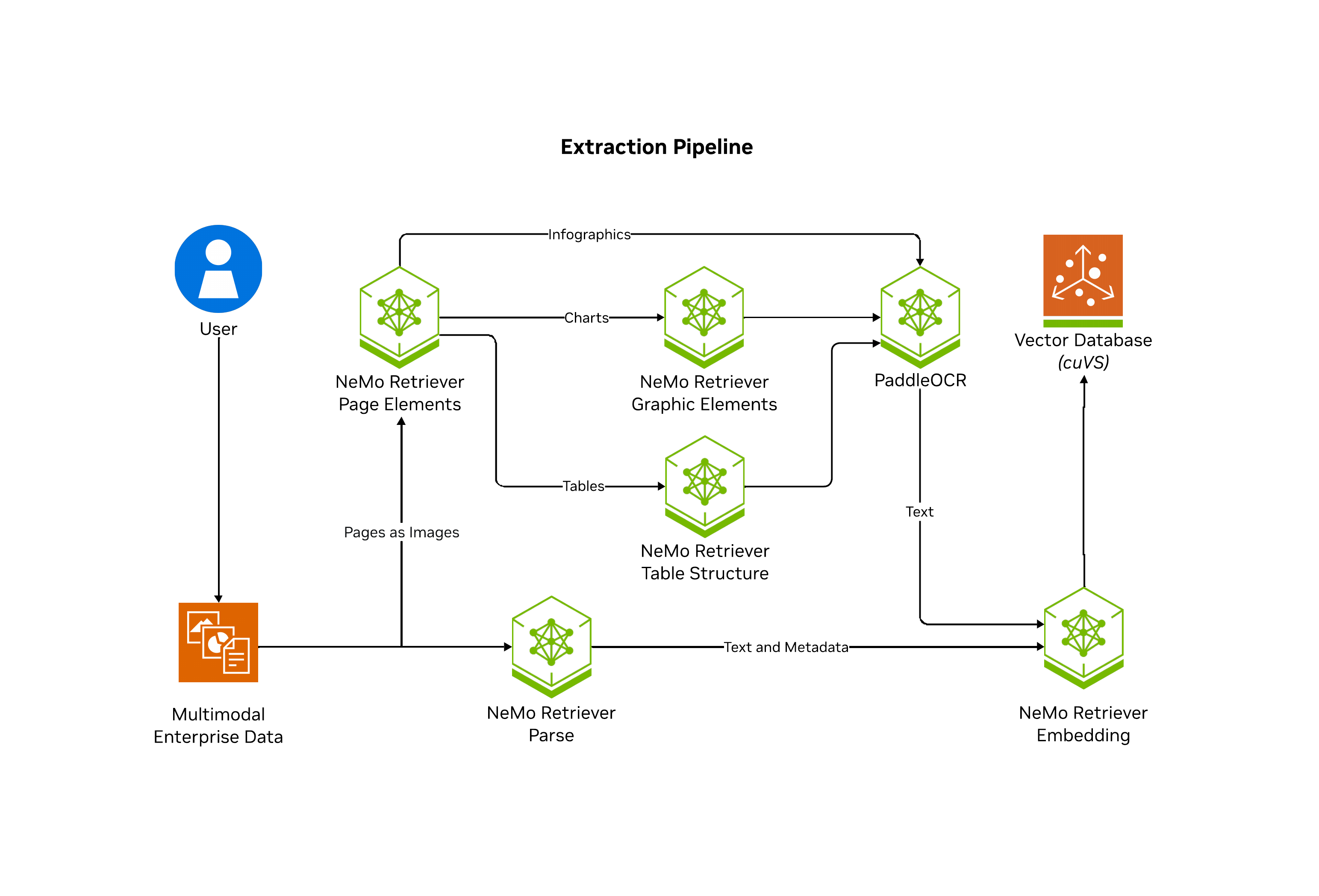
NeMo Retriever extraction is a scalable, performance-oriented document content and metadata extraction microservice. NeMo Retriever extraction uses specialized NVIDIA NIM microservices to find, contextualize, and extract text, tables, charts and infographics that you can use in downstream generative applications.
Note
NeMo Retriever extraction is also known as NVIDIA Ingest and nv-ingest.
NeMo Retriever extraction enables parallelization of splitting documents into pages where artifacts are classified (such as text, tables, charts, and infographics), extracted, and further contextualized through optical character recognition (OCR) into a well defined JSON schema. From there, NeMo Retriever extraction can optionally manage computation of embeddings for the extracted content, and optionally manage storing into a vector database Milvus.
Note
Cached and Deplot are deprecated. Instead, NeMo Retriever extraction now uses the yolox-graphic-elements NIM. With this change, you should now be able to run NeMo Retriever Extraction on a single 24GB A10G or better GPU. If you want to use the old pipeline, with Cached and Deplot, use the NeMo Retriever Extraction 24.12.1 release.
The following diagram shows the Nemo Retriever extraction pipeline.
NeMo Retriever Extraction is a library and microservice service that does the following:
- Accept a job specification that contains a document payload and a set of ingestion tasks to perform on that payload.
- Store the result of each job to retrieve later. The result is a dictionary that contains a list of metadata that describes the objects extracted from the base document, and processing annotations and timing/trace data.
- Support multiple methods of extraction for each document type to balance trade-offs between throughput and accuracy. For example, for .pdf documents, extraction is performed by using pdfium, nemoretriever-parse, Unstructured.io, and Adobe Content Extraction Services.
- Support various types of before and after processing operations, including text splitting and chunking, transform and filtering, embedding generation, and image offloading to storage.
NeMo Retriever Extraction supports the following file types:
bmpdocxhtml(converted to markdown format)jpegjson(treated as text)md(treated as text)pdfpngpptxsh(treated as text)tifftxt
NeMo Retriever extraction does not do the following:
- Run a static pipeline or fixed set of operations on every submitted document.
- Act as a wrapper for any specific document parsing library.
For more information, see the full NeMo Retriever Extraction documentation.
For production-level performance and scalability, we recommend that you deploy the pipeline and supporting NIMs by using Docker Compose or Kubernetes (helm charts). For more information, refer to prerequisites.
For small-scale workloads, such as workloads of fewer than 100 PDFs, you can use library mode setup. Library mode set up depends on NIMs that are already self-hosted, or, by default, NIMs that are hosted on build.nvidia.com.
Library mode deployment of nv-ingest requires:
- Linux operating systems (Ubuntu 22.04 or later recommended)
- Python 3.12
- We strongly advise using an isolated Python virtual env, such as provided by uv or conda
Create a fresh Python environment to install nv-ingest and dependencies.
uv venv --python 3.12 nvingest && \
source nvingest/bin/activate && \
uv pip install nv-ingest==25.9.0 nv-ingest-api==25.9.0 nv-ingest-client==25.9.0 milvus-lite==2.4.12Set your NVIDIA_API_KEY. If you don't have a key, you can get one on build.nvidia.com. For instructions, refer to Generate Your NGC Keys.
export NVIDIA_API_KEY=nvapi-...
You can submit jobs programmatically in Python.
To confirm that you have activated your Python environment, run which python and confirm that you see nvingest in the result. You can do this before any python command that you run.
which python
/home/dev/projects/nv-ingest/nvingest/bin/python
If you have a very high number of CPUs, and see the process hang without progress, we recommend that you use taskset to limit the number of CPUs visible to the process. Use the following code.
taskset -c 0-3 python your_ingestion_script.py
On a 4 CPU core low end laptop, the following code should take about 10 seconds.
import time
from nv_ingest.framework.orchestration.ray.util.pipeline.pipeline_runners import run_pipeline
from nv_ingest_client.client import Ingestor, NvIngestClient
from nv_ingest_api.util.message_brokers.simple_message_broker import SimpleClient
from nv_ingest_client.util.process_json_files import ingest_json_results_to_blob
# Start the pipeline subprocess for library mode
run_pipeline(block=False, disable_dynamic_scaling=True, run_in_subprocess=True)
client = NvIngestClient(
message_client_allocator=SimpleClient,
message_client_port=7671,
message_client_hostname="localhost",
)
# gpu_cagra accelerated indexing is not available in milvus-lite
# Provide a filename for milvus_uri to use milvus-lite
milvus_uri = "milvus.db"
collection_name = "test"
sparse = False
# do content extraction from files
ingestor = (
Ingestor(client=client)
.files("data/multimodal_test.pdf")
.extract(
extract_text=True,
extract_tables=True,
extract_charts=True,
extract_images=True,
table_output_format="markdown",
extract_infographics=True,
# extract_method="nemoretriever_parse", #Slower, but maximally accurate, especially for PDFs with pages that are scanned images
text_depth="page",
)
.embed()
.vdb_upload(
collection_name=collection_name,
milvus_uri=milvus_uri,
sparse=sparse,
# for llama-3.2 embedder, use 1024 for e5-v5
dense_dim=2048,
)
)
print("Starting ingestion..")
t0 = time.time()
# Return both successes and failures
# Use for large batches where you want successful chunks/pages to be committed, while collecting detailed diagnostics for failures.
results, failures = ingestor.ingest(show_progress=True, return_failures=True)
# Return only successes
# results = ingestor.ingest(show_progress=True)
t1 = time.time()
print(f"Total time: {t1 - t0} seconds")
# results blob is directly inspectable
if results:
print(ingest_json_results_to_blob(results[0]))
# (optional) Review any failures that were returned
if failures:
print(f"There were {len(failures)} failures. Sample: {failures[0]}")You can see the extracted text that represents the content of the ingested test document.
Starting ingestion..
Total time: 9.243880033493042 seconds
TestingDocument
A sample document with headings and placeholder text
Introduction
This is a placeholder document that can be used for any purpose. It contains some
headings and some placeholder text to fill the space. The text is not important and contains
no real value, but it is useful for testing. Below, we will have some simple tables and charts
that we can use to confirm Ingest is working as expected.
Table 1
This table describes some animals, and some activities they might be doing in specific
locations.
Animal Activity Place
Gira@e Driving a car At the beach
Lion Putting on sunscreen At the park
Cat Jumping onto a laptop In a home o@ice
Dog Chasing a squirrel In the front yard
Chart 1
This chart shows some gadgets, and some very fictitious costs.
... document extract continues ...To query for relevant snippets of the ingested content, and use them with an LLM to generate answers, use the following code.
import os
from openai import OpenAI
from nv_ingest_client.util.milvus import nvingest_retrieval
milvus_uri = "milvus.db"
collection_name = "test"
sparse = False
queries = ["Which animal is responsible for the typos?"]
retrieved_docs = nvingest_retrieval(
queries,
collection_name,
milvus_uri=milvus_uri,
hybrid=sparse,
top_k=1,
)
# simple generation example
extract = retrieved_docs[0][0]["entity"]["text"]
client = OpenAI(
base_url="https://integrate.api.nvidia.com/v1",
api_key=os.environ["NVIDIA_API_KEY"],
)
prompt = f"Using the following content: {extract}\n\n Answer the user query: {queries[0]}"
print(f"Prompt: {prompt}")
completion = client.chat.completions.create(
model="nvidia/llama-3.1-nemotron-70b-instruct",
messages=[{"role": "user", "content": prompt}],
)
response = completion.choices[0].message.content
print(f"Answer: {response}")Prompt: Using the following content: Table 1
| This table describes some animals, and some activities they might be doing in specific locations. | This table describes some animals, and some activities they might be doing in specific locations. | This table describes some animals, and some activities they might be doing in specific locations. |
| Animal | Activity | Place |
| Giraffe | Driving a car | At the beach |
| Lion | Putting on sunscreen | At the park |
| Cat | Jumping onto a laptop | In a home office |
| Dog | Chasing a squirrel | In the front yard |
Answer the user query: Which animal is responsible for the typos?
Answer: A clever query!
Based on the provided Table 1, I'd make an educated inference to answer your question. Since the activities listed are quite unconventional for the respective animals (e.g., a giraffe driving a car, a lion putting on sunscreen), it's likely that the table is using humor or hypothetical scenarios.
Given this context, the question "Which animal is responsible for the typos?" is probably a tongue-in-cheek inquiry, as there's no direct information in the table about typos or typing activities.
However, if we were to make a playful connection, we could look for an animal that's:
1. Typically found in a setting where typing might occur (e.g., an office).
2. Engaging in an activity that could potentially lead to typos (e.g., interacting with a typing device).
Based on these loose criteria, I'd jokingly point to:
**Cat** as the potential culprit, since it's:
* Located "In a home office"
* Engaged in "Jumping onto a laptop", which could theoretically lead to accidental keystrokes or typos if the cat were to start "walking" on the keyboard!
Please keep in mind that this response is purely humorous and interpretative, as the table doesn't explicitly mention typos or provide a straightforward answer to the question.Tip
Beyond inspecting the results, you can read them into things like llama-index or langchain retrieval pipelines.
Please also checkout our demo using a retrieval pipeline on build.nvidia.com to query over document content pre-extracted w/ NVIDIA Ingest.
The following is a description of the folders in the GitHub repository.
- .devcontainer — VSCode containers for local development
- .github — GitHub repo configuration files
- api — Core API logic shared across python modules
- ci — Scripts used to build the nv-ingest container and other packages
- client — Readme, examples, and source code for the nv-ingest-cli utility
- conda — Conda environment and packaging definitions
- config — Various .yaml files defining configuration for OTEL, Prometheus
- data — Sample PDFs for testing
- deploy — Brev.dev-hosted launchable
- docker — Scripts used by the nv-ingest docker container
- docs — Documentation for NV Ingest
- evaluation — Notebooks that demonstrate how to test recall accuracy
- examples — Notebooks, scripts, and tutorial content
- helm — Documentation for deploying nv-ingest to a Kubernetes cluster via Helm chart
- skaffold — Skaffold configuration
- src — Source code for the nv-ingest pipelines and service
- tests — Unit tests for nv-ingest
If configured to do so, this project will download and install additional third-party open source software projects. Review the license terms of these open source projects before use:
https://pypi.org/project/pdfservices-sdk/
INSTALL_ADOBE_SDK:- Description: If set to
true, the Adobe SDK will be installed in the container at launch time. This is required if you want to use the Adobe extraction service for PDF decomposition. Please review the license agreement for the pdfservices-sdk before enabling this option.
- Description: If set to
DOWNLOAD_LLAMA_TOKENIZER(Built With Llama)::- Description: The Split task uses the
meta-llama/Llama-3.2-1Btokenizer, which will be downloaded from HuggingFace at build time ifDOWNLOAD_LLAMA_TOKENIZERis set toTrue. Please review the license agreement for Llama 3.2 materials before using this. This is a gated model so you'll need to request access and setHF_ACCESS_TOKENto your HuggingFace access token in order to use it.
- Description: The Split task uses the
We require that all contributors "sign-off" on their commits. This certifies that the contribution is your original work, or you have rights to submit it under the same license, or a compatible license.
Any contribution which contains commits that are not signed off are not accepted.
To sign off on a commit, use the --signoff (or -s) option when you commit your changes as shown following.
$ git commit --signoff --message "Add cool feature."
This appends the following text to your commit message.
Signed-off-by: Your Name <[email protected]>
The following is the full text of the Developer Certificate of Origin (DCO)
Developer Certificate of Origin
Version 1.1
Copyright (C) 2004, 2006 The Linux Foundation and its contributors.
1 Letterman Drive
Suite D4700
San Francisco, CA, 94129
Everyone is permitted to copy and distribute verbatim copies of this license document, but changing it is not allowed.
Developer's Certificate of Origin 1.1
By making a contribution to this project, I certify that:
(a) The contribution was created in whole or in part by me and I have the right to submit it under the open source license indicated in the file; or
(b) The contribution is based upon previous work that, to the best of my knowledge, is covered under an appropriate open source license and I have the right under that license to submit that work with modifications, whether created in whole or in part by me, under the same open source license (unless I am permitted to submit under a different license), as indicated in the file; or
(c) The contribution was provided directly to me by some other person who certified (a), (b) or (c) and I have not modified it.
(d) I understand and agree that this project and the contribution are public and that a record of the contribution (including all personal information I submit with it, including my sign-off) is maintained indefinitely and may be redistributed consistent with this project or the open source license(s) involved.
- NeMo Retriever Extraction doesn't generate any code that may require sandboxing.
- NeMo Retriever Extraction is shared as a reference and is provided "as is". The security in the production environment is the responsibility of the end users deploying it. When deploying in a production environment, please have security experts review any potential risks and threats; define the trust boundaries, implement logging and monitoring capabilities, secure the communication channels, integrate AuthN & AuthZ with appropriate access controls, keep the deployment up to date, ensure the containers/source code are secure and free of known vulnerabilities.
- A frontend that handles AuthN & AuthZ should be in place as missing AuthN & AuthZ could provide ungated access to customer models if directly exposed to e.g. the internet, resulting in either cost to the customer, resource exhaustion, or denial of service.
- NeMo Retriever Extraction doesn't require any privileged access to the system.
- The end users are responsible for ensuring the availability of their deployment.
- The end users are responsible for building the container images and keeping them up to date.
- The end users are responsible for ensuring that OSS packages used by the developer blueprint are current.
- The logs from nginx proxy, backend, and demo app are printed to standard out. They can include input prompts and output completions for development purposes. The end users are advised to handle logging securely and avoid information leakage for production use cases.