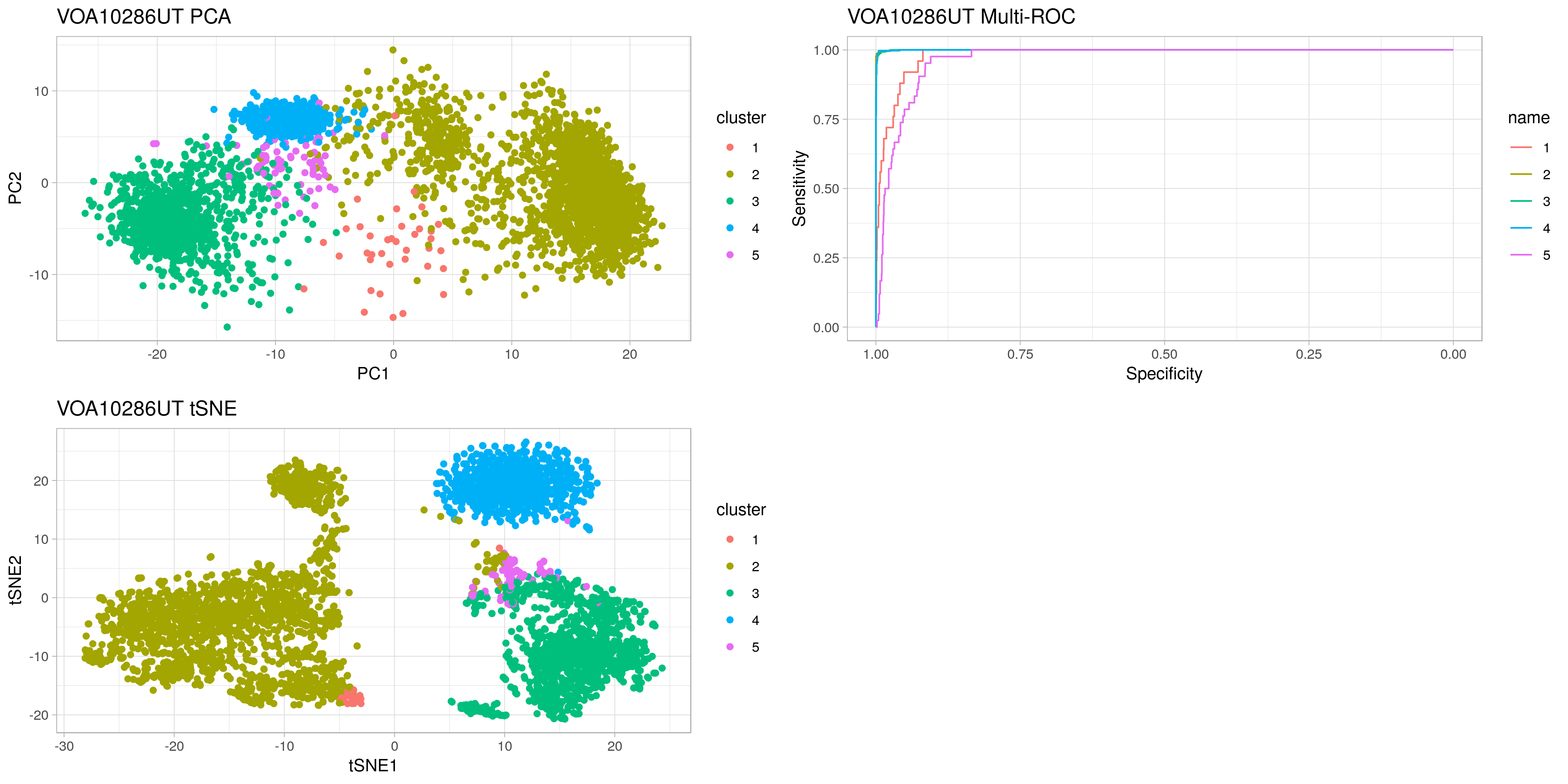
casc trains a logisitic regression model on provided single-cell RNAseq clusters. It creates an ROC curve for each cluster vs the others to help decide the appropriate number of clusters.
casc can be installed via devtools and github:
install.packages("devtools")
devtools::install_github("jamez-eh/casc")casc has the following parameters:
sce: ASingleCellExperimentwith normalized logcounts as an assay or a logcounts matrix with cells as columns and genes as rows.clusters: A list of clusterings to evaluate.alpha: A parameter for logistic regression. alpha = 1 represents the lasso penalty and alpha = 0 represents the ridge penalty.
registerDoSEQ()
sce_sim <- readRDS("~/sce_sim.rds")
casc_list <- casc(sce = sce_sim,
clusters = list(clustering_1, clustering_2, clustering_3, clustering_4, clustering_5),
alpha = 0.5)
James Hopkins