generated from databricks-industry-solutions/industry-solutions-blueprints
-
Notifications
You must be signed in to change notification settings - Fork 2
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
Showing
11 changed files
with
976 additions
and
566 deletions.
There are no files selected for viewing
This file was deleted.
Oops, something went wrong.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,236 @@ | ||
| # Databricks notebook source | ||
| # MAGIC %md | ||
| # MAGIC ## Parsing CSRD directive | ||
| # MAGIC In this section, we aim at programmatically extracting chapters / articles / paragraphs from the [CSRD document](https://eur-lex.europa.eu/legal-content/EN/TXT/HTML/?uri=CELEX:02013L0034-20240109&qid=1712714544806) (available as HTML) and provide users with solid data foundations to build advanced generative AI applications in the context of regulatory compliance. | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %run ./config/00_environment | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| import requests | ||
| act_url = 'https://eur-lex.europa.eu/legal-content/EN/TXT/HTML/?uri=CELEX:02013L0034-20240109&qid=1712714544806' | ||
| html_page = requests.get(act_url).text | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC We may apply different data strategies to extract chapters and articles from the CSRD directive. The simplest approach would be to extract raw content and extract chunks that could feed a vector database. Whilst this approach would certainly be the easiest route (and often times the preferred option for less material use cases), it might contribute to the notion of hallucination since most large language model would be tempted to "infer" missing words and generate content not 100% in line with regulatory articles, chapters and paragraphs. A model making new regulations up is probably something one may need to avoid... | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC Instead, we went down the "boring" and "outdated" approach of scraping documents manually. We assume that the efforts done upfront will pay off at a later stage when extracting facts around specific chapters, articles, paragraphs and citations. For that purpose, we make use of the [Beautiful soup](https://beautiful-soup-4.readthedocs.io/en/latest/) library against our HTML content that we previously inspected through a browser / developer tool as per screenshot below. | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC 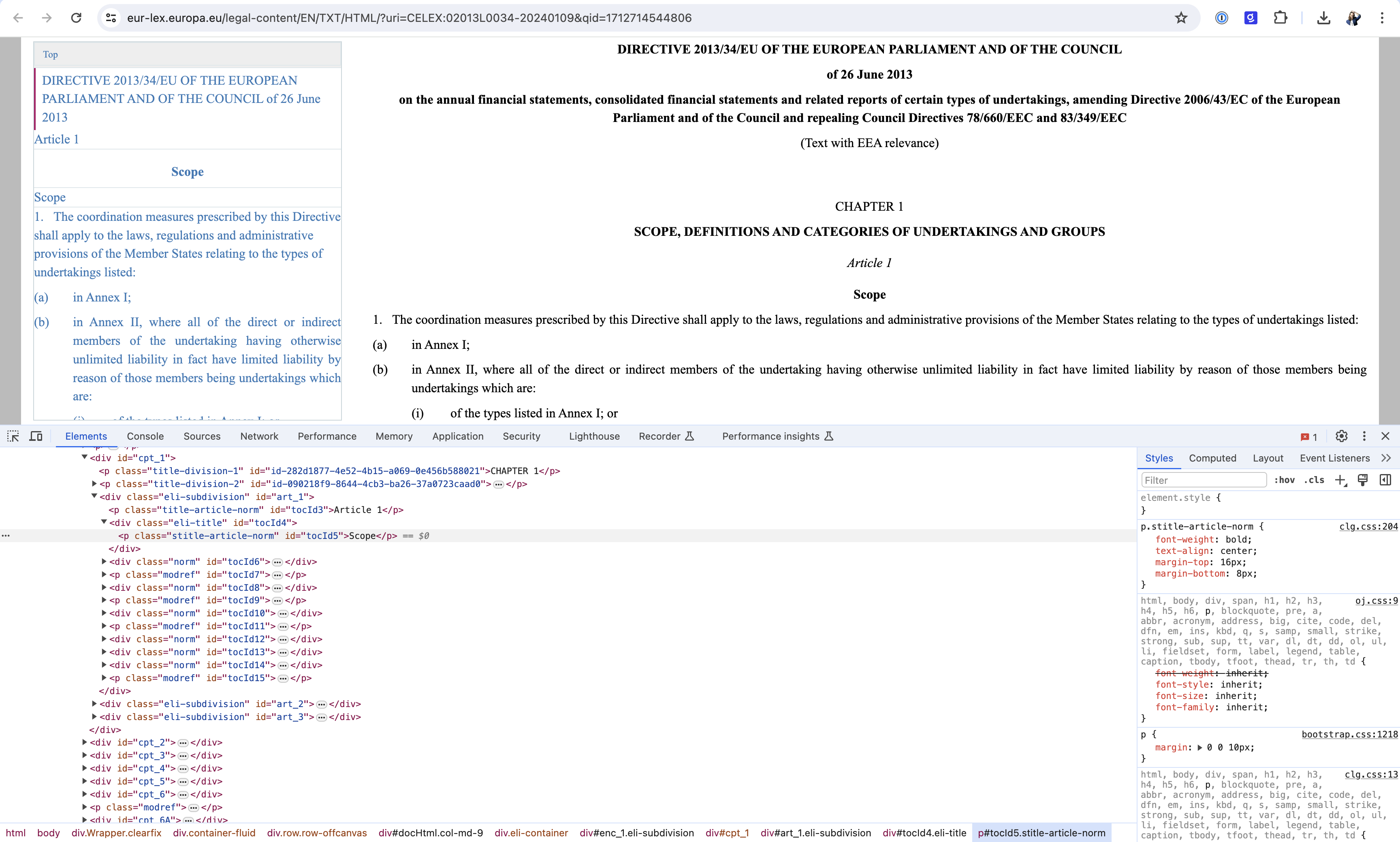 | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC Relatively complex, we could still observe delimiter tags / classes used in our scraping logic. | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| def get_directive_section(main_content): | ||
| return main_content.find('div', {'class': 'eli-main-title'}) | ||
|
|
||
| def get_content_section(main_content): | ||
| return main_content.find('div', {'class': 'eli-subdivision'}) | ||
|
|
||
| def get_chapter_sections(content_section): | ||
| return content_section.find_all('div', recursive=False) | ||
|
|
||
| def get_article_sections(chapter_section): | ||
| return chapter_section.find_all('div', {'class': 'eli-subdivision'}, recursive=False) | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| def get_directive_name(directive_section) -> str: | ||
| title_doc = directive_section.find_all('p', {'class': 'title-doc-first'}) | ||
| title_doc = ' '.join([t.text.strip() for t in title_doc]) | ||
| return title_doc | ||
|
|
||
| def get_chapter_name(chapter_section) -> str: | ||
| return chapter_section.find('p', {'class': 'title-division-2'}).text.strip().capitalize() | ||
|
|
||
| def get_chapter_id(chapter_section) -> str: | ||
| chapter_id = chapter_section.find('p', {'class': 'title-division-1'}).text.strip() | ||
| chapter_id = chapter_id.replace('CHAPTER', '').strip() | ||
| return chapter_id | ||
|
|
||
| def get_article_name(article_section) -> str: | ||
| return article_section.find('p', {'class': 'stitle-article-norm'}).text.strip() | ||
|
|
||
| def get_article_id(article_section) -> str: | ||
| article_id = article_section.find('p', {'class': 'title-article-norm'}).text.strip() | ||
| article_id = re.sub('\"?Article\s*', '', article_id).strip() | ||
| return article_id | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| from bs4.element import Tag | ||
| import re | ||
|
|
||
| def _clean_paragraph(txt): | ||
| # remove multiple break lines | ||
| txt = re.sub('\n+', '\n', txt) | ||
| # simplifies bullet points | ||
| txt = re.sub('(\([\d\w]+\)\s?)\n', r'\1\t', txt) | ||
| # simplifies quote | ||
| txt = re.sub('‘', '\'', txt) | ||
| # some weird references to other articles | ||
| txt = re.sub('\(\\n[\d\w]+\n\)', '', txt) | ||
| # remove spaces before punctuation | ||
| txt = re.sub(f'\s([\.;:])', r'\1', txt) | ||
| # remove reference links | ||
| txt = re.sub('▼\w+\n', '', txt) | ||
| # format numbers | ||
| txt = re.sub('(?<=\d)\s(?=\d)', '', txt) | ||
| # remove consecutive spaces | ||
| txt = re.sub('\s{2,}', ' ', txt) | ||
| # remove leading / trailing spaces | ||
| txt = txt.strip() | ||
| return txt | ||
|
|
||
| def get_paragraphs(article_section): | ||
| content = {} | ||
| paragraph_number = '0' | ||
| paragraph_content = [] | ||
| for child in article_section.children: | ||
| if isinstance(child, Tag): | ||
| if 'norm' in child.attrs.get('class'): | ||
| if child.name == 'p': | ||
| paragraph_content.append(child.text.strip()) | ||
| elif child.name == 'div': | ||
| content[paragraph_number] = _clean_paragraph('\n'.join(paragraph_content)) | ||
| paragraph_number = child.find('span', {'class': 'no-parag'}).text.strip().split('.')[0] | ||
| paragraph_content = [child.find('div', {'class': 'inline-element'}).text] | ||
| elif 'grid-container' in child.attrs.get('class'): | ||
| paragraph_content.append(child.text) | ||
| content[paragraph_number] = _clean_paragraph('\n'.join(paragraph_content)) | ||
| return {k:v for k, v in content.items() if len(v) > 0} | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC Finally, we could extract the full content hierarchy from the CSRD directive, from chapter to articles and paragraph. | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| from bs4 import BeautifulSoup | ||
|
|
||
| main_content = BeautifulSoup(html_page, 'html.parser') | ||
| directive_section = get_directive_section(main_content) | ||
| directive_name = get_directive_name(directive_section) | ||
| content_section = get_content_section(main_content) | ||
|
|
||
| for chapter_section in get_chapter_sections(content_section): | ||
| chapter_id = get_chapter_id(chapter_section) | ||
| chapter_name = get_chapter_name(chapter_section) | ||
| articles = len(get_article_sections(chapter_section)) | ||
| print(f'Chapter {chapter_id}: {chapter_name}') | ||
| print(f'{articles} article(s)') | ||
| print('') | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC ## Knowledge graph | ||
| # MAGIC Our content follows a tree structure where each chapter has multiple articles and each article has multiple paragraphs. A graph structure becomes a logical representation of our data. Let's create 2 dataframes representing both our nodes (article content) and edges (relationships). | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| import pandas as pd | ||
|
|
||
| nodes = [] | ||
| edges = [] | ||
|
|
||
| # root node, starting with Id 0 | ||
| nodes.append(['0', 'CSRD', directive_name, 'DIRECTIVE']) | ||
|
|
||
| for chapter_section in get_chapter_sections(content_section): | ||
|
|
||
| chapter_id = get_chapter_id(chapter_section) | ||
| chapter_name = get_chapter_name(chapter_section) | ||
|
|
||
| # level 1, chapter | ||
| # chapters are included in root node | ||
| nodes.append([ chapter_id, f'Chapter {chapter_id}', chapter_name, 'CHAPTER']) | ||
| edges.append(['0', f'{chapter_id}', 'CONTAINS']) | ||
|
|
||
| for article_section in get_article_sections(chapter_section): | ||
| article_id = get_article_id(article_section) | ||
| article_name = get_article_name(article_section) | ||
| article_paragraphs = get_paragraphs(article_section) | ||
|
|
||
| # level 2, article | ||
| # articles are included in chapters | ||
| nodes.append([f'{chapter_id}.{article_id}', f'Article {article_id}', article_name, 'ARTICLE']) | ||
| edges.append([chapter_id, f'{chapter_id}.{article_id}', 'CONTAINS']) | ||
|
|
||
| for paragraph_id, paragraph_text in article_paragraphs.items(): | ||
|
|
||
| # level 3, paragraph | ||
| # paragraphs are included in articles | ||
| nodes.append([f'{chapter_id}.{article_id}.{paragraph_id}', f'Article {article_id}({paragraph_id})', paragraph_text, 'PARAGRAPH']) | ||
| edges.append([f'{chapter_id}.{article_id}', f'{chapter_id}.{article_id}.{paragraph_id}', 'CONTAINS']) | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| nodes_df = pd.DataFrame(nodes, columns=['id', 'label', 'content', 'group']) | ||
| edges_df = pd.DataFrame(edges, columns=['src', 'dst', 'label']) | ||
| display(nodes_df) | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| _ = spark.createDataFrame(nodes_df).write.format('delta').mode('overwrite').saveAsTable(table_nodes) | ||
| _ = spark.createDataFrame(edges_df).write.format('delta').mode('overwrite').saveAsTable(table_edges) | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC Physically stored as delta tables, the same can be rendered in memory and visualized through the [NetworkX](https://networkx.org/) libary. | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| nodes_df = spark.read.table(table_nodes).toPandas() | ||
| edges_df = spark.read.table(table_edges).toPandas() | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| import networkx as nx | ||
|
|
||
| CSRD = nx.DiGraph() | ||
|
|
||
| for i, n in nodes_df.iterrows(): | ||
| CSRD.add_node(n['id'], label=n['label'], title=n['content'], group=n['group']) | ||
|
|
||
| for i, e in edges_df.iterrows(): | ||
| if e['label'] == 'CONTAINS': | ||
| CSRD.add_edge(e['src'], e['dst'], label=e['label']) | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC Our directive contains ~ 350 nodes where each node is connected to maximum 1 parent (this is expected from a tree structure), as represented below. Feel free to Zoom in and out, hovering some nodes to read their actual text content. | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| from scripts.graph import displayGraph | ||
| displayHTML(displayGraph(CSRD)) | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC Please note that we carefully designed our graph unique identifiers so that one can access a given paragraph through a simple reference, expressed in the form of `[chapter-article-paragraph]` coordinate (e.g. `3.9.7`). | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| from scripts.html_output import * | ||
| p_id = '3.9.7' | ||
| p = CSRD.nodes[p_id] | ||
| displayHTML(article_html(p['label'], p['title'])) |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,160 @@ | ||
| # Databricks notebook source | ||
| # MAGIC %md | ||
| # MAGIC ## Indexing content | ||
| # MAGIC Though representing our CSRD directive as a graph was visually compelling, it offers no semantic search capability. In this section, we will further index our graph data to offer search functionality for a given question / query that can be combined with Generative AI capabilities as part of a Retrieval Augmented Generation strategy (RAG). | ||
| # MAGIC | ||
| # MAGIC In production settings, we highly encourage users to leverage [Databricks vector store](https://docs.databricks.com/en/generative-ai/vector-search.html) capability, linking records to the actual binary file that may have been previously stored on your volume, hence part of a unified governance strategy. | ||
| # MAGIC | ||
| # MAGIC However, in the context of a solution accelerator, we decided to limit the infrastructure requirements (volume creation, table creation, DLT pipelines, etc.) and provide entry level capabilities, in memory, leveraging FAISS as our de facto vector store and leveraging out-of-the-box foundation models provided by Databricks. | ||
| # MAGIC | ||
| # MAGIC For more information on E2E applications, please refer to [DB Demo](https://www.databricks.com/resources/demos/tutorials/data-science-and-ai/lakehouse-ai-deploy-your-llm-chatbot). | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %run ./config/00_environment | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| nodes_df = spark.read.table(table_nodes).toPandas() | ||
| display(nodes_df) | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC We leverage the langchain framework for its ease of use, converting our graph content as a set of langchain documents that we can index on our vector store. Please note that chunking is no longer required since we have been able to extract granular information (at a paragraph level) in our previous notebook. | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
|
|
||
| from langchain.docstore.document import Document | ||
|
|
||
| documents = [] | ||
| for i, n in nodes_df[nodes_df['group'] == 'PARAGRAPH'].iterrows(): | ||
| metadata=n.to_dict() | ||
| page_content=metadata['content'] | ||
| del metadata['content'] | ||
| documents.append(Document( | ||
| page_content=page_content, | ||
| metadata=metadata | ||
| )) | ||
|
|
||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC For our embedding strategy, we will leverage foundational models provided out of the box through your development workspace environment. | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| from langchain_community.embeddings import DatabricksEmbeddings | ||
| embeddings = DatabricksEmbeddings(endpoint="databricks-bge-large-en") | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| from langchain.vectorstores import FAISS | ||
| CSRD_search = FAISS.from_documents(documents=documents, embedding=embeddings) | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC We can now retrieve content based on similarity search. Given a question, part of a text, or simple keywords, we retrieve specific facts, articles and paragraph that we can trace back to our CSRD directive. This will become the foundation to our RAG strategy later, answering specific regulatory compliance questions by citing existing chapters and articles. The example below returns the best matching paragraph with a relevance score given a user question. | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| from scripts.html_output import * | ||
|
|
||
| question = '''disclosing transactions between related parties, | ||
| transactions between related parties included in a consolidation | ||
| that are eliminated on consolidation shall not be included''' | ||
|
|
||
| search, score = CSRD_search.similarity_search_with_relevance_scores(question)[0] | ||
| displayHTML(vector_html(search.metadata['label'], search.page_content, '{}%'.format(int(score * 100)))) | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC We serialize our vector store for future use. Once again, we highly recommend leveraging vector store capability instead of local FAISS used for demo purpose only. | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| CSRD_search.save_local(faiss_output_dir) | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC ## Foundation model | ||
| # MAGIC It is expected that foundational models like DBRX, Llama 3, Mixtral or OpenAI that learned knowledge from trillion of tokens may already have acquired some knowledge for generic questions related to the CSRD directive. However, it would be cavalier to ask specific questions to our foundation model and solely rely on its built-in knowledge we could not link to exact articles. In the context of regulatory compliance, we cannot afford for a model to return information as "best guess", regardless of how convincing its answer might be. We leverage DBRX model (provided as a service on your databricks workspace). | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| from langchain_community.chat_models import ChatDatabricks | ||
| chat_model = ChatDatabricks(endpoint="databricks-dbrx-instruct", max_tokens = 300, temperature=0) | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| query = 'Which disclosures will be subject to assurance, and what level of assurance is required?' | ||
| displayHTML(llm_html(query, chat_model.invoke(query).content)) | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC The question above might yield a well formed answer that may seem convincing to the naked eye, but definitely lacks substance and sound quality required for regulatory compliance. Instead of an output we may have to take at its face value, it would certainly be more comfortable to cite actual facts and cross references to existing articles / paragraphs from a trusted source (such as the directive itself). | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC | ||
| # MAGIC ## RAG strategy | ||
| # MAGIC Instead, we should prompt a model to search for specific knowledge, knowledge that we acquired throughout the first part of our notebook. This creates our foundation for RAG. In this section, we will design a simple prompt and "chain" our vector store logic with actual model inference. | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| from langchain.chains import RetrievalQA | ||
| from langchain.prompts import PromptTemplate | ||
| from langchain_community.chat_models import ChatDatabricks | ||
| import json | ||
|
|
||
| TEMPLATE = """ | ||
| Context information is below. | ||
| --------------------- | ||
| {context} | ||
| --------------------- | ||
| Given the context information and not prior knowledge. | ||
| Answer compliance issue related to the CSRD directive only. | ||
| If the question is not related to regulatory compliance, kindly decline to answer. | ||
| If you don't know the answer, just say that you don't know, don't try to make up an answer. | ||
| Keep the answer as concise as possible, citing articles and chapters whenever applicable. | ||
| Please do not repeat the answer and do not add any additional information. | ||
| Question: {question} | ||
| Answer: | ||
| """ | ||
| prompt = PromptTemplate(template=TEMPLATE, input_variables=["context", "question"]) | ||
|
|
||
| chain = RetrievalQA.from_chain_type( | ||
| llm=chat_model, | ||
| chain_type="stuff", | ||
| retriever=CSRD_search.as_retriever(), | ||
| chain_type_kwargs={"prompt": prompt}, | ||
| return_source_documents = True | ||
| ) | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC Prompts must be designed in a way that provides user with better clarity and / or confidence as well as safeguarding model against malicious or offensive use. This, however, is not part of this solution. We invite users to explore our [DB Demo](https://www.databricks.com/resources/demos/tutorials/data-science-and-ai/lakehouse-ai-deploy-your-llm-chatbot) that covers the basics to most advanced use of RAG and prompt engineering. A good example would be to restrict our model to only answer questions that are CSRD related, possibly linking multiple chains together. | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| question = {"query": 'Which disclosures will be subject to assurance, and what level of assurance is required?'} | ||
| answer = chain.invoke(question) | ||
| displayHTML(rag_html(question['query'], answer['result'], answer['source_documents'])) | ||
|
|
||
| # COMMAND ---------- | ||
|
|
||
| # MAGIC %md | ||
| # MAGIC In this simple example, we have let our model formulate a point of view based on actual facts we know we can trust. For the purpose of that demo, we represented output as a form of an HTML notebook. In real life scenario, we should offer that capability as a chat interface to a set of non technologist user, hence requiring building a UI and application server outside of a notebook based environment (outside of the scope here). |
Oops, something went wrong.