-
Notifications
You must be signed in to change notification settings - Fork 4
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
Showing
25 changed files
with
1,885 additions
and
17 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1 +1,2 @@ | ||
| /build/ | ||
| *__pycache__* |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -27,3 +27,4 @@ if(BUILD_EXAMPLES) | |
| endif() | ||
|
|
||
| install(FILES LICENSE.md DESTINATION .) | ||
| install(DIRECTORY train DESTINATION .) | ||
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,3 +1,51 @@ | ||
| ## Annotation Guidelines | ||
| # Annotation Guidelines | ||
|
|
||
| Guidelines coming soon... | ||
| This document describes the data layout for building your own models with OpenEM. Training routines in OpenEM expect the following directory layout: | ||
|
|
||
| ```shell | ||
| your-top-level-directory | ||
| └── train | ||
| ├── cover.csv | ||
| ├── length.csv | ||
| ├── ruler_position.csv | ||
| └── videos | ||
| ├── 00WK7DR6FyPZ5u3A.mp4 | ||
| ├── 01wO3HNwawJYADQw.mp4 | ||
| ├── 02p3Yn87z0b5grhL.mp4 | ||
| ``` | ||
|
|
||
| Many of the annotations require video frame numbers. It is important to point out that most video players do not have frame level accuracy, so attempting to convert timestamps in a typical video player to frame numbers will likely be inaccurate. Therefore we recommend using the frame accurate video annotator [Tator][Tator], which guarantees frame level accuracy when seeking and allows line annotations which are useful for generating frame level fish length. | ||
|
|
||
| **videos** contains video files in mp4 format. The content of these videos should follow the [data collection guidelines][CollectionGuidelines]. We refer to the basename of each video file as the *video ID*, a unique identifier for each video. In the directory layout above, the video ID for the videos are 00WK7DR6FyPZ5u3A, 01wO3HNwawJYADQw, and 02p3Yn87z0b5grhL. | ||
|
|
||
| **length.csv** contains length annotations of fish in the videos. Each row corresponds to an individual fish, specifically the video frame containing the clearest view of each fish. This file is also used to train the counting algorithm, so exactly one frame should be annotated per individual fish. The columns of this file are: | ||
|
|
||
| * *video_id*: The basename of the video. | ||
| * *frame*: The zero-based frame number in the video. | ||
| * *x1, y1, x2, y2*: xy-coordinates of the tip and tail of the fish in pixels. | ||
| * *species_id*: The one-based index of the species as listed in the ini file, as described in the [tutorial][Tutorial]. If this value is zero, it indicates that no fish are present. While length.csv can be used to include no fish example frames, it is encouraged to instead include them in cover.csv. Both are used when training the detection model, but only cover.csv is used when training the classification model. | ||
|
|
||
| 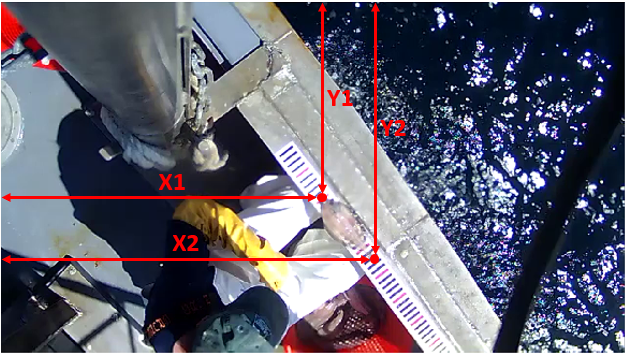 | ||
|
|
||
| **cover.csv** contains examples of frames that contain no fish, fish covered by a hand or other obstruction, and fish that can be clearly viewed. The columns of this file are: | ||
|
|
||
| * *video_id*: The basename of the video. | ||
| * *frame*: The zero-based frame number in the video. | ||
| * *cover*: 0 for no fish, 1 for covered fish, 2 for clear view of fish. | ||
|
|
||
|  | ||
|
|
||
|  | ||
|
|
||
|  | ||
|
|
||
| **ruler_position.csv** contains the ruler position in each video. The columns of this file are: | ||
|
|
||
| * *video_id*: The basename of the video. | ||
| * *x1, y1, x2, y2*: xy-coordinates of the ends of the ruler in pixels. | ||
|
|
||
| 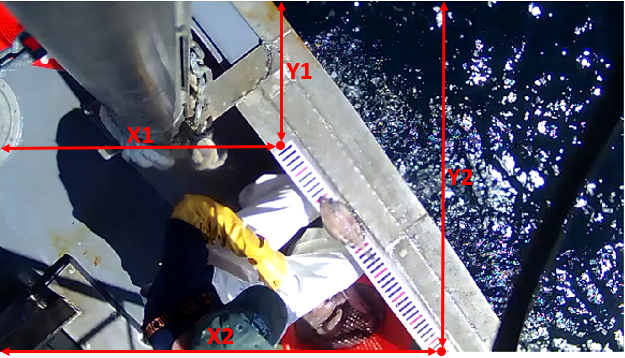 | ||
|
|
||
| [Tator]: https://github.com/cvisionai/Tator/releases | ||
| [CollectionGuidelines]: ./data_collection.md | ||
| [Tutorial]: ./tutorial.md |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,4 +1,4 @@ | ||
| ## Build and Test Instructions | ||
| # Build and Test Instructions | ||
|
|
||
| Choose your operating system: | ||
|
|
||
|
|
||
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,37 @@ | ||
| # Python Environment for Training Library | ||
|
|
||
| This document describes how to set up a python environment to use the training library. It specifies specific module versions that were used to test the training library, but more recent versions of these modules may also work. The training library has been tested on Windows 10 x64 and Ubuntu 18.04 LTS. | ||
|
|
||
| ## Installing Miniconda | ||
|
|
||
| [Miniconda][Miniconda] is a cross platform distribution of python that includes a utility for managing packages called conda. It allows for maintenance of multiple python environments that each have different modules or libraries installed. Download the version for python 3 and install it for your operating system of choice. | ||
|
|
||
| ## Creating a new environment (optional) | ||
|
|
||
| To create a new environment, use this command: | ||
|
|
||
| ```shell | ||
| conda create --name openem | ||
| ``` | ||
|
|
||
| This will create a new environment called openem. To start using it: | ||
|
|
||
| ```shell | ||
| source activate openem | ||
| ``` | ||
|
|
||
| If you do not do these steps then modules will be installed in the base environment, which is activated by default. This is fine if you do not plan to use Miniconda for other purposes than OpenEM training. | ||
|
|
||
| ## Install modules | ||
|
|
||
| Versions are included here for reference, different versions may work but have not been tested. | ||
|
|
||
| ```shell | ||
| conda install keras-gpu==2.2.4 | ||
| conda install pandas==0.23.4 | ||
| conda install opencv==3.4.2 | ||
| conda install scikit-image==0.14.0 | ||
| conda install scikit-learn==0.20.1 | ||
| ``` | ||
|
|
||
| [Miniconda]: https://conda.io/miniconda.html |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
Empty file.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,121 @@ | ||
| """Functions for training detection algorithm. | ||
| """ | ||
|
|
||
| import os | ||
| import glob | ||
| import numpy as np | ||
| from keras.optimizers import Adam | ||
| from keras.callbacks import ModelCheckpoint | ||
| from keras.callbacks import TensorBoard | ||
| from openem_train.ssd import ssd | ||
| from openem_train.ssd.ssd_training import MultiboxLoss | ||
| from openem_train.ssd.ssd_utils import BBoxUtility | ||
| from openem_train.ssd.ssd_dataset import SSDDataset | ||
| from openem_train.util.model_utils import keras_to_tensorflow | ||
|
|
||
| def _save_model(config, model): | ||
| """Loads best weights and converts to protobuf file. | ||
| # Arguments | ||
| config: ConfigInterface object. | ||
| model: Keras Model object. | ||
| """ | ||
| best = glob.glob(os.path.join(config.checkpoints_dir(), '*best*')) | ||
| latest = max(best, key=os.path.getctime) | ||
| model.load_weights(latest) | ||
| os.makedirs(config.detect_model_dir(), exist_ok=True) | ||
| keras_to_tensorflow(model, ['output_node0'], config.detect_model_path()) | ||
|
|
||
| def train(config): | ||
| """Trains detection model. | ||
| # Arguments | ||
| config: ConfigInterface object. | ||
| """ | ||
|
|
||
| # Create tensorboard and checkpoints directories. | ||
| os.makedirs(config.checkpoints_dir(), exist_ok=True) | ||
| os.makedirs(config.tensorboard_dir(), exist_ok=True) | ||
|
|
||
| # Build the ssd model. | ||
| model = ssd.ssd_model( | ||
| input_shape=(config.detect_height(), config.detect_width(), 3), | ||
| num_classes=config.num_classes()) | ||
|
|
||
| # Set trainable layers. | ||
| for layer in model.layers: | ||
| layer.trainable = True | ||
|
|
||
| # Set up loss and optimizer. | ||
| loss_obj = MultiboxLoss( | ||
| config.num_classes(), | ||
| neg_pos_ratio=2.0, | ||
| pos_cost_multiplier=1.1) | ||
| adam = Adam(lr=3e-5) | ||
|
|
||
| # Compile the model. | ||
| model.compile(loss=loss_obj.compute_loss, optimizer=adam) | ||
| model.summary() | ||
|
|
||
| # Get prior box layers from model. | ||
| prior_box_names = [ | ||
| 'conv4_3_norm_mbox_priorbox', | ||
| 'fc7_mbox_priorbox', | ||
| 'conv6_2_mbox_priorbox', | ||
| 'conv7_2_mbox_priorbox', | ||
| 'conv8_2_mbox_priorbox', | ||
| 'pool6_mbox_priorbox'] | ||
| priors = [] | ||
| for prior_box_name in prior_box_names: | ||
| layer = model.get_layer(prior_box_name) | ||
| if layer is not None: | ||
| priors.append(layer.prior_boxes) | ||
| priors = np.vstack(priors) | ||
|
|
||
| # Set up bounding box utility. | ||
| bbox_util = BBoxUtility(config.num_classes(), priors) | ||
|
|
||
| # Set up dataset interface. | ||
| dataset = SSDDataset( | ||
| config, | ||
| bbox_util=bbox_util, | ||
| preproc=lambda x: x) | ||
|
|
||
| # Set up keras callbacks. | ||
| checkpoint_best = ModelCheckpoint( | ||
| config.checkpoint_best(), | ||
| verbose=1, | ||
| save_weights_only=False, | ||
| save_best_only=True) | ||
|
|
||
| checkpoint_periodic = ModelCheckpoint( | ||
| config.checkpoint_periodic(), | ||
| verbose=1, | ||
| save_weights_only=False, | ||
| period=1) | ||
|
|
||
| tensorboard = TensorBoard( | ||
| config.tensorboard_dir(), | ||
| histogram_freq=0, | ||
| write_graph=True, | ||
| write_images=True) | ||
|
|
||
| # Fit the model. | ||
| batch_size = config.detect_batch_size() | ||
| val_batch_size = config.detect_val_batch_size() | ||
| model.fit_generator( | ||
| dataset.generate_ssd( | ||
| batch_size=batch_size, | ||
| is_training=True), | ||
| steps_per_epoch=dataset.nb_train_samples // batch_size, | ||
| epochs=config.detect_num_epochs(), | ||
| verbose=1, | ||
| callbacks=[checkpoint_best, checkpoint_periodic, tensorboard], | ||
| validation_data=dataset.generate_ssd( | ||
| batch_size=val_batch_size, | ||
| is_training=False), | ||
| validation_steps=dataset.nb_test_samples // val_batch_size, | ||
| initial_epoch=0) | ||
|
|
||
| # Load weights of the best model. | ||
| _save_model(config, model) |
Oops, something went wrong.