-
Notifications
You must be signed in to change notification settings - Fork 2
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Updated README.md to better match index.rst. Also, updated link to at…
…las images
- Loading branch information
1 parent
3a16166
commit 5cddc6c
Showing
11 changed files
with
115 additions
and
66 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,42 +1,90 @@ | ||
|  | ||
|
|
||
| ## UNRAVEL is a command line tool for: | ||
| * Voxel-wise analysis of fluorescent signals (e.g., c-Fos immunofluorescence) across the mouse brains in atlas space | ||
| * Validation of hot/cold spots via cell/label density quantification at cellular resolution | ||
|
|
||
| ## Publications: | ||
| * UNRAVELing the synergistic effects of psilocybin and environment on brain-wide immediate early gene expression in mice | ||
| * [Neuropsychopharmacology](https://www.nature.com/articles/s41386-023-01613-4) | ||
|
|
||
| ## IF/iDISCO+/LSFM guide: | ||
| [Heifets lab guide to immunofluorescence staining, iDISCO+, & lightsheet fluorescence microscopy](https://docs.google.com/document/d/16yowBhiBQWz8_VX2t9Rf6Xo3Ub4YPYD6qeJP6vJo6P4/edit?usp=sharing) | ||
|
|
||
| ## UNRAVEL guide: | ||
| * Example workflow for region-wise and voxel-wise c-Fos+ cell densities: | ||
| * prep_reg.py (To see help, run: prep_reg.py -h) | ||
| * copy_tifs.py | ||
| * brain_mask.py | ||
| * reg.py | ||
| * check_reg.py | ||
| * regional_cell_densities.py | ||
| * regional_cell_densities_summary.py | ||
| * prep_vstats.py | ||
| * z-score.py | ||
| * aggregate_files_from_sample_dirs.py | ||
| * whole_to_LR_avg.py | ||
| * vstats.py | ||
| * fdr_range.py | ||
| * fdr.py | ||
| * recursively_mirror_rev_cluster_indices.py | ||
| * ilastik_pixel_classification.py | ||
| * validate_clusters.py | ||
| * valid_clusters_summary.py | ||
|
|
||
| \ | ||
| Please send questions/suggestions to: | ||
| * Daniel Ryskamp Rijsketic ([email protected]) | ||
| * Austen Casey ([email protected]) | ||
| * Boris Heifets ([email protected]) | ||
|
|
||
| \ | ||
| For command line interface help, please review [Unix tutorials](https://andysbrainbook.readthedocs.io/en/latest/index.html) | ||
| [](https://b-heifets.github.io/UNRAVEL/) | ||
|
|
||
|
|
||
| <style> | ||
| video { | ||
| border: none; | ||
| outline: none; | ||
| } | ||
| </style> | ||
|
|
||
| <div style="display: flex; justify-content: space-between;"> | ||
| <div style="flex: 1; margin-right: 10px;"> | ||
| <a href="https://www.nature.com/articles/s41386-023-01613-4" target="_blank"> | ||
| <video width="100%" autoplay muted playsinline> | ||
| <source src="https://b-heifets.github.io/UNRAVEL/_static/psilocybin_up_valid_clusters.mp4" type="video/mp4"> | ||
| Your browser does not support the video tag. | ||
| </video> | ||
| </a> | ||
| </div> | ||
| <div style="flex: 1; margin-left: 10px;"> | ||
| <a href="https://www.nature.com/articles/s41386-023-01613-4" target="_blank"> | ||
| <video width="100%" autoplay muted playsinline> | ||
| <source src="https://b-heifets.github.io/UNRAVEL/_static/psilocybin_up_sunburst.mp4" type="video/mp4"> | ||
| Your browser does not support the video tag. | ||
| </video> | ||
| </a> | ||
| </div> | ||
| </div> | ||
|
|
||
| ### UN-biased high-Resolution Analysis and Validation of Ensembles using Light sheet images | ||
| * UNRAVEL is a [Python](https://www.python.org/) package & command line tool for the analysis of brain-wide imaging data, automating: | ||
| * Registration of brain-wide images to a common atlas space | ||
| * Quantification of cell/label densities across the brain | ||
| * Voxel-wise analysis of fluorescent signals and cluster correction | ||
| * Validation of hot/cold spots via cell/label density quantification at cellular resolution | ||
| * [UNRAVEL GitHub repository](https://github.com/b-heifets/UNRAVEL/tree/dev) | ||
| * [Initial UNRAVEL publication](https://www.nature.com/articles/s41386-023-01613-4) | ||
| * UNRAVEL was developed by [the Heifets lab](https://heifetslab.stanford.edu/) and [TensorAnalytics](https://sites.google.com/view/tensoranalytics/home?authuser=0) | ||
| * Additional support was provided by [the Shamloo lab](https://med.stanford.edu/neurosurgery/research/shamloo.html) | ||
|
|
||
|
|
||
|  | ||
|
|
||
| --- | ||
|
|
||
| ### *Please see [UNRAVEL documentation](https://b-heifets.github.io/UNRAVEL/) for guides on [installation](https://b-heifets.github.io/UNRAVEL/installation.html) and [anaysis](https://b-heifets.github.io/UNRAVEL/guide.html)* | ||
|
|
||
| --- | ||
|
|
||
| ### UNRAVEL visualizer | ||
| * [UNRAVEL visualizer](https://heifetslab-unravel.org/) is a web-based tool for visualizing and exploring 3D maps output from UNRAVEL | ||
| * [UNRAVEL visualizer GitHub repo](https://github.com/MetaCell/cfos-visualizer/) | ||
| * Developed by [MetaCell](https://metacell.us/) with support from the [Heifets lab](https://heifetslab.stanford.edu/) | ||
|
|
||
| 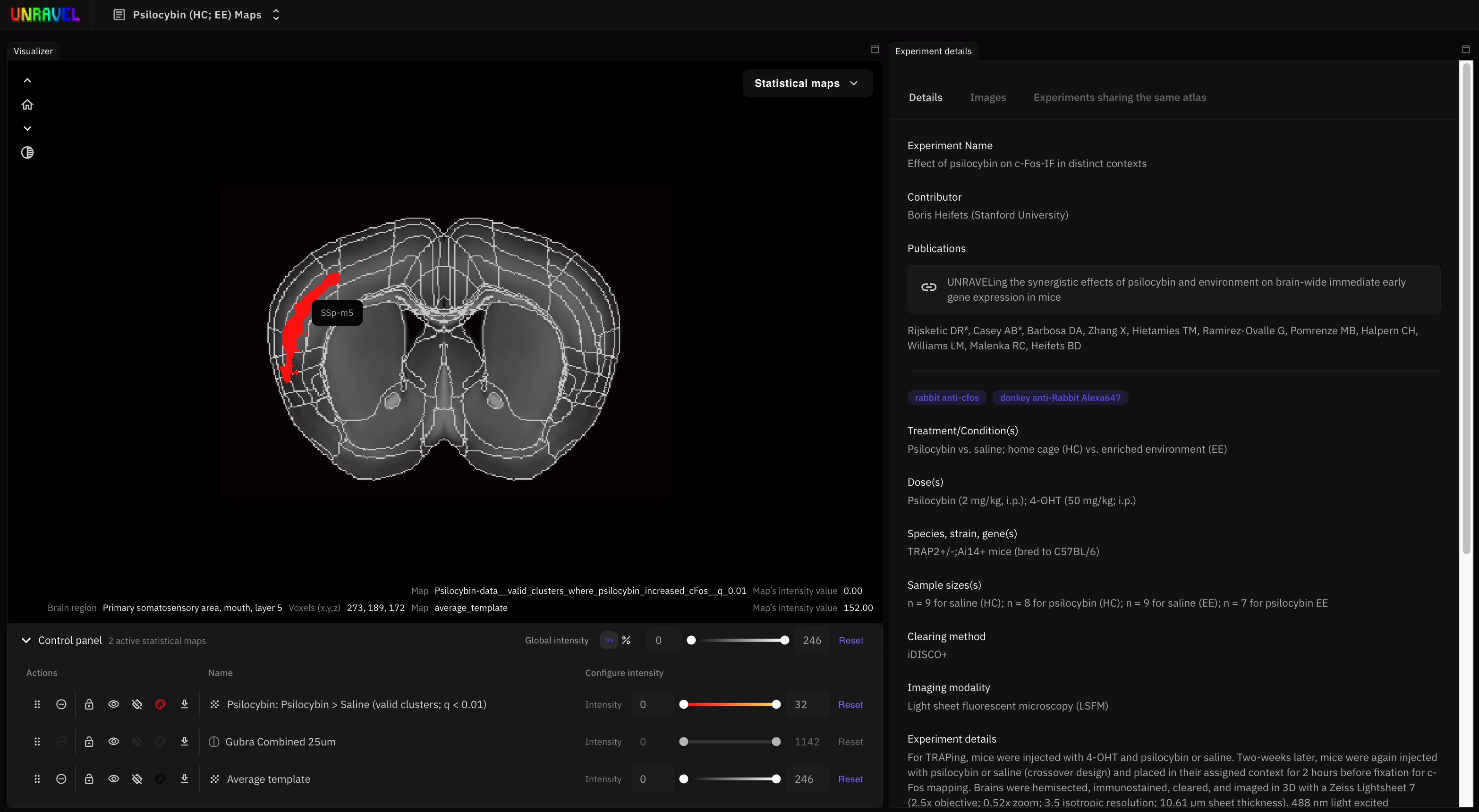 | ||
|
|
||
| ### Contact us | ||
| If you have any questions, suggestions, or are interested in collaborations and contributions, please reach out to us. | ||
|
|
||
| ### Developers | ||
| * **Daniel Ryskamp Rijsketic** (lead developer and maintainer) - [[email protected]](mailto:[email protected]) | ||
| * **Austen Casey** (developer) - [[email protected]](mailto:[email protected]) | ||
| * **MetaCell** (UNRAVEL visualizer developers) - [[email protected]](mailto:[email protected]) | ||
| * **Boris Heifets** (PI) - [[email protected]](mailto:[email protected]) | ||
|
|
||
| ### Additional contributions from | ||
| * **Mehrdad Shamloo** (PI) - [[email protected]](mailto:[email protected]) | ||
| * **Daniel Barbosa** (early contributer and guidance) - [[email protected]](mailto:[email protected]) | ||
| * **Wesley Zhao** (guidance) - [[email protected]](mailto:[email protected]) | ||
| * **Nick Gregory** (guidance) - [[email protected]](mailto:[email protected]) | ||
|
|
||
| ### Main dependencies | ||
| * [Allen Institute for Brain Science](https://portal.brain-map.org/) | ||
| * [FSL](https://fsl.fmrib.ox.ac.uk/fsl/fslwiki) | ||
| * [fslpy](https://git.fmrib.ox.ac.uk/fsl/fslpy) | ||
| * [ANTsPy](https://github.com/ANTsX/ANTsPy) | ||
| * [Ilastik](https://www.ilastik.org/) | ||
| * [nibabel](https://nipy.org/nibabel/) | ||
| * [numpy](https://numpy.org/) | ||
| * [scipy](https://www.scipy.org/) | ||
| * [pandas](https://pandas.pydata.org/) | ||
| * [cc3d](https://pypi.org/project/connected-components-3d/) | ||
| * Registration and warping workflows were inspired by [MIRACL](https://miracl.readthedocs.io/en/latest/) | ||
| * We adapted [LSFM/iDISCO+ atlases](https://pubmed.ncbi.nlm.nih.gov/33063286/) from [Gubra](https://www.gubra.dk/cro-services/3d-imaging/) | ||
|
|
||
| ### Support is welcome for | ||
| * Analysis of new datasets | ||
| * Development of new features | ||
| * Maintenance of the codebase | ||
| * Guidance of new users |
Binary file not shown.
Binary file not shown.
Binary file not shown.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -107,16 +107,16 @@ Developers | |
| * **Boris Heifets** (PI) - `[email protected] <mailto:[email protected]>`_ | ||
|
|
||
|
|
||
| Additional contributions from: | ||
| ------------------------------ | ||
| Additional contributions from | ||
| ----------------------------- | ||
| * **Mehrdad Shamloo** (PI) - `[email protected] <mailto:[email protected]>`_ | ||
| * **Daniel Barbosa** (early contributer and guidance) - `[email protected] <mailto:[email protected]>`_ | ||
| * **Wesley Zhao** (guidance) - `[email protected] <mailto:[email protected]>`_ | ||
| * **Nick Gregory** (guidance) - `[email protected] <mailto:[email protected]>`_ | ||
|
|
||
|
|
||
| Main dependencies: | ||
| ------------------------------------------------ | ||
| Main dependencies | ||
| ----------------- | ||
| * `Allen Institute for Brain Science <https://portal.brain-map.org/>`_ | ||
| * `FSL <https://fsl.fmrib.ox.ac.uk/fsl/fslwiki>`_ | ||
| * `fslpy <https://git.fmrib.ox.ac.uk/fsl/fslpy>`_ | ||
|
|
@@ -128,10 +128,10 @@ Main dependencies: | |
| * `pandas <https://pandas.pydata.org/>`_ | ||
| * `cc3d <https://pypi.org/project/connected-components-3d/>`_ | ||
| * Registration and warping workflows were inspired by `MIRACL <https://miracl.readthedocs.io/en/latest/>`_ | ||
| * We adapted `LSFM/iDISCO+ atlases <https://pubmed.ncbi.nlm.nih.gov/33063286/>`_ from `Gubra <https://www.gubra.dk/cro-services/3d-imaging/>`_ | ||
|
|
||
|
|
||
| Support is welcome for: | ||
| ----------------------- | ||
| Support is welcome for | ||
| ---------------------- | ||
| * Analysis of new datasets | ||
| * Development of new features | ||
| * Maintenance of the codebase | ||
|
|
||
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
Large diffs are not rendered by default.
Oops, something went wrong.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -107,16 +107,16 @@ Developers | |
| * **Boris Heifets** (PI) - `[email protected] <mailto:[email protected]>`_ | ||
|
|
||
|
|
||
| Additional contributions from: | ||
| ------------------------------ | ||
| Additional contributions from | ||
| ----------------------------- | ||
| * **Mehrdad Shamloo** (PI) - `[email protected] <mailto:[email protected]>`_ | ||
| * **Daniel Barbosa** (early contributer and guidance) - `[email protected] <mailto:[email protected]>`_ | ||
| * **Wesley Zhao** (guidance) - `[email protected] <mailto:[email protected]>`_ | ||
| * **Nick Gregory** (guidance) - `[email protected] <mailto:[email protected]>`_ | ||
|
|
||
|
|
||
| Main dependencies: | ||
| ------------------------------------------------ | ||
| Main dependencies | ||
| ----------------- | ||
| * `Allen Institute for Brain Science <https://portal.brain-map.org/>`_ | ||
| * `FSL <https://fsl.fmrib.ox.ac.uk/fsl/fslwiki>`_ | ||
| * `fslpy <https://git.fmrib.ox.ac.uk/fsl/fslpy>`_ | ||
|
|
@@ -128,10 +128,10 @@ Main dependencies: | |
| * `pandas <https://pandas.pydata.org/>`_ | ||
| * `cc3d <https://pypi.org/project/connected-components-3d/>`_ | ||
| * Registration and warping workflows were inspired by `MIRACL <https://miracl.readthedocs.io/en/latest/>`_ | ||
| * We adapted `LSFM/iDISCO+ atlases <https://pubmed.ncbi.nlm.nih.gov/33063286/>`_ from `Gubra <https://www.gubra.dk/cro-services/3d-imaging/>`_ | ||
|
|
||
|
|
||
| Support is welcome for: | ||
| ----------------------- | ||
| Support is welcome for | ||
| ---------------------- | ||
| * Analysis of new datasets | ||
| * Development of new features | ||
| * Maintenance of the codebase | ||
|
|
||
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters