SeqALib contains efficient implementation of sequence alignment algorithms from Bioinformatics.
Templated to allow for aligning of strings and custom objects by providing a matching function of your own design.
The algorithms currently provided by SeqALib are:
- Needleman-Wunsch
- Hirschberg
- Smith-Waterman
- Gotoh Local and Global
- Myers and Miller
- BLAST (pairwise)
- FOGSAA
- MUMMER
- Suffix Trees
AUTHOR: Rodrigo Rocha and Sean Stirling
See full example in the file: include/Test.cpp
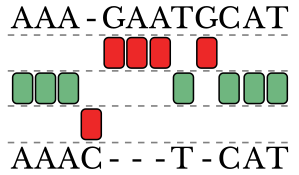
std::string seq1 = "AAAGAATGCAT";
std::string seq2 = "AAACTCAT";
NeedlemanWunschSA<std::string,char,'-'> SA(ScoringSystem(-1,2));
AlignedSequence<char,'-'> Alignment = SA.getAlignment(seq1,seq2);
// The resultng Alignment contains:
// AAA GAATGCAT
// ||| | |||
// AAAC T CATMultiple Sequence Alignment