-
Notifications
You must be signed in to change notification settings - Fork 0
Protein Annotations in PINT
The annotations that are automatically imported are UniProtKB and OMIM
-
UniProtKB The pint annotations module takes care of retrieving and storing at the server the protein annotations of all the proteins stored in the server.
These annotations will be stored at the server folder at: PINT_HOME_PATH/uniprot It will create a subfolder for each new release that is found, and it will do it automatically. -
OMIM The import of OMIM annotations is made using the API provided at (https://omim.org/api). But in order to use it, you have to register in OMIM to get an API key that you should include in the configuration.
Other external resources that are linked:
- PRIDE Cluster
- IntAct database
- ComplexPortal
First, define where are you going to store the uniprot annotations in your computer:
final File uniprotReleasesFolder = new File("C:\\Users\\salvador\\Desktop\\uniprotKB");Then, create an UniprotProteinLocalRetriever object using that location:
final UniprotProteinLocalRetriever uplr = new UniprotProteinLocalRetriever(uniprotReleasesFolder, true);Then, use that object to query your accession numbers of interest:
Collection<String> accs = # here is where I have my Uniprot Accession numbers
final Map<String, Entry> annotatedProteins = uplr.getAnnotatedProteins(null, accs);Note that you can query as many accessions as you want at once. It will make multiple remote queries to the EBI Uniprot service in parallel, with 200 proteins per query (maximum defined by EBI).
This will download the XML formatted Uniprot entries (i.e. P12345), and index it so that subsequent queries of the same accessions are retrieved fast.
Once the query has finished, you obtain a map in which the keys are the accessions of the proteins and the values are the uniprot Entry objects.
Note that in Uniprot proteins have multiple accession numbers pointing to the same entry. This query will do the same and for example, if you query the protein P12345, you will notice that it is also defined by the secondary accession G1SKL2. The map recovered will have the keys "P12345" and "G1SKL2" pointing to the same Entry object.
Then, the object Entry is the one representing the XML structure of the Uniprot entries.
However, you have multiple useful static methods in the class UniprotEntryUtil that you will find very handy, such as:
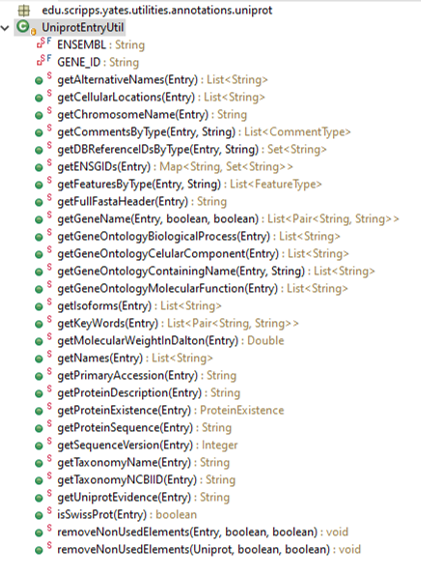
As an example:
// we have an Entry object retrieved using UniprotProteinLocalRetriever
final List<CommentType> commentsFunction = UniprotEntryUtil.getCommentsByType(entry, "Function");
String commentsFunctionsString = parseTextInFunctionComments(commentsFunction);
if ("".equals(commentsFunctionsString) || "N/A".equals(commentsFunctionsString)) {
// look into GO terms
final List<String> goProperties = UniprotEntryUtil.getGeneOntologyMolecularFunction(entry);
commentsFunctionsString = parseGOs(goProperties);
}final String proteinSequence = UniprotEntryUtil.getProteinSequence(entry);How to get the subcellular localization annotation of a protein and information about possible transmembrane regions:
final List<String> cellularLocations = UniprotEntryUtil.getCellularLocations(entry);
final List<FeatureType> transmembraneRegions = entry.getFeature().stream()
.filter(feature -> "transmembrane region".equalsIgnoreCase(feature.getType()))
.collect(Collectors.toList());The same module has an object that can be used to easily retrieve information from OMIM:
// at OmimRetriever class:
public static Map<String, List<OmimEntry>> getAssociatedOmimEntries(String omimAPIKey, Collection<String> uniprotAccs)Note that in order to this query to work you will need to register yourself to get a valid omimAPIKey at https://www.omim.org/api
This module contains a class called UniprotGeneMapping which allows you to map any uniprot accession to gene name and the other way around.
It will automatically go to Uniprot and download the appropriate Uniprot ID mapping file, which is species-specific. Therefore you will need to specify the species (one or more) you want to query. See example:
final String[] ORGANISMS = { "Rat", "Mouse", "Human" };
final boolean mapToGENESYNONIM = false;
final boolean mapToENSEMBL = false;
final boolean mapToGENENAME = true;
UniprotGeneMapping geneMapping = UniprotGeneMapping.getInstance(new File(uniprotReleasesFolder), ORGANISMS, mapToENSEMBL,
mapToGENENAME, mapToGENESYNONIM);
final Set<String> uniprotAccs = geneMapping.mapGeneToUniprotACC(geneName);
Final Set<String> geneNames = geneMapping.mapUniprotACCToGene(acc);Proteomics Yates Laboratory
Salvador Martínez-Bartolomé (salvador at scripps.edu)
Research Associate
The Scripps Research Institute
10550 North Torrey Pines Road
La Jolla, CA 92037
Git-Hub profile