砖厂很早就实现了 Z-Order,一直没明白是如何实现的,正好得知 Impala 4.0 有实现,找到对应的Commit(IMPALA-8755: Backend support for Z-ordering),了解了下原理。原来一直以为会对 Parquet 格式有所修改,但事实上 Impala 只是实现了一种新的比较函数。Spark 保存 DataFrame 时可以这么写:
df.write
.mode(SaveMode.Overwrite)
.sortBy("id", "FLD1", "FLD2")
.saveAsTable("TBL")概念上,接口这么修改即可支持 Z-Order:
df.write
.mode(SaveMode.Overwrite)
.zsortBy("id", "FLD1", "FLD2")
.saveAsTable("TBL")本质上,Z-order 曲线是把多维空间的点**==映射==到一维空间。采用的方法称之为==位交织==**,参见下图:
该图表示的是二维空间的转换,多维空间的转换一样。下图显示了二维坐标为 0≤x≤7、0≤y≤7(以十进制和二进制显示)的二维情况的 Z 值。 如图所示,对二进制坐标值进行交织会产生二进制 z 值。 按数值顺序连接 z 值会生成Z形递归曲线。
假设坐标是由 U = 2^w^ 界定的正整数。给定 n 维坐标中的点 p,其中第 i 维 pi 是二进制数 piw ... pi0,定义点 p 的 Z 值是二进制数:p1w ... pnw ... p11 ... pn1。上图中 W = 3,n = 2。
不一定要计算出真正的 Z 值,例如 W = 64,n = 4 时,位串为 256 位。注意,我们真正的需要比较大小 n 维坐标中两个点 Z 值的大小!
对于 n 维坐标中的两个点,计算这两个点每个维度值异或后的最高有效位,然后用最高有效位最大的维度来比较两个点,就以确定两个点 Z 值的大小。 用 Python 表示如下:
def cmp_zorder(lhs, rhs) -> bool:
"""Compare z-ordering."""
# Assume lhs and rhs array-like objects of indices.
assert len(lhs) == len(rhs)
# Will contain the most significant dimension.
msd = 0
# Loop over the other dimensions.
for dim in range(1, len(lhs)):
# Check if the current dimension is more significant
# by comparing the most significant bits.
if less_msb(lhs[msd] ^ rhs[msd], lhs[dim] ^ rhs[dim]):
msd = dim
return lhs[msd] < rhs[msd]异或的特点是相同位的异或值为 0,所以掩盖了两个维度值相同的高位,再基于位交织的事实,也就是说某个维度异或值越大,说明最先从该维度开始区分出大小。我们来比较(3,3)和(0,4):
y | x
--------------
011 | 011 // (3, 3)
000 | 100 // (0, 4)
XOR --------------
3 | 7
--------------
MSB 2 | 3
因为 y 异或后的最高有效位是 2,而 x 异或后的最高有效位是 3,因此用 x 的值来确定两个点的大小,所以(3,3)<(0,4)。比较最高有效位的一种方法是比较每个值以 2 为底对数的下限,less_msb(x, y) 实际上是比较
log2x 和 log2y
def less_msb(x: int, y: int) -> bool:
return x < y and x < (x ^ y)这里的关键点是如何自定义比较函数,Demo 可参考 Z-Ordering索引加速大数据查询 的实现,基于 RDD.sortBy,利用 Scala 的机制,传入定制的比较函数,这个实现是把 Z 值都计算出来了,要想办法改成不需要。
因为 Spark 的 DataFrameWriter 没有提供自定义比较函数的机制,要改 Spark。修改的思路可以参考 Impala,分为:
- 前端,增加 zorder 选项
- 后端,增加 zorder 比较函数。
后端的修改可从内部实现排序的类 UnsafeInMemorySorter 开始:
public UnsafeInMemorySorter(
final MemoryConsumer consumer,
final TaskMemoryManager memoryManager,
final RecordComparator recordComparator, // 比较函数,可定制为 zorder compare 函数
final PrefixComparator prefixComparator, // 基数排序
LongArray array,
boolean canUseRadixSort) {
// ....
if (recordComparator != null) {
//...
this.sortComparator = new SortComparator(recordComparator, prefixComparator, memoryManager);
}
// ...
}使用的地方在:
public UnsafeSorterIterator getSortedIterator() {
// ....
if (sortComparator != null) {
if (this.radixSortSupport != null) {
//...
} else {
//...
sorter.sort(array, 0, pos / 2, sortComparator); // 内部使用的是TimSort
}
}其他实现技巧可参考 Impala 的 Commit。
几个没想清楚的问题:
- 什么场景下使用?
- 如何做文件级别的裁剪?上述的实现,可以做 page 和 rowgroup 的裁剪。
The figure below shows the Z-values for the two dimensional case with integer coordinates 0 ≤ x ≤ 7, 0 ≤ y ≤ 7 (shown both in decimal and binary). Interleaving the binary coordinate values yields binary z-values as shown. Connecting the z-values in their numerical order produces the recursively Z-shaped curve. Two-dimensional Z-values are also called as quadkey ones.
下图显示了二维坐标为0≤x≤7、0≤y≤7(以十进制和二进制显示)的二维情况的Z值。 如图所示,对二进制坐标值进行交织会产生二进制z值。 按数值顺序连接z值会生成Z形递归曲线。 二维Z值也称为四键值。
The Z-ordering can be used to efficiently build a quadtree for a set of points.3 The basic idea is to sort the input set according to Z-order. Once sorted, the points can either be stored in a ==binary search tree== and used directly, which is called a linear quadtree,[4] or they can be used to build a pointer based quadtree.
Z-order 可用于为一组点有效地构建四叉树3。基本思想 是根据 Z-order 对输入集进行排序。 排序后,这些点可以存储在**==二叉搜索树==**中并直接使用,这称为线性四叉树[4](https://en.wikipedia.org/wiki/Z-order_curve#cite_note- 4),或它们可用于构建基于指针的四叉树。
The input points are usually scaled in each dimension to be positive integers, either as ==a fixed point representation== over the unit range [0, 1] or corresponding to the machine word size. Both representations are equivalent and allow for the highest order non-zero bit to be found in constant time. Each square in the quadtree has a side length which is a power of two, and corner coordinates which are multiples of the side length. Given any two points, the derived square for the two points is the smallest square covering both points. The interleaving of bits from the x and y components of each point is called the shuffle of x and y, and can be extended to higher dimensions.3
输入点通常在每个维度上缩放为正整数,以单位范围[0,1]上的**==定点表示形式==,或者对应于机器字的大小。两种表示形式都是等效的,并允许在恒定时间内找到最高阶的非零位。四叉树中的每个正方形的边长为2的幂,并且角坐标为边长的倍数。给定任意两个点,两个点的派生平方是覆盖两个点的最小平方。来自每个点的 x 和 y 分量的比特交织**称为 x 和 y 的混洗,并且可以扩展到更高的维度3。
Points can be sorted according to their shuffle without explicitly interleaving the bits. To do this, for each dimension, the most significant bit of the exclusive or of the coordinates of the two points for that dimension is examined. The dimension for which the most significant bit is largest is then used to compare the two points to determine their shuffle order.
可以根据点的混洗对点进行排序,而无需显式交织位。为此,对于每个维度,都要检查该维度的两个点的坐标的异或的最高有效位。 然后,最高有效位最大的维度用于比较两个点,以确定其混洗顺序。
The exclusive or operation masks off the higher order bits for which the two coordinates are identical. Since the ==shuffle== interleaves bits from higher order to lower order, identifying the coordinate with the ==largest most significant bit==, identifies the first bit in the shuffle order which differs, and that coordinate can be used to compare the two points.5 This is shown in the following Python code:
异或运算掩盖了两个坐标相同的高位。由于**==混洗==将位从高位到低位交织,因此以==最大的最高有效位==来标识坐标,==以混洗顺序中的第一位来标识不同的第一位==**,并且该坐标可用于比较两个点5。如下 Python 代码所示:
def cmp_zorder(lhs, rhs) -> bool:
"""Compare z-ordering."""
# Assume lhs and rhs array-like objects of indices.
assert len(lhs) == len(rhs)
# Will contain the most significant dimension.
msd = 0
# Loop over the other dimensions.
for dim in range(1, len(lhs)):
# Check if the current dimension is more significant
# by comparing the most significant bits.
if less_msb(lhs[msd] ^ rhs[msd], lhs[dim] ^ rhs[dim]):
msd = dim
return lhs[msd] < rhs[msd]One way to determine whether the most significant bit is smaller is to compare the floor of the base-2 logarithm of each point. It turns out the following operation is equivalent, and only requires exclusive or operations:5
确定最高有效位是否较小的一种方法是比较每个点的以 2 为底的对数的下限。下面的操作是等价的,只需要异或操作:
def less_msb(x: int, y: int) -> bool:
return x < y and x < (x ^ y)It is also possible to compare floating point numbers using the same technique. The less_msb function is modified to first compare the exponents. Only when they are equal is the standard less_msb function used on the mantissas.[6]
Once the points are in sorted order, two properties make it easy to build a quadtree: The first is that the points contained in a square of the quadtree form a contiguous interval in the sorted order. The second is that if more than one child of a square contains an input point, the square is the derived square for two adjacent points in the sorted order.
For each adjacent pair of points, the derived square is computed and its side length determined. For each derived square, the interval containing it is bounded by the first larger square to the right and to the left in sorted order.[3] Each such interval corresponds to a square in the quadtree. The result of this is a compressed quadtree, where only nodes containing input points or two or more children are present. A non-compressed quadtree can be built by restoring the missing nodes, if desired.
Rather than building a pointer based quadtree, the points can be maintained in sorted order in a data structure such as a binary search tree. This allows points to be added and deleted in O(log n) time. Two quadtrees can be merged by merging the two sorted sets of points, and removing duplicates. Point location can be done by searching for the points preceding and following the query point in the sorted order. If the quadtree is compressed, the predecessor node found may be an arbitrary leaf inside the compressed node of interest. In this case, it is necessary to find the predecessor of the least common ancestor of the query point and the leaf found.[7]
The CP algorithm can be stated completely in one paragraph. Assume coordinates are positive integers bounded by U=2^w^. Given a point p in a constant dimension d where the i-th coordinate pi is the number piw ... pi0 in the binary,
- define its shuffle
$\sigma(p)$ to be the number p1w... pdw... p10... pd0in binary, and - define its shifts
$\tau(p)=$ for$i = 0,...,d$ , assuming w.lo.g. that d is even
One objection is that we can't directly compute
i = 1;
for j = 2, ...., d do
if | pi XOR qi | < | pj XOR qj | then i = j;
return pi < qi
Here, XOR denotes bitwise excusive-or and
if x > y then return false else return x < x XOR y
So, using comparsion-base sorting, the entire algorithm can be implemented in
The general use-case for these features is to improve the performance of ==needle-in-the-haystack kind of queries== against huge data sets. The typical RDBMS solution, namely secondary indexes, is not practical in a big data context due to scalability reasons.
If you’re familiar with big data systems (be it Apache Spark, Hive, Impala, Vertica, etc.), you might already be thinking: (horizontal) partitioning.
Quick reminder: In Spark, just like Hive, partitioning 1 works by having one subdirectory for every distinct value of the partition column(s). Queries with filters on the partition column(s) can then benefit from partition pruning, i.e., avoid scanning any partition that doesn’t satisfy those filters.
The main question is: What columns do you partition by? And the typical answer is: The ones you’re most likely to filter by in time-sensitive queries. But… What if there are multiple (say 4+), equally relevant columns?
The problem, in that case, is that you end up with a huge number of unique combinations of values, which means a huge number of partitions and therefore files. Having data split across many small files brings up the following main issues:
- Metadata becomes as large as the data itself, causing performance issues for various driver-side operations
- In particular, file listing is affected, becoming very slow
- Compression effectiveness is compromised, leading to wasted space and slower IO
So while data partitioning in Spark generally works great for dates or categorical columns, it is not well suited for high-cardinality columns and, in practice, it is usually limited to one or two columns at most.
这些功能的一般用例是提高针对大数据集的==大海捞针式查询==的性能。由于可伸缩性原因,RDBMS 的典型解决方案(即辅助索引)在大数据环境中不切实际。
如果熟悉大数据系统(如Apache Spark,Hive,Impala,Vertica等),可能已经开始考虑(水平)分区。
快速提醒:在Spark中,就像Hive一样,分区[1]的工作原理是,每个分区列的不同值都有一个子目录。 这样,分区列上具有过滤器的查询就可以从分区裁剪中受益,即避免扫描任何不满足这些过滤条件的分区。
主要问题是:按什么列进行分区? 通常的答案是:在对时间敏感的查询中,最可能过滤那些利。 但是... 如果有多个(例如4个以上),同等相关的列怎么办?
在这种情况下,问题在于最终会产生大量的值的唯一组合,这意味着会有大量的分区,因此文件也很多。 将数据拆分为许多小文件会带来以下主要问题:
- 元数据变得和数据本身一样大,从而导致各种 ==driver== 端的性能问题
- 特别是枚举文件将受到影响,变得非常慢
- 压缩效率受到损害,导致空间浪费和 IO 变慢
因此,尽管 Spark 中的数据分区通常适用于日期或分类列,但它不适用于高基数列,并且在实践中,通常最多限制为一列或两列。
Apart from partition pruning, another common technique that’s used in the data warehousing world, but which Spark currently lacks, is I/O pruning based on Small Materialized Aggregates. In short, the idea is to:
- Keep track of simple statistics such as minimum and maximum values at a certain granularity that’s correlated with I/O granularity.
- Leverage those statistics at query planning time in order to avoid unnecessary I/O.
This is exactly what Databricks Delta’s data skipping feature is about. As new data is inserted into a Databricks Delta table, file-level min/max statistics are collected for all columns (including nested ones) of supported types. Then, when there’s a lookup query against the table, Databricks Delta first consults these statistics in order to determine which files can safely be skipped. But, as they say, a GIF is worth a thousand words, so here you go:
On the one hand, this is a lightweight and flexible (the granularity can be tuned) technique that is easy to implement and reason about. It’s also completely orthogonal to partitioning: it works great alongside it, but doesn’t depend on it. On the other hand, it’s a probabilistic indexing approach which, like bloom filters, may give false-positives, especially when data is not clustered. Which brings us to our next technique.
除了分区修剪之外,数据仓库世界中使用的另一种常见技术(但Spark目前还缺少这种技术)是基于小型物化聚合的 I/O 修剪。 简而言之,该想法是:
- 跟踪与 I/O 粒度相关的特定粒度下的简单统计信息,例如最小值和最大值。
- 在查询计划时利用这些统计信息以避免不必要的I/O。
这正是 Databricks Delta 数据跳过功能的含义。将新数据插入Databricks Delta 表后,将为受支持类型的所有列(包括嵌套列)收集文件级最小/最大统计信息。然后,当针对该表进行查找查询时,Databricks Delta首先会查询这些统计信息,以确定可以安全地跳过哪些文件。但是,正如他们所说,GIF 值千言,所以:
一方面,这是一种轻量级和灵活(粒度可以调整)的技术,易于实现和推理。它也完全与分区正交:它与分区并行工作,但并不依赖于分区。另一方面,它是一种概率索引方法,与bloom过滤器一样,可能会产生误报,尤其是在数据没有聚集的情况下。这就引出了我们的下一项技术。
For I/O pruning to be effective data needs to be clustered so that min-max ranges are narrow and, ideally, non-overlapping. That way, for a given point lookup, the number of min-max range hits is minimized, i.e. skipping is maximized.
Sometimes, data just happens to be naturally clustered: monotonically increasing IDs, columns that are correlated with insertion time (e.g., dates / timestamps) or the partition key (e.g., pk_brand_name – model_name). When that’s not the case, you can still enforce clustering by explicitly sorting or range-partitioning your data before insertions.
But, again, suppose your workload consists of equally frequent/relevant single-column predicates on (e.g. n = 4) different columns.
In that case, “linear” a.k.a. “lexicographic” or “major-minor” sorting by all of the n columns will strongly favor the first one that’s specified, clustering its values perfectly. However, it won’t do much, if anything at all (depending on how many duplicate values there are on that first column) for the second one, and so on. Therefore, in all likelihood, there will be no clustering on the nth column and therefore no skipping possible for lookups involving it.
So how can we do better? More precisely, how can we achieve similar skipping effectiveness along every individual dimension?
If we think about it, what we’re looking for is a way of assigning n-dimensional data points to data files, such that points assigned to the same file are also close to each other along each of the n dimensions individually. In other words, we want to map multi-dimensional points to one-dimensional values in a way that preserves locality.
This is a well-known problem, encountered not only in the database world, but also in domains such as computer graphics and geohashing. The answer is: locality-preserving space-filling curves, the most commonly used ones being the Z-order and Hilbert curves.
Below is a simple illustration of how Z-ordering can be applied for improving data layout with regard to data skipping effectiveness. Legend:
- Gray dot = data point e.g., chessboard square coordinates
- Gray box = data file; in this example, we aim for files of 4 points each
- Yellow box = data file that’s read for the given query
- Green dot = data point that passes the query’s filter and answers the query
- Red dot = data point that’s read, but doesn’t satisfy the filter; “false positive”
Okay, enough theory, let’s get back to the Spark + AI Summit keynote and see how Databricks Delta can be used for real-time cybersecurity threat response.
Say you’re using Bro, the popular open-source network traffic analyzer, which produces real-time, comprehensive network activity information2. The more popular your product is, the more heavily your services get used and, therefore, the more data Bro starts outputting. Writing this data at a fast enough pace to persistent storage in a more structured way for future processing is the first big data challenge you’ll face.
This is exactly what Databricks Delta was designed for in the first place, making this task easy and reliable. What you could do is use structured streaming to pipe your Bro conn data into a date-partitioned Databricks Delta table, which you’ll periodically run OPTIMIZE on so that your log records end up evenly distributed across reasonably-sized data files. But that’s not the focus of this blog post, so, for illustration purposes, let’s keep it simple and use a non-streaming, non-partitioned Databricks Delta table consisting of uniformly distributed random data.
Faced with a potential cyber-attack threat, the kind of ad-hoc data analysis you’ll want to run is a series of interactive “point lookups” against the logged network connection data. For example, “find all recent network activity involving this suspicious IP address.” We’ll model this workload by assuming it’s made out of basic lookup queries with single-column equality filters, using both random and sampled IPs and ports. Such simple queries are IO-bound, i.e. their runtime depends linearly on the amount of data scanned.
These lookup queries will typically turn into full table scans that might run for hours, depending on how much data you’re storing and how far back you’re looking. Your end goal is likely to minimize the total amount of time spent on running these queries, but, for illustration purposes, let’s instead define our cost function as the total number of records scanned. This metric should be a good approximation of total runtime and has the benefit of being well defined and deterministic, allowing interested readers to easily and reliably reproduce our experiments.
So here we go, this is what we’ll work with, concretely:
case class ConnRecord(src_ip: String, src_port: Int, dst_ip: String, dst_port: Int)
def randomIPv4(r: Random) = Seq.fill(4)(r.nextInt(256)).mkString(".")
def randomPort(r: Random) = r.nextInt(65536)
def randomConnRecord(r: Random) = ConnRecord(
src_ip = randomIPv4(r), src_port = randomPort(r),
dst_ip = randomIPv4(r), dst_port = randomPort(r))
case class TestResult(numFilesScanned: Long, numRowsScanned: Long, numRowsReturned: Long)
def testFilter(table: String, filter: String): TestResult = {
val query = s"SELECT COUNT(*) FROM $table WHERE $filter"
val(result, metrics) = collectWithScanMetrics(sql(query).as[Long])
TestResult(
numFilesScanned = metrics("filesNum"),
numRowsScanned = metrics.get("numOutputRows").getOrElse(0L),
numRowsReturned = result.head)
}
// Runs testFilter() on all given filters and returns the percent of rows skipped
// on average, as a proxy for Data Skipping effectiveness: 0 is bad, 1 is good
def skippingEffectiveness(table: String, filters: Seq[String]): Double = { ... }
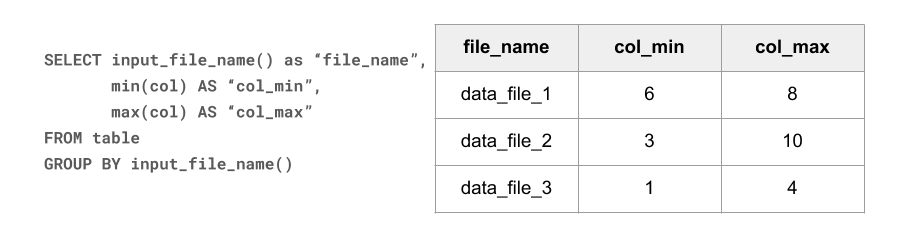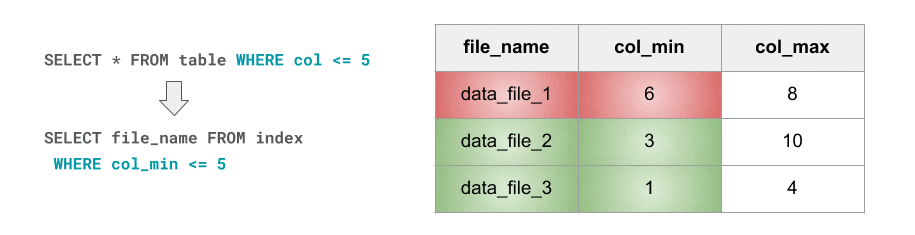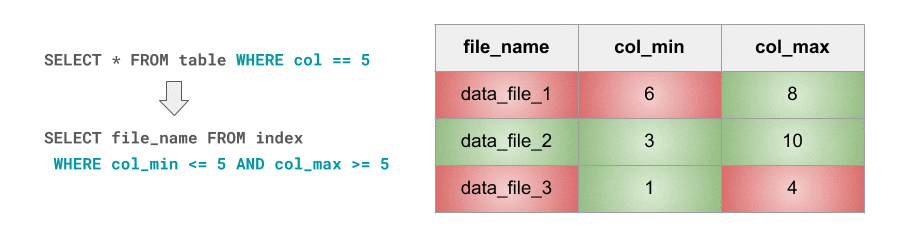
Here’s how a randomly generated table of 100 files, 1K random records each, might look like:
SELECT row_number() OVER (ORDER BY file) AS file_id,
count(*) as numRecords, min(src_ip), max(src_ip), min(src_port),
max(src_port), min(dst_ip), max(dst_ip), min(dst_port), max(dst_port)
FROM (
SELECT input_file_name() AS file, * FROM conn_random)
GROUP BY file
Seeing how every file’s min-max ranges cover almost the entire domain of values, it is easy to predict that there will be very little opportunity for file skipping. Our evaluation function confirms that:
skippingEffectiveness(connRandom, singleColumnFilters)
Ok, that’s expected, as our data is randomly generated and so there are no correlations. So let’s try explicitly sorting data before writing it.
spark.read.table(connRandom)
.repartitionByRange($"src_ip", $"src_port", $"dst_ip", $"dst_port")
// or just .sort($"src_ip", $"src_port", $"dst_ip", $"dst_port")
.write.format("delta").saveAsTable(connSorted)
skippingEffectiveness(connRandom, singleColumnFilters)
Hmm, we have indeed improved our metric, but 25% is still not great. Let’s take a closer look:
val src_ip_eff = skippingEffectiveness(connSorted, srcIPv4Filters)
val src_port_eff = skippingEffectiveness(connSorted, srcPortFilters)
val dst_ip_eff = skippingEffectiveness(connSorted, dstIPv4Filters)
val dst_port_eff = skippingEffectiveness(connSorted, dstPortFilters)
Turns out src_ip lookups are really fast but all others are basically just full table scans. Again, that’s no surprise. As explained earlier, that’s what you get with linear sorting: the resulting data is clustered perfectly along the first dimension (src_ip in our case), but almost not at all along further dimensions.
So how can we do better? By enforcing ZORDER clustering.
spark.read.table(connRandom)
.write.format("delta").saveAsTable(connZorder)
sql(s"OPTIMIZE $connZorder ZORDER BY (src_ip, src_port, dst_ip, dst_port)")
skippingEffectiveness(connZorder, singleColumnFilters)
Quite a bit better than the 0.25 obtained by linear sorting, right? Also, here’s the breakdown:
val src_ip_eff = skippingEffectiveness(connZorder, srcIPv4Filters)
val src_port_eff = skippingEffectiveness(connZorder, srcPortFilters)
val dst_ip_eff = skippingEffectiveness(connZorder, dstIPv4Filters)
val dst_port_eff = skippingEffectiveness(connZorder, dstPortFilters)
A couple of observations worth noting:
- It is expected that skipping effectiveness on src_ip is now lower than with linear ordering, as the latter would ensure perfect clustering, unlike z-ordering. However, the other columns’ score is now almost just as good, unlike before when it was 0.
- It is also expected that the more columns you z-order by, the lower the effectiveness.
For example,
ZORDER BY (src_ip, dst_ip)achieves 0.82. So it is up to you to decide what filters you care about the most.
In the real-world use case presented at the Spark + AI summit, the skipping effectiveness on a typical WHERE src_ip = x AND dst_ip = y query was even higher. In a data set of 504 terabytes (over 11 trillion rows), only 36.5 terabytes needed to be scanned thanks to data skipping. That’s a significant reduction of 92.4% in the number of bytes and 93.2% in the number of rows.
Using Databricks Delta’s built-in data skipping and ZORDER clustering features, large cloud data lakes can be queried in a matter of seconds by skipping files not relevant to the query. In a real-world cybersecurity analysis use case, 93.2% of the records in a 504 terabytes dataset were skipped for a typical query, reducing query times by up to two orders of magnitude.
In other words, Databricks Delta can speed up your queries by as much as 100X.
Note: Data skipping has been offered as an independent option outside of Databricks Delta in the past as a separate preview. That option will be deprecated in the near future. We highly recommend you move to Databricks Delta to take advantage of the data skipping capability.