-
Notifications
You must be signed in to change notification settings - Fork 2
Stage 2 – ICA & source localization
For ICA decomposition, the AMICA algorithm is used, which has been shown to result in the greater mutual information reduction and dipolarity than competing algorithms for EEG data (Palmer et al., 2011). For source localization, EEGLAB's DIPFIT plugin is used for equivalent dipole fitting of IC scalp projections (Oostenveld et al., 2011).
Data preprocessing for optimized ICA decomposition differs from what is optimal for experimental analysis. This is why the ICA (stage 2) and final (stage 3) preprocessing stages are run separately.
Stage 2 is implemented in the function procICAdipfit.m, which is meant to be called by its wrapper, procICAdipfit_wrapper.m. The wrapper contains all parameters used in the main function, and should be edited to match user preferences and directory structure.
- The top of
procICAdipfit_wrapper.mcontains path and directory parameters. The wrapper takes subject # as an input, and assumes that your directory and filename structure is based on subject #. This can be changed in the code.- Edit the path directory to match the location of your custom path file
- Edit the folders to match your directory structure
- Input the directory of your PREP file (
procPREP.mshould've made a PREP subdir in the subject dir) - Input the directory where the ICA & final datasets will be saved (separate subdirs will be made here)
- Input the directory of your PREP file (
- The channel parameters are optimized for CMU Psych's BioSemi. Epoch parmeters depend on your paradigm
-
p.epEvents- events (trigger numbers) to make epochs around -
p.epMin- epoch start (seconds before trigger) -
p.epMax- epoch end (seconds after trigger)
-
Load the output from procPREP.m. The processing done in PREP has been shown to improve ICA decomposition, especially it's scalp-average robust referencing procedure (Bigdely-Shamlo et al., 2015). The included functions require data that has been run through PREP.
Data should be high-pass filtered at 1–2Hz for optimal ICA decomposition, as ICA is very sensitive to the non-linearity and non-stationarity present at infraslow frequencies (Winkler et al., 2015). However, high-pass filtering over 0.1 Hz has been shown to induce distortions in ERP data (Rousselet et al., 2012); one of the reasons why stage 2 and stage 3 are run separately. Data should be high-passed prior to epoching.
I prefer data to be epoched to the experimental trials for ICA. This way, the vast majority of the data fed into ICA is recorded during the neurocognitive processes of interest. Furthermore, much "artifactual" activity (e.g. blinking, stretching, and mindwandering) occurs between trials, runs, and blocks. Thus, in my experience, ICA performed on epoched data (vs. continuous data) appears to better discerns the neural processes of interest (e.g. produces ICs that represent the distinct neural processes that underlie a cognitive phenomenon), and will likely devote less ICs to artifactual activity.
Do not perform baseline correction prior to ICA, as this can adversely impact decomposition quality (Groppe et al., 2010).
In this pipeline, channel removal is highly recommended before ICA for several reasons listed below.
- They do not add to data rank, and can adversely affect ICA. These can be re-interpolated after ICA and artifactual IC subtraction
- They don't add much to the dataset since they're not used as a reference, nor have standardized electrode locations. The more channels there are, the more data is needed for ICA. However, if mastoid locations are recorded, they can improve source localization – don't remove if so.
- The "robust referencing" (scalp-average) used in PREP reduces data rank by one; ICA requires the number of channels and data rank to correspond.
- Rank is reduced by removing a scalp channel. Cz is removed by default. The data recorded by Cz is least "irreplacable" as it is surrounded by other electrodes at all sides, which also makes it easily interpolated. If Cz has been interpolated, the function will remove a channel adjacent to Cz. The removed channel is interpolated after ICA & IC subtraction.
- Alternatively, PCA can be used to reduce data dimensionality to match rank. However, PCA reduces ICA decomposition quality by increasing non-linearity in the data. If only 1 dimension needs to be reduced, I prefer removing a channel over PCA. If 2 or more dimensions need to be reduced, I prefer PCA. You will have to play with the code (look at the AMICA functions) to perform PCA instead of channel removal.
ICA is performed twice for the best possible decomposition. The first ICA run is to identify sections of data that result in non-stereotyped artifactual ICs. After these sections are removed, a ICA is run a second time, producing the final ICA weight matrices to be used for equivalent dipole fitting and Stage 3. For faster preprocessing, ICA can be run just once.
A detailed tutorial on ICA for EEGLAB can be found in documentation.
- Both ICA runs have separate sets of parameters: AMICA (ICA algorithm), FASTER (statistical artifactual IC detection), MARA (classification-based artifactual IC detection). Included in the function are my preferred values for each ICA run, tailored for CMU Psy's BioSemi.
- AMICA parameters (in wrapper):
- kVal = number of (time) frames required per IC (explained in detail below)
- threads = number of CPU threads to use (Psych-O nodes have 72–128 threads)
- iter = number of iterations before completion
- rej = enable/disable rejection of data that doesn't fit AMICA model (further params described in wrapper)
- Pure ICA produces the same number of ICs as data dimensions (channels). However, more dimensions require exponentially longer (temporally) data. CMU Psy's BioSemi requires a ~40min recording to support full-dimension ICA decomposition.
- The number of ICs the data length supports can be calculated by:
maxIC = floor(sqrt(size(EEG.data(:, :), 2) / kVal));
- The number of (time) frames required per IC is represented by the parameter kVal. This number increases exponentially as the number of channels/dimensions increases. For CMU Psy's BioSemi, I recommend 30, but more would be needed for, say, a 256ch system.
- If the data length is too short to support full-dimension decomposition, PCA dimension reduction is performed to reduce dimensionality to maxIC
- The first AMICA run is to identify sections of data that produce non-stereotyped "one-of-a-kind" artifactual ICs (e.g. sudden impedance shifts, extreme muscle activity). This run should be performed on uncleaned data. AMICA data rejection should not be enabled.
- The initial AMICA run's ICs are evaluated for stereotyped artifactual ICs by calculating their mutual information with the EOG and ECG channels. ICs with mutual information ≥ 3 SDs above average are identified as stereotyped ICs (can be changed in wrapper params).
- ICs representing stereotyped artifacts will not be used for epoch rejection. Epochs containing stereotyped artifacts should not be removed, as they are often numerous, and can be easily cleaned. Keeping these epochs improves the decomposition quality of the second ICA run, as stereotyped artifacts are represented by a relatively small number of ICs, yet account for the vast majority of artifacts.
- Activity of the initial AMICA run's ICs are used for epoch rejection, aside from the aforementioned stereotyped ICs. Epoch rejection uses statistical thresholding for the following parameters. These are evaluated, using different parameter values, for individual ICs and all ICs. The all-IC thresholds are very liberal, since the goal is to find data that "generate" specific artifactual ICs.
- Max and min amplitude (absolute threshold)
- Amplitude improbability
- Temporal kurtosis
- Spectral power of ECG-associated frequencies (disabled; muscle noise is stereotyped)
- AMICA is run a final time after epoch rejection. AMICA also has its own data rejection scheme, is used in the pipeline for further data cleaning. In the pipeline, data that did not fit AMICA's model (≥ 5 SDs) are removed at early iterations (every 3 iterations starting at iteration 4). These values can be changed in the wrapper. Similar methods were used in Rissling et al. (2014). The resulting ICA weights matrices will be saved to a separate .mat file and used for source localization & all further processing/analyses (Stage 3).
Hopefully there are less bad ICs in the final versus initial AMICA run. It's best to identify ICs prior to equivalent dipole fitting. Dipole fitting is computationally intensive and there's no point in localizing artifactual ICs. Change parameters to be more liberal or conservative depending on your needs.
- MARA is a linear classifier that identifies artifactual ICs (Winkler et al., 2011). It extracts EEG features in the spectral, spatial, and temporal domains to use for classification. Binary classification or posterior probability can be used (changeable in wrapper).
- MARA was trained on EEG systems with fewer electrodes, so often some artifactual ICs are missed. FASTER uses z-score thresholds to identify bad ICs (Nolan et al., 2010). Features used are: median gradient, spatial kurtosis, Hurst exponent, and EOG/ECG correlation. The included z-score thresholds are very conservative as MARA has already been performed.
The sources of the remaining ICs can now be found using DIPFIT, which uses FieldTrip (Oostenveld et al., 2011). DIPFIT the scalp projections of ICs to fit their equivalent dipoles in source-space.
Source localization can be fully- or semi-automated. Procedures that can be done manually are channel position co-registration and identification of dual-dipole ICs.
- The following should be performed prior to running DIPFIT. A detailed tutorial on DIPFIT can be found here. As always, parameters for DIPFIT can be found in the wrapper.
- The pipeline uses a 4-shell spherical head model by default. This is less accurate but more stable (e.g. less sensitive to discrepancies in electrode location and individual head anatomy) than the more sophisticated head models available. Custom head models can be made from anatomical MRIs of subjects. The use of custom head models and accurate electrode location digitization can result in accuracy of up to ~12mm. EEG systems with more electrodes also results in greater localization accuracy (Acar & Makeig, 2013)
- If you recorded (digitized) the location of subject electrode positions, you should co-register them with the standard electrode positions. This is explained in detail in the DIPFIT guide linked above.
- If you've set up DIPFIT correctly in the wrapper prior to running Stage 2, the DIPFIT will run fully-automated. If you've enabled manual identification of dual-dipoles (on by default, explained below), the DIPFIT model will not be saved to a separate .mat file. If manual identification is disabled, the DIPFIT model will be saved in a .mat file, ready to be imported in Stage 3.
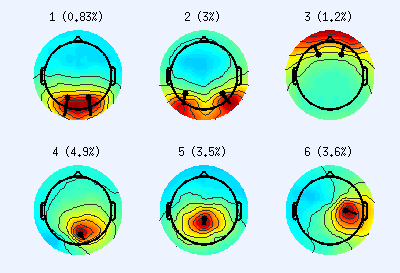
Scalp projections of neurogenic ICs. Top row ICs appear to originate from bilateral dual-dipoles, while bottom row ICs appear to originate from single dipoles. Colors indicate IC weights. Dots represent fitted dipole locations, and their lines represent dipole moment direction. Note the low residual variance (%), suggesting high localization accuracy- In a decomposition, some ICs produce scalp projections that resemble dual-dipoles (almost always bilateral). By default, DIPFIT only fits ICs for single dipoles. Dual-dipole ICs can be identified manually after the initial DIPFIT run (this is the default in the pipeline):
- Open the dataset produced after DIPFIT
*_DIPFIT.set. - Plot the IC scalp projections:
Plot > Component maps > In 2-D. - Identify ICs with scalp maps that appear to originate from dual-dipoles
- Manually re-fit these ICs using two dipoles:
Tools > Locate dipoles using DIPFIT > Fine fit (iterative)- This is explained in detail here
- After re-fitting, save the
EEG.dipfitstruct to a .mat file, this will be imported during Stage 3. - Save the whole dataset:
File > Save current dataset as
- Open the dataset produced after DIPFIT
- Alternatively, dual-dipole ICs can be automatically identified using the fitTwoDipoles plugin (Piazza et al., 2016). This is not yet implemented in the code; feel free to edit it to do so and add a branch to the git repo.
For faster results, ICA be performed just once using the parameters tailored for the final ICA run.