Pytaxon: A Python software package for the identification and correction of errors in the taxonomic data of biodiversity species
We present pytaxon, a Python software designed to resolve and correct taxonomic names in biodiversity data by leveraging the Global Names Verifier (GNV) API and employing fuzzy matching techniques to suggest corrections for discrepancies and nomenclatural inconsistencies. The pytaxon offers both a Command Line Interface (CLI) and a Graphical User Interface (GUI), ensuring accessibility to users with different levels of computing expertise.
- Listed at requirements.txt
Install the package from PyPI:
$ pip install pytaxon
To download the Pytaxon GUI .exe:
| Windows | Linux | |
|---|---|---|
| .zip | Link | Link |
| .rar | Link | Link |
Firstly, you will want to check your spreadsheet for errors, then the program will return you and Excel file (.xlsx) containing all the incorrect data depending on the selected data source.
Then, you may select which data are to be corrected with the "Change" column, after this, you may run the second command to correct automatically the original spreadsheet with the checked spreadsheet.
$ pytaxon -r <column names> -os <path to original spreadsheet> -ss <name of suggestion spreadsheet> -si <source id>
$ pytaxon -os <path to original spreadsheet> -cs <path of checked spreadsheet> -o <name of corrected spreadsheet>
Explore the options for these commands with the --help flag.
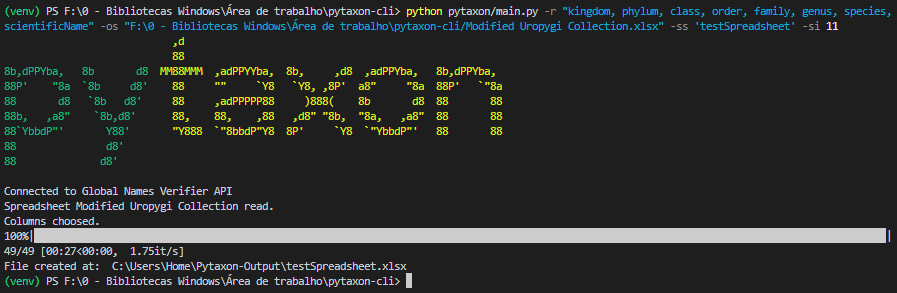
Pytaxon CLI running on the Visual Studio Code terminal (Powershell) with a modified version of the Uropygi dataset
The to correct spreadsheet of the modified Uropygi dataset
Pytaxon GUI application running with a modified version of the Uropygi dataset
Pytaxon's CLI and GUI workflow
If you use the source code of Pytaxon in any form, please cite the following manuscript (we encorage citing Global Names Resolver as well):
Proença Neto MA, De Sousa MPA (2025) Pytaxon: A Python software for resolving and correcting taxonomic names in biodiversity data. Biodiversity Data Journal 13: e138257. https://doi.org/10.3897/BDJ.13.e138257
We thank the following institutions, which contributed to ensuring the success of our work:
Museu Paraense Emílio Goeldi (MPEG)
Centro Universitário do Estado do Pará (CESUPA)
This research was supported by Centro Universitário do Pará - CESUPA with the PIBICT scientific initiation scholarship project.
Marco Aurélio Proença Neto
Marcos Paulo Alves de Sousa
Dr. Marcos Paulo Alves de Sousa (Project leader)
Email: [email protected]
Grupo de Estudos Temático em Computação Aplicada (GET-COM)
Centro Universitário do Pará - CESUPA
Av. Perimetral 1901. CEP 66077- 530. Belém, Pará, Brazil.