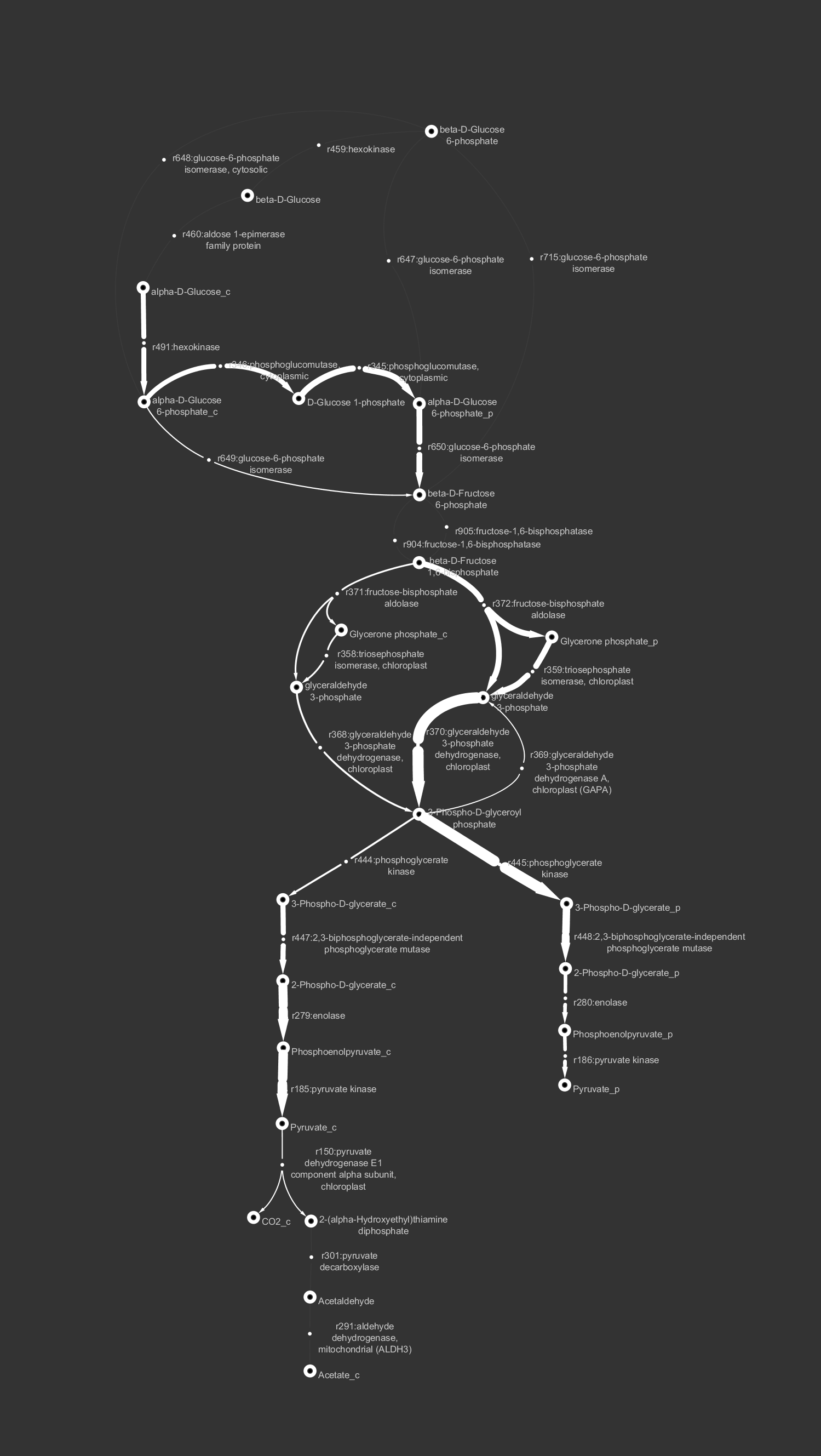
Create Cytoscape network with thickness of arrows representing fluxes or rates of reactions
Example: The glycolysis/gluconeogenesis pathway in AraGEM (a genome-scale model of Arabidopsis thaliana) 3 files are generated:
- network.sif network for Cytoscape
- edge.txt table to import (match interaction to network)
- node.txt table to import (match name to network) Use cytoscape_style_GEM00.xml