A graphical backend module to visualize, edit and export haplotype networks.
This is not a standalone application. For an implementation that visualizes sequence files, visit Hapsolutely.
Lay out the initial graph using a modified spring algorithm:
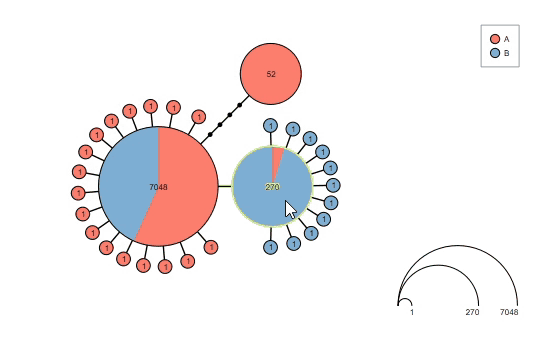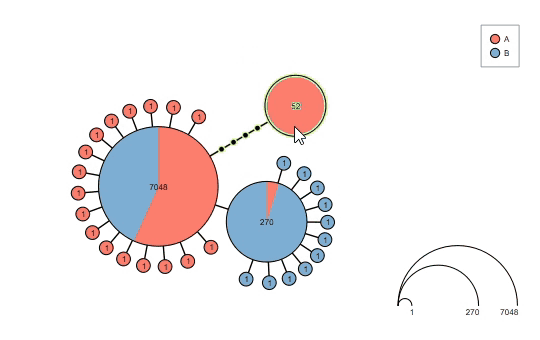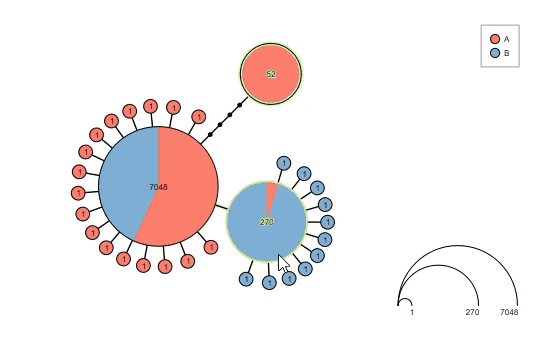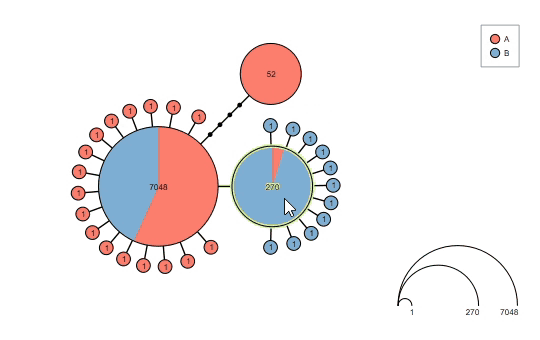
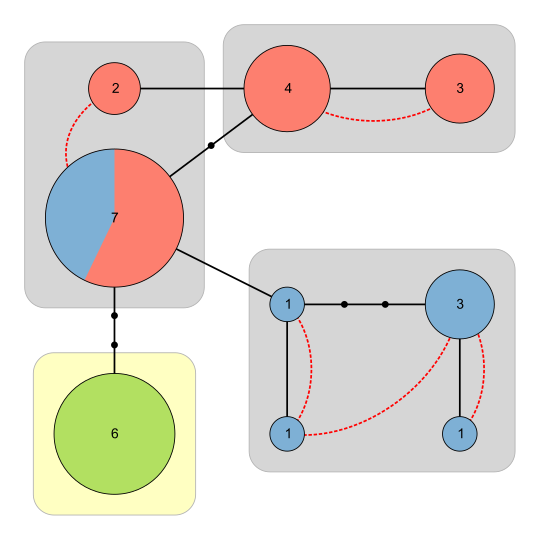
Interact with the graph before saving the results:
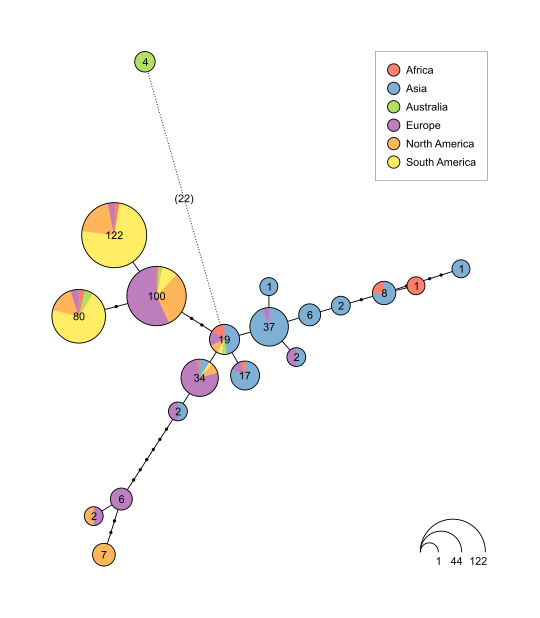
Supports haploweb visualization:
Haplodemo is available on PyPI. You can install it through pip:
pip install itaxotools-haplodemo
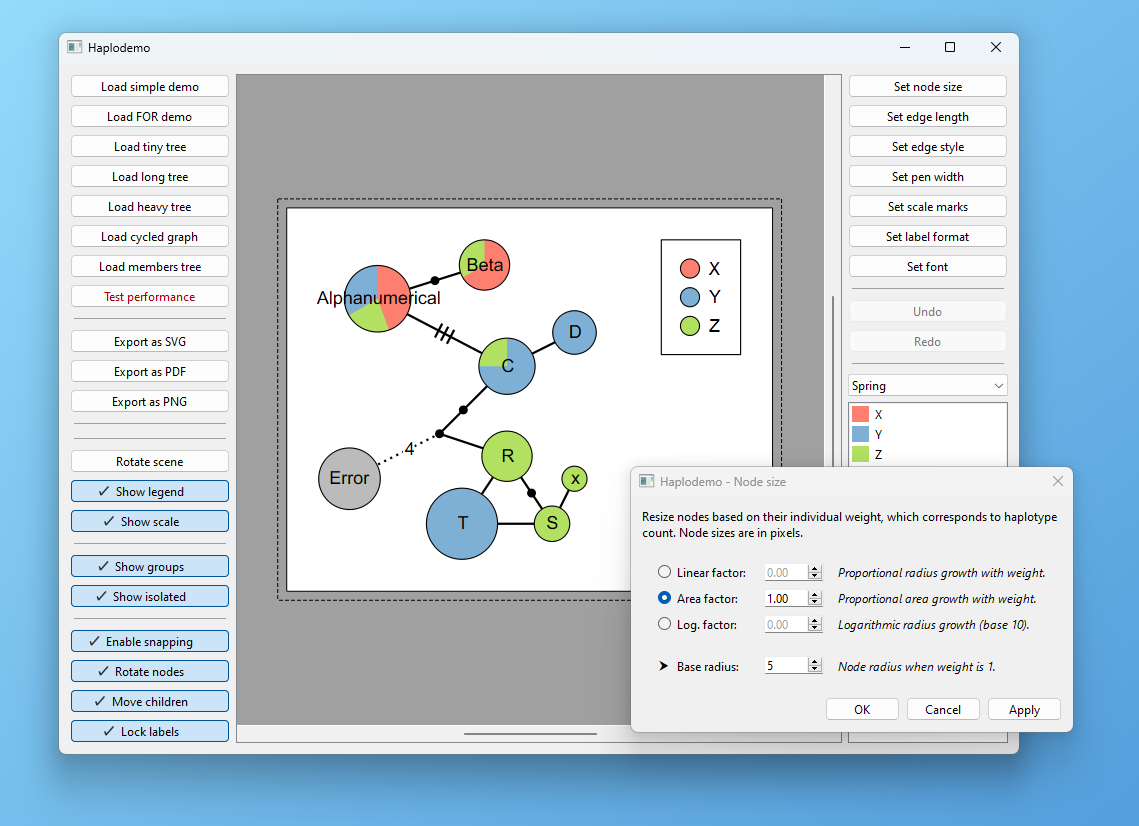
Standalone executables are included for demonstrating the library capabilities.
It is not possible to open custom haplotype networks with the demo program.
Launch the demo application to get an overview of the features: haplodemo
To get started on instantiating the scene, view and controls, look at window.py.
For some examples of data visualization, look at demos.py.
For a comprehensive example of a network in YAML format, look at members_graph.yaml.
The network can be given in tree or graph format using the HaploTreeNode and HaploGraph types.
It is also possible to load a network from a properly formatted YAML file, or directly from a Python dictionary.
Alternatively, populate the scene manually with nodes and edges.