-
Notifications
You must be signed in to change notification settings - Fork 0
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
profiling package and results for pandas extensions #11
Conversation
* profilers for copying dataframe to database * profilers moved to subpackage * profiler group tagging * profilers executed in subprocess to ensure memory measurements * automatic plotting of memory & time results for each profiler group * randomize profiler order in group * run multiple trials * filter by profiler function name and/or by group tag * removed in-development artifacts (txt, yaml, png) * made management command wrapper handle arguments and errors correctly
prof/util.py
Outdated
|
|
||
|
|
||
| def histogram(stream): | ||
| counts = collections.defaultdict(int) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Is histogram here just reimplementing collections.Counter? Although I usually don't use it this way it appears it can take an iterable in the constructor and I would guess does exactly what histogram is doing here.
https://docs.python.org/3/library/collections.html#collections.Counter
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Cool, thanks – I didn't see this added in Python 3. (It's not hard to do, but it did seem like something that belonged there.)
| return f"contextmanager({self.func})" | ||
|
|
||
|
|
||
| class contextmanager(_ComposingMixin, Wrapper): |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Can this explain why it's a better contextmanager?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yep, that was an oversight – documentation added (here in util and in tool).
|
It feels like your gigantic PR introductory text belongs in something more persistently visible, like a markdown file in the repo linked to from the README. Technically you are describing changes, but also so much more! I think PR introductory text works better for things that are useful context to the reviewer, but the audience for this text seems like potential users |
This profiler permits a proper comparision to pg_copy_to, (and exposes potential opportunities for improvement).
|
@thcrock I agree! And I fully intend to publish these results in some such way. I just want to be sure of what we have, here, first. |
|
All right I have:
The results of the latter are interesting. As we saw before, the "generic" version, which uses The novelty of the new profiler, which goes through a Pandas extension – What's interesting is that the StringIO accessor is as fast as it is. It's just using So …
|
|
I've added an issue, #12, for exploring speed improvement in And we already have #4 for exploring an I just kicked off another profiling run, in hopes of getting nice clean plots for I should probably do the same for (Unfortunately, as fast as my laptop is, it'll take a while. Of course, this is mostly the fault of Pandas |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The code looks good to me. There's probably a better way to brand these in the future. You now have multiple 'like a python standard library but more advanced' things put into Ohio. Some of them are hard to find here if somebody were to want to use elsewhere (who goes combing through util.py files?)
You could:
- Go the toolz route and make boring 'itertoolz' 'functoolz' in the same repo (maybe it wouldn't be called ohio in that case) https://github.com/pytoolz/toolz
- Milk the 'ohio' thing into ones that make no sense: 'ohcontext', 'ohfunc'
- Expend each of them into its own thing: 'contextliberal', 'funktools'
- Something better than any of these
But this works and is fine for now.
|
@thcrock Hah sure. I was thinking the same, it might be nice to put some of these "toolz" elsewhere; but, I'm not too worried about that for the moment. Certainly I won't just put stuff unrelated to Perhaps the profiling framework should be split out, such that what's here is more of a stub; but, same story. Once we want to reuse this stuff, there's work to do. But that's a welcome alternative to restarting from scratch. I think better to just make use of it here first, and see, (than to make library atomicity a prerequisite). Anyway, thanks 👍 |
basic documentation **Changes** * rounded out library docstrings, such that the heart of the (API) documentation is there * integrated with Sphinx (and autodoc) * such that standard documentation can be generated from docstrings, (tho HTML currently unused) * submitted patches to extension [sphinx-contrib.restbuilder](sphinx-contrib/restbuilder#8), such that Sphinx can also build RST docs – (it's a little surprising that Sphinx can't on its own or otherwise its build process can't be intercepted to retrieve RST formatted output before it's parsed into an abstract tree, considering its input is RST, albeit Sphinx- and autodoc-extended RST; but, nonetheless, this appeared to be the easiest way forward ... though markdown is another obvious target, it doesn't cleanly support all the directives that you get from RST ... and Github won't render README.html). * scripted a development command to generate HTML docs and README.rst In the end, I don't love [how the auto-generated README.rst looks](https://github.com/dssg/ohio/tree/docs), at least not as Github renders it; but, for something like this, I think it's fine, and keeps API documentation in the code. A discussion of [benchmarking the Pandas extension](#11) is included in `doc/index.rst` (and from there included in built documentation). resolves #1
Branch summary
The below package was used to generate the following performance results for the Ohio-Pandas integration extensions to
DataFrame–pg_copy_to&pg_copy_from.Results summary
In general, all solutions involving the Ohio extensions require significantly less RAM; and, (except for the
buffer_size=1case, which is no longer the default), Ohio extensions are also significantly faster.Unlike Pandas built-in methods
to_sqlandread_sql, doing a custom PostgreSQLCOPYviaio.StringIOis competitive, but still significantly slower and more RAM-intensive than Ohio extensions. (And Ohio extensions have the added benefit of being one-liners, simple method invocations, and "built-in" toDataFrame, once the integration is loaded.)Profiling
The profiling package,
prof, is executable as a package, (like a module):I added a management command wrapper largely for added documentation and cleanliness:
Generally, I used variations of the following command:
I.e.:
timereports on that.Results
These take a really long time to run, and so while you could just run it as above, and get plots for each group, I generally filtered down to the one group of the two I was trying to test.
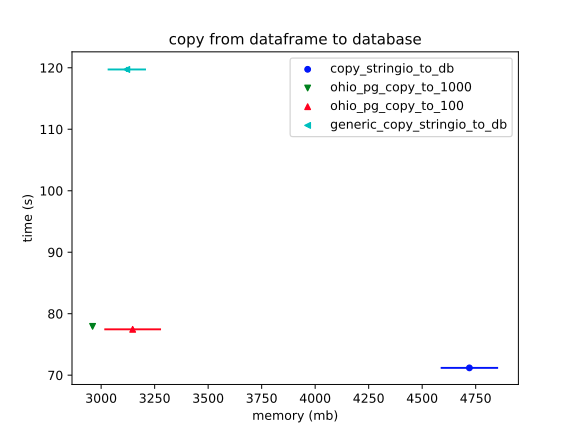
Copy from DataFrame to database table
Basically,
to_sqlis a lost cause, (even playing with themultiparameter). However, theStringIOmethod comes close; and, it's unclear how close, at this scale.So, ignoring
to_sql:Now we can see more clearly what was apparent in the previous plot: the
StringIOversion is significantly slower. (This makes sense, since it has to encode all the data into a string before beginning toCOPYit into the database.)The error bars on RAM usage are a mess, however. It doesn't help that I'm comparing a ton of different configurations of Ohio, (including unreleased ones, which this profiling has almost certainly proven to be unnecessary).
These RAM usage measurements were tough to make seem "true." Running all profilers, by default, in a subprocess made a huge difference; but, for whatever reason, you can still get a wide variance – perhaps because I'm just running these on my laptop, and even at three trials, a weird one can throw things off.
In conclusion, I'd say that Ohio – especially with the current default configuration – does fair better on RAM usage, as well. And, that's just with this particular dataset. Unlike the
StringIOversion, the overhead of Ohio's solution shouldn't increase linearly.Copy from database table to DataFrame
Pandas –
read_sql– is again a huge outlier in slowness and RAM usage.Unlike in the previous context, here
StringIOdoes much worse on RAM. And, again, it's significantly slower, (if not tremendously so).The Ohio solutions all do about the same, (and it's unclear which of them is definitely best).
All that said, I'm happy to run more trials. And, with this package, others are welcome to do the same! 😸