-
Notifications
You must be signed in to change notification settings - Fork 2
Filtering datasets
By clicking on the 'Filters' menu, you will obtain several option menus:
- Define filters: this will open the filters definition window
- And then, there is one option menu for each one of the available filters, which will open the filters definition window for that specific filter:
Note: Clicking on an individual filter menu when that filter was already defined, will not open the filter definition window, and it will just enable (or disable it). If the filter was not defined before, the filter definition window will open when clicking on the individual filter menu. Note 2: When a filter is active, you will see a tick mark on the individual filter menu. In order to deactivate the filter, just click again on the individual filter menu, or click on the 'Define filters' menu and click on the corresponding checkbox to disable the filter you want.
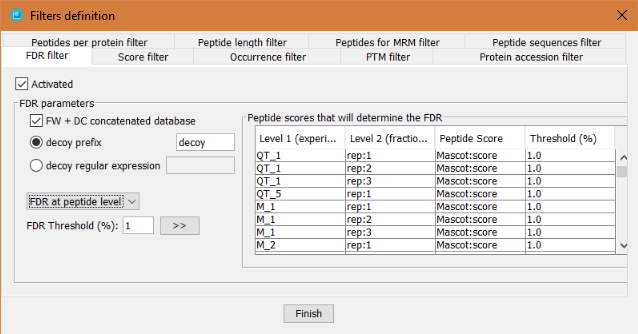
This filter will perform a False Discovery Rate (FDR) filter by calculating the FDR at PSM, peptide or protein level. Important notes about FDR in PACOM:
- The FDR is applied at level 2 of the comparison project tree.
- The FDR is applied using the peptide score, even for FDR at protein level. In case of FDR at protein level, the best peptide score for each protein is used to rank the proteins.
We are aware that this is not the most accurate way to do it, specially for protein level FDR, but due to the heterogeneity of the data that can be analyzed by PACOM, we didn't want to be stuck with a particular solution and we offer this general one. For a more accurate FDR filter, either import your data with a p-value or any other score that can be used to sort the dataset, or just import the data from an already filtered dataset.
In order to define the FDR filter, the user has to:
- Click on ?Activated? checkbox
- Define how to identify the decoy hits: by a regular expression or by a prefix.
- Define which peptide score will be taken into account to sort the peptides for level 2 nodes.
- Define which threshold will be applied. The threshold value can be different for each level 2 node.
Contact person for any suggestion or error report:
Salvador Martínez-Bartolomé (salvador at scripps.edu)
Senior Research Associate
The Scripps Research Institute
10550 North Torrey Pines Road
La Jolla, CA 92037
Git-Hub profile