-
Notifications
You must be signed in to change notification settings - Fork 0
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
LDSC rg between FinnGen and UKB #27
Comments
|
We have computation started. While we can run this preliminary analysis, this approach is not scalable. There are 1801 FinnGen summary statistics. When focusing on This means > 1M (= 1801 * 580) comparison. Each of them takes ~ 5 min... |
|
A pilot analysis of 5000 comparisons is finished. It seems like some comparison dumped NA due to low N or h2 in the input trait. To reduce the number of comparisons, we can run LDSC h2 first for both UKB and FinnGen, drop phenotypes with NA as the h2 estimates, and impose a min h2 threshold. |
|
The heritability filter reduced the combinations. FinnGen: 1801 --> 1482
--> 20% reduction. |
|
171k comparison is done by now. |
|
295,616 / 775,086 by now. ~980 jobs are running using Sherlock's owners queue. Hopefully, we will have an initial results by the weekend. |
|
580,450/775,086 |
|
753,824/775,086 almost there |
|
This initial batch is now finished. Here we have some commands that I used to check the progress of the computation. 523 * 1482 = 775086 1-100000 |
|
Tabulate the results into a table
This would take some time... |
|
773,617 comparisons in It turned out that there are 775265 LDSC rg log files but some of them are empty files (presumably due to the owners job). |
|
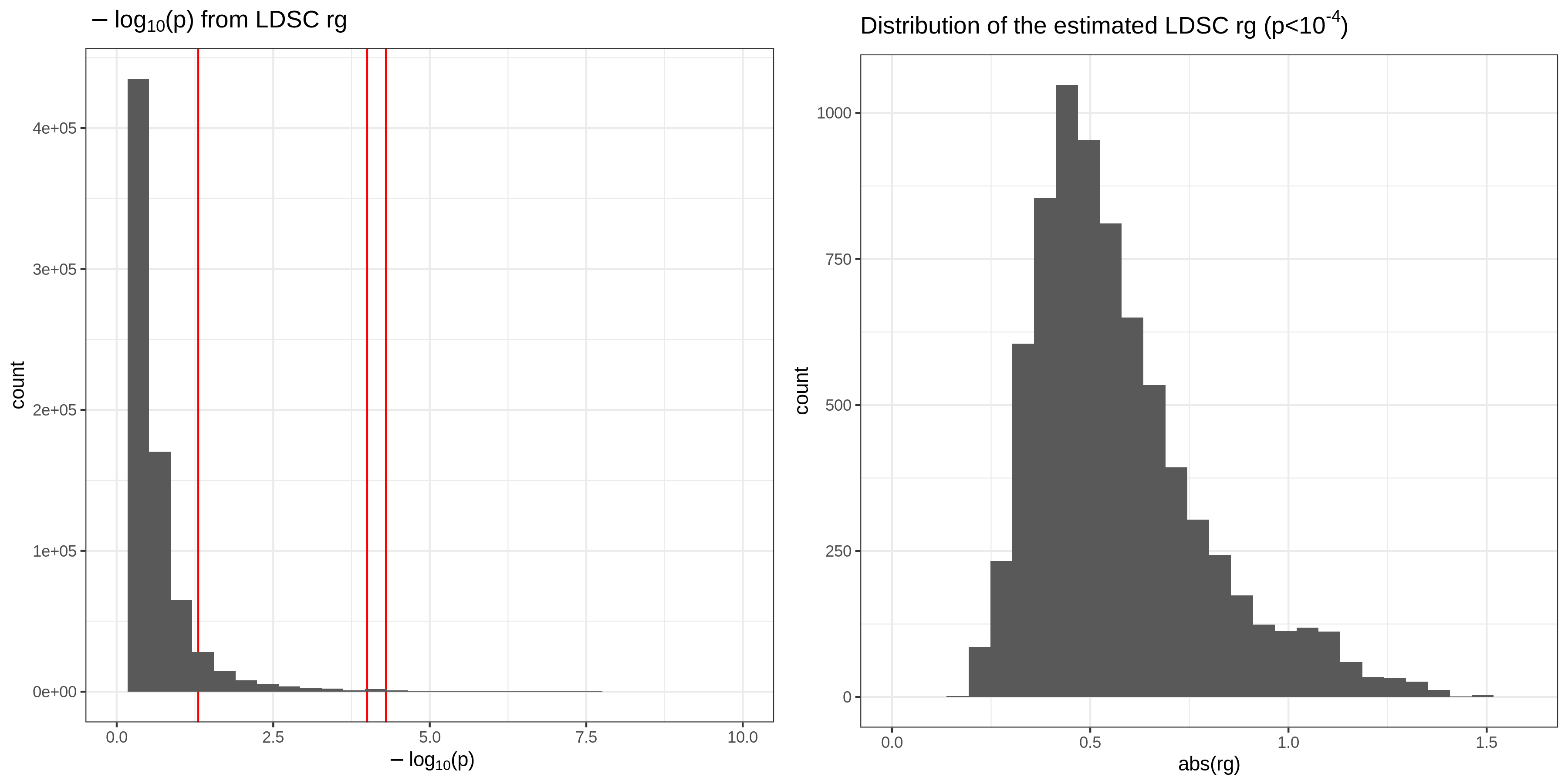
We computed the LDSC rg between FinnGen sumstats (estimated heritability > 0) and UKB WB sumstats (HC phenotypes with estimated heritability > 0). We investigated the distribution of p-value. Because there are ~580 UKB traits, we put p-value threshold of 1e-4 and focused on those significant associations. We also checked the distribution of rg. After imposing p < 1e-4 filter, there are 3,597 rg estimates across 282 FinnGen phenotypes and 158 UKB HC phenotypes. We sorted the table by FinnGen phenocode and p-value of rg and uploaded to a Google Spreadsheet. https://docs.google.com/spreadsheets/d/1ul4hr00KKZy0JRUW2ZW5-LORWEyeBCt7B3pAiNKRj5g/edit?usp=sharing Analysis repo: https://github.com/rivas-lab/ukbb-tools/tree/master/04_gwas/extras/finngen_r3#ldsc-rg |
|
To finish up the missing files (files of size 0), we resubmitted the jobs. |
|
Commands to check the empty files: |
|
Empty files were recomputed. |
|
Yet, we don't have comparison for cancer phenotypes. Let's compute rg for UKB cancer phenotypes. |
We don't have heritability estimates for cancer phenotypes. |
|
Let's make some progress on #21 first... |
update the LDSC rg analysis with cancer and BIN_FC phenotypes (and a few updated HC phenotypes)UKB GWAS is now updated. Let's generate the list of UKB (and Finngen) traits and see how many phenotypes do we have in This is a good increase from the previous run. We have With this, we have the input files for the rg computation. We updated the script and tested its behavior There are |
|
Let's check the jobs again. 1483 * 991 < 1470 * 1000 |
LDSC rg table (full)We used out scripts to aggregate LDSC rg results. This resulted in:
|
|
We computed the LDSC rg between FinnGen sumstats (estimated heritability > 0) and UKB WB sumstats (HC phenotypes with estimated heritability > 0). We investigated the distribution of p-value. Because there are ~990 UKB traits, we put a p-value threshold of 5e-5 and focused on those significant associations. After imposing |
|
The corresponding directory in the codebase: https://github.com/rivas-lab/ukbb-tools/blob/master/04_gwas/extras/finngen_r3/README.md |

To generate
GBE_IDmapping between FinnGen and UKB, we apply LDSC rg between UKB and FinnGen.We prepared FinnGen in LDSC munge format here.
We are also preparing UKB in LDSC munge format in issue #26.
We use WB sum stats for this rg analysis.
The text was updated successfully, but these errors were encountered: