WORK IN PROGRESS
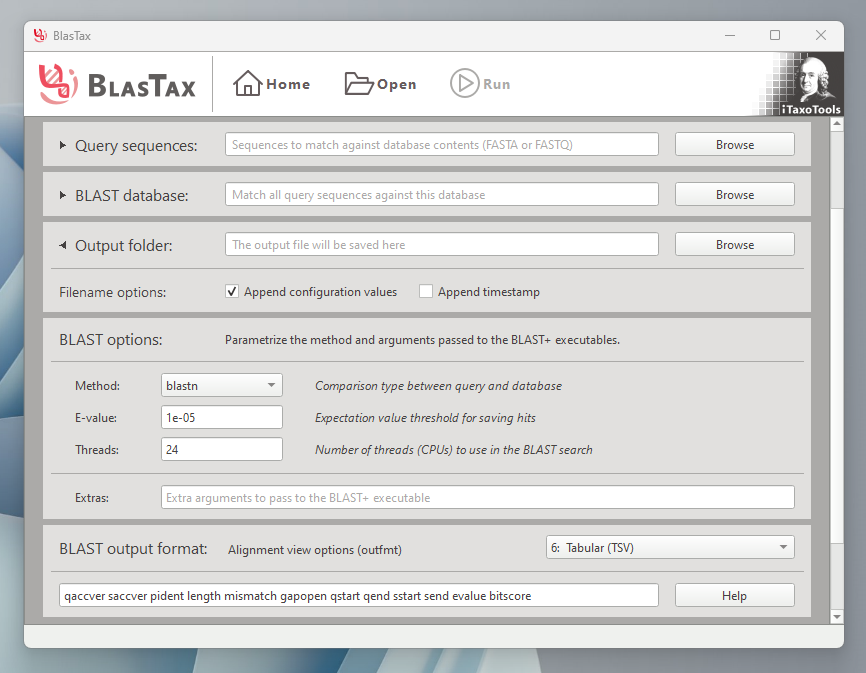
A graphical user interface to run BLAST and parse hits.
- Make BLAST database: Create a BLAST database from a sequence file
- Regular BLAST: Find regions of similarity between sequences in a query file and a BLAST database
- BLAST-Append: Append the aligned part of matching sequences to the original query sequences
- BLAST-Append-X: Like BLAST-Append, but appends nucleotides c orresponding to the protein database
- Decontaminate: Remove contaminants from query sequences based on two ingroup and outgroup databases
- Museoscript: Create sequence files from BLAST matches
- FastaSeqRename: Rename FASTA sequence identifiers in preparation for BLAST analysis
Input sequences must be in the FASTA or FASTQ file formats.
Download and run the standalone executables without installing Python or BLAST+.
First clone the repository and install all dependencies.
git clone https://github.com/iTaxoTools/BlasTax.git
cd BlasTax
pip install -r requirements.txt
Then fetch the BLAST+ binaries if not already installed on the system:
python tools/get_blast_binaries.py
Finally run BlasTax using:
python gui.py
BlasTax was developed in the framework of the iTaxoTools project:
Vences M. et al. (2021): iTaxoTools 0.1: Kickstarting a specimen-based software toolkit for taxonomists. - Megataxa 6: 77-92.
Code by Nikita Kulikov, Anja-Kristina Schulz and Stefanos Patmanidis.
BlasTax integrates the BLAST+ suite from NCBI:
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., and Madden, T.L. 2009. BLAST+: architecture and applications. BMC Bioinformatics, 10, 421.
Museoscript recoded following original concept Linux bash script:
Rancilhac, L., Bruy, T., Scherz, M. D., Pereira, E. A., Preick, M., Straube, N., Lyra, M. L., Ohler, A., Streicher, J. W., Andreone, F., Crottini, A., Hutter, C. R., Randrianantoandro,J. C., Rokotoarison, A., Glaw, F., Hofreiter, M. & Vences, M. (2020). Target-enriched DNA sequencing from historical type material enables a partial revision of the Madagascar giant stream frogs (genus Mantidactylus). Journal of Natural History, 1-32.