Developed by Tashrif Billah and Sylvain Bouix, Brigham and Women's Hospital (Harvard Medical School).
Table of Contents created by gh-md-toc
This tutorial assumes you have made yourself familiar with how Luigi works. In brief, each Luigi task executes its prerequisite tasks before executing the task itself. If expected output of a task exist, that will not rerun. Progress of the pipelines can be viewed in http://cmu166.research.partners.org:8082.
Another assumption is--you have organized your data according to BIDS convention. In the following, we shall explain how to run pipelines on DIAGNOSE-CTE data.
The Luigi workflows accept input in two ways-- through the command line and through a config file.
The motivation for that compartmentalization is to differentiate between high-level (former) and low-level (latter) inputs.
High-level inputs facilitate launching workflows while low-level inputs specify parameters pertinent to tasks in the workflow,
some of which are optional. Task specific parameters are provided in a configuration file defined via
environment variable LUIGI_CONFIG_PATH.
usage: ExecuteTask.py [-h] --bids-data-dir BIDS_DATA_DIR -c C -s S
[--dwi-template DWI_TEMPLATE]
[--t1-template T1_TEMPLATE] [--t2-template T2_TEMPLATE]
--task
{StructMask,Freesurfer,CnnMask,PnlEddy,FslEddy,TopupEddy,EddyEpi,Ukf,Fs2Dwi,Wmql,Wmqlqc,TractMeasures}
[--num-workers NUM_WORKERS]
[--derivatives-name DERIVATIVES_NAME]
pnlpipe glued together using Luigi, optional parameters can be set by
environment variable LUIGI_CONFIG_PATH, see luigi-pnlpipe/scripts/params/*.cfg
as examples
optional arguments:
-h, --help show this help message and exit
--bids-data-dir BIDS_DATA_DIR
/path/to/bids/data/directory
-c C a single case ID or a .txt file where each line is a
case ID
-s S a single session ID or a .txt file where each line is
a session ID (default: 1)
--dwi-template DWI_TEMPLATE
dwi pipeline: glob bids-data-dir/dwi-template to find
input data e.g. sub-*/ses-*/dwi/*_dwi.nii.gz, fs2dwi
pipeline: glob bids-data-dir/derivatives/derivatives-
name/dwi-template to find input data (default:
sub-*/dwi/*_dwi.nii.gz)
--t1-template T1_TEMPLATE
glob bids-data-dir/t1-template to find input data e.g.
sub-*/ses-*/anat/*_T1w.nii.gz (default:
sub-*/anat/*_T1w.nii.gz)
--t2-template T2_TEMPLATE
glob bids-data-dir/t2-template to find input data
(default: None)
--task {StructMask,Freesurfer,CnnMask,PnlEddy,FslEddy,TopupEddy,EddyEpi,Ukf,Fs2Dwi,Wmql,Wmqlqc,TractMeasures}
number of Luigi workers
--num-workers NUM_WORKERS
number of Luigi workers (default: 1)
--derivatives-name DERIVATIVES_NAME
relative name of bids derivatives directory,
translates to bids-data-dir/derivatives/derivatives-
name (default: pnlpipe)In the following, we shall provide both mandatory and optional settings for each workflow. Before running any workflow,
you should set up your terminal so that various prerequisite software i.e. FSL, FreeSurfer, ANTs etc. are found.
In addition, define PYTHONPATH:
cd ~/luigi-pnlpipe
export PYTHONPATH=`pwd`/scripts:`pwd`/workflows:$PYTHONPATH
# in case you have space shortage in /tmp or it is inaccessible to you
export PNLPIPE_TMPDIR=~/tmp
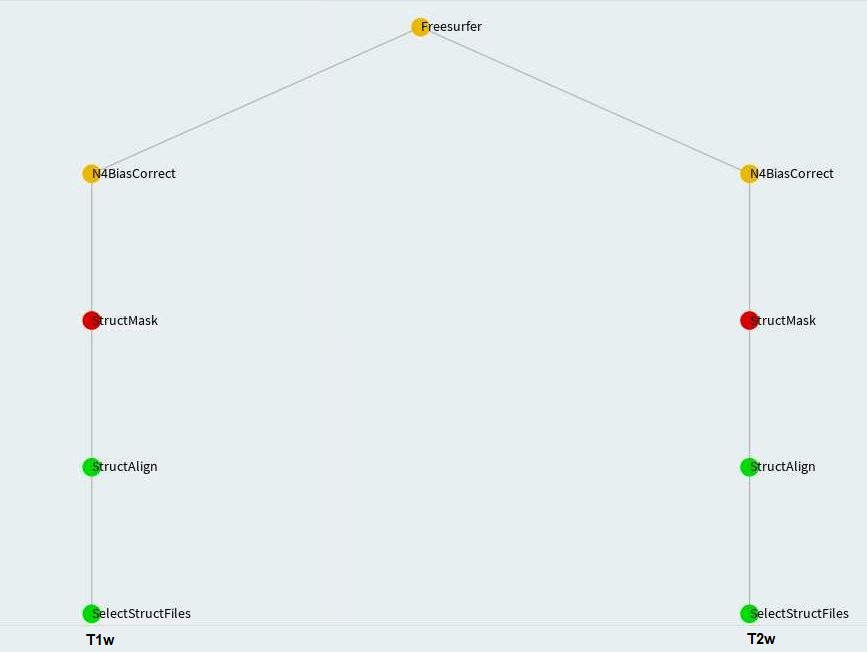
The goal of structural pipeline is to perform FreeSurfer segmentation. To be able to do that, we need mask for structural images-- T1w and maybe T2w.
MABS (Multi Atlas Brain Segmentation) remains as the acceptable technique to create mask for structural images. We shall create a MABS mask for one modality only and then quality check it manually. We shall warp that quality checked mask to obtain mask for other modalities. This approach minimizes the human effort required to quality check masks for all modalities. Nevertheless, you can create MABS mask for all modalities and quality check them manually.
MABS requires training data which are specified through csvFile below. StructMask task is capable of waiting until
quality checking of automated mask is complete, an option which can be invoked by setting mask_qc: True.
However, you can set mask_qc: False, obtain all masks, and then quality check at a later time.
So the content of LUIGI_CONFIG_PATH is following:
[StructMask]
mabs_mask_nproc: 8
fusion:
debug: False
reg_method:
slicer_exec:
mask_qc: False
csvFile: /path/to/trainingDataT2Masks.csv
ref_img:
ref_mask:Finally, save the above configuration in mabs_mask_params.cfg and run StructMask task as follows:
export LUIGI_CONFIG_PATH=/path/to/mabs_mask_params.cfg
# individual
# MABS masking of T2w image for case 1001
exec/ExecuteTask --task StructMask \
--bids-data-dir /data/pnl/DIAGNOSE_CTE_U01/rawdata -c 1001 --t2-template sub-*/ses-01/anat/*_T2w.nii.gz
# group
# MABS masking of T2w image for all cases
exec/ExecuteTask --task StructMask \
--bids-data-dir /data/pnl/DIAGNOSE_CTE_U01/rawdata -c /path/to/caselist.txt --t2-template sub-*/anat/*_T2w.nii.gz \
--num-workers 8Quality checked mask must be saved with Qc suffix in the desc field for its integration with later part of
the structural pipeline. Example:
Mabs mask : sub-1001/ses-01/anat/sub-1001_ses-01_desc-T2wXcMabs_mask.nii.gz
Quality checked : sub-1001/ses-01/anat/sub-1001_ses-01_desc-T2wXcMabsQc_mask.nii.gz
A couple of points to note regarding the above examples:
- For executing task through LSF, you should be using the individual example.
- When using group example, depending on the number of cases and CPUs in your machine, you should provide
a suitable
--num-workersso optimal number of cases are processed parallelly allowing room for other users in a shared cluster environment. - Although we provided both individual and group examples in the above, we shall be providing only individual examples in the rest of the tutorial. You can follow the above group example as a model for the rest.
- Each relevant configuration snippet should be saved in a
.cfgfile and defined inLUIGI_CONFIG_PATHenvironment variable. - If there is more than one task under a workflow, which is generally the case, a
[DEFAULT]section should be used in their configuration file to allow parameter sharing with top level-tasks. EddyEpi section explains this requirement in detail.
Tips:
lscpucommand will show you the number of processors available. As a rule of thumb, you can choose--num-workersto be half the number of available processors on a shared environment.vmstat -s -S Mwill show you the RAM available
Recently, we have adopted a better brain masking tool in Luigi pipeline: https://github.com/MIC-DKFZ/HD-BET. Unlike the CSV file with a list of training masks used by MABS, it uses a pre-trained deep learning model. To use this tool, use the following configuration:
[StructMask]
mask_method: HD-BETAs simple as that! However, it is advisable to run this program on a GPU enabled device. It can also be
run on a CPU enabled device but quite slowly. To do that, you need an additional parameter hdbet_device: cpu in
the above configuration.
For DIAG-CTE data, we have created MABS mask for T2w images. Then we have warped them to obtain mask for T1w and AXT2
images. Alternatively, you can create masks for T1w images first and then use them to create masks
for T2w and AXT2 images. Once again, we need a configuration to be provided via LUIGI_CONFIG_PATH:
[StructMask]
mabs_mask_nproc: 8
fusion:
debug: False
reg_method: rigid
slicer_exec:
mask_qc: False
csvFile:
ref_img: *_desc-Xc_T2w.nii.gz
ref_mask: *_desc-T2wXcMabsQc_mask.nii.gzNotice the values of ref_img and ref_mask beginning with asterisk (*). The asterisk (*) is important.
These are the patterns with which output directory is searched to obtain structural image and associated MABS mask.
The structural image is used to register to target space, in this case T1w or AXT2 space. Finally, the associated MABS
mask is warped to target space.
Other important parameters are reg_method and mask_qc. reg_method takes a value of either rigid or SyN
indicating the type of antsRegistration you would like to perform. On the other hand, since MABS mask was already
quality checked, mask_qc is set to False. Notice the csvFile field that must be kept empty otherwise MABS masking
procedure will be triggered.
Putting them all together:
export LUIGI_CONFIG_PATH=/path/to/mask_warping_params.cfg
exec/ExecuteTask --task StructMask \
--bids-data-dir /data/pnl/DIAGNOSE_CTE_U01/rawdata -c 1001 --t1-template sub-*/ses-01/anat/*_T1w.nii.gzNOTE Creating T1w and AXT2 masks separately is not required. They can be generated as part of
Freesurfer and FslEddyEpi tasks respectively.
We use T1w and/or T2w images to perform FreeSurfer segmentation. T1w and/or T2w images are multiplied by
their respective masks and passed through N4BiasFieldCorrection. This intermediate step does not require
any user intervention. On the other hand, since the method for obtaining T1w masks is
dependent upon T2w MABS masks, the latter have to be generated separately before running Freesurfer. The Freesurfer
task will generate T1w masks to fulfill its requirement.
[DEFAULT]
## [StructMask] ##
mabs_mask_nproc: 8
fusion:
debug: False
reg_method: rigid
slicer_exec:
## [Freesurfer] ##
t1_csvFile:
t1_ref_img: *_desc-Xc_T2w.nii.gz
t1_ref_mask: *_desc-T2wXcMabsQc_mask.nii.gz
t1_mask_qc: False
freesurfer_nproc: 4
expert_file:
no_hires: True
no_skullstrip: True
subfields: True
fs_dirname: freesurfer
[StructMask]
[Freesurfer]Notice the introduction of t1_ prefix preceding parameters defined for StructMask task itself.
The purpose of this introduction would be clear in the following section. Run Freesurfer task as follows:
export LUIGI_CONFIG_PATH=/path/to/fs_with_t1.cfg
exec/ExecuteTask --task Freesurfer \
--bids-data-dir /data/pnl/DIAGNOSE_CTE_U01/rawdata -c 1001 --t1-template sub-*/ses-01/anat/*_T1w.nii.gz[DEFAULT]
## [StructMask] ##
mabs_mask_nproc: 8
fusion:
debug: False
reg_method: rigid
slicer_exec:
## [Freesurfer] ##
t1_csvFile:
t1_ref_img: *_desc-Xc_T2w.nii.gz
t1_ref_mask: *_desc-T2wXcMabsQc_mask.nii.gz
t1_mask_qc: False
t2_csvFile: /path/to/trainingDataT2Masks.csv
t2_ref_img:
t2_ref_mask:
t2_mask_qc: True
freesurfer_nproc: 4
expert_file:
no_hires: True
no_skullstrip: True
subfields: True
fs_dirname: freesurfer
[StructMask]
[Freesurfer]Now, let's look at the purpose of using t1_ and t2_ prefix preceding parameters defined for StructMask task itself.
T2w masks are product of MABS while T1w masks are product of warping. Having same values for StructMask task parameters
would not allow this differentiation. Moreover, notice t1_mask_qc: False but t2_mask_qc: True, the latter telling
the pipeline to look for T2w masks with Qc suffix in desc field.
Finally, to tell Freesurfer task to use both T1w and T2w images, we shall provide both templates:
export LUIGI_CONFIG_PATH=/path/to/fs_with_both_t1_t2.cfg
exec/ExecuteTask --task Freesurfer \
--bids-data-dir /data/pnl/DIAGNOSE_CTE_U01/rawdata -c 1001 \
--t1-template sub-*/ses-01/anat/*_T1w.nii.gz \
--t2-template sub-*/ses-01/anat/*_T2w.nii.gzAny automated mask should be quality checked visually. To reiterate, the pipelines are equipped with the capability to wait until an automatically created mask is quality checked and progress from there. Yet, we explain the means of generating all the masks and quality checking them in two discrete steps.
[CnnMask]
slicer_exec:
dwi_mask_qc: False
model_folder: /data/pnl/soft/pnlpipe3/CNN-Diffusion-MRIBrain-Segmentation/model_folder
percentile: 97
filter:Run CnnMask task as follows:
export LUIGI_CONFIG_PATH=/path/to/dwi_pipe_params.cfg
exec/ExecuteTask --task CnnMask \
--bids-data-dir /data/pnl/DIAGNOSE_CTE_U01/rawdata -c 1001 --dwi-template sub-*/ses-01/dwi/*_dwi.nii.gzAs it was done for saving quality checked structural masks, quality checked dwi masks must be saved
with Qc suffix in the desc field for its integration with later part of the dwi pipeline. Example:
Cnn mask : sub-1001/ses-01/dwi/sub-1001_ses-01_desc-dwiXcCNN_mask.nii.gz
Quality checked : sub-1001/ses-01/dwi/sub-1001_ses-01_desc-dwiXcCNNQc_mask.nii.gz
NOTE To clean up the CNN generated mask, assign scipy or mrtrix to filter field. See here for details.
Different methods for eddy and epi corrections are described below:
- PnlEddy + PnlEpi (EddyEpi)
- FslEddy + PnlEpi (EddyEpi)
- SynB0
- TopupEddy
- HcpPipe
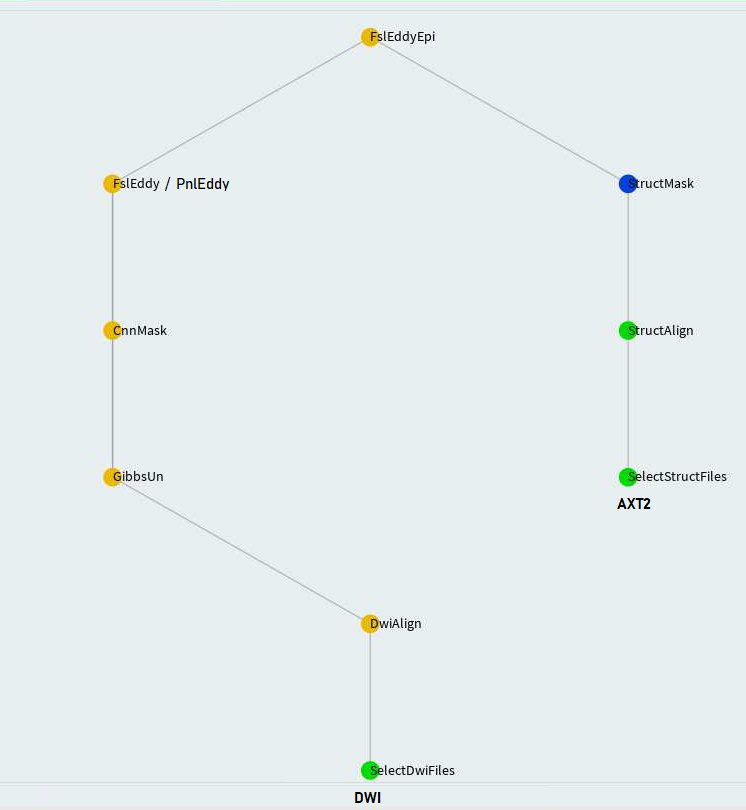
Epi correction method at PNL makes use of an axial T2w image acquired in the same plane of dwi,
identified by _AXT2.nii.gz suffix. This method can be applied on top of either PNL eddy or FSL eddy. The task
that implements this method is called EddyEpi. The configuration file used for this task can be found at dwi_pipe_params.cfg.
In the configuration, one has to define eddy_task parameter properly as eddy_task: PnlEddy or
eddy_task: FslEddy based on what method they have chosen for eddy correction.
Notice the use of [DEFAULT] section that allows sharing of parameters across various tasks. Refer to the flowchart of
FslEddyEpi (now called EddyEpi):
FslEddyEpi (now called EddyEpi) task directly depends upon FslEddy and StructMask tasks. So the parameters of the latter two also become
the parameters of the top-level task. On the other hand, defining task specific parameters under FslEddy and StructMask
in the configuration would not make them visible to the top-level task. Hence, the right thing is to define them under
[DEFAULT] section so they are visible to any top-level tasks.
Finally, run the big dwi pipeline as follows:
export LUIGI_CONFIG_PATH=/path/to/dwi_pipe_params.cfg
exec/ExecuteTask --task EddyEpi \
--bids-data-dir /data/pnl/DIAGNOSE_CTE_U01/rawdata -c 1001 \
--dwi-template sub-*/ses-01/dwi/*_dwi.nii.gz \
--t2-template sub-*/ses-01/anat/*_AXT2.nii.gzA few things to note here:
- The
--t2-templateis different than what we used forFreesurfertask since we need to use an in-plane T2w image for EPI correction, not the structural one. - The parameters for
StructMasktask echo those explained in Warped mask dwi_mask_qc: Truetells the program to look for quality checked dwi masks whilemask_qc: Falsetells it to look for non quality checked mask. As mentioned before, since a MABS mask was already quality checked, we should not require to quality check it again after warp assumingantsRegistration, eitherrigidorSyN, did a good job and not introduce any new artifact in the warped mask.- Notice that we used computation intensive
SyNregistration to obtain AXT2 mask compared torigidregistration we used for obtaining T2w mask.
This section serves as an example about running FslEddy/PnlEddy separately. When you run EddyEpi, it will run the former
already to fulfill its requirement.
Configuration:
The configuration file used for FslEddy/PnlEddy tasks is the same as above dwi_pipe_params.cfg.
FslEddy requires the following parameters definition in addition to the ones already defined for PnlEddy:
## [FslEddy] ##
acqp: /data/pnl/DIAGNOSE_CTE_U01/acqp.txt
index: /data/pnl/DIAGNOSE_CTE_U01/index.txt
config: /data/pnl/DIAGNOSE_CTE_U01/eddy_config.txtRun it:
export LUIGI_CONFIG_PATH=/path/to/dwi_pipe_params.cfg
exec/ExecuteTask --task FslEddy \
--bids-data-dir /data/pnl/DIAGNOSE_CTE_U01/rawdata -c 1001 --dwi-template sub-*/ses-01/dwi/*_dwi.nii.gzThe mask and baseline image provided to FslEddy are approximated as eddy corrected mask and baseline image that can
be fed into later task FslEddyEpi.
When an axial T2w image is absent and there is only AP or PA acquisition, we make use of Synb0-DISCO program to perform EPI distortion correction. The Synb0-DISCO program is a replacement of topup. It creates a distortion corrected B0 from a single AP or PA acquisition. It also provides the usual topup correction parameters that are used to perform eddy correction later. However, Synb0-DISCO requires a T1w image.
The configuration file used for SynB0 task is same as the above dwi_pipe_params.cfg. Specifically, SynB0 requires
the following parameters definition:
## [SynB0] ##
acqp: /path/to/acqp.txt
index: /path/to/index.txtWhere acqp.txt just:
0 1 0 0.05
0 1 0 0
Run it:
export LUIGI_CONFIG_PATH=/path/to/dwi_pipe_params.cfg
exec/ExecuteTask --task SynB0 \
--bids-data-dir /data/pnl/Collaborators/EDCRP/1110/1110_ses-002_T1_nii/rawdata/ \
-c ne00181 -s 002 \
--dwi-template sub-*/ses-*/dwi/*_dwi.nii.gz \
--t1-template sub-*/ses-*/anat/*_T1w.nii.gzNote that, SynB0 task is run in coordination with _synb0_eddy.sh .
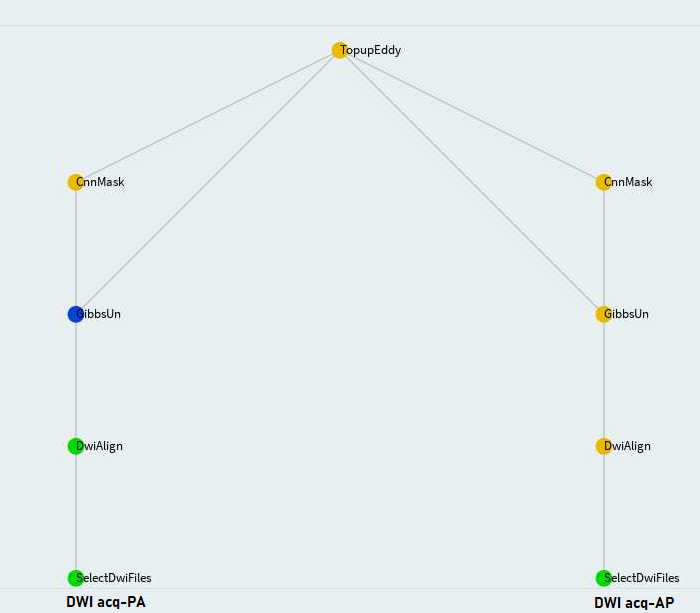
When two opposing acquisitions--AP and PA are available such as in Human Connectom Project (HCP), eddy+epi correction can be done in a more sophisticated way through FSL topup and eddy. In that way, AP and PA acquisitions are processed independently until TopupEddy:
Configuration:
Once again, the configuration file is dwi_pipe_params.cfg. Like FslEddy, TopupEddy requires
the following parameters definition:
## [TopupEddy] ##
acqp: /data/pnl/U01_HCP_Psychosis/acqp.txt
index: /data/pnl/U01_HCP_Psychosis/index.txt
config: /data/pnl/U01_HCP_Psychosis/eddy_config.txt
TopupOutDir: topup_eddy_107_dir
numb0: 1
whichVol: 1
scale: 2Run it:
export LUIGI_CONFIG_PATH=/path/to/dwi_pipe_params.cfg
exec/ExecuteTask --task TopupEddy \
--bids-data-dir /data/pnl/DIAGNOSE_CTE_U01/rawdata -c 1001 -s BWH01 \
--dwi-template sub-*/ses-*/dwi/*_acq-AP_*_dwi.nii.gz,sub-*/ses-*/dwi/*_acq-PA_*_dwi.nii.gzNotice that --dwi-template is followed by two templates, one for AP and the other for PA acquisition. Output prefix
for corrected data is determined as follows:
# correct only the first volume (whichVol: 1)
# preserve _acq-*_ and _dir-*_
_acq-AP_*_dir-107_*
# correct both volumes (whichVol: 1,2)
# omit _acq-*_ and double _dir-*_
*_dir-214_*
This task supersedes TopupEddy task. The HcpPipe way of processing opposing pair of DWIs is inherited from Washington-University/HCPpipelines. PNL introduced some improvements in that pipeline including:
- Use CNN brain masking program to generate topup mask
- Replace outliers (
--repolflag) for >500 b-shells
Since external processing is involved in this task, it is run via a separate script in three steps:
- Gibbs unringing of opposing pair of DWIs via Luigi
- HCP pipeline via shell scripts
- Creation of soft links in
sub-*/ses-*/dwi/directory according to BIDS convention via Luigi
This HcpPipe task has been used to process HCP-EP data.
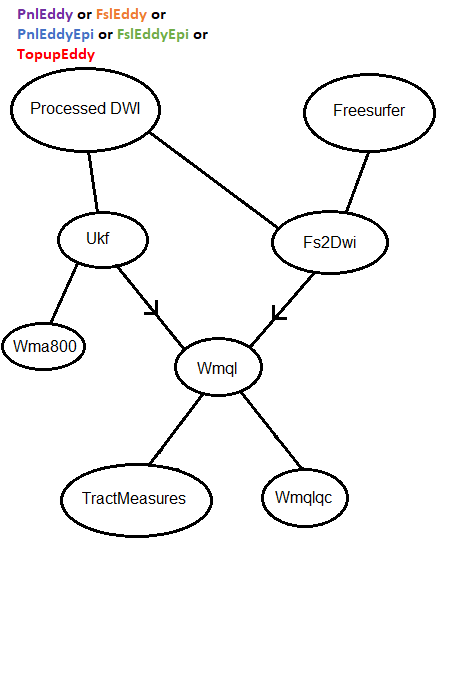
UKFTractography can be performed after any of the methods of eddy and epi correction. To make it aware of what methods were used, two parameters are needed in dwi_pipe_params.cfg:
eddy_task: # one of {FslEddy,PnlEddy}
eddy_epi_task: # one of {FslEddy,PnlEddy,EddyEpi,TopupEddy}
In addition, as of now, UKFTractography is performed on b-shells<=2000. This default can be adjusted through:
bhigh: 2000
Run it:
export LUIGI_CONFIG_PATH=/path/to/dwi_pipe_params.cfg
exec/ExecuteTask --task Ukf \
--bids-data-dir /data/pnl/DIAGNOSE_CTE_U01/rawdata -c 1001 --dwi-template sub-*/ses-01/dwi/*_dwi.nii.gzWe use SlicerDMRI/whitematteranalysis provided
wm_apply_ORG_atlas_to_subject.sh script and ORG-800FC-100HCP white matter atlas
for performing whole-brain tractography parcellation. The outputs of that script include:
- a parcellation of the entire white matter into 800 fiber clusters
- a parcellation of anatomical fiber tracts organized according to the brain lobes they connect
Here is an abridged snapshot of the output directory (you should examine the *csv files):
wma800/
└── sub-1122_ses-1_dir-416
├── AnatomicalTracts
│ └── diffusion_measurements_anatomical_tracts.csv
├── FiberClustering
│ ├── InitialClusters
│ ├── OutlierRemovedClusters
│ ├── SeparatedClusters
│ │ ├── diffusion_measurements_commissural.csv
│ │ ├── diffusion_measurements_left_hemisphere.csv
│ │ ├── diffusion_measurements_right_hemisphere.csv
│ │ ├── tracts_commissural
│ │ ├── tracts_left_hemisphere
│ │ └── tracts_right_hemisphere
│ └── TransformedClusters
└── TractRegistration
└── sub-1122_ses-1_dir-416
└── output_tractographyConfiguration:
Usability of this script imposes a number of dependencies on your software environment--3D Slicer executable,
SlicerDMRI extension, and xvfb-run server. Those dependencies of wm_apply_ORG_atlas_to_subject.sh
are specified in dwi_pipe_params.cfg as follows:
slicer_exec: /path/to/Slicer-4.10.2-linux-amd64/SlicerWithExtensions.sh
FiberTractMeasurements: /path/to/Slicer-4.10.2-linux-amd64/SlicerWithExtensions.sh --launch /path/to/Slicer-4.10.2-linux-amd64/.config/NA-MIC/Extensions-28257/SlicerDMRI/lib/Slicer-4.10/cli-modules/FiberTractMeasurements
atlas: /path/to/ORG-Atlases-1.1.1
wma_nproc: 4
xvfb: 1
wma_cleanup: 0Consult wm_apply_ORG_atlas_to_subject.sh --help to know the details of these parameters.
Run it:
export LUIGI_CONFIG_PATH=/path/to/dwi_pipe_params.cfg
exec/ExecuteTask --task Wma800 \
--bids-data-dir /data/pnl/DIAGNOSE_CTE_U01/rawdata -c 1001 --dwi-template sub-*/ses-01/dwi/*_dwi.nii.gzNEW_SOFT_DIR=/rfanfs/pnl-zorro/software/pnlpipe3/
source ${NEW_SOFT_DIR}/bashrc3
wm_apply_ORG_atlas_to_subject.sh \
-i /path/to/your.vtk \
-o /path/to/output/dir/ \
-a ${NEW_SOFT_DIR}/ORG-Atlases-1.2 \
-s ${NEW_SOFT_DIR}/Slicer-4.10.2-linux-amd64/SlicerWithExtensions.sh \
-m "${NEW_SOFT_DIR}/Slicer-4.10.2-linux-amd64/SlicerWithExtensions.sh --launch FiberTractMeasurements" \
-n 8 -c 2 -d 1
The same command applies for running it on PNL's GRX node: dna007.partners.org except a change in the software directory:
NEW_SOFT_DIR=/data/pnl/soft/pnlpipe3/
You should consult SlicerDMRI/whitematteranalysis repository for meaning of the above arguments.
Just append the -x 1 flag to the above command to enable virtual X server:
wm_apply_ORG_atlas_to_subject.sh \
...
-n 8 -c 2 -d 1 -x 1
However, running it through SSH requires availability of xvfb-run on that server--may it be a PNL workstation or a GRX node.
The good news is that we have already installed xorg-x11-server-Xvfb in most servers. So you should be all set.
But if you find xvfb-run's absence, reach out to PNL engineers for help.
Lastly, you can use the same -x 1 flag to run it through bsub on pri_pnl queue in ERIS cluster.
After the completion of structural and diffusion pipelines, Fs2Dwi pipeline can proceed as follows:
The tasks defined in Fs2Dwi pipeline are the ones downstream from Fs2Dwi node. The configuration file is fs2dwi_pipe_params.cfg.
The parameter of most interest is fs2dwi.py registration mode. The default is mode: direct. However, you can choose
to do through T2w registration by mode: witht2.
Run it:
export LUIGI_CONFIG_PATH=/path/to/fs2dwi_pipe_params.cfg
# direct fs2dwi.py
workflows/ExecuteTask.py --task Wmql --bids-data-dir $HOME/CTE/rawdata -c 1004 -s 01 \
--dwi-template sub-*/ses-*/dwi/*EdEp_dwi.nii.gz
# witht2 fs2dwi.py
workflows/ExecuteTask.py --task Wmql --bids-data-dir $HOME/CTE/rawdata -c 1004 -s 01 \
--dwi-template sub-*/ses-*/dwi/*EdEp_dwi.nii.gz --t2-template sub-*/ses-*/anat/*_T2w.nii.gzNOTE In Fs2Dwi pipeline, the --dwi-template is used to search bids-data-dir/derivatives/derivatives-name/dwi-template
to find input data. Note that the search directory is different than that of DWI pipeline where --dwi-template was used to search
rawdata directory i.e. bids-data-dir/rawdata/dwi-template. The reason behind this discrepancy is--Fs2Dwi pipeline starts
from processed dwi data, not rawdata. However, once a dwi is found, the associated mask is found according to
the knowledge of outputs of different nodes from the DWI pipeline. There is a one to one mapping between dwi and mask
of each pipeline node as written here.
You can run Fs2Dwi, TractMeasures, and Wmqlqc tasks in the same way as above. Note that, Wmqlqc task
would require GUI capability so it is better to attempt that through a virtual desktop e.g. NoMachine.