-
Notifications
You must be signed in to change notification settings - Fork 1
Simple Systematic Point Simulation
Laura Marshall edited this page Feb 6, 2017
·
11 revisions
This simulation sets up a square study area of 1000 by 1000 km. The population is a fixed size of 2000 individuals and has an even density throughout the study region. The detectability of the individuals is based on a half-normal detection function with sigma / scale parameter of 20km and sightings are truncated at 60km (a rather optomistic detection function!). The survey design uses a systematic grid of point transect spaced at 100 km intervals. The resulting data are analysed by fitting a half-normal detection function.
#Load DSsim
library(DSsim)
# Create coordinates for a 1000 by 1000 km study region
coords <- list()
coords[[1]] <- list(data.frame(x = c(0, 0 , 1000, 1000, 0),
y = c(0, 1000, 1000, 0, 0)))
# Create survey region
region <- make.region(coords = coords, units = "km")
plot(region)
# Create flat density surface
density <- make.density(region)
# Plotting density grid as points (the default style)
plot(density)
plot(region, add = TRUE)
# Create a population of 2000, need to give it non default region and density
pop.desc <- make.population.description(region, density, N = 2000)
# Define detectability, by default a half normal (hn) detection function will be used
detect <- make.detectability(scale.param = 20, truncation = 60)
# specify design
# default for point is a systematic placement with a spacing of 100
design <- make.design("point")
# Define analysis
analyses <- make.ddf.analysis.list(dsmodel = list(~cds(key = "hn", formula = ~1)),
criteria = "AIC",
truncation = 60)
# Put everything together as a simulation
sim <- make.simulation(999,
region.obj = region,
population.description.obj = pop.desc,
detectability.obj = detect,
design.obj = design,
ddf.analyses.list = analyses)
# Let's set a seed to check our results match
set.seed(735)
# Check the setup
check.sim.setup(sim)
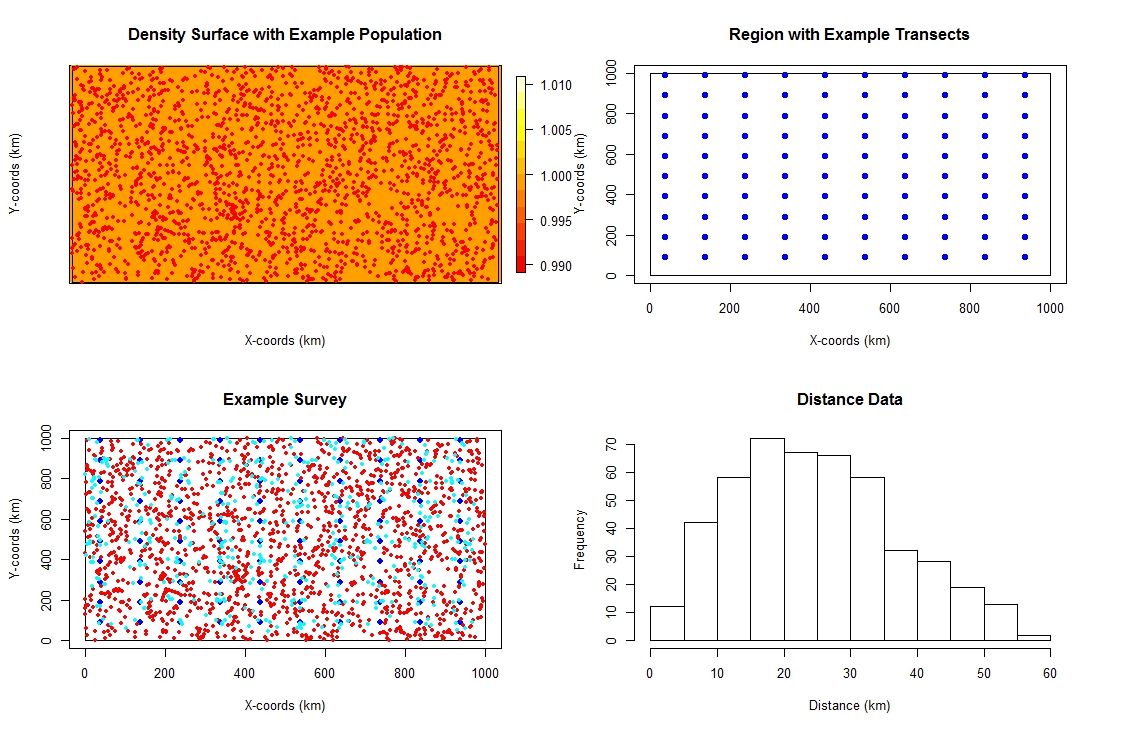
# Run the simulation
sim <- run(sim, run.parallel = TRUE, max.cores = 4)
# View the results
summary(sim)
GLOSSARY
--------
Summary of Simulation Output
Region : the region name.
No. Repetitions : the number of times the simulation was repeated.
No. Failures : the number of times the simulation failed (too
few sightings, model fitting failure etc.)
Summary for Individuals
Summary Statistics:
mean.Cover.Area : mean covered across simulation.
mean.Effort : mean effort across simulation.
mean.n : mean number of observed objects across
simulation.
no.zero.n : number of surveys in simulation where
nothing was detected.
mean.ER : mean encounter rate across simulation.
mean.se.ER : mean standard error of the encounter rates
across simulation.
sd.mean.ER : standard deviation of the encounter rates
across simulation.
Estimates of Abundance:
Truth : true population size, (or mean of true
population sizes across simulation for Poisson N.
mean.Estimate : mean estimate of abundance across simulation.
percent.bias : the percentage of bias in the estimates.
RMSE : root mean squared error/no.successful reps
CI.coverage.prob : proportion of times the 95% confidence interval
contained the true value.
mean.se : the mean standard error of the estimates of
abundance
sd.of.means : the standard deviation of the estimates
Estimates of Density:
Truth : true average density.
mean.Estimate : mean estimate of density across simulation.
percent.bias : the percentage of bias in the estimates.
RMSE : root mean squared error/no.successful reps
CI.coverage.prob : proportion of times the 95% confidence interval
contained the true value.
mean.se : the mean standard error of the estimates.
sd.of.means : the standard deviation of the estimates.
Detection Function Values
mean.observed.Pa : mean proportion of animals observed in the covered
region.
mean.estimte.Pa : mean estimate of the proportion of animals observed
in the covered region.
sd.estimate.Pa : standard deviation of the mean estimates of the
proportion of animals observed in the covered region.
mean.ESW : mean estimated strip width.
sd.ESW : standard deviation of the mean estimated strip widths.
Region: region
No. Repetitions: 999
No. Failures: 0
Design: Systematic Point Transect
design.axis = 0
spacing = 100
Population Detectability Summary:
key.function = hn
scale.param = 20
truncation = 60
Analysis Summary:
Candidate Models:
Model 1 : ~ cds(key = "hn", formula = ~1) was selected 999 time(s).
criteria = AIC
truncation = 60
Summary for Individuals
Summary Statistics
mean.Cover.Area mean.Effort mean.n no.zero.n mean.ER mean.se.ER sd.mean.ER
1 1130973 100 481.7728 0 4.817728 0.224798 0.2161754
Estimates of Abundance (N)
Truth mean.Estimate percent.bias RMSE CI.coverage.prob mean.se sd.of.means
1 2000 1978.28 -1.09 135.26 0.95 134.01 133.57
Estimates of Density (D)
Truth mean.Estimate percent.bias RMSE CI.coverage.prob mean.se sd.of.means
1 0.002 0.001978284 -1.085784 0.0001352564 0.9469469 0.0001340121 0.0001335687
Detection Function Values
mean.observed.Pa mean.estimate.Pa sd.estimate.Pa mean.ESW sd.ESW
1 0.22 0.22 0.01 12.95 0.65