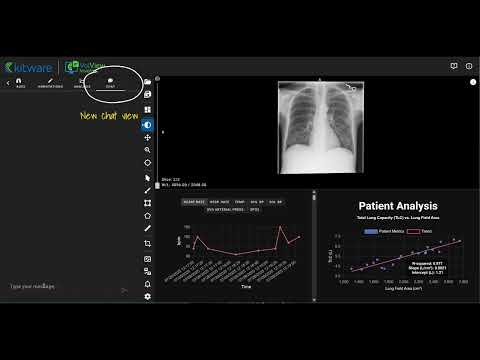
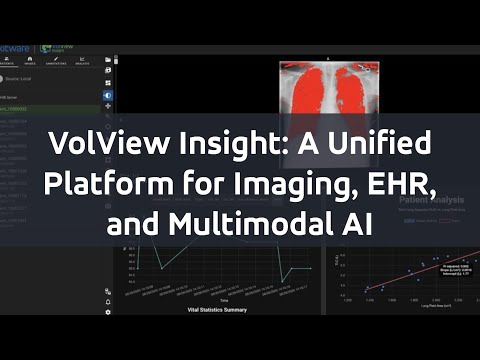
VolView Insight is an open-source platform that unifies medical imaging and clinical data into one extensible interface. It provides researchers with a realistic testbed for developing, testing, and extending multimodal models in settings that mirror clinical reality.
Highlights
- 🔬 Unified access to imaging data (DICOM/DICOMWeb) and clinical records (FHIR/EHR).
- 🧠 Extensible backend for AI/ML pipelines, including deep learning inference.
- ⚡ Web-based UI built on VolView.
- 🔌 Pluggable integrations — Orthanc, HAPI on FHIR, and MedGemma are examples. Users can bring their own DICOM/FHIR servers or multimodal pipelines.
Install the following:
- Node.js (use
nvmto manage versions) - Python ≥
3.10(tested with 3.10) - Poetry
2.1.2 - Docker
28.0.04 - A DICOMWeb server (e.g., Orthanc)
- A FHIR server (e.g., HAPI on FHIR)
git clone https://github.com/KitwareMedical/volview-insight.git
cd volview-insight
git submodule update --init# All platforms
cat ./core-volview-patches/VOLVIEW_BACKEND.patch | git -C core/VolView apply
# macOS only
cat ./core-volview-patches/MACOS_COMPATIBILITY.patch | git -C core/VolView applyYou can use any DICOMWeb server. Below are example setups for Orthanc.
# Run Orthanc with DICOMWeb plugin and NO authentication (dev only)
docker run --rm -p 8042:8042 -p 4242:4242 \
-e ORTHANC__AUTHENTICATION_ENABLED=false \
-e DICOM_WEB_PLUGIN_ENABLED=true \
orthancteam/orthancVerify at: http://localhost:8042/
- Download Orthanc: https://www.orthanc-server.com/static.php?page=download-mac
- Run
startOrthanc.command - Verify at:
http://localhost:8042/
You can use any FHIR R4 server. Example with SMART on FHIR HAPI image:
docker pull smartonfhir/hapi-5:r4-empty
docker run -dp 3000:8080 smartonfhir/hapi-5:r4-emptyVerify at: http://localhost:3000/hapi-fhir-jpaserver/fhir/Patient
The Python backend executes multimodal pipelines. You can extend it with your own.
cd volview-insight/server
poetry env use /path/to/bin/python3.10
poetry install
poetry run python -m volview_server -P 4014 -H 0.0.0.0 volview_insight_methods.pyIf you encounter issues, remove
-P 4014.
- MedGemma: Hugging Face gated models (requires account + access token).
- Segmentation: Example lung segmentation model:
curl https://data.kitware.com/api/v1/file/65bd8c2f03c3115909f73dd7/download --output segmentLungsModel-v1.0.ckptnvm use 18.17.1
npm install --force
npm run setup-project
npm run build
npm run previewNow open: http://localhost:4173/
cd volview-insight/orthanc-proxy
nvm use 23.10.0
npm install
npm run dev
⚠️ Must use Node.js 23.10.0 for the proxy. Runs at port5173.
- DICOMWeb: Replace Orthanc with your own server.
- FHIR/EHR: Replace HAPI with your own server.
- Pipelines: Extend the Python backend with your custom multimodal methods.
We welcome contributions!
- Add new pipelines or models
- Add integrations with other EHR/DICOM backends
- Report bugs and propose features
This software is provided solely for research and educational purposes.
It is a proof-of-concept research platform and is not intended for clinical use.
- This software has not been reviewed or approved by the U.S. Food and Drug Administration (FDA) or any other regulatory authority.
- It must not be used for diagnosis, treatment, or any clinical decision-making.
- No warranties or guarantees of performance, safety, or fitness for medical purposes are provided.
By using this software, you acknowledge that it is for non-clinical, investigational research only.
Apache 2.0 © Kitware