-Title: Next-generation file formats (NGFF) -Shortname: ome-ngff -Level: 1 -Status: w3c/CG-FINAL -TR: https://ngff.openmicroscopy.org/0.1/ -URL: https://ngff.openmicroscopy.org/0.1/ -Repository: https://github.com/ome/ngff -Issue Tracking: Forums https://forum.image.sc/tag/ome-ngff -Logo: http://www.openmicroscopy.org/img/logos/ome-logomark.svg -Local Boilerplate: header yes, copyright yes -Boilerplate: style-darkmode off -Markup Shorthands: markdown yes -Editor: Josh Moore, Open Microscopy Environment (OME) https://www.openmicroscopy.org -Editor: Sébastien Besson, Open Microscopy Environment (OME) https://www.openmicroscopy.org -Abstract: This document contains next-generation file format (NGFF) -Abstract: specifications for storing bioimaging data in the cloud. -Abstract: All specifications are submitted to the https://image.sc community for review. -Status Text: This is the 0.1 release of this specification. Migration scripts -Status Text: will be provided between numbered versions. Data written with the latest version -Status Text: (an "editor's draft") will not necessarily be supported. -- -Introduction {#intro} -===================== - -Bioimaging science is at a crossroads. Currently, the drive to acquire more, -larger, preciser spatial measurements is unfortunately at odds with our ability -to structure and share those measurements with others. During a global pandemic -more than ever, we believe fervently that global, collaborative discovery as -opposed to the post-publication, "data-on-request" mode of operation is the -path forward. Bioimaging data should be shareable via open and commercial cloud -resources without the need to download entire datasets. - -At the moment, that is not the norm. The plethora of data formats produced by -imaging systems are ill-suited to remote sharing. Individual scientists -typically lack the infrastructure they need to host these data themselves. When -they acquire images from elsewhere, time-consuming translations and data -cleaning are needed to interpret findings. Those same costs are multiplied when -gathering data into online repositories where curator time can be the limiting -factor before publication is possible. Without a common effort, each lab or -resource is left building the tools they need and maintaining that -infrastructure often without dedicated funding. - -This document defines a specification for bioimaging data to make it possible -to enable the conversion of proprietary formats into a common, cloud-ready one. -Such next-generation file formats layout data so that individual portions, or -"chunks", of large data are reference-able eliminating the need to download -entire datasets. - - -Why "NGFF"? {#why-ngff} -------------------------------------------------------------------------------------------------- - -A short description of what is needed for an imaging format is "a hierarchy -of n-dimensional (dense) arrays with metadata". This combination of features -is certainly provided by HDF5 -from the HDF Group, which a number of -bioimaging formats do use. HDF5 and other larger binary structures, however, -are ill-suited for storage in the cloud where accessing individual chunks -of data by name rather than seeking through a large file is at the heart of -parallelization. - -As a result, a number of formats have been developed more recently which provide -the basic data structure of an HDF5 file, but do so in a more cloud-friendly way. -In the [PyData](https://pydata.org/) community, the Zarr [[zarr]] format was developed -for easily storing collections of [NumPy](https://numpy.org/) arrays. In the -[ImageJ](https://imagej.net/) community, N5 [[n5]] was developed to work around -the limitations of HDF5 ("N5" was originally short for "Not-HDF5"). -Both of these formats permit storing individual chunks of data either locally in -separate files or in cloud-based object stores as separate keys. - -A [current effort](https://zarr-specs.readthedocs.io/en/core-protocol-v3.0-dev/protocol/core/v3.0.html) -is underway to unify the two similar specifications to provide a single binary -specification. The editor's draft will soon be entering a [request for comments (RFC)](https://github.com/zarr-developers/zarr-specs/issues/101) phase with the goal of having a first version early in 2021. As that -process comes to an end, this document will be updated. - -OME-NGFF {#ome-ngff} --------------------- - -The conventions and specifications defined in this document are designed to -enable next-generation file formats to represent the same bioimaging data -that can be represented in \[OME-TIFF](http://www.openmicroscopy.org/ome-files/) -and beyond. However, the conventions will also be usable by HDF5 and other sufficiently advanced -binary containers. Eventually, we hope, the moniker "next-generation" will no longer be -applicable, and this will simply be the most efficient, common, and useful representation -of bioimaging data, whether during acquisition or sharing in the cloud. - -Note: The following text makes use of OME-Zarr [[ome-zarr-py]], the current prototype implementation, -for all examples. - -On-disk (or in-cloud) layout {#on-disk} -======================================= - -An overview of the layout of an OME-Zarr fileset should make -understanding the following metadata sections easier. The hierarchy -is represented here as it would appear locally but could equally -be stored on a web server to be accessed via HTTP or in object storage -like S3 or GCS. - -Images {#image-layout} ----------------------- - -The following layout describes the expected Zarr hierarchy for images with -multiple levels of resolutions and optionally associated labels. - -``` -. # Root folder, potentially in S3, -│ # with a flat list of images by image ID. -│ -├── 123.zarr # One image (id=123) converted to Zarr. -│ -└── 456.zarr # Another image (id=456) converted to Zarr. - │ - ├── .zgroup # Each image is a Zarr group, or a folder, of other groups and arrays. - ├── .zattrs # Group level attributes are stored in the .zattrs file and include - │ # "multiscales" and "omero" below) - │ - ├── 0 # Each multiscale level is stored as a separate Zarr array, - │ ... # which is a folder containing chunk files which compose the array. - ├── n # The name of the array is arbitrary with the ordering defined by - │ │ # by the "multiscales" metadata, but is often a sequence starting at 0. - │ │ - │ ├── .zarray # All image arrays are 5-dimensional - │ │ # with dimension order (t, c, z, y, x). - │ │ - │ ├── 0.0.0.0.0 # Chunks are stored with the flat directory layout. - │ │ ... # Each dotted component of the chunk file represents - │ └── t.c.z.y.x # a "chunk coordinate", where the maximum coordinate - │ # will be `dimension_size / chunk_size`. - │ - └── labels - │ - ├── .zgroup # The labels group is a container which holds a list of labels to make the objects easily discoverable - │ - ├── .zattrs # All labels will be listed in `.zattrs` e.g. `{ "labels": [ "original/0" ] }` - │ # Each dimension of the label `(t, c, z, y, x)` should be either the same as the - │ # corresponding dimension of the image, or `1` if that dimension of the label - │ # is irrelevant. - │ - └── original # Intermediate folders are permitted but not necessary and currently contain no extra metadata. - │ - └── 0 # Multiscale, labeled image. The name is unimportant but is registered in the "labels" group above. - ├── .zgroup # Zarr Group which is both a multiscaled image as well as a labeled image. - ├── .zattrs # Metadata of the related image and as well as display information under the "image-label" key. - │ - ├── 0 # Each multiscale level is stored as a separate Zarr array, as above, but only integer values - │ ... # are supported. - └── n -``` - -High-content screening {#hcs-layout} ------------------------------------- - -The following specification defines the hierarchy for a high-content screening -dataset. Three groups must be defined above the images: - -- the group above the images defines the well and MUST implement the - [well specification](#well-md). All images contained in a well are fields - of view of the same well -- the group above the well defines a row of wells -- the group above the well row defines an entire plate i.e. a two-dimensional - collection of wells organized in rows and columns. It MUST implement the - [plate specification](#plate-md) - - -``` -. # Root folder, potentially in S3, -│ -└── 5966.zarr # One plate (id=5966) converted to Zarr - ├── .zgroup - ├── .zattrs # Implements "plate" specification - ├── A # First row of the plate - │ ├── .zgroup - │ │ - │ ├── 1 # First column of row A - │ │ ├── .zgroup - │ │ ├── .zattrs # Implements "well" specification - │ │ │ - │ │ ├── 0 # First field of view of well A1 - │ │ │ │ - │ │ │ ├── .zgroup - │ │ │ ├── .zattrs # Implements "multiscales", "omero" - │ │ │ ├── 0 - │ │ │ │ ... # Resolution levels - │ │ │ ├── n - │ │ │ └── labels # Labels (optional) - │ │ ├── ... # Fields of view - │ │ └── m - │ ├── ... # Columns - │ └── 12 - ├── ... # Rows - └── H -``` - -Metadata {#metadata} -==================== - -The various `.zattrs` files throughout the above array hierarchy may contain metadata -keys as specified below for discovering certain types of data, especially images. - -"multiscales" metadata {#multiscale-md} ---------------------------------------- - -Metadata about the multiple resolution representations of the image can be -found under the "multiscales" key in the group-level metadata. -The specification for the multiscale (i.e. "resolution") metadata is provided -in [zarr-specs#50](https://github.com/zarr-developers/zarr-specs/issues/50). -If only one multiscale is provided, use it. Otherwise, the user can choose by -name, using the first multiscale as a fallback: - -```python -datasets = [] -for named in multiscales: - if named["name"] == "3D": - datasets = [x["path"] for x in named["datasets"]] - break -if not datasets: - # Use the first by default. Or perhaps choose based on chunk size. - datasets = [x["path"] for x in multiscales[0]["datasets"]] -``` - -The subresolutions in each multiscale are ordered from highest-resolution -to lowest. - -"omero" metadata {#omero-md} ----------------------------- - -Information specific to the channels of an image and how to render it -can be found under the "omero" key in the group-level metadata: - -```json -"id": 1, # ID in OMERO -"name": "example.tif", # Name as shown in the UI -"version": "0.1", # Current version -"channels": [ # Array matching the c dimension size - { - "active": true, - "coefficient": 1, - "color": "0000FF", - "family": "linear", - "inverted": false, - "label": "LaminB1", - "window": { - "end": 1500, - "max": 65535, - "min": 0, - "start": 0 - } - } -], -"rdefs": { - "defaultT": 0, # First timepoint to show the user - "defaultZ": 118, # First Z section to show the user - "model": "color" # "color" or "greyscale" -} -``` - -See https://docs.openmicroscopy.org/omero/5.6.1/developers/Web/WebGateway.html#imgdata -for more information. - -"labels" metadata {#labels-md} ------------------------------- - -The special group "labels" found under an image Zarr contains the key `labels` containing -the paths to label objects which can be found underneath the group: - -```json -{ - "labels": [ - "orphaned/0" - ] -} -``` - -Unlisted groups MAY be labels. - -"image-label" metadata {#label-md} ----------------------------------- - -Groups containing the `image-label` dictionary represent an image segmentation -in which each unique pixel value represents a separate segmented object. -`image-label` groups MUST also contain `multiscales` metadata and the two -"datasets" series MUST have the same number of entries. - -The `colors` key defines a list of JSON objects describing the unique label -values. Each entry in the list MUST contain the key "label-value" with the -pixel value for that label. Additionally, the "rgba" key MAY be present, the -value for which is an RGBA unsigned-int 4-tuple: `[uint8, uint8, uint8, uint8]` -All `label-value`s must be unique. Clients who choose to not throw an error -should ignore all except the _last_ entry. - -Some implementations may represent overlapping labels by using a specially assigned -value, for example the highest integer available in the pixel range. - -The `source` key is an optional dictionary which contains information on the -image the label is associated with. If included it MAY include a key `image` -whose value is the relative path to a Zarr image group. The default value is -"../../" since most labels are stored under a subgroup named "labels/" (see -above). - - -```json -"image-label": - { - "version": "0.1", - "colors": [ - { - "label-value": 1, - "rgba": [255, 255, 255, 0] - }, - { - "label-value": 4, - "rgba": [0, 255, 255, 128] - }, - ... - ] - }, - "source": { - "image": "../../" - } -] -``` - -"plate" metadata {#plate-md} ----------------------------- - -For high-content screening datasets, the plate layout can be found under the -custom attributes of the plate group under the `plate` key. - -
-
-
- acquisitions -
- An optional list of JSON objects defining the acquisitions for a given - plate. Each acquisition object MUST contain an `id` key providing an - unique identifier within the context of the plate to which fields of - view can refer to. It SHOULD contain a `name` key identifying the name - of the acquisition. It SHOULD contain a `maximumfieldcount` key - indicating the maximum number of fields of view for the acquisition. It - MAY contain a `description` key providing a description for the - acquisition. It MAY contain a `startime` and/or `endtime` key specifying - the start and/or end timestamp of the acquisition using an epoch - string. -
- columns -
- A list of JSON objects defining the columns of the plate. Each column - object defines the properties of the column at the index of the object - in the list. If not empty, it MUST contain a `name` key specifying the - column name. -
- field_count -
- An integer defining the maximum number of fields per view across all - wells. -
- name -
- A string defining the name of the plate. -
- rows -
- A list of JSON objects defining the rows of the plate. Each row object - defines the properties of the row at the index of the object in the - list. If not empty, it MUST contain a `name` key specifying the row - name. -
- version -
- A string defining the version of the specification. -
- wells -
- A list of JSON objects defining the wells of the plate. Each well object - MUST contain a `path` key identifying the path to the well subgroup. -
-
-
- images -
- A list of JSON objects defining the fields of views for a given well. - Each object MUST contain a `path` key identifying the path to the - field of view. If multiple acquisitions were performed in the plate, it - SHOULD contain an `acquisition` key identifying the id of the - acquisition which must match one of acquisition JSON objects defined in - the plate metadata. -
- version -
- A string defining the version of the specification. -
-
-
-
- [omero-ms-zarr](https://github.com/ome/omero-ms-zarr) -
- A microservice for OMERO.server that converts images stored in OMERO to OME Zarr files on the fly, served via a web API. - -
- [idr-zarr-tools](https://github.com/IDR/idr-zarr-tools) -
- A full workflow demonstrating the conversion of IDR images to OME Zarr images on S3. - -
- [OMERO CLI Zarr plugin](https://github.com/ome/omero-cli-zarr) -
- An OMERO CLI plugin that converts images stored in OMERO.server into a local Zarr file. - -
- [ome-zarr-py](https://github.com/ome/ome-zarr-py) -
- A napari plugin for reading ome-zarr files. - -
- [bioformats2raw](https://github.com/glencoesoftware/bioformats2raw) -
- A performant, Bio-Formats image file format converter. - -
- [vizarr](https://github.com/hms-dbmi/vizarr/) -
- A minimal, purely client-side program for viewing Zarr-based images with Viv & ImJoy. - -
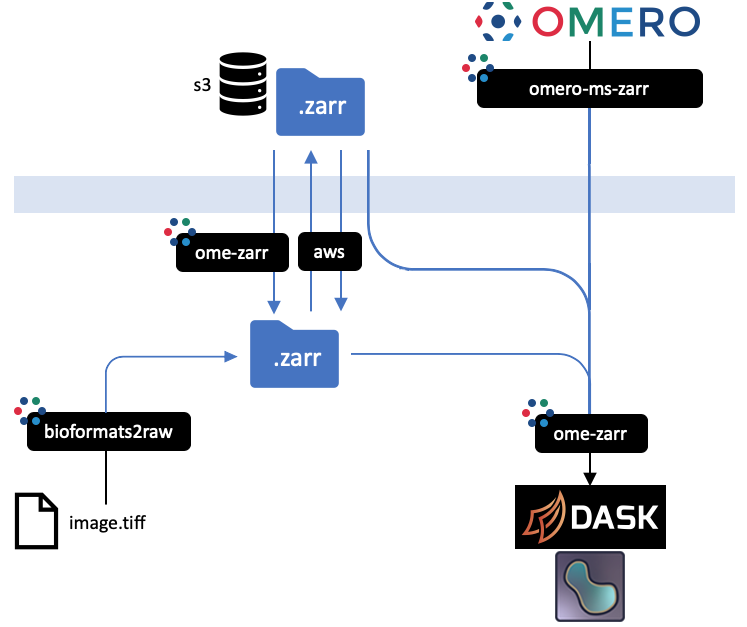 -
-All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
-version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
-Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
-
-Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
-
-Citing {#citing}
-================
-
-[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.1)
-J. Moore, *et al*. Open Microscopy Environment Consortium, 20 November 2020.
-This edition of the specification is [https://ngff.openmicroscopy.org/0.1/](https://ngff.openmicroscopy.org/0.1/]).
-The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
-[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
-
-Version History {#history}
-==========================
-
-
-
-All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
-version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
-Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
-
-Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
-
-Citing {#citing}
-================
-
-[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.1)
-J. Moore, *et al*. Open Microscopy Environment Consortium, 20 November 2020.
-This edition of the specification is [https://ngff.openmicroscopy.org/0.1/](https://ngff.openmicroscopy.org/0.1/]).
-The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
-[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
-
-Version History {#history}
-==========================
-
-| Revision | -Date | -Description | -
| 0.1.4 | -2020-11-26 | -Add HCS specification | -
| 0.1.3 | -2020-09-14 | -Add labels specification | -
| 0.1.2 | -2020-05-07 | -Add description of "omero" metadata | -
| 0.1.1 | -2020-05-06 | -Add info on the ordering of resolutions | -
| 0.1.0 | -2020-04-20 | -First version for internal demo | -
-{
- "blogNov2020": {
- "href": "https://blog.openmicroscopy.org/file-formats/community/2020/11/04/zarr-data/",
- "title": "Public OME-Zarr data (Nov. 2020)",
- "authors": [
- "OME Team"
- ],
- "status": "Informational",
- "publisher": "OME",
- "id": "blogNov2020",
- "date": "04 November 2020"
- },
- "imagesc26952": {
- "href": "https://forum.image.sc/t/ome-s-position-regarding-file-formats/26952",
- "title": "OME’s position regarding file formats",
- "authors": [
- "OME Team"
- ],
- "status": "Informational",
- "publisher": "OME",
- "id": "imagesc26952",
- "date": "19 June 2020"
- },
- "n5": {
- "id": "n5",
- "href": "https://github.com/saalfeldlab/n5/issues/62",
- "title": "N5---a scalable Java API for hierarchies of chunked n-dimensional tensors and structured meta-data",
- "status": "Informational",
- "authors": [
- "John A. Bogovic",
- "Igor Pisarev",
- "Philipp Hanslovsky",
- "Neil Thistlethwaite",
- "Stephan Saalfeld"
- ],
- "date": "2020"
- },
- "ome-zarr-py": {
- "id": "ome-zarr-py",
- "href": "https://doi.org/10.5281/zenodo.4113931",
- "title": "ome-zarr-py: Experimental implementation of next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.",
- "status": "Informational",
- "publisher": "Zenodo",
- "authors": [
- "OME",
- "et al"
- ],
- "date": "06 October 2020"
- },
- "zarr": {
- "id": "zarr",
- "href": "https://doi.org/10.5281/zenodo.4069231",
- "title": "Zarr: An implementation of chunked, compressed, N-dimensional arrays for Python.",
- "status": "Informational",
- "publisher": "Zenodo",
- "authors": [
- "Alistair Miles",
- "et al"
- ],
- "date": "06 October 2020"
- }
-}
-
diff --git a/0.1/schemas/image.schema b/0.1/schemas/image.schema
deleted file mode 100644
index 5a97f3f2..00000000
--- a/0.1/schemas/image.schema
+++ /dev/null
@@ -1,112 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/0.1/schemas/image.schema",
- "title": "NGFF Image",
- "description": "JSON from OME-NGFF .zattrs",
- "type": "object",
- "properties": {
- "multiscales": {
- "description": "The multiscale datasets for this image",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- },
- "datasets": {
- "type": "array",
- "minItems": 1,
- "items": {
- "type": "object",
- "properties": {
- "path": {
- "type": "string"
- }
- },
- "required": ["path"]
- }
- },
- "version": {
- "type": "string",
- "enum": [
- "0.1"
- ]
- },
- "metadata": {
- "type": "object",
- "properties": {
- "method": {
- "type": "string"
- },
- "version": {
- "type": "string"
- }
- }
- }
- },
- "required": [
- "datasets"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "omero": {
- "type": "object",
- "properties": {
- "channels": {
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "window": {
- "type": "object",
- "properties": {
- "end": {
- "type": "number"
- },
- "max": {
- "type": "number"
- },
- "min": {
- "type": "number"
- },
- "start": {
- "type": "number"
- }
- },
- "required": [
- "start",
- "min",
- "end",
- "max"
- ]
- },
- "label": {
- "type": "string"
- },
- "family": {
- "type": "string"
- },
- "color": {
- "type": "string"
- },
- "active": {
- "type": "boolean"
- }
- },
- "required": [
- "window",
- "color"
- ]
- }
- }
- },
- "required": [
- "channels"
- ]
- }
- },
- "required": [ "multiscales" ]
-}
diff --git a/0.1/schemas/plate.schema b/0.1/schemas/plate.schema
deleted file mode 100644
index e4f4f6f7..00000000
--- a/0.1/schemas/plate.schema
+++ /dev/null
@@ -1,112 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/0.1/schemas/plate.schema",
- "title": "OME-NGFF plate schema",
- "description": "JSON from OME-NGFF Plate .zattrs",
- "type": "object",
- "properties": {
- "plate": {
- "type": "object",
- "properties": {
- "version": {
- "type": "string",
- "enum": [
- "0.1"
- ]
- },
- "name": {
- "type": "string"
- },
- "columns": {
- "description": "Columns of the Plate grid",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- }
- },
- "required": [
- "name"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "rows": {
- "description": "Rows of the Plate grid",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- }
- },
- "required": [
- "name"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "wells": {
- "description": "Rows of the Plate grid",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "path": {
- "type": "string"
- }
- },
- "required": [
- "path"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "field_count": {
- "description": "Maximum number of fields per view across all wells."
- },
- "acquisitions": {
- "description": "Rows of the Plate grid",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "id": {
- "type": "number"
- },
- "maximumfieldcount": {
- "type": "number"
- },
- "name": {
- "type": "string"
- },
- "description": {
- "type": "string"
- },
- "starttime": {
- "type": "number"
- }
- },
- "required": [
- "id"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- }
- },
- "required": [
- "version", "columns", "rows", "wells"
- ]
- }
- },
- "required": [
- "plate"
- ]
-}
\ No newline at end of file
diff --git a/0.1/schemas/strict_image.schema b/0.1/schemas/strict_image.schema
deleted file mode 100644
index ac375073..00000000
--- a/0.1/schemas/strict_image.schema
+++ /dev/null
@@ -1,19 +0,0 @@
-{
- "$id": "https://ngff.openmicroscopy.org/0.1/schemas/strict_image.schema",
- "allOf": [
- {
- "$ref": "https://ngff.openmicroscopy.org/0.1/schemas/image.schema"
- },
- {
- "properties": {
- "multiscales": {
- "items": {
- "required": [
- "version", "metadata", "type", "name"
- ]
- }
- }
- }
- }
- ]
-}
\ No newline at end of file
diff --git a/0.1/schemas/well.schema b/0.1/schemas/well.schema
deleted file mode 100644
index 02475934..00000000
--- a/0.1/schemas/well.schema
+++ /dev/null
@@ -1,47 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/0.1/schemas/well.schema",
- "title": "OME-NGFF well schema",
- "description": "JSON from OME-NGFF .zattrs",
- "type": "object",
- "properties": {
- "well": {
- "type": "object",
- "properties": {
- "images": {
- "description": "The fields of view for this well",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "acquisition": {
- "description": "A unique identifier within the context of the plate",
- "type": "integer"
- },
- "path": {
- "description": "The path for this field of view subgroup",
- "type": "string",
- "pattern": "^[A-Za-z0-9]+$"
- }
- },
- "required": [
- "path"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "version": {
- "description": "The version of the specification",
- "type": "string",
- "enum": [
- "0.1"
- ]
- }
- },
- "required": [
- "images"
- ]
- }
- }
-}
diff --git a/0.1/tests/test_validation.py b/0.1/tests/test_validation.py
deleted file mode 100644
index 21d0e826..00000000
--- a/0.1/tests/test_validation.py
+++ /dev/null
@@ -1,97 +0,0 @@
-import json
-import os
-import glob
-
-import pytest
-
-from jsonschema import validate, RefResolver
-from jsonschema.validators import validator_for
-from jsonschema.exceptions import ValidationError
-
-
-IMAGE_SCHEMA_KEY = "https://ngff.openmicroscopy.org/0.1/schemas/image.schema"
-
-
-def files():
- return list(glob.glob(f"examples/*/valid/*.json")) + \
- list(glob.glob(f"examples/*/invalid/*.json"))
-
-
-def strict():
- return list(glob.glob(f"examples/image/valid/*.json"))
-
-
-def ids(paths):
- return [str(x).split("/")[-1][0:-5] for x in paths]
-
-
-@pytest.mark.parametrize("testfile", files(), ids=ids(files()))
-def test_json(testfile):
-
- test_json, schema = load_instance_and_schema(testfile)
-
- if "invalid" in testfile:
- with pytest.raises(ValidationError):
- validate(instance=test_json, schema=schema)
- else:
- validate(instance=test_json, schema=schema)
-
-
-class LocalRefResolver(RefResolver):
-
- def resolve_remote(self, url):
- # Use remote URL to generate local path
- url = url.replace("https://ngff.openmicroscopy.org/0.1/", "")
- # Load local document and cache it
- document = load_json(url)
- self.store[url] = document
- return document
-
-
-@pytest.mark.parametrize("testfile", strict(), ids=ids(strict()))
-def test_strict_rules(testfile):
-
- test_json, schema = load_instance_and_schema(testfile, strict=True)
-
- # Check for all validation errors without throwing exception
- cls = validator_for(schema)
- cls.check_schema(schema)
-
- # Use our local resolver subclass to resolve local documents
- localResolver = LocalRefResolver.from_schema(schema)
- validator = cls(schema, localResolver)
-
- warnings = list(validator.iter_errors(test_json))
- for warning in warnings:
- print("WARNING", warning.message)
- # ONLY the complete example has no warnings in strict mode
- if "complete" not in testfile:
- assert len(warnings) > 0
-
-
-def load_instance_and_schema(path, strict=False):
- # Load the correct schema
- test_json = load_json(path)
- # we don't have @type in this version
- if "multiscales" in test_json:
- schema_name = "image.schema"
- elif "plate" in test_json:
- schema_name = "plate.schema"
- elif "well" in test_json:
- schema_name = "well.schema"
- else:
- raise Exception("No schema found")
-
- schema = load_json('schemas/' + schema_name)
-
- if strict and schema_name == "image.schema":
- strict_path = 'schemas/strict_' + schema_name
- schema = load_json(strict_path)
-
- return (test_json, schema)
-
-
-def load_json(path):
- with open(path) as f:
- json_data = json.loads(f.read())
- return json_data
diff --git a/0.1/tox.ini b/0.1/tox.ini
deleted file mode 100644
index 2de32a96..00000000
--- a/0.1/tox.ini
+++ /dev/null
@@ -1,10 +0,0 @@
-[tox]
-envlist = v01
-skipsdist = True
-
-[testenv]
-deps =
- pytest
- jsonschema
-commands =
- pytest tests --color=yes --basetemp={envtmpdir} {posargs:-v}
diff --git a/0.2/copyright.include b/0.2/copyright.include
deleted file mode 100644
index f0def708..00000000
--- a/0.2/copyright.include
+++ /dev/null
@@ -1,4 +0,0 @@
-Copyright © 2020-[YEAR]
-OME®
-(U. Dundee).
-OME trademark rules apply.
diff --git a/0.2/examples/image/invalid/invalid_channels_color.json b/0.2/examples/image/invalid/invalid_channels_color.json
deleted file mode 100644
index 56f9f7f2..00000000
--- a/0.2/examples/image/invalid/invalid_channels_color.json
+++ /dev/null
@@ -1,31 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.2",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- }
- ]
- }
- ],
- "omero": {
- "channels": [
- {
- "active": true,
- "coefficient": 1.0,
- "color": 255,
- "family": "linear",
- "label": "1234",
- "window": {
- "end": 1765.0,
- "max": 2555.0,
- "min": 5.0,
- "start": 0.0
- }
- }
- ]
- }
-}
diff --git a/0.2/examples/image/invalid/invalid_channels_window.json b/0.2/examples/image/invalid/invalid_channels_window.json
deleted file mode 100644
index 534b6eb6..00000000
--- a/0.2/examples/image/invalid/invalid_channels_window.json
+++ /dev/null
@@ -1,31 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.2",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- }
- ]
- }
- ],
- "omero": {
- "channels": [
- {
- "active": true,
- "coefficient": 1.0,
- "color": "ff0000",
- "family": "linear",
- "label": "1234",
- "window": {
- "end": "100",
- "max": 2555.0,
- "min": 5.0,
- "start": 0.0
- }
- }
- ]
- }
-}
diff --git a/0.2/examples/image/invalid/invalid_path.json b/0.2/examples/image/invalid/invalid_path.json
deleted file mode 100644
index 0ea1f2d9..00000000
--- a/0.2/examples/image/invalid/invalid_path.json
+++ /dev/null
@@ -1,17 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.2",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- },
- {
- "path": 0
- }
- ]
- }
- ]
-}
diff --git a/0.2/examples/image/invalid/missing_datasets.json b/0.2/examples/image/invalid/missing_datasets.json
deleted file mode 100644
index 6604968d..00000000
--- a/0.2/examples/image/invalid/missing_datasets.json
+++ /dev/null
@@ -1,9 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.2",
- "name": "example"
- }
- ]
-}
diff --git a/0.2/examples/image/invalid/missing_path.json b/0.2/examples/image/invalid/missing_path.json
deleted file mode 100644
index 487a4266..00000000
--- a/0.2/examples/image/invalid/missing_path.json
+++ /dev/null
@@ -1,17 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.2",
- "name": "example",
- "datasets": [
- {
- "foo": "path/to/0"
- },
- {
- "path": "1"
- }
- ]
- }
- ]
-}
diff --git a/0.2/examples/image/invalid/no_datasets.json b/0.2/examples/image/invalid/no_datasets.json
deleted file mode 100644
index 839a60aa..00000000
--- a/0.2/examples/image/invalid/no_datasets.json
+++ /dev/null
@@ -1,15 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.2",
- "name": "example",
- "datasets": [],
- "axes": [
- "z",
- "y",
- "x"
- ]
- }
- ]
-}
diff --git a/0.2/examples/image/invalid/no_multiscales.json b/0.2/examples/image/invalid/no_multiscales.json
deleted file mode 100644
index d6dbfb33..00000000
--- a/0.2/examples/image/invalid/no_multiscales.json
+++ /dev/null
@@ -1,4 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": []
-}
diff --git a/0.2/examples/image/valid/image.json b/0.2/examples/image/valid/image.json
deleted file mode 100644
index 2c1e7d27..00000000
--- a/0.2/examples/image/valid/image.json
+++ /dev/null
@@ -1,14 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.2",
- "name": "example",
- "datasets": [
- {"path": "path/to/0"},
- {"path": "1"},
- {"path": "2"}
- ]
- }
- ]
-}
diff --git a/0.2/examples/image/valid/image_metadata.json b/0.2/examples/image/valid/image_metadata.json
deleted file mode 100644
index 3ca5cd47..00000000
--- a/0.2/examples/image/valid/image_metadata.json
+++ /dev/null
@@ -1,28 +0,0 @@
-{
- "@id": "top",
- "@type": "ngff:Image",
- "multiscales": [
- {
- "@id": "inner",
- "version": "0.2",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- }
- ],
- "type": "gaussian",
- "metadata": {
- "method": "skimage.transform.pyramid_gaussian",
- "version": "0.16.1",
- "args": [
- "true",
- "false"
- ],
- "kwargs": {
- "multichannel": true
- }
- }
- }
- ]
-}
diff --git a/0.2/examples/image/valid/image_omero.json b/0.2/examples/image/valid/image_omero.json
deleted file mode 100644
index 20b3b892..00000000
--- a/0.2/examples/image/valid/image_omero.json
+++ /dev/null
@@ -1,58 +0,0 @@
-{
- "@id": "#my-image",
- "@type": "ngff:Image",
- "multiscales": [
- {
- "@id": "#my-pyramid",
- "version": "0.2",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- },
- {
- "path": "1"
- },
- {
- "path": "2"
- }
- ],
- "type": "gaussian",
- "metadata": {
- "method": "skimage.transform.pyramid_gaussian",
- "version": "0.16.1",
- "args": [
- "true",
- "false"
- ],
- "kwargs": {
- "multichannel": true
- }
- }
- }
- ],
- "omero": {
- "id": 1,
- "version": "0.2",
- "channels": [
- {
- "active": true,
- "color": "0000FF",
- "family": "linear",
- "inverted": false,
- "label": "1234",
- "window": {
- "end": 1765.0,
- "max": 2555.0,
- "min": 5.0,
- "start": 0.0
- }
- }
- ],
- "rdefs": {
- "defaultZ": 0,
- "defaultT": 0,
- "model": "color"
- }
- }
-}
diff --git a/0.2/examples/image/valid/missing_name.json b/0.2/examples/image/valid/missing_name.json
deleted file mode 100644
index 3942a543..00000000
--- a/0.2/examples/image/valid/missing_name.json
+++ /dev/null
@@ -1,13 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.2",
- "datasets": [
- {
- "path": "path/to/0"
- }
- ]
- }
- ]
-}
diff --git a/0.2/examples/image/valid/missing_version.json b/0.2/examples/image/valid/missing_version.json
deleted file mode 100644
index c278ddef..00000000
--- a/0.2/examples/image/valid/missing_version.json
+++ /dev/null
@@ -1,13 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- }
- ]
- }
- ]
-}
diff --git a/0.2/examples/plate/valid/plate.json b/0.2/examples/plate/valid/plate.json
deleted file mode 100644
index 0712bad8..00000000
--- a/0.2/examples/plate/valid/plate.json
+++ /dev/null
@@ -1,40 +0,0 @@
-{
- "plate": {
- "columns": [
- {
- "name": "1"
- },
- {
- "name": "2"
- },
- {
- "name": "3"
- }
- ],
- "field_count": 1,
- "name": "plate name",
- "rows": [
- {
- "name": "A"
- },
- {
- "name": "B"
- }
- ],
- "version": "0.2",
- "wells": [
- {
- "path": "A/3"
- },
- {
- "path": "B/2"
- },
- {
- "path": "A/1"
- },
- {
- "path": "B/3"
- }
- ]
- }
-}
\ No newline at end of file
diff --git a/0.2/examples/well/valid/well.json b/0.2/examples/well/valid/well.json
deleted file mode 100644
index fe6edd09..00000000
--- a/0.2/examples/well/valid/well.json
+++ /dev/null
@@ -1,15 +0,0 @@
-{
- "well": {
- "images": [
- {
- "acquisition": 0,
- "path": "0"
- },
- {
- "acquisition": 3,
- "path": "1"
- }
- ],
- "version": "0.2"
- }
-}
\ No newline at end of file
diff --git a/0.2/header.include b/0.2/header.include
deleted file mode 100644
index 28305fe4..00000000
--- a/0.2/header.include
+++ /dev/null
@@ -1,34 +0,0 @@
-
-
-
-
-
-
-
-
-  -
-
-
-
-
-
-
-
- -
-
-
- [TITLE]
-[LONGSTATUS], -
- - - --
Status of this document
- - - - --Title: Next-generation file formats (NGFF) -Shortname: ome-ngff -Level: 1 -Status: w3c/CG-FINAL -TR: https://ngff.openmicroscopy.org/0.2/ -URL: https://ngff.openmicroscopy.org/0.2/ -Repository: https://github.com/ome/ngff -Issue Tracking: Forums https://forum.image.sc/tag/ome-ngff -Logo: http://www.openmicroscopy.org/img/logos/ome-logomark.svg -Local Boilerplate: header yes, copyright yes -Boilerplate: style-darkmode off -Markup Shorthands: markdown yes -Editor: Josh Moore, Open Microscopy Environment (OME) https://www.openmicroscopy.org -Editor: Sébastien Besson, Open Microscopy Environment (OME) https://www.openmicroscopy.org -Abstract: This document contains next-generation file format (NGFF) -Abstract: specifications for storing bioimaging data in the cloud. -Abstract: All specifications are submitted to the https://image.sc community for review. -Status Text: This is the 0.2 release of this specification. Migration scripts -Status Text: will be provided between numbered versions. Data written with the latest version -Status Text: (an "editor's draft") will not necessarily be supported. -- -Introduction {#intro} -===================== - -Bioimaging science is at a crossroads. Currently, the drive to acquire more, -larger, preciser spatial measurements is unfortunately at odds with our ability -to structure and share those measurements with others. During a global pandemic -more than ever, we believe fervently that global, collaborative discovery as -opposed to the post-publication, "data-on-request" mode of operation is the -path forward. Bioimaging data should be shareable via open and commercial cloud -resources without the need to download entire datasets. - -At the moment, that is not the norm. The plethora of data formats produced by -imaging systems are ill-suited to remote sharing. Individual scientists -typically lack the infrastructure they need to host these data themselves. When -they acquire images from elsewhere, time-consuming translations and data -cleaning are needed to interpret findings. Those same costs are multiplied when -gathering data into online repositories where curator time can be the limiting -factor before publication is possible. Without a common effort, each lab or -resource is left building the tools they need and maintaining that -infrastructure often without dedicated funding. - -This document defines a specification for bioimaging data to make it possible -to enable the conversion of proprietary formats into a common, cloud-ready one. -Such next-generation file formats layout data so that individual portions, or -"chunks", of large data are reference-able eliminating the need to download -entire datasets. - - -Why "NGFF"? {#why-ngff} -------------------------------------------------------------------------------------------------- - -A short description of what is needed for an imaging format is "a hierarchy -of n-dimensional (dense) arrays with metadata". This combination of features -is certainly provided by HDF5 -from the HDF Group, which a number of -bioimaging formats do use. HDF5 and other larger binary structures, however, -are ill-suited for storage in the cloud where accessing individual chunks -of data by name rather than seeking through a large file is at the heart of -parallelization. - -As a result, a number of formats have been developed more recently which provide -the basic data structure of an HDF5 file, but do so in a more cloud-friendly way. -In the [PyData](https://pydata.org/) community, the Zarr [[zarr]] format was developed -for easily storing collections of [NumPy](https://numpy.org/) arrays. In the -[ImageJ](https://imagej.net/) community, N5 [[n5]] was developed to work around -the limitations of HDF5 ("N5" was originally short for "Not-HDF5"). -Both of these formats permit storing individual chunks of data either locally in -separate files or in cloud-based object stores as separate keys. - -A [current effort](https://zarr-specs.readthedocs.io/en/core-protocol-v3.0-dev/protocol/core/v3.0.html) -is underway to unify the two similar specifications to provide a single binary -specification. The editor's draft will soon be entering a [request for comments (RFC)](https://github.com/zarr-developers/zarr-specs/issues/101) phase with the goal of having a first version early in 2021. As that -process comes to an end, this document will be updated. - -OME-NGFF {#ome-ngff} --------------------- - -The conventions and specifications defined in this document are designed to -enable next-generation file formats to represent the same bioimaging data -that can be represented in \[OME-TIFF](http://www.openmicroscopy.org/ome-files/) -and beyond. However, the conventions will also be usable by HDF5 and other sufficiently advanced -binary containers. Eventually, we hope, the moniker "next-generation" will no longer be -applicable, and this will simply be the most efficient, common, and useful representation -of bioimaging data, whether during acquisition or sharing in the cloud. - -Note: The following text makes use of OME-Zarr [[ome-zarr-py]], the current prototype implementation, -for all examples. - -On-disk (or in-cloud) layout {#on-disk} -======================================= - -An overview of the layout of an OME-Zarr fileset should make -understanding the following metadata sections easier. The hierarchy -is represented here as it would appear locally but could equally -be stored on a web server to be accessed via HTTP or in object storage -like S3 or GCS. - -Images {#image-layout} ----------------------- - -The following layout describes the expected Zarr hierarchy for images with -multiple levels of resolutions and optionally associated labels. - -``` -. # Root folder, potentially in S3, -│ # with a flat list of images by image ID. -│ -├── 123.zarr # One image (id=123) converted to Zarr. -│ -└── 456.zarr # Another image (id=456) converted to Zarr. - │ - ├── .zgroup # Each image is a Zarr group, or a folder, of other groups and arrays. - ├── .zattrs # Group level attributes are stored in the .zattrs file and include - │ # "multiscales" and "omero" below) - │ - ├── 0 # Each multiscale level is stored as a separate Zarr array, - │ ... # which is a folder containing chunk files which compose the array. - ├── n # The name of the array is arbitrary with the ordering defined by - │ │ # by the "multiscales" metadata, but is often a sequence starting at 0. - │ │ - │ ├── .zarray # All image arrays are 5-dimensional - │ │ # with dimension order (t, c, z, y, x). - │ │ - │ └─ t # Chunks are stored with the nested directory layout. - │ └─ c # All but the last chunk element are stored as directories. - │ └─ z # The terminal chunk is a file. Together the directory and file names - │ └─ y # provide the "chunk coordinate" (t, c, z, y, x), where the maximum coordinate - │ └─ x # will be `dimension_size / chunk_size`. - │ - └── labels - │ - ├── .zgroup # The labels group is a container which holds a list of labels to make the objects easily discoverable - │ - ├── .zattrs # All labels will be listed in `.zattrs` e.g. `{ "labels": [ "original/0" ] }` - │ # Each dimension of the label `(t, c, z, y, x)` should be either the same as the - │ # corresponding dimension of the image, or `1` if that dimension of the label - │ # is irrelevant. - │ - └── original # Intermediate folders are permitted but not necessary and currently contain no extra metadata. - │ - └── 0 # Multiscale, labeled image. The name is unimportant but is registered in the "labels" group above. - ├── .zgroup # Zarr Group which is both a multiscaled image as well as a labeled image. - ├── .zattrs # Metadata of the related image and as well as display information under the "image-label" key. - │ - ├── 0 # Each multiscale level is stored as a separate Zarr array, as above, but only integer values - │ ... # are supported. - └── n -``` - -High-content screening {#hcs-layout} ------------------------------------- - -The following specification defines the hierarchy for a high-content screening -dataset. Three groups must be defined above the images: - -- the group above the images defines the well and MUST implement the - [well specification](#well-md). All images contained in a well are fields - of view of the same well -- the group above the well defines a row of wells -- the group above the well row defines an entire plate i.e. a two-dimensional - collection of wells organized in rows and columns. It MUST implement the - [plate specification](#plate-md) - - -``` -. # Root folder, potentially in S3, -│ -└── 5966.zarr # One plate (id=5966) converted to Zarr - ├── .zgroup - ├── .zattrs # Implements "plate" specification - ├── A # First row of the plate - │ ├── .zgroup - │ │ - │ ├── 1 # First column of row A - │ │ ├── .zgroup - │ │ ├── .zattrs # Implements "well" specification - │ │ │ - │ │ ├── 0 # First field of view of well A1 - │ │ │ │ - │ │ │ ├── .zgroup - │ │ │ ├── .zattrs # Implements "multiscales", "omero" - │ │ │ ├── 0 - │ │ │ │ ... # Resolution levels - │ │ │ ├── n - │ │ │ └── labels # Labels (optional) - │ │ ├── ... # Fields of view - │ │ └── m - │ ├── ... # Columns - │ └── 12 - ├── ... # Rows - └── H -``` - -Metadata {#metadata} -==================== - -The various `.zattrs` files throughout the above array hierarchy may contain metadata -keys as specified below for discovering certain types of data, especially images. - -"multiscales" metadata {#multiscale-md} ---------------------------------------- - -Metadata about the multiple resolution representations of the image can be -found under the "multiscales" key in the group-level metadata. - -"multiscales" contains a list of dictionaries where each entry describes a multiscale image. - -Each dictionary contained in the list MUST contain the field "datasets", which is a list of dictionaries describing -the arrays storing the individual resolution levels. -Each dictionary in "datasets" MUST contain the field "path", whose value contains the path to the array for this resolution relative -to the current zarr group. The "path"s MUST be ordered from largest (i.e. highest resolution) to smallest. - -It SHOULD contain the field "name". - -It SHOULD contain the field "version", which indicates the version of the -multiscale metadata of this image (current version is 0.2). - -It SHOULD contain the field "type", which gives the type of downscaling method used to generate the multiscale image pyramid. - -It SHOULD contain the field "metadata", which contains a dictionary with additional information about the downscaling method. - -```json -{ - "multiscales": [ - { - "version": "0.2", - "name": "example", - "datasets": [ - {"path": "0"}, - {"path": "1"}, - {"path": "2"} - ], - "type": "gaussian", - "metadata": { # the fields in metadata depend on the downscaling implementation - "method": "skimage.transform.pyramid_gaussian", # here, the parameters passed to the skimage function are given - "version": "0.16.1", - "args": "[true]", - "kwargs": {"multichannel": true} - } - } - ] -} -``` - -If only one multiscale is provided, use it. Otherwise, the user can choose by -name, using the first multiscale as a fallback: - -```python -datasets = [] -for named in multiscales: - if named["name"] == "3D": - datasets = [x["path"] for x in named["datasets"]] - break -if not datasets: - # Use the first by default. Or perhaps choose based on chunk size. - datasets = [x["path"] for x in multiscales[0]["datasets"]] -``` - -"omero" metadata {#omero-md} ----------------------------- - -Information specific to the channels of an image and how to render it -can be found under the "omero" key in the group-level metadata: - -```json -"id": 1, # ID in OMERO -"name": "example.tif", # Name as shown in the UI -"version": "0.2", # Current version -"channels": [ # Array matching the c dimension size - { - "active": true, - "coefficient": 1, - "color": "0000FF", - "family": "linear", - "inverted": false, - "label": "LaminB1", - "window": { - "end": 1500, - "max": 65535, - "min": 0, - "start": 0 - } - } -], -"rdefs": { - "defaultT": 0, # First timepoint to show the user - "defaultZ": 118, # First Z section to show the user - "model": "color" # "color" or "greyscale" -} -``` - -See https://docs.openmicroscopy.org/omero/5.6.1/developers/Web/WebGateway.html#imgdata -for more information. - -"labels" metadata {#labels-md} ------------------------------- - -The special group "labels" found under an image Zarr contains the key `labels` containing -the paths to label objects which can be found underneath the group: - -```json -{ - "labels": [ - "orphaned/0" - ] -} -``` - -Unlisted groups MAY be labels. - -"image-label" metadata {#label-md} ----------------------------------- - -Groups containing the `image-label` dictionary represent an image segmentation -in which each unique pixel value represents a separate segmented object. -`image-label` groups MUST also contain `multiscales` metadata and the two -"datasets" series MUST have the same number of entries. - -The `colors` key defines a list of JSON objects describing the unique label -values. Each entry in the list MUST contain the key "label-value" with the -pixel value for that label. Additionally, the "rgba" key MAY be present, the -value for which is an RGBA unsigned-int 4-tuple: `[uint8, uint8, uint8, uint8]` -All `label-value`s must be unique. Clients who choose to not throw an error -should ignore all except the _last_ entry. - -Some implementations may represent overlapping labels by using a specially assigned -value, for example the highest integer available in the pixel range. - -The `properties` key defines a list of JSON objects which also describes the unique -label values. Each entry in the list MUST contain the key "label-value" with the -pixel value for that label. Additionally, an arbitrary number of key-value pairs -MAY be present for each label value denoting associated metadata. Not all label -values must share the same key-value pairs within the properties list. - -The `source` key is an optional dictionary which contains information on the -image the label is associated with. If included it MAY include a key `image` -whose value is the relative path to a Zarr image group. The default value is -"../../" since most labels are stored under a subgroup named "labels/" (see -above). - - -```json -"image-label": - { - "version": "0.2", - "colors": [ - { - "label-value": 1, - "rgba": [255, 255, 255, 0] - }, - { - "label-value": 4, - "rgba": [0, 255, 255, 128] - }, - ... - ], - "properties": [ - { - "label-value": 1, - "area (pixels)": 1200, - "class": "foo" - - }, - { - "label-value": 4, - "area (pixels)": 1650 - }, - ... - ] - }, - "source": { - "image": "../../" - } -] -``` - -"plate" metadata {#plate-md} ----------------------------- - -For high-content screening datasets, the plate layout can be found under the -custom attributes of the plate group under the `plate` key. - -
-
-
- acquisitions -
- An optional list of JSON objects defining the acquisitions for a given - plate. Each acquisition object MUST contain an `id` key providing an - unique identifier within the context of the plate to which fields of - view can refer to. It SHOULD contain a `name` key identifying the name - of the acquisition. It SHOULD contain a `maximumfieldcount` key - indicating the maximum number of fields of view for the acquisition. It - MAY contain a `description` key providing a description for the - acquisition. It MAY contain a `startime` and/or `endtime` key specifying - the start and/or end timestamp of the acquisition using an epoch - string. -
- columns -
- A list of JSON objects defining the columns of the plate. Each column - object defines the properties of the column at the index of the object - in the list. If not empty, it MUST contain a `name` key specifying the - column name. -
- field_count -
- An integer defining the maximum number of fields per view across all - wells. -
- name -
- A string defining the name of the plate. -
- rows -
- A list of JSON objects defining the rows of the plate. Each row object - defines the properties of the row at the index of the object in the - list. If not empty, it MUST contain a `name` key specifying the row - name. -
- version -
- A string defining the version of the specification. -
- wells -
- A list of JSON objects defining the wells of the plate. Each well object - MUST contain a `path` key identifying the path to the well subgroup. -
-
-
- images -
- A list of JSON objects defining the fields of views for a given well. - Each object MUST contain a `path` key identifying the path to the - field of view. If multiple acquisitions were performed in the plate, it - SHOULD contain an `acquisition` key identifying the id of the - acquisition which must match one of acquisition JSON objects defined in - the plate metadata. -
- version -
- A string defining the version of the specification. -
-
-
-
- [bigdataviewer-ome-zarr](https://github.com/mobie/bigdataviewer-ome-zarr) -
- Fiji-plugin for reading OME-Zarr. - -
- [bioformats2raw](https://github.com/glencoesoftware/bioformats2raw) -
- A performant, Bio-Formats image file format converter. - -
- [omero-ms-zarr](https://github.com/ome/omero-ms-zarr) -
- A microservice for OMERO.server that converts images stored in OMERO to OME-Zarr files on the fly, served via a web API. - -
- [idr-zarr-tools](https://github.com/IDR/idr-zarr-tools) -
- A full workflow demonstrating the conversion of IDR images to OME-Zarr images on S3. - -
- [OMERO CLI Zarr plugin](https://github.com/ome/omero-cli-zarr) -
- An OMERO CLI plugin that converts images stored in OMERO.server into a local Zarr file. - -
- [ome-zarr-py](https://github.com/ome/ome-zarr-py) -
- A napari plugin for reading ome-zarr files. - -
- [vizarr](https://github.com/hms-dbmi/vizarr/) -
- A minimal, purely client-side program for viewing Zarr-based images with Viv & ImJoy. - -
 -
-All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
-version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
-Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
-
-Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
-
-Citing {#citing}
-================
-
-[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.2)
-J. Moore, *et al*. Open Microscopy Environment Consortium, 29 March, 2021.
-This edition of the specification is [https://ngff.openmicroscopy.org/0.2/](https://ngff.openmicroscopy.org/0.2/]).
-The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
-[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
-
-Version History {#history}
-==========================
-
-
-
-All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
-version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
-Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
-
-Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
-
-Citing {#citing}
-================
-
-[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.2)
-J. Moore, *et al*. Open Microscopy Environment Consortium, 29 March, 2021.
-This edition of the specification is [https://ngff.openmicroscopy.org/0.2/](https://ngff.openmicroscopy.org/0.2/]).
-The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
-[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
-
-Version History {#history}
-==========================
-
-| Revision | -Date | -Description | -
| 0.2.0 | -2021-03-29 | -Change chunk dimension separator to "/" | -
| 0.1.4 | -2020-11-26 | -Add HCS specification | -
| 0.1.3 | -2020-09-14 | -Add labels specification | -
| 0.1.2 | -2020-05-07 | -Add description of "omero" metadata | -
| 0.1.1 | -2020-05-06 | -Add info on the ordering of resolutions | -
| 0.1.0 | -2020-04-20 | -First version for internal demo | -
-{
- "blogNov2020": {
- "href": "https://blog.openmicroscopy.org/file-formats/community/2020/11/04/zarr-data/",
- "title": "Public OME-Zarr data (Nov. 2020)",
- "authors": [
- "OME Team"
- ],
- "status": "Informational",
- "publisher": "OME",
- "id": "blogNov2020",
- "date": "04 November 2020"
- },
- "imagesc26952": {
- "href": "https://forum.image.sc/t/ome-s-position-regarding-file-formats/26952",
- "title": "OME’s position regarding file formats",
- "authors": [
- "OME Team"
- ],
- "status": "Informational",
- "publisher": "OME",
- "id": "imagesc26952",
- "date": "19 June 2020"
- },
- "n5": {
- "id": "n5",
- "href": "https://github.com/saalfeldlab/n5/issues/62",
- "title": "N5---a scalable Java API for hierarchies of chunked n-dimensional tensors and structured meta-data",
- "status": "Informational",
- "authors": [
- "John A. Bogovic",
- "Igor Pisarev",
- "Philipp Hanslovsky",
- "Neil Thistlethwaite",
- "Stephan Saalfeld"
- ],
- "date": "2020"
- },
- "ome-zarr-py": {
- "id": "ome-zarr-py",
- "href": "https://doi.org/10.5281/zenodo.4113931",
- "title": "ome-zarr-py: Experimental implementation of next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.",
- "status": "Informational",
- "publisher": "Zenodo",
- "authors": [
- "OME",
- "et al"
- ],
- "date": "06 October 2020"
- },
- "zarr": {
- "id": "zarr",
- "href": "https://doi.org/10.5281/zenodo.4069231",
- "title": "Zarr: An implementation of chunked, compressed, N-dimensional arrays for Python.",
- "status": "Informational",
- "publisher": "Zenodo",
- "authors": [
- "Alistair Miles",
- "et al"
- ],
- "date": "06 October 2020"
- }
-}
-
diff --git a/0.2/schemas/image.schema b/0.2/schemas/image.schema
deleted file mode 100644
index 6eb26274..00000000
--- a/0.2/schemas/image.schema
+++ /dev/null
@@ -1,101 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "http://localhost:8000/image.schema",
- "title": "NGFF Image",
- "description": "JSON from OME-NGFF .zattrs",
- "type": "object",
- "properties": {
- "multiscales": {
- "description": "The multiscale datasets for this image",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- },
- "datasets": {
- "type": "array",
- "minItems": 1,
- "items": {
- "type": "object",
- "properties": {
- "path": {
- "type": "string"
- }
- },
- "required": ["path"]
- }
- },
- "version": {
- "type": "string",
- "enum": [
- "0.2"
- ]
- }
- },
- "required": [
- "datasets"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "omero": {
- "type": "object",
- "properties": {
- "channels": {
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "window": {
- "type": "object",
- "properties": {
- "end": {
- "type": "number"
- },
- "max": {
- "type": "number"
- },
- "min": {
- "type": "number"
- },
- "start": {
- "type": "number"
- }
- },
- "required": [
- "start",
- "min",
- "end",
- "max"
- ]
- },
- "label": {
- "type": "string"
- },
- "family": {
- "type": "string"
- },
- "color": {
- "type": "string"
- },
- "active": {
- "type": "boolean"
- }
- },
- "required": [
- "window",
- "color"
- ]
- }
- }
- },
- "required": [
- "channels"
- ]
- }
- },
- "required": [ "multiscales" ]
-}
diff --git a/0.2/schemas/plate.schema b/0.2/schemas/plate.schema
deleted file mode 100644
index e30b1662..00000000
--- a/0.2/schemas/plate.schema
+++ /dev/null
@@ -1,119 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/0.2/schemas/plate.schema",
- "title": "OME-NGFF plate schema",
- "description": "JSON from OME-NGFF Plate .zattrs",
- "type": "object",
- "properties": {
- "plate": {
- "type": "object",
- "properties": {
- "version": {
- "type": "string",
- "enum": [
- "0.2"
- ]
- },
- "name": {
- "type": "string"
- },
- "columns": {
- "description": "Columns of the Plate grid",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- }
- },
- "required": [
- "name"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "rows": {
- "description": "Rows of the Plate grid",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- }
- },
- "required": [
- "name"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "wells": {
- "description": "Rows of the Plate grid",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "path": {
- "type": "string"
- }
- },
- "required": [
- "path"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "field_count": {
- "description": "Maximum number of fields per view across all wells."
- },
- "acquisitions": {
- "description": "Rows of the Plate grid",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "id": {
- "type": "integer"
- },
- "maximumfieldcount": {
- "type": "integer"
- },
- "name": {
- "type": "string"
- },
- "description": {
- "type": "string"
- },
- "starttime": {
- "description": "The start timestamp of the acquisition, expressed as epoch time i.e. the number seconds since the Epoch",
- "type": "integer",
- "minimum": 0
- },
- "endtime": {
- "description": "The end timestamp of the acquisition, expressed as epoch time i.e. the number seconds since the Epoch",
- "type": "integer",
- "minimum": 0
- }
- },
- "required": [
- "id"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- }
- },
- "required": [
- "version", "columns", "rows", "wells"
- ]
- }
- },
- "required": [
- "plate"
- ]
-}
\ No newline at end of file
diff --git a/0.2/schemas/well.schema b/0.2/schemas/well.schema
deleted file mode 100644
index 8dca3290..00000000
--- a/0.2/schemas/well.schema
+++ /dev/null
@@ -1,47 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/0.2/schemas/well.schema",
- "title": "OME-NGFF well schema",
- "description": "JSON from OME-NGFF .zattrs",
- "type": "object",
- "properties": {
- "well": {
- "type": "object",
- "properties": {
- "images": {
- "description": "The fields of view for this well",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "acquisition": {
- "description": "A unique identifier within the context of the plate",
- "type": "integer"
- },
- "path": {
- "description": "The path for this field of view subgroup",
- "type": "string",
- "pattern": "^[A-Za-z0-9]+$"
- }
- },
- "required": [
- "path"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "version": {
- "description": "The version of the specification",
- "type": "string",
- "enum": [
- "0.2"
- ]
- }
- },
- "required": [
- "images"
- ]
- }
- }
-}
diff --git a/0.2/tests/test_validation.py b/0.2/tests/test_validation.py
deleted file mode 100644
index 80decdf4..00000000
--- a/0.2/tests/test_validation.py
+++ /dev/null
@@ -1,44 +0,0 @@
-import json
-import glob
-
-import pytest
-
-from jsonschema import validate
-from jsonschema.exceptions import ValidationError
-
-
-def files():
- return list(glob.glob(f"examples/*/valid/*.json")) + \
- list(glob.glob(f"examples/*/invalid/*.json"))
-
-def ids():
- return [str(x).split("/")[-1][0:-5] for x in files()]
-
-
-@pytest.mark.parametrize("testfile", files(), ids=ids())
-def test_json(testfile):
-
- if "invalid" in testfile:
- with pytest.raises(ValidationError):
- json_schema(testfile)
- else:
- json_schema(testfile)
-
-
-def json_schema(path):
- # Load the correct schema
- with open(path) as f:
- test_json = json.loads(f.read())
- # we don't have @type in this version
- if "multiscales" in test_json:
- schema_name = "image.schema"
- elif "plate" in test_json:
- schema_name = "plate.schema"
- elif "well" in test_json:
- schema_name = "well.schema"
- else:
- raise Exception("No schema found")
-
- with open('schemas/' + schema_name) as f:
- schema = json.loads(f.read())
- validate(instance=test_json, schema=schema)
diff --git a/0.2/tox.ini b/0.2/tox.ini
deleted file mode 100644
index 5f9175d3..00000000
--- a/0.2/tox.ini
+++ /dev/null
@@ -1,10 +0,0 @@
-[tox]
-envlist = v02
-skipsdist = True
-
-[testenv]
-deps =
- pytest
- jsonschema
-commands =
- pytest tests --color=yes --basetemp={envtmpdir} {posargs:-v}
diff --git a/0.3/copyright.include b/0.3/copyright.include
deleted file mode 100644
index f0def708..00000000
--- a/0.3/copyright.include
+++ /dev/null
@@ -1,4 +0,0 @@
-Copyright © 2020-[YEAR]
-OME®
-(U. Dundee).
-OME trademark rules apply.
diff --git a/0.3/examples/image/invalid/invalid_axes.json b/0.3/examples/image/invalid/invalid_axes.json
deleted file mode 100644
index 74fb909d..00000000
--- a/0.3/examples/image/invalid/invalid_axes.json
+++ /dev/null
@@ -1,22 +0,0 @@
-{
- "@id": "#my-image",
- "@type": "ngff:Image",
- "multiscales": [
- {
- "@id": "#my-pyramid",
- "version": "0.3",
- "name": "example",
- "datasets": [
- {
- "@id": "#my-full-resolution",
- "path": "path/to/0"
- }
- ],
- "axes": [
- "z",
- "y",
- "ct"
- ]
- }
- ]
-}
diff --git a/0.3/examples/image/invalid/invalid_axes_count.json b/0.3/examples/image/invalid/invalid_axes_count.json
deleted file mode 100644
index b79d5ee2..00000000
--- a/0.3/examples/image/invalid/invalid_axes_count.json
+++ /dev/null
@@ -1,20 +0,0 @@
-{
- "@id": "#my-image",
- "@type": "ngff:Image",
- "multiscales": [
- {
- "@id": "#my-pyramid",
- "version": "0.3",
- "name": "example",
- "datasets": [
- {
- "@id": "#my-full-resolution",
- "path": "path/to/0"
- }
- ],
- "axes": [
- "y"
- ]
- }
- ]
-}
\ No newline at end of file
diff --git a/0.3/examples/image/invalid/invalid_axes_order.json b/0.3/examples/image/invalid/invalid_axes_order.json
deleted file mode 100644
index d27e3e8a..00000000
--- a/0.3/examples/image/invalid/invalid_axes_order.json
+++ /dev/null
@@ -1,22 +0,0 @@
-{
- "@id": "#my-image",
- "@type": "ngff:Image",
- "multiscales": [
- {
- "@id": "#my-pyramid",
- "version": "0.3",
- "name": "example",
- "datasets": [
- {
- "@id": "#my-full-resolution",
- "path": "path/to/0"
- }
- ],
- "axes": [
- "y",
- "t",
- "c"
- ]
- }
- ]
-}
\ No newline at end of file
diff --git a/0.3/examples/image/invalid/invalid_channels_color.json b/0.3/examples/image/invalid/invalid_channels_color.json
deleted file mode 100644
index 6c6990c0..00000000
--- a/0.3/examples/image/invalid/invalid_channels_color.json
+++ /dev/null
@@ -1,36 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.3",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- }
- ],
- "axes": [
- "z",
- "y",
- "x"
- ]
- }
- ],
- "omero": {
- "channels": [
- {
- "active": true,
- "coefficient": 1.0,
- "color": 255,
- "family": "linear",
- "label": "1234",
- "window": {
- "end": 1765.0,
- "max": 2555.0,
- "min": 5.0,
- "start": 0.0
- }
- }
- ]
- }
-}
diff --git a/0.3/examples/image/invalid/invalid_channels_window.json b/0.3/examples/image/invalid/invalid_channels_window.json
deleted file mode 100644
index 9c198f02..00000000
--- a/0.3/examples/image/invalid/invalid_channels_window.json
+++ /dev/null
@@ -1,36 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.3",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- }
- ],
- "axes": [
- "z",
- "y",
- "x"
- ]
- }
- ],
- "omero": {
- "channels": [
- {
- "active": true,
- "coefficient": 1.0,
- "color": "ff0000",
- "family": "linear",
- "label": "1234",
- "window": {
- "end": "100",
- "max": 2555.0,
- "min": 5.0,
- "start": 0.0
- }
- }
- ]
- }
-}
diff --git a/0.3/examples/image/invalid/invalid_path.json b/0.3/examples/image/invalid/invalid_path.json
deleted file mode 100644
index 1c517fb8..00000000
--- a/0.3/examples/image/invalid/invalid_path.json
+++ /dev/null
@@ -1,22 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.3",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- },
- {
- "path": 0
- }
- ],
- "axes": [
- "z",
- "y",
- "x"
- ]
- }
- ]
-}
diff --git a/0.3/examples/image/invalid/invalid_version.json b/0.3/examples/image/invalid/invalid_version.json
deleted file mode 100644
index ba995783..00000000
--- a/0.3/examples/image/invalid/invalid_version.json
+++ /dev/null
@@ -1,25 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "invalid",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- },
- {
- "path": "1"
- },
- {
- "path": "2"
- }
- ],
- "axes": [
- "z",
- "y",
- "x"
- ]
- }
- ]
-}
\ No newline at end of file
diff --git a/0.3/examples/image/invalid/missing_axes.json b/0.3/examples/image/invalid/missing_axes.json
deleted file mode 100644
index 5c1ebc4b..00000000
--- a/0.3/examples/image/invalid/missing_axes.json
+++ /dev/null
@@ -1,14 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.3",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- }
- ]
- }
- ]
-}
\ No newline at end of file
diff --git a/0.3/examples/image/invalid/missing_datasets.json b/0.3/examples/image/invalid/missing_datasets.json
deleted file mode 100644
index bcf96e46..00000000
--- a/0.3/examples/image/invalid/missing_datasets.json
+++ /dev/null
@@ -1,14 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.3",
- "name": "example",
- "axes": [
- "z",
- "y",
- "x"
- ]
- }
- ]
-}
diff --git a/0.3/examples/image/invalid/missing_path.json b/0.3/examples/image/invalid/missing_path.json
deleted file mode 100644
index 36439fa2..00000000
--- a/0.3/examples/image/invalid/missing_path.json
+++ /dev/null
@@ -1,22 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.3",
- "name": "example",
- "datasets": [
- {
- "foo": "path/to/0"
- },
- {
- "path": "1"
- }
- ],
- "axes": [
- "z",
- "y",
- "x"
- ]
- }
- ]
-}
diff --git a/0.3/examples/image/invalid/no_axes.json b/0.3/examples/image/invalid/no_axes.json
deleted file mode 100644
index 7bc397bb..00000000
--- a/0.3/examples/image/invalid/no_axes.json
+++ /dev/null
@@ -1,15 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.3",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- }
- ],
- "axes": []
- }
- ]
-}
diff --git a/0.3/examples/image/invalid/no_datasets.json b/0.3/examples/image/invalid/no_datasets.json
deleted file mode 100644
index 31537f03..00000000
--- a/0.3/examples/image/invalid/no_datasets.json
+++ /dev/null
@@ -1,15 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.3",
- "name": "example",
- "datasets": [],
- "axes": [
- "z",
- "y",
- "x"
- ]
- }
- ]
-}
diff --git a/0.3/examples/image/invalid/no_multiscales.json b/0.3/examples/image/invalid/no_multiscales.json
deleted file mode 100644
index d6dbfb33..00000000
--- a/0.3/examples/image/invalid/no_multiscales.json
+++ /dev/null
@@ -1,4 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": []
-}
diff --git a/0.3/examples/image/valid/image.json b/0.3/examples/image/valid/image.json
deleted file mode 100644
index ebcb882d..00000000
--- a/0.3/examples/image/valid/image.json
+++ /dev/null
@@ -1,15 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "datasets": [
- {"path": "path/to/0"},
- {"path": "1"},
- {"path": "2"}
- ],
- "axes": [
- "z", "y", "x"
- ]
- }
- ]
-}
diff --git a/0.3/examples/image/valid/image_metadata.json b/0.3/examples/image/valid/image_metadata.json
deleted file mode 100644
index 7f1e2ffd..00000000
--- a/0.3/examples/image/valid/image_metadata.json
+++ /dev/null
@@ -1,33 +0,0 @@
-{
- "@id": "top",
- "@type": "ngff:Image",
- "multiscales": [
- {
- "@id": "inner",
- "version": "0.3",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- }
- ],
- "type": "gaussian",
- "metadata": {
- "method": "skimage.transform.pyramid_gaussian",
- "version": "0.16.1",
- "args": [
- "true",
- "false"
- ],
- "kwargs": {
- "multichannel": true
- }
- },
- "axes": [
- "z",
- "y",
- "x"
- ]
- }
- ]
-}
diff --git a/0.3/examples/image/valid/image_omero.json b/0.3/examples/image/valid/image_omero.json
deleted file mode 100644
index 775dc2dc..00000000
--- a/0.3/examples/image/valid/image_omero.json
+++ /dev/null
@@ -1,63 +0,0 @@
-{
- "@id": "#my-image",
- "@type": "ngff:Image",
- "multiscales": [
- {
- "@id": "#my-pyramid",
- "version": "0.3",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- },
- {
- "path": "1"
- },
- {
- "path": "2"
- }
- ],
- "type": "gaussian",
- "metadata": {
- "method": "skimage.transform.pyramid_gaussian",
- "version": "0.16.1",
- "args": [
- "true",
- "false"
- ],
- "kwargs": {
- "multichannel": true
- }
- },
- "axes": [
- "z",
- "y",
- "x"
- ]
- }
- ],
- "omero": {
- "id": 1,
- "version": "0.3",
- "channels": [
- {
- "active": true,
- "color": "0000FF",
- "family": "linear",
- "inverted": false,
- "label": "1234",
- "window": {
- "end": 1765.0,
- "max": 2555.0,
- "min": 5.0,
- "start": 0.0
- }
- }
- ],
- "rdefs": {
- "defaultZ": 0,
- "defaultT": 0,
- "model": "color"
- }
- }
-}
diff --git a/0.3/examples/image/valid/missing_name.json b/0.3/examples/image/valid/missing_name.json
deleted file mode 100644
index 22697e44..00000000
--- a/0.3/examples/image/valid/missing_name.json
+++ /dev/null
@@ -1,30 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.3",
- "datasets": [
- {
- "path": "path/to/0"
- }
- ],
- "type": "gaussian",
- "metadata": {
- "method": "skimage.transform.pyramid_gaussian",
- "version": "0.16.1",
- "args": [
- "true",
- "false"
- ],
- "kwargs": {
- "multichannel": true
- }
- },
- "axes": [
- "z",
- "y",
- "x"
- ]
- }
- ]
-}
diff --git a/0.3/examples/image/valid/missing_version.json b/0.3/examples/image/valid/missing_version.json
deleted file mode 100644
index df165311..00000000
--- a/0.3/examples/image/valid/missing_version.json
+++ /dev/null
@@ -1,18 +0,0 @@
-{
- "@type": "ngff:Image",
- "multiscales": [
- {
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0"
- }
- ],
- "axes": [
- "z",
- "y",
- "x"
- ]
- }
- ]
-}
diff --git a/0.3/examples/plate/valid/plate.json b/0.3/examples/plate/valid/plate.json
deleted file mode 100644
index d06552f3..00000000
--- a/0.3/examples/plate/valid/plate.json
+++ /dev/null
@@ -1,40 +0,0 @@
-{
- "plate": {
- "columns": [
- {
- "name": "1"
- },
- {
- "name": "2"
- },
- {
- "name": "3"
- }
- ],
- "field_count": 1,
- "name": "plate name",
- "rows": [
- {
- "name": "A"
- },
- {
- "name": "B"
- }
- ],
- "version": "0.3",
- "wells": [
- {
- "path": "A/3"
- },
- {
- "path": "B/2"
- },
- {
- "path": "A/1"
- },
- {
- "path": "B/3"
- }
- ]
- }
-}
\ No newline at end of file
diff --git a/0.3/examples/well/valid/well.json b/0.3/examples/well/valid/well.json
deleted file mode 100644
index e4efac11..00000000
--- a/0.3/examples/well/valid/well.json
+++ /dev/null
@@ -1,15 +0,0 @@
-{
- "well": {
- "images": [
- {
- "acquisition": 0,
- "path": "0"
- },
- {
- "acquisition": 3,
- "path": "1"
- }
- ],
- "version": "0.3"
- }
-}
\ No newline at end of file
diff --git a/0.3/index.bs b/0.3/index.bs
deleted file mode 100644
index f8b3550e..00000000
--- a/0.3/index.bs
+++ /dev/null
@@ -1,700 +0,0 @@
--Title: Next-generation file formats (NGFF) -Shortname: ome-ngff -Level: 1 -Status: w3c/CG-FINAL -TR: https://ngff.openmicroscopy.org/0.3/ -URL: https://ngff.openmicroscopy.org/0.3/ -Repository: https://github.com/ome/ngff -Issue Tracking: Forums https://forum.image.sc/tag/ome-ngff -Logo: http://www.openmicroscopy.org/img/logos/ome-logomark.svg -Local Boilerplate: header yes, copyright yes -Boilerplate: style-darkmode off -Markup Shorthands: markdown yes -Editor: Josh Moore, Open Microscopy Environment (OME) https://www.openmicroscopy.org -Editor: Sébastien Besson, Open Microscopy Environment (OME) https://www.openmicroscopy.org -Editor: Constantin Pape, European Molecular Biology Laboratory (EMBL) https://www.embl.org/sites/heidelberg/ -Abstract: This document contains next-generation file format (NGFF) -Abstract: specifications for storing bioimaging data in the cloud. -Abstract: All specifications are submitted to the https://image.sc community for review. -Status Text: This is the 0.3 release of this specification. Migration scripts -Status Text: will be provided between numbered versions. Data written with the latest version -Status Text: (an "editor's draft") will not necessarily be supported. -- - -Introduction {#intro} -===================== - -Bioimaging science is at a crossroads. Currently, the drive to acquire more, -larger, preciser spatial measurements is unfortunately at odds with our ability -to structure and share those measurements with others. During a global pandemic -more than ever, we believe fervently that global, collaborative discovery as -opposed to the post-publication, "data-on-request" mode of operation is the -path forward. Bioimaging data should be shareable via open and commercial cloud -resources without the need to download entire datasets. - -At the moment, that is not the norm. The plethora of data formats produced by -imaging systems are ill-suited to remote sharing. Individual scientists -typically lack the infrastructure they need to host these data themselves. When -they acquire images from elsewhere, time-consuming translations and data -cleaning are needed to interpret findings. Those same costs are multiplied when -gathering data into online repositories where curator time can be the limiting -factor before publication is possible. Without a common effort, each lab or -resource is left building the tools they need and maintaining that -infrastructure often without dedicated funding. - -This document defines a specification for bioimaging data to make it possible -to enable the conversion of proprietary formats into a common, cloud-ready one. -Such next-generation file formats layout data so that individual portions, or -"chunks", of large data are reference-able eliminating the need to download -entire datasets. - - -Why "NGFF"? {#why-ngff} -------------------------------------------------------------------------------------------------- - -A short description of what is needed for an imaging format is "a hierarchy -of n-dimensional (dense) arrays with metadata". This combination of features -is certainly provided by HDF5 -from the HDF Group, which a number of -bioimaging formats do use. HDF5 and other larger binary structures, however, -are ill-suited for storage in the cloud where accessing individual chunks -of data by name rather than seeking through a large file is at the heart of -parallelization. - -As a result, a number of formats have been developed more recently which provide -the basic data structure of an HDF5 file, but do so in a more cloud-friendly way. -In the [PyData](https://pydata.org/) community, the Zarr [[zarr]] format was developed -for easily storing collections of [NumPy](https://numpy.org/) arrays. In the -[ImageJ](https://imagej.net/) community, N5 [[n5]] was developed to work around -the limitations of HDF5 ("N5" was originally short for "Not-HDF5"). -Both of these formats permit storing individual chunks of data either locally in -separate files or in cloud-based object stores as separate keys. - -A [current effort](https://zarr-specs.readthedocs.io/en/core-protocol-v3.0-dev/protocol/core/v3.0.html) -is underway to unify the two similar specifications to provide a single binary -specification. The editor's draft will soon be entering a [request for comments (RFC)](https://github.com/zarr-developers/zarr-specs/issues/101) phase with the goal of having a first version early in 2021. As that -process comes to an end, this document will be updated. - -OME-NGFF {#ome-ngff} --------------------- - -The conventions and specifications defined in this document are designed to -enable next-generation file formats to represent the same bioimaging data -that can be represented in \[OME-TIFF](http://www.openmicroscopy.org/ome-files/) -and beyond. However, the conventions will also be usable by HDF5 and other sufficiently advanced -binary containers. Eventually, we hope, the moniker "next-generation" will no longer be -applicable, and this will simply be the most efficient, common, and useful representation -of bioimaging data, whether during acquisition or sharing in the cloud. - -Note: The following text makes use of OME-Zarr [[ome-zarr-py]], the current prototype implementation, -for all examples. - -On-disk (or in-cloud) layout {#on-disk} -======================================= - -An overview of the layout of an OME-Zarr fileset should make -understanding the following metadata sections easier. The hierarchy -is represented here as it would appear locally but could equally -be stored on a web server to be accessed via HTTP or in object storage -like S3 or GCS. - -Images {#image-layout} ----------------------- - -The following layout describes the expected Zarr hierarchy for images with -multiple levels of resolutions and optionally associated labels. - -``` -. # Root folder, potentially in S3, -│ # with a flat list of images by image ID. -│ -├── 123.zarr # One image (id=123) converted to Zarr. -│ -└── 456.zarr # Another image (id=456) converted to Zarr. - │ - ├── .zgroup # Each image is a Zarr group, or a folder, of other groups and arrays. - ├── .zattrs # Group level attributes are stored in the .zattrs file and include - │ # "multiscales" and "omero" (see below). In addition, the group level attributes - │ # must also contain "_ARRAY_DIMENSIONS" if this group directly contains multi-scale arrays. - │ - ├── 0 # Each multiscale level is stored as a separate Zarr array, - │ ... # which is a folder containing chunk files which compose the array. - ├── n # The name of the array is arbitrary with the ordering defined by - │ │ # by the "multiscales" metadata, but is often a sequence starting at 0. - │ │ - │ ├── .zarray # All image arrays must be up to 5-dimensional - │ │ # with dimension order (t, c, z, y, x). - │ │ - │ └─ t # Chunks are stored with the nested directory layout. - │ └─ c # All but the last chunk element are stored as directories. - │ └─ z # The terminal chunk is a file. Together the directory and file names - │ └─ y # provide the "chunk coordinate" (t, c, z, y, x), where the maximum coordinate - │ └─ x # will be `dimension_size / chunk_size`. - │ - └── labels - │ - ├── .zgroup # The labels group is a container which holds a list of labels to make the objects easily discoverable - │ - ├── .zattrs # All labels will be listed in `.zattrs` e.g. `{ "labels": [ "original/0" ] }` - │ # Each dimension of the label `(t, c, z, y, x)` should be either the same as the - │ # corresponding dimension of the image, or `1` if that dimension of the label - │ # is irrelevant. - │ - └── original # Intermediate folders are permitted but not necessary and currently contain no extra metadata. - │ - └── 0 # Multiscale, labeled image. The name is unimportant but is registered in the "labels" group above. - ├── .zgroup # Zarr Group which is both a multiscaled image as well as a labeled image. - ├── .zattrs # Metadata of the related image and as well as display information under the "image-label" key. - │ - ├── 0 # Each multiscale level is stored as a separate Zarr array, as above, but only integer values - │ ... # are supported. - └── n -``` - -High-content screening {#hcs-layout} ------------------------------------- - -The following specification defines the hierarchy for a high-content screening -dataset. Three groups must be defined above the images: - -- the group above the images defines the well and MUST implement the - [well specification](#well-md). All images contained in a well are fields - of view of the same well -- the group above the well defines a row of wells -- the group above the well row defines an entire plate i.e. a two-dimensional - collection of wells organized in rows and columns. It MUST implement the - [plate specification](#plate-md) - - -``` -. # Root folder, potentially in S3, -│ -└── 5966.zarr # One plate (id=5966) converted to Zarr - ├── .zgroup - ├── .zattrs # Implements "plate" specification - ├── A # First row of the plate - │ ├── .zgroup - │ │ - │ ├── 1 # First column of row A - │ │ ├── .zgroup - │ │ ├── .zattrs # Implements "well" specification - │ │ │ - │ │ ├── 0 # First field of view of well A1 - │ │ │ │ - │ │ │ ├── .zgroup - │ │ │ ├── .zattrs # Implements "multiscales", "omero" - │ │ │ ├── 0 - │ │ │ │ ... # Resolution levels - │ │ │ ├── n - │ │ │ └── labels # Labels (optional) - │ │ ├── ... # Fields of view - │ │ └── m - │ ├── ... # Columns - │ └── 12 - ├── ... # Rows - └── H -``` - -Metadata {#metadata} -==================== - -The various `.zattrs` files throughout the above array hierarchy may contain metadata -keys as specified below for discovering certain types of data, especially images. - -"multiscales" metadata {#multiscale-md} ---------------------------------------- - -Metadata about the multiple resolution representations of the image can be -found under the "multiscales" key in the group-level metadata. - -"multiscales" contains a list of dictionaries where each entry describes a multiscale image. - -Each dictionary contained in the list MUST contain the field "datasets", which is a list of dictionaries describing -the arrays storing the individual resolution levels. -Each dictionary in "datasets" MUST contain the field "path", whose value contains the path to the array for this resolution relative -to the current zarr group. The "path"s MUST be ordered from largest (i.e. highest resolution) to smallest. - -It MUST contain the field "axes", which is a list of dimension names of the axes. -The values MUST be unique and one of `{"t", "c", "z", "y", "x"}`. -The number of values MUST be the same as the number of dimensions of the arrays corresponding to this image. -In addition, the "axes" values MUST be repeated in the field "_ARRAY_DIMENSIONS" of all scale groups -(i.e. groups containing arrays with the multiscale data). -This ensures compatibility with the [xarray zarr encoding](http://xarray.pydata.org/en/stable/internals/zarr-encoding-spec.html#zarr-encoding). - -It SHOULD contain the field "name". - -It SHOULD contain the field "version", which indicates the version of the -multiscale metadata of this image (current version is 0.3). - -It SHOULD contain the field "type", which gives the type of downscaling method used to generate the multiscale image pyramid. - -It SHOULD contain the field "metadata", which contains a dictionary with additional information about the downscaling method. - -```json -{ - "multiscales": [ - { - "version": "0.3", - "name": "example", - "datasets": [ - {"path": "0"}, - {"path": "1"}, - {"path": "2"} - ], - "axes": [ - "t", "c", "z", "y", "x" - ], - "type": "gaussian", - "metadata": { # the fields in metadata depend on the downscaling implementation - "method": "skimage.transform.pyramid_gaussian", # here, the parameters passed to the skimage function are given - "version": "0.16.1", - "args": "[true]", - "kwargs": {"multichannel": true} - } - } - ] -} -``` - -If only one multiscale is provided, use it. Otherwise, the user can choose by -name, using the first multiscale as a fallback: - -```python -datasets = [] -for named in multiscales: - if named["name"] == "3D": - datasets = [x["path"] for x in named["datasets"]] - break -if not datasets: - # Use the first by default. Or perhaps choose based on chunk size. - datasets = [x["path"] for x in multiscales[0]["datasets"]] -``` - -"omero" metadata {#omero-md} ----------------------------- - -Information specific to the channels of an image and how to render it -can be found under the "omero" key in the group-level metadata: - -```json -"id": 1, # ID in OMERO -"name": "example.tif", # Name as shown in the UI -"version": "0.3", # Current version -"channels": [ # Array matching the c dimension size - { - "active": true, - "coefficient": 1, - "color": "0000FF", - "family": "linear", - "inverted": false, - "label": "LaminB1", - "window": { - "end": 1500, - "max": 65535, - "min": 0, - "start": 0 - } - } -], -"rdefs": { - "defaultT": 0, # First timepoint to show the user - "defaultZ": 118, # First Z section to show the user - "model": "color" # "color" or "greyscale" -} -``` - -See https://docs.openmicroscopy.org/omero/5.6.1/developers/Web/WebGateway.html#imgdata -for more information. - -"labels" metadata {#labels-md} ------------------------------- - -The special group "labels" found under an image Zarr contains the key `labels` containing -the paths to label objects which can be found underneath the group: - -```json -{ - "labels": [ - "orphaned/0" - ] -} -``` - -Unlisted groups MAY be labels. - -"image-label" metadata {#label-md} ----------------------------------- - -Groups containing the `image-label` dictionary represent an image segmentation -in which each unique pixel value represents a separate segmented object. -`image-label` groups MUST also contain `multiscales` metadata and the two -"datasets" series MUST have the same number of entries. - -The `colors` key defines a list of JSON objects describing the unique label -values. Each entry in the list MUST contain the key "label-value" with the -pixel value for that label. Additionally, the "rgba" key MAY be present, the -value for which is an RGBA unsigned-int 4-tuple: `[uint8, uint8, uint8, uint8]` -All `label-value`s must be unique. Clients who choose to not throw an error -should ignore all except the _last_ entry. - -Some implementations may represent overlapping labels by using a specially assigned -value, for example the highest integer available in the pixel range. - -The `properties` key defines a list of JSON objects which also describes the unique -label values. Each entry in the list MUST contain the key "label-value" with the -pixel value for that label. Additionally, an arbitrary number of key-value pairs -MAY be present for each label value denoting associated metadata. Not all label -values must share the same key-value pairs within the properties list. - -The `source` key is an optional dictionary which contains information on the -image the label is associated with. If included it MAY include a key `image` -whose value is the relative path to a Zarr image group. The default value is -"../../" since most labels are stored under a subgroup named "labels/" (see -above). - - -```json -"image-label": - { - "version": "0.3", - "colors": [ - { - "label-value": 1, - "rgba": [255, 255, 255, 0] - }, - { - "label-value": 4, - "rgba": [0, 255, 255, 128] - }, - ... - ], - "properties": [ - { - "label-value": 1, - "area (pixels)": 1200, - "class": "foo" - - }, - { - "label-value": 4, - "area (pixels)": 1650 - }, - ... - ] - }, - "source": { - "image": "../../" - } -] -``` - -"plate" metadata {#plate-md} ----------------------------- - -For high-content screening datasets, the plate layout can be found under the -custom attributes of the plate group under the `plate` key. - -
-
-
- acquisitions -
- An optional list of JSON objects defining the acquisitions for a given - plate. Each acquisition object MUST contain an `id` key providing an - unique identifier within the context of the plate to which fields of - view can refer to. It SHOULD contain a `name` key identifying the name - of the acquisition. It SHOULD contain a `maximumfieldcount` key - indicating the maximum number of fields of view for the acquisition. It - MAY contain a `description` key providing a description for the - acquisition. It MAY contain a `startime` and/or `endtime` key specifying - the start and/or end timestamp of the acquisition using an epoch - string. -
- columns -
- A list of JSON objects defining the columns of the plate. Each column - object defines the properties of the column at the index of the object - in the list. If not empty, it MUST contain a `name` key specifying the - column name. -
- field_count -
- An integer defining the maximum number of fields per view across all - wells. -
- name -
- A string defining the name of the plate. -
- rows -
- A list of JSON objects defining the rows of the plate. Each row object - defines the properties of the row at the index of the object in the - list. If not empty, it MUST contain a `name` key specifying the row - name. -
- version -
- A string defining the version of the specification. -
- wells -
- A list of JSON objects defining the wells of the plate. Each well object - MUST contain a `path` key identifying the path to the well subgroup. -
-
-
- images -
- A list of JSON objects defining the fields of views for a given well. - Each object MUST contain a `path` key identifying the path to the - field of view. If multiple acquisitions were performed in the plate, it - SHOULD contain an `acquisition` key identifying the id of the - acquisition which must match one of acquisition JSON objects defined in - the plate metadata. -
- version -
- A string defining the version of the specification. -
-
-
-
- [omero-ms-zarr](https://github.com/ome/omero-ms-zarr) -
- A microservice for OMERO.server that converts images stored in OMERO to OME Zarr files on the fly, served via a web API. - -
- [idr-zarr-tools](https://github.com/IDR/idr-zarr-tools) -
- A full workflow demonstrating the conversion of IDR images to OME Zarr images on S3. - -
- [OMERO CLI Zarr plugin](https://github.com/ome/omero-cli-zarr) -
- An OMERO CLI plugin that converts images stored in OMERO.server into a local Zarr file. - -
- [ome-zarr-py](https://github.com/ome/ome-zarr-py) -
- A napari plugin for reading ome-zarr files. - -
- [bioformats2raw](https://github.com/glencoesoftware/bioformats2raw) -
- A performant, Bio-Formats image file format converter. - -
- [vizarr](https://github.com/hms-dbmi/vizarr/) -
- A minimal, purely client-side program for viewing Zarr-based images with Viv & ImJoy. - -
 -
-All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
-version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
-Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
-
-Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
-
-Citing {#citing}
-================
-
-[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.3)
-J. Moore, *et al*. Open Microscopy Environment Consortium, 20 November 2020.
-This edition of the specification is [https://ngff.openmicroscopy.org/0.3/](https://ngff.openmicroscopy.org/0.3/]).
-The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
-[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
-
-Version History {#history}
-==========================
-
-
-
-All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
-version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
-Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
-
-Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
-
-Citing {#citing}
-================
-
-[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.3)
-J. Moore, *et al*. Open Microscopy Environment Consortium, 20 November 2020.
-This edition of the specification is [https://ngff.openmicroscopy.org/0.3/](https://ngff.openmicroscopy.org/0.3/]).
-The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
-[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
-
-Version History {#history}
-==========================
-
-| Revision | -Date | -Description | -
| 0.3.0 | -2021-08-24 | -Add axes field to multiscale metadata | -
| 0.2.0 | -2021-03-29 | -Change chunk dimension separator to "/" | -
| 0.1.4 | -2020-11-26 | -Add HCS specification | -
| 0.1.3 | -2020-09-14 | -Add labels specification | -
| 0.1.2 | -2020-05-07 | -Add description of "omero" metadata | -
| 0.1.1 | -2020-05-06 | -Add info on the ordering of resolutions | -
| 0.1.0 | -2020-04-20 | -First version for internal demo | -
-{
- "blogNov2020": {
- "href": "https://blog.openmicroscopy.org/file-formats/community/2020/11/04/zarr-data/",
- "title": "Public OME-Zarr data (Nov. 2020)",
- "authors": [
- "OME Team"
- ],
- "status": "Informational",
- "publisher": "OME",
- "id": "blogNov2020",
- "date": "04 November 2020"
- },
- "imagesc26952": {
- "href": "https://forum.image.sc/t/ome-s-position-regarding-file-formats/26952",
- "title": "OME’s position regarding file formats",
- "authors": [
- "OME Team"
- ],
- "status": "Informational",
- "publisher": "OME",
- "id": "imagesc26952",
- "date": "19 June 2020"
- },
- "n5": {
- "id": "n5",
- "href": "https://github.com/saalfeldlab/n5/issues/62",
- "title": "N5---a scalable Java API for hierarchies of chunked n-dimensional tensors and structured meta-data",
- "status": "Informational",
- "authors": [
- "John A. Bogovic",
- "Igor Pisarev",
- "Philipp Hanslovsky",
- "Neil Thistlethwaite",
- "Stephan Saalfeld"
- ],
- "date": "2020"
- },
- "ome-zarr-py": {
- "id": "ome-zarr-py",
- "href": "https://doi.org/10.5281/zenodo.4113931",
- "title": "ome-zarr-py: Experimental implementation of next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.",
- "status": "Informational",
- "publisher": "Zenodo",
- "authors": [
- "OME",
- "et al"
- ],
- "date": "06 October 2020"
- },
- "zarr": {
- "id": "zarr",
- "href": "https://doi.org/10.5281/zenodo.4069231",
- "title": "Zarr: An implementation of chunked, compressed, N-dimensional arrays for Python.",
- "status": "Informational",
- "publisher": "Zenodo",
- "authors": [
- "Alistair Miles",
- "et al"
- ],
- "date": "06 October 2020"
- }
-}
-
diff --git a/0.3/schemas/image.schema b/0.3/schemas/image.schema
deleted file mode 100644
index afc2bd16..00000000
--- a/0.3/schemas/image.schema
+++ /dev/null
@@ -1,109 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/0.3/schemas/image.schema",
- "title": "NGFF Image",
- "description": "JSON from OME-NGFF .zattrs",
- "type": "object",
- "properties": {
- "multiscales": {
- "description": "The multiscale datasets for this image",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- },
- "datasets": {
- "type": "array",
- "minItems": 1,
- "items": {
- "type": "object",
- "properties": {
- "path": {
- "type": "string"
- }
- },
- "required": ["path"]
- }
- },
- "version": {
- "type": "string",
- "enum": [
- "0.3"
- ]
- },
- "axes": {
- "type": "array",
- "minItems": 2,
- "items": {
- "type": "string",
- "pattern": "^[xyzct]$"
- }
- }
- },
- "required": [
- "datasets", "axes"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "omero": {
- "type": "object",
- "properties": {
- "channels": {
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "window": {
- "type": "object",
- "properties": {
- "end": {
- "type": "number"
- },
- "max": {
- "type": "number"
- },
- "min": {
- "type": "number"
- },
- "start": {
- "type": "number"
- }
- },
- "required": [
- "start",
- "min",
- "end",
- "max"
- ]
- },
- "label": {
- "type": "string"
- },
- "family": {
- "type": "string"
- },
- "color": {
- "type": "string"
- },
- "active": {
- "type": "boolean"
- }
- },
- "required": [
- "window",
- "color"
- ]
- }
- }
- },
- "required": [
- "channels"
- ]
- }
- },
- "required": [ "multiscales" ]
-}
diff --git a/0.3/schemas/jsonld/context.json b/0.3/schemas/jsonld/context.json
deleted file mode 100644
index 0341b30f..00000000
--- a/0.3/schemas/jsonld/context.json
+++ /dev/null
@@ -1,6 +0,0 @@
-{
- "@context": {
- "@vocab" : "http://localhost:8000/context.json#",
- "ngff" : "http://localhost:8000/context.json#"
- }
-}
diff --git a/0.3/schemas/jsonld/frame.json b/0.3/schemas/jsonld/frame.json
deleted file mode 100644
index 092ff3f6..00000000
--- a/0.3/schemas/jsonld/frame.json
+++ /dev/null
@@ -1,42 +0,0 @@
-{
- "@context": {
- "@vocab" : "http://localhost:8000/context.json#",
- "xsd": "http://www.w3.org/2001/XMLSchema#",
- "ngff" : "http://localhost:8000/context.json#",
- "ngff:multiscales": {
- "@container": "@set"
- },
- "ngff:datasets": {
- "@container": "@set"
- },
- "ngff:end": {
- "@type": "xsd:decimal"
- }
- },
- "@type": "Image",
- "multiscales": {
- "@type": {
- "@default": "Multiscale"
- },
- "datasets" : {
- "@type": {
- "@default": "Dataset"
- }
- }
- },
- "omero": {
- "@type": {
- "@default": "omero"
- },
- "channels": {
- "@type": {
- "@default": "Channel"
- },
- "window": {
- "@type": {
- "@default": "Window"
- }
- }
- }
- }
-}
diff --git a/0.3/schemas/jsonld/shacl.ttl b/0.3/schemas/jsonld/shacl.ttl
deleted file mode 100644
index 02cb213b..00000000
--- a/0.3/schemas/jsonld/shacl.ttl
+++ /dev/null
@@ -1,210 +0,0 @@
-@prefix xsd:
-
-
-  -
-
-
-
-
-
-
-
- -
-
-
- [TITLE]
-[LONGSTATUS], -
- - - --
Status of this document
- - - - --Title: Next-generation file formats (NGFF) -Shortname: ome-ngff -Level: 1 -Status: w3c/CG-FINAL -TR: https://ngff.openmicroscopy.org/0.4/ -URL: https://ngff.openmicroscopy.org/0.4/ -Repository: https://github.com/ome/ngff -Issue Tracking: Forums https://forum.image.sc/tag/ome-ngff -Logo: http://www.openmicroscopy.org/img/logos/ome-logomark.svg -Local Boilerplate: header yes, copyright yes -Boilerplate: style-darkmode off -Markup Shorthands: markdown yes -Editor: Josh Moore, University of Dundee (UoD) https://www.dundee.ac.uk, https://orcid.org/0000-0003-4028-811X -Editor: Sébastien Besson, University of Dundee (UoD) https://www.dundee.ac.uk, https://orcid.org/0000-0001-8783-1429 -Editor: Constantin Pape, European Molecular Biology Laboratory (EMBL) https://www.embl.org/sites/heidelberg/, https://orcid.org/0000-0001-6562-7187 -Abstract: This document contains next-generation file format (NGFF) -Abstract: specifications for storing bioimaging data in the cloud. -Abstract: All specifications are submitted to the https://image.sc community for review. -Status Text: This is the 0.4 release of this specification. Migration scripts -Status Text: will be provided between numbered versions. Data written with the latest version -Status Text: (an "editor's draft") will not necessarily be supported. -- -Introduction {#intro} -===================== - -Bioimaging science is at a crossroads. Currently, the drive to acquire more, -larger, preciser spatial measurements is unfortunately at odds with our ability -to structure and share those measurements with others. During a global pandemic -more than ever, we believe fervently that global, collaborative discovery as -opposed to the post-publication, "data-on-request" mode of operation is the -path forward. Bioimaging data should be shareable via open and commercial cloud -resources without the need to download entire datasets. - -At the moment, that is not the norm. The plethora of data formats produced by -imaging systems are ill-suited to remote sharing. Individual scientists -typically lack the infrastructure they need to host these data themselves. When -they acquire images from elsewhere, time-consuming translations and data -cleaning are needed to interpret findings. Those same costs are multiplied when -gathering data into online repositories where curator time can be the limiting -factor before publication is possible. Without a common effort, each lab or -resource is left building the tools they need and maintaining that -infrastructure often without dedicated funding. - -This document defines a specification for bioimaging data to make it possible -to enable the conversion of proprietary formats into a common, cloud-ready one. -Such next-generation file formats layout data so that individual portions, or -"chunks", of large data are reference-able eliminating the need to download -entire datasets. - - -Why "NGFF"? {#why-ngff} -------------------------------------------------------------------------------------------------- - -A short description of what is needed for an imaging format is "a hierarchy -of n-dimensional (dense) arrays with metadata". This combination of features -is certainly provided by HDF5 -from the HDF Group, which a number of -bioimaging formats do use. HDF5 and other larger binary structures, however, -are ill-suited for storage in the cloud where accessing individual chunks -of data by name rather than seeking through a large file is at the heart of -parallelization. - -As a result, a number of formats have been developed more recently which provide -the basic data structure of an HDF5 file, but do so in a more cloud-friendly way. -In the [PyData](https://pydata.org/) community, the Zarr [[zarr]] format was developed -for easily storing collections of [NumPy](https://numpy.org/) arrays. In the -[ImageJ](https://imagej.net/) community, N5 [[n5]] was developed to work around -the limitations of HDF5 ("N5" was originally short for "Not-HDF5"). -Both of these formats permit storing individual chunks of data either locally in -separate files or in cloud-based object stores as separate keys. - -A [current effort](https://zarr-specs.readthedocs.io/en/core-protocol-v3.0-dev/protocol/core/v3.0.html) -is underway to unify the two similar specifications to provide a single binary -specification. The editor's draft will soon be entering a [request for comments (RFC)](https://github.com/zarr-developers/zarr-specs/issues/101) phase with the goal of having a first version early in 2021. As that -process comes to an end, this document will be updated. - -OME-NGFF {#ome-ngff} --------------------- - -The conventions and specifications defined in this document are designed to -enable next-generation file formats to represent the same bioimaging data -that can be represented in \[OME-TIFF](http://www.openmicroscopy.org/ome-files/) -and beyond. However, the conventions will also be usable by HDF5 and other sufficiently advanced -binary containers. Eventually, we hope, the moniker "next-generation" will no longer be -applicable, and this will simply be the most efficient, common, and useful representation -of bioimaging data, whether during acquisition or sharing in the cloud. - -Note: The following text makes use of OME-Zarr [[ome-zarr-py]], the current prototype implementation, -for all examples. - -Document conventions --------------------- - -The key words “MUST”, “MUST NOT”, “REQUIRED”, “SHALL”, “SHALL NOT”, “SHOULD”, “SHOULD NOT”, -“RECOMMENDED”, “MAY”, and “OPTIONAL” are to be interpreted as described in -[RFC 2119](https://tools.ietf.org/html/rfc2119). - -
-Transitional metadata is added to the specification with the -intention of removing it in the future. Implementations may be expected (MUST) or -encouraged (SHOULD) to support the reading of the data, but writing will usually -be optional (MAY). Examples of transitional metadata include custom additions by -implementations that are later submitted as a formal specification. (See [[#bf2raw]]) -
- -Some of the JSON examples in this document include comments. However, these are only for -clarity purposes and comments MUST NOT be included in JSON objects. - -On-disk (or in-cloud) layout {#on-disk} -======================================= - -An overview of the layout of an OME-Zarr fileset should make -understanding the following metadata sections easier. The hierarchy -is represented here as it would appear locally but could equally -be stored on a web server to be accessed via HTTP or in object storage -like S3 or GCS. - -OME-Zarr is an implementation of the OME-NGFF specification using the Zarr -format. Arrays MUST be defined and stored in a hierarchical organization as -defined by the -[version 2 of the Zarr specification ](https://zarr.readthedocs.io/en/stable/spec/v2.html). -OME-NGFF metadata MUST be stored as attributes in the corresponding Zarr -groups. - -Images {#image-layout} ----------------------- - -The following layout describes the expected Zarr hierarchy for images with -multiple levels of resolutions and optionally associated labels. -Note that the number of dimensions is variable between 2 and 5 and that axis names are arbitrary, see [[#multiscale-md]] for details. -For this example we assume an image with 5 dimensions and axes called `t,c,z,y,x`. - -
-. # Root folder, potentially in S3,
-│ # with a flat list of images by image ID.
-│
-├── 123.zarr # One image (id=123) converted to Zarr.
-│
-└── 456.zarr # Another image (id=456) converted to Zarr.
- │
- ├── .zgroup # Each image is a Zarr group, or a folder, of other groups and arrays.
- ├── .zattrs # Group level attributes are stored in the .zattrs file and include
- │ # "multiscales" and "omero" (see below). In addition, the group level attributes
- │ # may also contain "_ARRAY_DIMENSIONS" for compatibility with xarray if this group directly contains multi-scale arrays.
- │
- ├── 0 # Each multiscale level is stored as a separate Zarr array,
- │ ... # which is a folder containing chunk files which compose the array.
- ├── n # The name of the array is arbitrary with the ordering defined by
- │ │ # by the "multiscales" metadata, but is often a sequence starting at 0.
- │ │
- │ ├── .zarray # All image arrays must be up to 5-dimensional
- │ │ # with the axis of type time before type channel, before spatial axes.
- │ │
- │ └─ t # Chunks are stored with the nested directory layout.
- │ └─ c # All but the last chunk element are stored as directories.
- │ └─ z # The terminal chunk is a file. Together the directory and file names
- │ └─ y # provide the "chunk coordinate" (t, c, z, y, x), where the maximum coordinate
- │ └─ x # will be `dimension_size / chunk_size`.
- │
- └── labels
- │
- ├── .zgroup # The labels group is a container which holds a list of labels to make the objects easily discoverable
- │
- ├── .zattrs # All labels will be listed in `.zattrs` e.g. `{ "labels": [ "original/0" ] }`
- │ # Each dimension of the label `(t, c, z, y, x)` should be either the same as the
- │ # corresponding dimension of the image, or `1` if that dimension of the label
- │ # is irrelevant.
- │
- └── original # Intermediate folders are permitted but not necessary and currently contain no extra metadata.
- │
- └── 0 # Multiscale, labeled image. The name is unimportant but is registered in the "labels" group above.
- ├── .zgroup # Zarr Group which is both a multiscaled image as well as a labeled image.
- ├── .zattrs # Metadata of the related image and as well as display information under the "image-label" key.
- │
- ├── 0 # Each multiscale level is stored as a separate Zarr array, as above, but only integer values
- │ ... # are supported.
- └── n
-
-
-
-
-High-content screening {#hcs-layout}
-------------------------------------
-
-The following specification defines the hierarchy for a high-content screening
-dataset. Three groups MUST be defined above the images:
-
-- the group above the images defines the well and MUST implement the
- [well specification](#well-md). All images contained in a well are fields
- of view of the same well
-- the group above the well defines a row of wells
-- the group above the well row defines an entire plate i.e. a two-dimensional
- collection of wells organized in rows and columns. It MUST implement the
- [plate specification](#plate-md)
-
-A well row group SHOULD NOT be present if there are no images in the well row.
-A well group SHOULD NOT be present if there are no images in the well.
-
-
--. # Root folder, potentially in S3, -│ -└── 5966.zarr # One plate (id=5966) converted to Zarr - ├── .zgroup - ├── .zattrs # Implements "plate" specification - ├── A # First row of the plate - │ ├── .zgroup - │ │ - │ ├── 1 # First column of row A - │ │ ├── .zgroup - │ │ ├── .zattrs # Implements "well" specification - │ │ │ - │ │ ├── 0 # First field of view of well A1 - │ │ │ │ - │ │ │ ├── .zgroup - │ │ │ ├── .zattrs # Implements "multiscales", "omero" - │ │ │ ├── 0 - │ │ │ │ ... # Resolution levels - │ │ │ ├── n - │ │ │ └── labels # Labels (optional) - │ │ ├── ... # Fields of view - │ │ └── m - │ ├── ... # Columns - │ └── 12 - ├── ... # Rows - └── H -- -Metadata {#metadata} -==================== - -The various `.zattrs` files throughout the above array hierarchy may contain metadata -keys as specified below for discovering certain types of data, especially images. - -"axes" metadata {#axes-md} --------------------------- - -"axes" describes the dimensions of a physical coordinate space. It is a list of dictionaries, where each dictionary describes a dimension (axis) and: -- MUST contain the field "name" that gives the name for this dimension. The values MUST be unique across all "name" fields. -- SHOULD contain the field "type". It SHOULD be one of "space", "time" or "channel", but MAY take other string values for custom axis types that are not part of this specification yet. -- SHOULD contain the field "unit" to specify the physical unit of this dimension. The value SHOULD be one of the following strings, which are valid units according to UDUNITS-2. - - Units for "space" axes: 'angstrom', 'attometer', 'centimeter', 'decimeter', 'exameter', 'femtometer', 'foot', 'gigameter', 'hectometer', 'inch', 'kilometer', 'megameter', 'meter', 'micrometer', 'mile', 'millimeter', 'nanometer', 'parsec', 'petameter', 'picometer', 'terameter', 'yard', 'yoctometer', 'yottameter', 'zeptometer', 'zettameter' - - Units for "time" axes: 'attosecond', 'centisecond', 'day', 'decisecond', 'exasecond', 'femtosecond', 'gigasecond', 'hectosecond', 'hour', 'kilosecond', 'megasecond', 'microsecond', 'millisecond', 'minute', 'nanosecond', 'petasecond', 'picosecond', 'second', 'terasecond', 'yoctosecond', 'yottasecond', 'zeptosecond', 'zettasecond' - -If part of [[#multiscale-md]], the length of "axes" MUST be equal to the number of dimensions of the arrays that contain the image data. - -"bioformats2raw.layout" (transitional) {#bf2raw} ------------------------------------------------- - -[=Transitional=] "bioformats2raw.layout" metadata identifies a group which implicitly describes a series of images. -The need for the collection stems from the common "multi-image file" scenario in microscopy. Parsers like Bio-Formats -define a strict, stable ordering of the images in a single container that can be used to refer to them by other tools. - -In order to capture that information within an OME-NGFF dataset, `bioformats2raw` internally introduced a wrapping layer. -The bioformats2raw layout has been added to v0.4 as a transitional specification to specify filesets that already exist -in the wild. An upcoming NGFF specification will replace this layout with explicit metadata. - -
Layout
- -Typical Zarr layout produced by running `bioformats2raw` on a fileset that contains more than one image (series > 1): - --series.ome.zarr # One converted fileset from bioformats2raw - ├── .zgroup - ├── .zattrs # Contains "bioformats2raw.layout" metadata - ├── OME # Special group for containing OME metadata - │ ├── .zgroup - │ ├── .zattrs # Contains "series" metadata - │ └── METADATA.ome.xml # OME-XML file stored within the Zarr fileset - ├── 0 # First image in the collection - ├── 1 # Second image in the collection - └── ... -- -
Attributes
- -The top-level `.zattrs` file must contain the `bioformats2raw.layout` key: --path: examples/bf2raw/image.json -highlight: json -- -If the top-level group represents a plate, the `bioformats2raw.layout` metadata will be present but -the "plate" key MUST also be present, takes precedence and parsing of such datasets should follow [[#plate-md]]. It is not -possible to mix collections of images with plates at present. - -
-path: examples/bf2raw/plate.json -highlight: json -- -The `.zattrs` file within the OME group may contain the "series" key: - -
-path: examples/ome/series-2.json -highlight: json -- -
Details
- -Conforming groups: - -- MUST have the value "3" for the "bioformats2raw.layout" key in their `.zattrs` metadata at the top of the hierarchy; -- SHOULD have OME metadata representing the entire collection of images in a file named "OME/METADATA.ome.xml" which: - - MUST adhere to the OME-XML specification but - - MUST use `| `identity` | identity transformation, is the default transformation and is typically not explicitly defined - | ||||
|---|---|---|---|---|---|
| `translation` | one of: `"translation":List[float]`, `"path":str` | translation vector, stored either as a list of floats (`"translation"`) or as binary data at a location in this container (`path`). The length of vector defines number of dimensions. | - | |||
| `scale` | one of: `"scale":List[float]`, `"path":str` | scale vector, stored either as a list of floats (`scale`) or as binary data at a location in this container (`path`). The length of vector defines number of dimensions. |
-
- | type | fields | description
- | |
-path: examples/multiscales_strict/multiscales_example.json -highlight: json -- - -If only one multiscale is provided, use it. Otherwise, the user can choose by -name, using the first multiscale as a fallback: - -```python -datasets = [] -for named in multiscales: - if named["name"] == "3D": - datasets = [x["path"] for x in named["datasets"]] - break -if not datasets: - # Use the first by default. Or perhaps choose based on chunk size. - datasets = [x["path"] for x in multiscales[0]["datasets"]] -``` - -"omero" metadata (transitional) {#omero-md} -------------------------------------------- - -[=Transitional=] information specific to the channels of an image and how to render it -can be found under the "omero" key in the group-level metadata: - -```json -"id": 1, # ID in OMERO -"name": "example.tif", # Name as shown in the UI -"version": "0.4", # Current version -"channels": [ # Array matching the c dimension size - { - "active": true, - "coefficient": 1, - "color": "0000FF", - "family": "linear", - "inverted": false, - "label": "LaminB1", - "window": { - "end": 1500, - "max": 65535, - "min": 0, - "start": 0 - } - } -], -"rdefs": { - "defaultT": 0, # First timepoint to show the user - "defaultZ": 118, # First Z section to show the user - "model": "color" # "color" or "greyscale" -} -``` - -See https://docs.openmicroscopy.org/omero/5.6.1/developers/Web/WebGateway.html#imgdata -for more information. - -The "omero" metadata is optional, but if present it MUST contain the field "channels", which is an array of dictionaries describing the channels of the image. -Each dictionary in "channels" MUST contain the field "color", which is a string of 6 hexadecimal digits specifying the color of the channel in RGB format. -Each dictionary in "channels" MUST contain the field "window", which is a dictionary describing the windowing of the channel. -The field "window" MUST contain the fields "min" and "max", which are the minimum and maximum values of the window, respectively. -It MUST also contain the fields "start" and "end", which are the start and end values of the window, respectively. - -"labels" metadata {#labels-md} ------------------------------- - -The special group "labels" found under an image Zarr contains the key `labels` containing -the paths to label objects which can be found underneath the group: - -```json -{ - "labels": [ - "orphaned/0" - ] -} -``` - -Unlisted groups MAY be labels. - -"image-label" metadata {#label-md} ----------------------------------- - -Groups containing the `image-label` dictionary represent an image segmentation -in which each unique pixel value represents a separate segmented object. -`image-label` groups MUST also contain `multiscales` metadata and the two -"datasets" series MUST have the same number of entries. - -The `image-label` dictionary SHOULD contain a `colors` key whose value MUST be a -list of JSON objects describing the unique label values. Each color object MUST -contain the `label-value` key whose value MUST be an integer specifying the -pixel value for that label. It MAY contain an `rgba` key whose value MUST be an array -of four integers between 0 and 255 `[uint8, uint8, uint8, uint8]` specifying the label -color as RGBA. All the values under the `label-value` key MUST be unique. Clients -who choose to not throw an error SHOULD ignore all except the _last_ entry. - -Some implementations MAY represent overlapping labels by using a specially assigned -value, for example the highest integer available in the pixel range. - -The `image-label` dictionary MAY contain a `properties` key whose value MUST be a -list of JSON objects which also describes the unique label values. Each property object -MUST contain the `label-value` key whose value MUST be an integer specifying the pixel -value for that label. Additionally, an arbitrary number of key-value pairs -MAY be present for each label value denoting associated metadata. Not all label -values must share the same key-value pairs within the properties list. - -The `image-label` dictionary MAY contain a `source` key whose value MUST be a JSON -object containing information on the image the label is associated with. If included, -it MAY include a key `image` whose value MUST be a string specifying the relative -path to a Zarr image group. The default value is "../../" since most labels are stored -under a subgroup named "labels/" (see above). - -The `image-label` dictionary SHOULD contain a `version` key whose value MUST be a string -specifying the version of the image-label specification. - -
-path: examples/label_strict/colors_properties.json -highlight: json -- -"plate" metadata {#plate-md} ----------------------------- - -For high-content screening datasets, the plate layout can be found under the -custom attributes of the plate group under the `plate` key in the group-level metadata. - -The `plate` dictionary MAY contain an `acquisitions` key whose value MUST be a list of -JSON objects defining the acquisitions for a given plate to which wells can refer to. Each -acquisition object MUST contain an `id` key whose value MUST be an unique integer identifier -greater than or equal to 0 within the context of the plate to which fields of view can refer -to (see #well-md). -Each acquisition object SHOULD contain a `name` key whose value MUST be a string identifying -the name of the acquisition. Each acquisition object SHOULD contain a `maximumfieldcount` -key whose value MUST be a positive integer indicating the maximum number of fields of view for the -acquisition. Each acquisition object MAY contain a `description` key whose value MUST be a -string specifying a description for the acquisition. Each acquisition object MAY contain -a `starttime` and/or `endtime` key whose values MUST be integer epoch timestamps specifying -the start and/or end timestamp of the acquisition. - -The `plate` dictionary MUST contain a `columns` key whose value MUST be a list of JSON objects -defining the columns of the plate. Each column object defines the properties of -the column at the index of the object in the list. Each column in the physical plate -MUST be defined, even if no wells in the column are defined. Each column object MUST -contain a `name` key whose value is a string specifying the column name. The `name` MUST -contain only alphanumeric characters, MUST be case-sensitive, and MUST NOT be a duplicate of any -other `name` in the `columns` list. Care SHOULD be taken to avoid collisions on -case-insensitive filesystems (e.g. avoid using both `Aa` and `aA`). - -The `plate` dictionary SHOULD contain a `field_count` key whose value MUST be a positive integer -defining the maximum number of fields per view across all wells. - -The `plate` dictionary SHOULD contain a `name` key whose value MUST be a string defining the -name of the plate. - -The `plate` dictionary MUST contain a `rows` key whose value MUST be a list of JSON objects -defining the rows of the plate. Each row object defines the properties of -the row at the index of the object in the list. Each row in the physical plate -MUST be defined, even if no wells in the row are defined. Each defined row MUST -contain a `name` key whose value MUST be a string defining the row name. The `name` MUST -contain only alphanumeric characters, MUST be case-sensitive, and MUST NOT be a duplicate of any -other `name` in the `rows` list. Care SHOULD be taken to avoid collisions on -case-insensitive filesystems (e.g. avoid using both `Aa` and `aA`). - -The `plate` dictionary SHOULD contain a `version` key whose value MUST be a string specifying the -version of the plate specification. - -The `plate` dictionary MUST contain a `wells` key whose value MUST be a list of JSON objects -defining the wells of the plate. Each well object MUST contain a `path` key whose value MUST -be a string specifying the path to the well subgroup. The `path` MUST consist of a `name` in -the `rows` list, a file separator (`/`), and a `name` from the `columns` list, in that order. -The `path` MUST NOT contain additional leading or trailing directories. -Each well object MUST contain both a `rowIndex` key whose value MUST be an integer identifying -the index into the `rows` list and a `columnIndex` key whose value MUST be an integer identifying -the index into the `columns` list. `rowIndex` and `columnIndex` MUST be 0-based. The -`rowIndex`, `columnIndex`, and `path` MUST all refer to the same row/column pair. - -For example the following JSON object defines a plate with two acquisitions and -6 wells (2 rows and 3 columns), containing up to 2 fields of view per acquisition. - -
-path: examples/plate_strict/plate_6wells.json -highlight: json -- -The following JSON object defines a sparse plate with one acquisition and -2 wells in a 96 well plate, containing one field of view per acquisition. - -
-path: examples/plate_strict/plate_2wells.json -highlight: json -- -"well" metadata {#well-md} --------------------------- - -For high-content screening datasets, the metadata about all fields of views -under a given well can be found under the "well" key in the attributes of the -well group. - -The `well` dictionary MUST contain an `images` key whose value MUST be a list of JSON objects -specifying all fields of views for a given well. Each image object MUST contain a -`path` key whose value MUST be a string specifying the path to the field of view. The `path` -MUST contain only alphanumeric characters, MUST be case-sensitive, and MUST NOT be a duplicate -of any other `path` in the `images` list. If multiple acquisitions were performed in the plate, -it MUST contain an `acquisition` key whose value MUST be an integer identifying the acquisition -which MUST match one of the acquisition JSON objects defined in the plate metadata (see #plate-md). - -The `well` dictionary SHOULD contain a `version` key whose value MUST be a string specifying the -version of the well specification. - -For example the following JSON object defines a well with four fields of -view. The first two fields of view were part of the first acquisition while -the last two fields of view were part of the second acquisition. - -
-path: examples/well_strict/well_4fields.json -highlight: json -- -The following JSON object defines a well with two fields of view in a plate with -four acquisitions. The first field is part of the first acquisition, and the second -field is part of the last acquisition. - -
-path: examples/well_strict/well_2fields.json -highlight: json -- -Specification naming style {#naming-style} -========================================== - -Multi-word keys in this specification should use the `camelCase` style. -NB: some parts of the specification don't obey this convention as they -were added before this was adopted, but they should be updated in due course. - -Implementations {#implementations} -================================== - -Projects which support reading and/or writing OME-NGFF data include: - -
-
-
-
- [bigdataviewer-ome-zarr](https://github.com/mobie/bigdataviewer-ome-zarr) -
- Fiji-plugin for reading OME-Zarr. - -
- [bioformats2raw](https://github.com/glencoesoftware/bioformats2raw) -
- A performant, Bio-Formats image file format converter. - -
- [omero-ms-zarr](https://github.com/ome/omero-ms-zarr) -
- A microservice for OMERO.server that converts images stored in OMERO to OME-Zarr files on the fly, served via a web API. - -
- [idr-zarr-tools](https://github.com/IDR/idr-zarr-tools) -
- A full workflow demonstrating the conversion of IDR images to OME-Zarr images on S3. - -
- [OMERO CLI Zarr plugin](https://github.com/ome/omero-cli-zarr) -
- An OMERO CLI plugin that converts images stored in OMERO.server into a local Zarr file. - -
- [ome-zarr-py](https://github.com/ome/ome-zarr-py) -
- A napari plugin for reading ome-zarr files. - -
- [vizarr](https://github.com/hms-dbmi/vizarr/) -
- A minimal, purely client-side program for viewing Zarr-based images with Viv & ImJoy. - -
 -
-All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
-version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
-Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
-
-Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
-
-Citing {#citing}
-================
-
-[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.4)
-J. Moore, *et al*. Open Microscopy Environment Consortium, 8 February 2022.
-This edition of the specification is [https://ngff.openmicroscopy.org/0.4/](https://ngff.openmicroscopy.org/0.4/]).
-The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
-[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
-
-Version History {#history}
-==========================
-
-
-
-All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
-version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
-Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
-
-Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
-
-Citing {#citing}
-================
-
-[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.4)
-J. Moore, *et al*. Open Microscopy Environment Consortium, 8 February 2022.
-This edition of the specification is [https://ngff.openmicroscopy.org/0.4/](https://ngff.openmicroscopy.org/0.4/]).
-The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
-[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
-
-Version History {#history}
-==========================
-
-| Revision | -Date | -Description | -
| 0.4.1 | -2022-09-26 | -transitional metadata for image collections ("bioformats2raw.layout") | -
| 0.4.0 | -2022-02-08 | -multiscales: add axes type, units and coordinateTransformations | -
| 0.4.0 | -2022-02-08 | -plate: add rowIndex/columnIndex | -
| 0.3.0 | -2021-08-24 | -Add axes field to multiscale metadata | -
| 0.2.0 | -2021-03-29 | -Change chunk dimension separator to "/" | -
| 0.1.4 | -2020-11-26 | -Add HCS specification | -
| 0.1.3 | -2020-09-14 | -Add labels specification | -
| 0.1.2 | -2020-05-07 | -Add description of "omero" metadata | -
| 0.1.1 | -2020-05-06 | -Add info on the ordering of resolutions | -
| 0.1.0 | -2020-04-20 | -First version for internal demo | -
-{
- "blogNov2020": {
- "href": "https://blog.openmicroscopy.org/file-formats/community/2020/11/04/zarr-data/",
- "title": "Public OME-Zarr data (Nov. 2020)",
- "authors": [
- "OME Team"
- ],
- "status": "Informational",
- "publisher": "OME",
- "id": "blogNov2020",
- "date": "04 November 2020"
- },
- "imagesc26952": {
- "href": "https://forum.image.sc/t/ome-s-position-regarding-file-formats/26952",
- "title": "OME’s position regarding file formats",
- "authors": [
- "OME Team"
- ],
- "status": "Informational",
- "publisher": "OME",
- "id": "imagesc26952",
- "date": "19 June 2020"
- },
- "n5": {
- "id": "n5",
- "href": "https://github.com/saalfeldlab/n5/issues/62",
- "title": "N5---a scalable Java API for hierarchies of chunked n-dimensional tensors and structured meta-data",
- "status": "Informational",
- "authors": [
- "John A. Bogovic",
- "Igor Pisarev",
- "Philipp Hanslovsky",
- "Neil Thistlethwaite",
- "Stephan Saalfeld"
- ],
- "date": "2020"
- },
- "ome-zarr-py": {
- "id": "ome-zarr-py",
- "href": "https://doi.org/10.5281/zenodo.4113931",
- "title": "ome-zarr-py: Experimental implementation of next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.",
- "status": "Informational",
- "publisher": "Zenodo",
- "authors": [
- "OME",
- "et al"
- ],
- "date": "06 October 2020"
- },
- "zarr": {
- "id": "zarr",
- "href": "https://doi.org/10.5281/zenodo.4069231",
- "title": "Zarr: An implementation of chunked, compressed, N-dimensional arrays for Python.",
- "status": "Informational",
- "publisher": "Zenodo",
- "authors": [
- "Alistair Miles",
- "et al"
- ],
- "date": "06 October 2020"
- }
-}
-
diff --git a/0.4/schemas/bf2raw.schema b/0.4/schemas/bf2raw.schema
deleted file mode 100644
index 834aee24..00000000
--- a/0.4/schemas/bf2raw.schema
+++ /dev/null
@@ -1,14 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/latest/schemas/bf2raw.schema",
- "title": "NGFF container produced by bioformats2raw",
- "description": "JSON from OME-NGFF .zattrs",
- "type": "object",
- "properties": {
- "bioformats2raw.layout": {
- "description": "The top-level identifier metadata added by bioformats2raw",
- "type": "number",
- "enum": [3]
- }
- }
-}
diff --git a/0.4/schemas/image.schema b/0.4/schemas/image.schema
deleted file mode 100644
index aec5db5f..00000000
--- a/0.4/schemas/image.schema
+++ /dev/null
@@ -1,233 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/0.4/schemas/image.schema",
- "title": "NGFF Image",
- "description": "JSON from OME-NGFF .zattrs",
- "type": "object",
- "properties": {
- "multiscales": {
- "description": "The multiscale datasets for this image",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- },
- "datasets": {
- "type": "array",
- "minItems": 1,
- "items": {
- "type": "object",
- "properties": {
- "path": {
- "type": "string"
- },
- "coordinateTransformations": {
- "$ref": "#/$defs/coordinateTransformations"
- }
- },
- "required": ["path", "coordinateTransformations"]
- }
- },
- "version": {
- "type": "string",
- "enum": [
- "0.4"
- ]
- },
- "axes": {
- "$ref": "#/$defs/axes"
- },
- "coordinateTransformations": {
- "$ref": "#/$defs/coordinateTransformations"
- }
- },
- "required": [
- "datasets", "axes"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "omero": {
- "type": "object",
- "properties": {
- "channels": {
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "window": {
- "type": "object",
- "properties": {
- "end": {
- "type": "number"
- },
- "max": {
- "type": "number"
- },
- "min": {
- "type": "number"
- },
- "start": {
- "type": "number"
- }
- },
- "required": [
- "start",
- "min",
- "end",
- "max"
- ]
- },
- "label": {
- "type": "string"
- },
- "family": {
- "type": "string"
- },
- "color": {
- "type": "string"
- },
- "active": {
- "type": "boolean"
- }
- },
- "required": [
- "window",
- "color"
- ]
- }
- }
- },
- "required": [
- "channels"
- ]
- }
- },
- "required": [ "multiscales" ],
-
- "$defs": {
- "axes": {
- "type": "array",
- "uniqueItems": true,
- "minItems": 2,
- "maxItems": 5,
- "contains": {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- },
- "type": {
- "type": "string",
- "enum": ["space"]
- },
- "unit": {
- "type": "string"
- }
- }
- },
- "minContains": 2,
- "maxContains": 3,
- "items": {
- "oneOf": [
- {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- },
- "type": {
- "type": "string",
- "enum": ["channel", "time", "space"]
- }
- },
- "required": ["name", "type"]
- },
- {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- },
- "type": {
- "type": "string",
- "not": {
- "enum": ["space", "time", "channel"]
- }
- }
- },
- "required": ["name"]
- }
- ]
- }
- },
- "coordinateTransformations": {
- "type": "array",
- "minItems": 1,
- "contains": {
- "type": "object",
- "properties": {
- "type": {
- "type": "string",
- "enum": [
- "scale"
- ]
- },
- "scale": {
- "type": "array",
- "minItems": 2,
- "items": {
- "type": "number"
- }
- }
- }
- },
- "maxContains": 1,
- "items": {
- "oneOf": [
- {
- "type": "object",
- "properties": {
- "type": {
- "type": "string",
- "enum": [
- "scale"
- ]
- },
- "scale": {
- "type": "array",
- "minItems": 2,
- "items": {
- "type": "number"
- }
- }
- },
- "required": ["type", "scale"]
- },
- {
- "type": "object",
- "properties": {
- "type": {
- "type": "string",
- "enum": [
- "translation"
- ]
- },
- "translation": {
- "type": "array",
- "minItems": 2,
- "items": {
- "type": "number"
- }
- }
- },
- "required": ["type", "translation"]
- }
- ]
- }
- }
- }
-}
diff --git a/0.4/schemas/label.schema b/0.4/schemas/label.schema
deleted file mode 100644
index 1629d0f9..00000000
--- a/0.4/schemas/label.schema
+++ /dev/null
@@ -1,77 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/0.4/schemas/label.schema",
- "title": "OME-NGFF labelled image schema",
- "description": "JSON from OME-NGFF .zattrs",
- "type": "object",
- "properties": {
- "image-label": {
- "type": "object",
- "properties": {
- "colors": {
- "description": "The colors for this label image",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "label-value": {
- "description": "The value of the label",
- "type": "number"
- },
- "rgba": {
- "description": "The RGBA color stored as an array of four integers between 0 and 255",
- "type": "array",
- "items": {
- "type": "integer",
- "minimum": 0,
- "maximum": 255
- },
- "minItems": 4,
- "maxItems": 4
- }
- },
- "required": [
- "label-value"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "properties": {
- "description": "The properties for this label image",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "label-value": {
- "description": "The pixel value for this label",
- "type": "integer"
- }
- },
- "required": [
- "label-value"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "source": {
- "description": "The source of this label image",
- "type": "object",
- "properties": {
- "image": {
- "type": "string"
- }
- }
- },
- "version": {
- "description": "The version of the specification",
- "type": "string",
- "enum": [
- "0.4"
- ]
- }
- }
- }
- }
-}
diff --git a/0.4/schemas/ome.schema b/0.4/schemas/ome.schema
deleted file mode 100644
index bd600a2a..00000000
--- a/0.4/schemas/ome.schema
+++ /dev/null
@@ -1,17 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/latest/schemas/ome.schema",
- "title": "NGFF group produced by bioformats2raw to contain OME metadata",
- "description": "JSON from OME-NGFF OME/.zattrs linked to an OME-XML file",
- "type": "object",
- "properties": {
- "series": {
- "description": "An array of the same length and the same order as the images defined in the OME-XML",
- "type": "array",
- "items": {
- "type": "string"
- },
- "minContains": 1
- }
- }
-}
diff --git a/0.4/schemas/plate.schema b/0.4/schemas/plate.schema
deleted file mode 100644
index df7cfad0..00000000
--- a/0.4/schemas/plate.schema
+++ /dev/null
@@ -1,140 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/0.4/schemas/plate.schema",
- "title": "OME-NGFF plate schema",
- "description": "JSON from OME-NGFF .zattrs",
- "type": "object",
- "properties": {
- "plate": {
- "type": "object",
- "properties": {
- "acquisitions": {
- "description": "The acquisitions for this plate",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "id": {
- "description": "A unique identifier within the context of the plate",
- "type": "integer",
- "minimum": 0
- },
- "maximumfieldcount": {
- "description": "The maximum number of fields of view for the acquisition",
- "type": "integer",
- "exclusiveMinimum": 0
- },
- "name": {
- "description": "The name of the acquisition",
- "type": "string"
- },
- "description": {
- "description": "The description of the acquisition",
- "type": "string"
- },
- "starttime": {
- "description": "The start timestamp of the acquisition, expressed as epoch time i.e. the number seconds since the Epoch",
- "type": "integer",
- "minimum": 0
- },
- "endtime": {
- "description": "The end timestamp of the acquisition, expressed as epoch time i.e. the number seconds since the Epoch",
- "type": "integer",
- "minimum": 0
- }
- },
- "required": [
- "id"
- ]
- }
- },
- "version": {
- "description": "The version of the specification",
- "type": "string",
- "enum": [
- "0.4"
- ]
- },
- "field_count": {
- "description": "The maximum number of fields per view across all wells",
- "type": "integer",
- "exclusiveMinimum": 0
- },
- "name": {
- "description": "The name of the plate",
- "type": "string"
- },
- "columns": {
- "description": "The columns of the plate",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "name": {
- "description": "The column name",
- "type": "string",
- "pattern": "^[A-Za-z0-9]+$"
- }
- },
- "required": [
- "name"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "rows": {
- "description": "The rows of the plate",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "name": {
- "description": "The row name",
- "type": "string",
- "pattern": "^[A-Za-z0-9]+$"
- }
- },
- "required": [
- "name"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "wells": {
- "description": "The wells of the plate",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "path": {
- "description": "The path to the well subgroup",
- "type": "string",
- "pattern": "^[A-Za-z0-9]+/[A-Za-z0-9]+$"
- },
- "rowIndex": {
- "description": "The index of the well in the rows list",
- "type": "integer",
- "minimum": 0
- },
- "columnIndex": {
- "description": "The index of the well in the columns list",
- "type": "integer",
- "minimum": 0
- }
- },
- "required": [
- "path", "rowIndex", "columnIndex"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- }
- },
- "required": [
- "columns", "rows", "wells"
- ]
- }
- }
-}
diff --git a/0.4/schemas/strict_image.schema b/0.4/schemas/strict_image.schema
deleted file mode 100644
index 87d0d6b5..00000000
--- a/0.4/schemas/strict_image.schema
+++ /dev/null
@@ -1,19 +0,0 @@
-{
- "$id": "https://ngff.openmicroscopy.org/0.4/schemas/strict_image.schema",
- "allOf": [
- {
- "$ref": "https://ngff.openmicroscopy.org/0.4/schemas/image.schema"
- },
- {
- "properties": {
- "multiscales": {
- "items": {
- "required": [
- "version", "metadata", "type", "name"
- ]
- }
- }
- }
- }
- ]
-}
\ No newline at end of file
diff --git a/0.4/schemas/strict_label.schema b/0.4/schemas/strict_label.schema
deleted file mode 100644
index af9090da..00000000
--- a/0.4/schemas/strict_label.schema
+++ /dev/null
@@ -1,18 +0,0 @@
-{
- "$id": "https://ngff.openmicroscopy.org/0.4/schemas/strict_label.schema",
- "allOf": [
- {
- "$ref": "https://ngff.openmicroscopy.org/0.4/schemas/label.schema"
- },
- {
- "properties": {
- "image-label": {
- "required": [
- "version",
- "colors"
- ]
- }
- }
- }
- ]
-}
diff --git a/0.4/schemas/strict_plate.schema b/0.4/schemas/strict_plate.schema
deleted file mode 100644
index 5a88ab72..00000000
--- a/0.4/schemas/strict_plate.schema
+++ /dev/null
@@ -1,28 +0,0 @@
-{
- "$id": "https://ngff.openmicroscopy.org/0.4/schemas/strict_plate.schema",
- "allOf": [
- {
- "$ref": "https://ngff.openmicroscopy.org/0.4/schemas/plate.schema"
- },
- {
- "properties": {
- "plate": {
- "properties": {
- "acquisitions": {
- "items": {
- "required": [
- "name",
- "maximumfieldcount"
- ]
- }
- }
- },
- "required": [
- "name",
- "version"
- ]
- }
- }
- }
- ]
-}
diff --git a/0.4/schemas/strict_well.schema b/0.4/schemas/strict_well.schema
deleted file mode 100644
index 1e200294..00000000
--- a/0.4/schemas/strict_well.schema
+++ /dev/null
@@ -1,17 +0,0 @@
-{
- "$id": "https://ngff.openmicroscopy.org/0.4/schemas/strict_well.schema",
- "allOf": [
- {
- "$ref": "https://ngff.openmicroscopy.org/0.4/schemas/well.schema"
- },
- {
- "properties": {
- "well": {
- "required": [
- "version"
- ]
- }
- }
- }
- ]
-}
diff --git a/0.4/schemas/well.schema b/0.4/schemas/well.schema
deleted file mode 100644
index 971a0102..00000000
--- a/0.4/schemas/well.schema
+++ /dev/null
@@ -1,47 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/0.4/schemas/well.schema",
- "title": "OME-NGFF well schema",
- "description": "JSON from OME-NGFF .zattrs",
- "type": "object",
- "properties": {
- "well": {
- "type": "object",
- "properties": {
- "images": {
- "description": "The fields of view for this well",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "acquisition": {
- "description": "A unique identifier within the context of the plate",
- "type": "integer"
- },
- "path": {
- "description": "The path for this field of view subgroup",
- "type": "string",
- "pattern": "^[A-Za-z0-9]+$"
- }
- },
- "required": [
- "path"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "version": {
- "description": "The version of the specification",
- "type": "string",
- "enum": [
- "0.4"
- ]
- }
- },
- "required": [
- "images"
- ]
- }
- }
-}
diff --git a/0.4/tests/image_suite.json b/0.4/tests/image_suite.json
deleted file mode 100644
index 42906985..00000000
--- a/0.4/tests/image_suite.json
+++ /dev/null
@@ -1,1148 +0,0 @@
-{
- "description": "TBD",
- "schema": {
- "id": "schemas/image.schema"
- },
- "tests": [
- {
- "formerly": "valid/mismatch_axes_units.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "t",
- "type": "time",
- "unit": "micrometer"
- },
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 0.13,
- 0.13
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid/untyped_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "angle"
- },
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid/missing_version.json",
- "description": "TBD",
- "data": {
- "@type": "ngff:Image",
- "multiscales": [
- {
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0",
- "coordinateTransformations": [
- {
- "type": "scale",
- "scale": [
- 1,
- 1
- ]
- }
- ]
- }
- ],
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ]
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid/invalid_axis_units.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micron"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 0.13,
- 0.13
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid/missing_name.json",
- "description": "TBD",
- "data": {
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.4",
- "datasets": [
- {
- "path": "path/to/0",
- "coordinateTransformations": [
- {
- "type": "scale",
- "scale": [
- 1,
- 1
- ]
- }
- ]
- }
- ],
- "type": "gaussian",
- "metadata": {
- "method": "skimage.transform.pyramid_gaussian",
- "version": "0.16.1",
- "args": [
- "true",
- "false"
- ],
- "kwargs": {
- "multichannel": true
- }
- },
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ]
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid/custom_type_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "angle",
- "type": "custom"
- },
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "invalid/duplicate_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_space_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_transformation_type.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "translation"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_scale.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "translation": [
- 1,
- 1
- ],
- "type": "translation"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/too_many_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "angle",
- "type": "custom"
- },
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "z",
- "type": "space"
- },
- {
- "name": "y",
- "type": "space"
- },
- {
- "name": "x",
- "type": "space"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1,
- 1,
- 1,
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_channels_color.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ],
- "omero": {
- "channels": [
- {
- "active": true,
- "coefficient": 1.0,
- "color": 255,
- "family": "linear",
- "label": "1234",
- "window": {
- "end": 1765.0,
- "max": 2555.0,
- "min": 5.0,
- "start": 0.0
- }
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_axes_name.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "type": "space",
- "unit": "micron"
- },
- {
- "type": "space",
- "unit": "micron"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 0.13,
- 0.13
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_axes_count.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/one_space_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "x",
- "type": "space"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_path.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": 0,
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_multiscales_transformations.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "coordinateTransformations": [
- {
- "scale": [
- "invalid"
- ],
- "type": "scale"
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_transformations.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0"
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/no_datasets.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_datasets.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_version.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.3"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_axis_type.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "invalid",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/duplicate_scale.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- },
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/no_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/too_many_space_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "X",
- "type": "space"
- },
- {
- "name": "z",
- "type": "space"
- },
- {
- "name": "y",
- "type": "space"
- },
- {
- "name": "x",
- "type": "space"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1,
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/no_multiscales.json",
- "description": "TBD",
- "data": {
- "@type": "ngff:Image",
- "multiscales": []
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_channels_window.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ],
- "omero": {
- "channels": [
- {
- "active": true,
- "coefficient": 1.0,
- "color": "ff0000",
- "family": "linear",
- "label": "1234",
- "window": {
- "end": "100",
- "max": 2555.0,
- "min": 5.0,
- "start": 0.0
- }
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "invalid/empty_transformations.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": []
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_path.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4"
- }
- ]
- },
- "valid": false
- }
- ]
-}
diff --git a/0.4/tests/label_suite.json b/0.4/tests/label_suite.json
deleted file mode 100644
index 634225c8..00000000
--- a/0.4/tests/label_suite.json
+++ /dev/null
@@ -1,131 +0,0 @@
-{
- "description": "Tests for the image-label JSON schema",
- "schema": {
- "id": "schemas/label.schema"
- },
- "tests": [
- {
- "formerly": "image-label/minimal",
- "data": {
- "image-label": {
- "colors": [
- {
- "label-value": 1,
- "rgba": [0, 0, 0, 0]
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "image-label/minimal_properties",
- "data": {
- "image-label": {
- "colors": [
- {
- "label-value": 1,
- "rgba": [0, 0, 0, 0]
- }
- ],
- "properties": [
- {
- "label-value": 1
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "image-label/empty_colors",
- "data": {
- "image-label": {
- "colors": []
- }
- },
- "valid": false
- },
- {
- "formerly": "image-label/empty_properties",
- "data": {
- "image-label": {
- "properties": []
- }
- },
- "valid": false
- },
- {
- "formerly": "image-label/colors_no_label_value",
- "data": {
- "image-label": {
- "colors": [
- {
- "rgba": [0, 0, 0, 0]
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "image-label/properties_no_label_value",
- "data": {
- "image-label": {
- "properties": [
- {
- "value": "foo"
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "image-label/colors_rgba_length",
- "data": {
- "image-label": {
- "colors": [
- {
- "label-value": 1,
- "rgba": [0, 0, 0]
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "image-label/colors_rgba_type",
- "data": {
- "image-label": {
- "colors": [
- {
- "label-value": 1,
- "rgba": [0, 0, 0, 500]
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "image-label/colors_duplicate",
- "data": {
- "image-label": {
- "colors": [
- {
- "label-value": 1,
- "rgba": [0, 0, 0, 0]
- },
- {
- "label-value": 1,
- "rgba": [0, 0, 0, 0]
- }
- ]
- }
- },
- "valid": false
- }
- ]
-}
diff --git a/0.4/tests/plate_suite.json b/0.4/tests/plate_suite.json
deleted file mode 100644
index c19ad60e..00000000
--- a/0.4/tests/plate_suite.json
+++ /dev/null
@@ -1,819 +0,0 @@
-{
- "description": "Tests for the plate JSON schema",
- "schema": {
- "id": "schemas/plate.schema"
- },
- "tests": [
- {
- "formerly": "plate/minimal_no_acquisitions",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "plate/minimal_acquisitions",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": 0
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "plate/missing_rows",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/empty_rows",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/duplicate_rows",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- },
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_columns",
- "data": {
- "plate": {
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/empty_columns",
- "data": {
- "plate": {
- "columns": [],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/duplicate_columns",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- },
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_wells",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/empty_wells",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": {}
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/duplicate_rows",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- },
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- },
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_column_name",
- "data": {
- "plate": {
- "columns": [
- {
- "concentration": 10
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_row_name",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "concentration": 10
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_well_path",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_well_rowIndex",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_well_columnIndex",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/well_1group",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A1",
- "rowIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/well_3groups",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "plate/A/1",
- "rowIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/invalid_version",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ],
- "version": "foo"
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/non_alphanumeric_column",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A-1"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A-1/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/non_alphanumeric_row",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "A1"
- }
- ],
- "wells": [
- {
- "path": "A/A1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "plate/missing_acquisition_id",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "maximumfieldcount": 1
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/non_integer_acquisition_id",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": "0"
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/negative_acquisition_id",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": -1
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/non_integer_acquisition_maximumfieldcount",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": 0,
- "maximumfieldcount": "0"
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/acquisition_zero_maximumfieldcount",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": 0,
- "maximumfieldcount": 0
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/acquisition_noninteger_starttime",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": 0,
- "starttime": "2022-05-13T13:48:06+00:00"
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/acquisition_negative_starttime",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": 0,
- "starttime": -1
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/acquisition_noninteger_endtime",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": 0,
- "endtime": "2022-05-13T13:48:06+00:00"
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/negative_endtime",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": 0,
- "endtime": -1
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/zero_field_count",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "field_count": 0,
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- }
- ]
-}
diff --git a/0.4/tests/strict_image_suite.json b/0.4/tests/strict_image_suite.json
deleted file mode 100644
index ec6295d3..00000000
--- a/0.4/tests/strict_image_suite.json
+++ /dev/null
@@ -1,388 +0,0 @@
-{
- "description": "TBD",
- "schema": {
- "id": "schemas/strict_image.schema"
- },
- "tests": [
- {
- "formerly": "valid_strict/multiscales_example.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "version": "0.4",
- "name": "example",
- "axes": [
- {
- "name": "t",
- "type": "time",
- "unit": "millisecond"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "z",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "type": "scale",
- "scale": [
- 1.0,
- 1.0,
- 0.5,
- 0.5,
- 0.5
- ]
- }
- ]
- },
- {
- "path": "1",
- "coordinateTransformations": [
- {
- "type": "scale",
- "scale": [
- 1.0,
- 1.0,
- 1.0,
- 1.0,
- 1.0
- ]
- }
- ]
- },
- {
- "path": "2",
- "coordinateTransformations": [
- {
- "type": "scale",
- "scale": [
- 1.0,
- 1.0,
- 2.0,
- 2.0,
- 2.0
- ]
- }
- ]
- }
- ],
- "coordinateTransformations": [
- {
- "type": "scale",
- "scale": [
- 0.1,
- 1.0,
- 1.0,
- 1.0,
- 1.0
- ]
- }
- ],
- "type": "gaussian",
- "metadata": {
- "description": "the fields in metadata depend on the downscaling implementation. Here, the parameters passed to the skimage function are given",
- "method": "skimage.transform.pyramid_gaussian",
- "version": "0.16.1",
- "args": "[true]",
- "kwargs": {
- "multichannel": true
- }
- }
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid_strict/multiscales_transformations.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "coordinateTransformations": [
- {
- "scale": [
- 10,
- 10
- ],
- "type": "scale"
- }
- ],
- "version": "0.4",
- "name": "image_with_coordinateTransformations",
- "type": "foo",
- "metadata": {
- "key": "value"
- }
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid_strict/image_metadata.json",
- "description": "TBD",
- "data": {
- "@id": "top",
- "@type": "ngff:Image",
- "multiscales": [
- {
- "@id": "inner",
- "version": "0.4",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0",
- "coordinateTransformations": [
- {
- "type": "scale",
- "scale": [
- 1,
- 1
- ]
- }
- ]
- }
- ],
- "type": "gaussian",
- "metadata": {
- "method": "skimage.transform.pyramid_gaussian",
- "version": "0.16.1",
- "args": [
- "true",
- "false"
- ],
- "kwargs": {
- "multichannel": true
- }
- },
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ]
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid_strict/image.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4",
- "name": "simple_image",
- "type": "foo",
- "metadata": {
- "key": "value"
- }
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid_strict/image_omero.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "z",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1,
- 0.5,
- 0.13,
- 0.13
- ],
- "type": "scale"
- },
- {
- "translation": [
- 0,
- 9,
- 0.5,
- 25.74,
- 21.58
- ],
- "type": "translation"
- }
- ]
- },
- {
- "path": "1",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1,
- 1,
- 0.26,
- 0.26
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.4",
- "name": "image_with_omero_metadata",
- "type": "foo",
- "metadata": {
- "key": "value"
- }
- }
- ],
- "omero": {
- "channels": [
- {
- "active": true,
- "coefficient": 1.0,
- "color": "00FF00",
- "family": "linear",
- "inverted": false,
- "label": "FITC",
- "window": {
- "end": 813.0,
- "max": 870.0,
- "min": 102.0,
- "start": 82.0
- }
- },
- {
- "active": true,
- "coefficient": 1.0,
- "color": "FF0000",
- "family": "linear",
- "inverted": false,
- "label": "RD-TR-PE",
- "window": {
- "end": 815.0,
- "max": 441.0,
- "min": 129.0,
- "start": 78.0
- }
- }
- ],
- "id": 1,
- "rdefs": {
- "defaultT": 0,
- "defaultZ": 2,
- "model": "color"
- },
- "version": "0.4"
- }
- },
- "valid": true
- }
- ]
-}
diff --git a/0.4/tests/strict_label_suite.json b/0.4/tests/strict_label_suite.json
deleted file mode 100644
index a4ccf01d..00000000
--- a/0.4/tests/strict_label_suite.json
+++ /dev/null
@@ -1,31 +0,0 @@
-{
- "description": "Tests for the strict image-label JSON schema",
- "schema": {
- "id": "schemas/strict_label.schema"
- },
- "tests": [
- {
- "formerly": "image-label/no_version",
- "data": {
- "image-label": {
- "colors": [
- {
- "label-value": 1,
- "rgba": [0, 0, 0, 0]
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "image-label/no_colors",
- "data": {
- "image-label": {
- "version": "0.4"
- }
- },
- "valid": false
- }
- ]
-}
diff --git a/0.4/tests/strict_plate_suite.json b/0.4/tests/strict_plate_suite.json
deleted file mode 100644
index 49eeb3fc..00000000
--- a/0.4/tests/strict_plate_suite.json
+++ /dev/null
@@ -1,187 +0,0 @@
-{
- "description": "Tests for the strict plate JSON schema",
- "schema": {
- "id": "schemas/strict_plate.schema"
- },
- "tests": [
- {
- "formerly": "plate/strict_no_acquisitions",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "name": "test plate",
- "rows": [
- {
- "name": "1"
- }
- ],
- "version": "0.4",
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "plate/missing_name",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "version": "0.4",
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_version",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "name": "test plate",
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/strict_acquisitions",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": 0,
- "name": "0",
- "maximumfieldcount": 1
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "name": "test plate",
- "rows": [
- {
- "name": "1"
- }
- ],
- "version": "0.4",
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "plate/missing_acquisition_name",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": 0,
- "maximumfieldcount": 1
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "name": "test plate",
- "rows": [
- {
- "name": "1"
- }
- ],
- "version": "0.4",
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_acquisition_maximumfieldcount",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": 0,
- "name": "0"
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "name": "test plate",
- "rows": [
- {
- "name": "1"
- }
- ],
- "version": "0.4",
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- }
- ]
-}
diff --git a/0.4/tests/strict_well_suite.json b/0.4/tests/strict_well_suite.json
deleted file mode 100644
index b4f8fe71..00000000
--- a/0.4/tests/strict_well_suite.json
+++ /dev/null
@@ -1,50 +0,0 @@
-{
- "description": "Tests for the strict well JSON schema",
- "schema": {
- "id": "schemas/strict_well.schema"
- },
- "tests": [
- {
- "formerly": "well/strict_no_acquisitions",
- "data": {
- "well": {
- "images": [
- {
- "path": "0"
- }
- ],
- "version": "0.4"
- }
- },
- "valid": true
- },
- {
- "formerly": "plate/missing_version",
- "data": {
- "well": {
- "images": [
- {
- "path": "0"
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/strict_acquisitions",
- "data": {
- "well": {
- "images": [
- {
- "acquisition": 0,
- "path": "0"
- }
- ],
- "version": "0.4"
- }
- },
- "valid": true
- }
- ]
-}
diff --git a/0.4/tests/test_validation.py b/0.4/tests/test_validation.py
deleted file mode 100644
index 82efd90d..00000000
--- a/0.4/tests/test_validation.py
+++ /dev/null
@@ -1,91 +0,0 @@
-import json
-import glob
-import os
-
-from dataclasses import dataclass
-from typing import List
-
-import pytest
-
-from jsonschema import RefResolver, Draft202012Validator as Validator
-from jsonschema.exceptions import ValidationError
-
-schema_store = {}
-for schema_filename in glob.glob("schemas/*"):
- with open(schema_filename) as f:
- schema = json.load(f)
- schema_store[schema["$id"]] = schema
-
-@dataclass
-class Suite:
- schema: dict
- data: dict
- valid: bool = True
-
- def validate(self, validator) -> None:
- if not self.valid:
- with pytest.raises(ValidationError):
- validator.validate(self.data)
- else:
- validator.validate(self.data)
-
-
-def pytest_generate_tests(metafunc):
- """
- Generates tests for the examples/ as well as tests/ subdirectories.
-
- Examples:
- These tests evaluate all of the files under the examples/ directory
- using the configuration in the provided config file in order detect
- what should be run. It is assumed that all files are valid and complete
- so that they can be wholly included into the specification. The
- .config.json file in each directory defines which schema will be used.
-
- Validation:
- These test consumes https://github.com/json-schema-org/JSON-Schema-Test-Suite#structure-of-a-test
- styled JSON tests. Metadata in each test defines which schema is used
- and whether or not the block is considered valid.
- """
- if "suite" in metafunc.fixturenames:
- suites: List[Schema] = []
- ids: List[str] = []
-
- # Validation
- for filename in glob.glob("tests/*.json"):
- with open(filename) as o:
- suite = json.load(o)
- schema = suite["schema"]
- with open(schema["id"]) as f:
- schema = json.load(f)
- for test in suite["tests"]:
- ids.append("validate_" + str(test["formerly"]).split("/")[-1][0:-5])
- suites.append(Suite(schema, test["data"], test["valid"]))
-
- # Examples
- for config_filename in glob.glob("examples/*/.config.json"):
- with open(config_filename) as o:
- data = json.load(o)
- schema = data["schema"]
- with open(schema) as f:
- schema = json.load(f)
- example_folder = os.path.dirname(config_filename)
- for filename in glob.glob(f"{example_folder}/*.json"):
- with open(filename) as f:
- # Strip comments
- data = ''.join(line for line in f if not line.lstrip().startswith('//'))
- data = json.loads(data)
- ids.append("example_" + str(filename).split("/")[-1][0:-5])
- suites.append(Suite(schema, data, True)) # Assume true
-
- metafunc.parametrize("suite", suites, ids=ids, indirect=True)
-
-
-@pytest.fixture
-def suite(request):
- return request.param
-
-
-def test_run(suite):
- resolver = RefResolver.from_schema(suite.schema, store=schema_store)
- validator = Validator(suite.schema, resolver=resolver)
- suite.validate(validator)
diff --git a/0.4/tests/well_suite.json b/0.4/tests/well_suite.json
deleted file mode 100644
index 0f752551..00000000
--- a/0.4/tests/well_suite.json
+++ /dev/null
@@ -1,88 +0,0 @@
-{
- "description": "Tests for the well JSON schema",
- "schema": {
- "id": "schemas/well.schema"
- },
- "tests": [
- {
- "formerly": "well/minimal_no_acquisition",
- "data": {
- "well": {
- "images": [
- {
- "path": "0"
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "well/minimal_acquisitions",
- "data": {
- "well": {
- "images": [
- {
- "acquisition": 1,
- "path": "0"
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "well/empty_images",
- "data": {
- "well": {
- "images": []
- }
- },
- "valid": false
- },
- {
- "formerly": "well/duplicate_images",
- "data": {
- "well": {
- "images": [
- {
- "path": "0"
- },
- {
- "path": "0"
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "well/invalid_version",
- "data": {
- "well": {
- "images": [
- {
- "path": "0"
- }
- ],
- "version": "foo"
- }
- },
- "valid": false
- },
- {
- "formerly": "well/non_integer_acquisition_id",
- "data": {
- "well": {
- "images": [
- {
- "acquisition": "0",
- "path": "0"
- }
- ]
- }
- },
- "valid": false
- }
- ]
-}
diff --git a/0.5/copyright.include b/0.5/copyright.include
deleted file mode 100644
index f0def708..00000000
--- a/0.5/copyright.include
+++ /dev/null
@@ -1,4 +0,0 @@
-Copyright © 2020-[YEAR]
-OME®
-(U. Dundee).
-OME trademark rules apply.
diff --git a/0.5/examples/bf2raw/.config.json b/0.5/examples/bf2raw/.config.json
deleted file mode 100644
index 2525328c..00000000
--- a/0.5/examples/bf2raw/.config.json
+++ /dev/null
@@ -1,3 +0,0 @@
-{
- "schema": "schemas/bf2raw.schema"
-}
diff --git a/0.5/examples/label_strict/.config.json b/0.5/examples/label_strict/.config.json
deleted file mode 100644
index e7329dc9..00000000
--- a/0.5/examples/label_strict/.config.json
+++ /dev/null
@@ -1,3 +0,0 @@
-{
- "schema": "schemas/strict_label.schema"
-}
diff --git a/0.5/examples/multiscales_strict/.config.json b/0.5/examples/multiscales_strict/.config.json
deleted file mode 100644
index b0469538..00000000
--- a/0.5/examples/multiscales_strict/.config.json
+++ /dev/null
@@ -1,3 +0,0 @@
-{
- "schema": "schemas/strict_image.schema"
-}
diff --git a/0.5/examples/ome/.config.json b/0.5/examples/ome/.config.json
deleted file mode 100644
index 8a611ccf..00000000
--- a/0.5/examples/ome/.config.json
+++ /dev/null
@@ -1,3 +0,0 @@
-{
- "schema": "schemas/ome.schema"
-}
diff --git a/0.5/examples/plate_strict/.config.json b/0.5/examples/plate_strict/.config.json
deleted file mode 100644
index a49b1743..00000000
--- a/0.5/examples/plate_strict/.config.json
+++ /dev/null
@@ -1,3 +0,0 @@
-{
- "schema": "schemas/strict_plate.schema"
-}
diff --git a/0.5/examples/well_strict/.config.json b/0.5/examples/well_strict/.config.json
deleted file mode 100644
index 129ac69c..00000000
--- a/0.5/examples/well_strict/.config.json
+++ /dev/null
@@ -1,3 +0,0 @@
-{
- "schema": "schemas/strict_well.schema"
-}
diff --git a/0.5/header.include b/0.5/header.include
deleted file mode 100644
index e98bf6fe..00000000
--- a/0.5/header.include
+++ /dev/null
@@ -1,34 +0,0 @@
-
-
-
-
-
-
-
-
-  -
-
-
-
-
-
-
-
- -
-
-
- [TITLE]
-[LONGSTATUS], -
- - - --