You signed in with another tab or window. Reload to refresh your session.You signed out in another tab or window. Reload to refresh your session.You switched accounts on another tab or window. Reload to refresh your session.Dismiss alert
Copy file name to clipboardExpand all lines: docs/Pipelines/PET_Introduction.md
+21Lines changed: 21 additions & 0 deletions
Display the source diff
Display the rich diff
Original file line number
Diff line number
Diff line change
@@ -35,6 +35,27 @@ bids
35
35
└─ sub-CLNC001_ses-M000_trc-18FFDG_pet.nii.gz
36
36
```
37
37
38
+
## Reconstruction methods
39
+
40
+
PET data can be reconstructed into the spatial distribution of the injected radio-tracer through various methods.
41
+
The specification of the method used to reconstruct a given image is supported by the [BIDS standard](https://bids-specification.readthedocs.io/en/stable/04-modality-specific-files/09-positron-emission-tomography.html#pet-recording-data).
42
+
43
+
There exists four reserved values:
44
+
45
+
-`acdyn`: for reconstructions with attenuation correction of dynamic data
46
+
-`acstat`: for reconstructions with attenuation correction of static data
47
+
-`nacdyn`: for reconstructions without attenuation correction of dynamic data
48
+
-`nacstat`: for reconstructions without attenuation correction of static data
49
+
50
+
In addition to these values, when working with ADNI data, the following values are possible:
51
+
52
+
-`coregdyn`: Corresponds to "ADNI processing steps 1"
53
+
-`coregavg`: Corresponds to "ADNI processing steps 2"
54
+
-`coregstd`: Corresponds to "ADNI processing steps 3"
55
+
-`coregiso`: Corresponds to "ADNI processing steps 4"
56
+
57
+
See [this page](https://adni.loni.usc.edu/data-samples/adni-data/neuroimaging/pet/) for more information.
58
+
38
59
## Reference regions used for intensity normalization
39
60
40
61
In neurology, an approach widely used to allow inter- and intra-subject comparison of [PET](../glossary.md#pet) images is to compute standardized uptake value ratio ([SUVR](../glossary.md#suvr)) maps.
Copy file name to clipboardExpand all lines: docs/Pipelines/PET_Linear.md
+26-34Lines changed: 26 additions & 34 deletions
Display the source diff
Display the rich diff
Original file line number
Diff line number
Diff line change
@@ -4,23 +4,17 @@
4
4
This pipeline performs spatial normalization to the MNI space and intensity normalization of [PET](../glossary.md#pet) images.
5
5
Its steps include:
6
6
7
-
- affine registration to the [MNI152NLin2009cSym](https://bids-specification.readthedocs.io/en/stable/99-appendices/08-coordinate-systems.html#template-based-coordinate-systems)
8
-
template [Fonov et al., [2011](https://doi.org/10.1016/j.neuroimage.2010.07.033),
9
-
[2009](https://doi.org/10.1016/S1053-8119(09)70884-5)] in MNI space with the
10
-
SyN algorithm [[Avants et al., 2008](https://doi.org/10.1016/j.media.2007.06.004)]
11
-
from the [ANTs](http://stnava.github.io/ANTs/) software package
12
-
[[Avants et al., 2014](https://doi.org/10.3389/fninf.2014.00044)];
13
-
- intensity normalization using the average [PET](../glossary.md#pet) uptake in reference regions
14
-
resulting in a standardized uptake value ratio ([SUVR](../glossary.md#suvr)) map;
15
-
- cropping of the registered images to remove the background.
7
+
- affine registration to the [MNI152NLin2009cSym](https://bids-specification.readthedocs.io/en/stable/99-appendices/08-coordinate-systems.html#template-based-coordinate-systems) template [Fonov et al., [2011](https://doi.org/10.1016/j.neuroimage.2010.07.033), [2009](https://doi.org/10.1016/S1053-8119(09)70884-5)] in MNI space with the SyN algorithm [[Avants et al., 2008](https://doi.org/10.1016/j.media.2007.06.004)] from the [ANTs](http://stnava.github.io/ANTs/) software package [[Avants et al., 2014](https://doi.org/10.3389/fninf.2014.00044)]
8
+
- intensity normalization using the average [PET](../glossary.md#pet) uptake in reference regions resulting in a standardized uptake value ratio ([SUVR](../glossary.md#suvr)) map
9
+
- cropping of the registered images to remove the background
16
10
17
11
!!! note "Clinica & BIDS specifications for PET modality"
18
12
Since Clinica `v0.6`, PET data following the official specifications in BIDS version 1.6.0 are now compatible with Clinica.
19
13
See [BIDS](../../BIDS) page for more information.
20
14
21
15
## Prerequisites
22
16
23
-
You need to have performed the [`t1-linear`](../T1_Linear) pipeline on your T1-weighted MR images.
17
+
You need to have performed the [`t1-linear`](./T1_Linear.md) pipeline on your T1-weighted MR images.
-`BIDS_DIRECTORY` is the input folder containing the dataset in a
41
-
[BIDS](../../BIDS) hierarchy;
42
-
-`CAPS_DIRECTORY` is the output folder containing the results in a
43
-
[CAPS](../../CAPS/Introduction) hierarchy;
44
-
-`ACQ_LABEL` is the label given to the PET acquisition, specifying the tracer
45
-
used (`trc-<acq_label>`). It can be for instance '18FFDG' for
46
-
<sup>18</sup>F-fluorodeoxyglucose or '18FAV45' for <sup>18</sup>F-florbetapir;
47
-
- The reference region is used to perform intensity normalization (i.e.
48
-
dividing each voxel of the image by the average uptake in this region)
49
-
resulting in a standardized uptake value ratio ([SUVR](../glossary.md#suvr)) map. It can be
50
-
`cerebellumPons` or `cerebellumPons2` (used for amyloid tracers) and `pons`
51
-
or `pons2` (used for FDG). See [PET introduction](./PET_Introduction.md) for
52
-
more details about masks versions.
53
-
54
-
By default, cropped images (matrix size 169×208×179, 1 mm isotropic voxels) are
55
-
generated to reduce the computing power required when training deep learning
56
-
models. Use the `--uncropped_image` option if you do not want to crop the image.
57
-
58
-
The pipeline also offers the possibility to save the [PET](../glossary.md#pet) image in the T1w space
59
-
after rigid transformation using the `--save_pet_in_t1w_space` option.
34
+
-`BIDS_DIRECTORY` is the input folder containing the dataset in a [BIDS](../BIDS.md) hierarchy
35
+
-`CAPS_DIRECTORY` is the output folder containing the results in a [CAPS](../CAPS/Introduction.md) hierarchy
36
+
-`ACQ_LABEL` is the label given to the PET acquisition, specifying the tracer used (`trc-<acq_label>`). It can be for instance '18FFDG' for <sup>18</sup>F-fluorodeoxyglucose or '18FAV45' for <sup>18</sup>F-florbetapir
37
+
- The reference region is used to perform intensity normalization (i.e. dividing each voxel of the image by the average uptake in this region) resulting in a standardized uptake value ratio ([SUVR](../glossary.md#suvr)) map.
38
+
It can be `cerebellumPons` or `cerebellumPons2` (used for amyloid tracers) and `pons` or `pons2` (used for FDG).
39
+
See [PET introduction](./PET_Introduction.md) for more details about masks versions.
40
+
41
+
By default, cropped images (matrix size 169×208×179, 1 mm isotropic voxels) are generated to reduce the computing power required when training deep learning models.
42
+
Use the `--uncropped_image` option if you do not want to crop the image.
43
+
44
+
It is possible to select only images based on a [specific reconstruction method](./PET_Introduction.md#reconstruction-methods) with the `--reconstruction_method` option.
45
+
46
+
!!! warning
47
+
It can happen that a [BIDS](../BIDS.md) dataset contains several [PET](../glossary.md#pet) scans for a given subject and session.
48
+
In this situation, these images will differ through at least one [BIDS](../BIDS.md) entity like the tracer or the reconstruction method.
49
+
When running the `pet-linear` pipeline, clinica will raise an error if more than one image matches the criteria provided through the command line.
50
+
To avoid that, it is important to specify values for these options such that a single image is selected per subject and session.
51
+
52
+
The pipeline also offers the possibility to save the [PET](../glossary.md#pet) image in the T1w space after rigid transformation using the `--save_pet_in_t1w_space` option.
60
53
61
54
!!! note
62
-
The arguments common to all Clinica pipelines are described in [Interacting with Clinica](../../InteractingWithClinica).
55
+
The arguments common to all Clinica pipelines are described in [Interacting with Clinica](../Software/InteractingWithClinica.md).
63
56
64
57
!!! tip
65
58
Do not hesitate to type `clinica run pet-linear --help` to see the full list of parameters.
66
59
67
60
## Outputs
68
61
69
-
Results are stored in the following folder of the [CAPS hierarchy](../../CAPS/Specifications/#pet-imaging-data): `subjects/<participant_id>/<session_id>/pet_linear`.
62
+
Results are stored in the following folder of the [CAPS hierarchy](../CAPS/Specifications.md#pet-imaging-data): `subjects/<participant_id>/<session_id>/pet_linear`.
70
63
71
64
The main output files are:
72
65
@@ -100,5 +93,4 @@ You can now use the [ClinicaDL framework](https://clinicadl.readthedocs.io/) pre
100
93
resulting in images of size 169×208×179 with 1 mm isotropic voxels.
Copy file name to clipboardExpand all lines: docs/Pipelines/PET_Surface.md
+10-16Lines changed: 10 additions & 16 deletions
Display the source diff
Display the rich diff
Original file line number
Diff line number
Diff line change
@@ -1,8 +1,7 @@
1
1
<!-- markdownlint-disable MD033 MD046 -->
2
2
# `pet-surface` - Surface-based processing of PET images
3
3
4
-
This pipeline performs several processing steps for the analysis of PET data on the cortical surface
5
-
[[Marcoux et al., 2018](https://doi.org/10.3389/fninf.2018.00094)]:
4
+
This pipeline performs several processing steps for the analysis of PET data on the cortical surface [[Marcoux et al., 2018](https://doi.org/10.3389/fninf.2018.00094)]:
6
5
7
6
- co-registration of PET and T1-weighted MR images;
8
7
- intensity normalization;
@@ -19,7 +18,7 @@ This pipeline relies mainly on tools from **[FreeSurfer](https://surfer.nmr.mgh.
19
18
20
19
## Prerequisite
21
20
22
-
You need to have performed the [`t1-freesurfer`](../T1_FreeSurfer) pipeline on your T1-weighted MR images.
21
+
You need to have performed the [`t1-freesurfer`](./T1_FreeSurfer.md) pipeline on your T1-weighted MR images.
-`BIDS_DIRECTORY` is the input folder containing the dataset in a [BIDS](../../BIDS) hierarchy.
42
-
-`CAPS_DIRECTORY` acts both as an input folder (where the results of the `t1-freesurfer` pipeline are stored) and as the output folder containing the results in a [CAPS](../../CAPS/Introduction) hierarchy.
40
+
-`BIDS_DIRECTORY` is the input folder containing the dataset in a [BIDS](../BIDS.md) hierarchy.
41
+
-`CAPS_DIRECTORY` acts both as an input folder (where the results of the `t1-freesurfer` pipeline are stored) and as the output folder containing the results in a [CAPS](../CAPS/Introduction.md) hierarchy.
43
42
-`ACQ_LABEL` is the label given to the PET acquisition, specifying the tracer used (`trc-<acq_label>`).
44
43
- The reference region is used to perform intensity normalization (i.e. dividing each voxel of the image by the average uptake in this region) resulting in a standardized uptake value ratio (SUVR) map.
45
-
It can be `cerebellumPons` or `cerebellumPons2` (used for amyloid tracers) or `pons` or `pons2` (used for FDG).
46
-
-`PVC_PSF_TSV` is the TSV file containing the `psf_x`, `psf_y` and `psf_z` of the PSF for each PET image. It is expected to be generated by the user.
47
-
More explanation is given in [PET Introduction](../PET_Introduction) page.
44
+
It can be `cerebellumPons` or `cerebellumPons2` (used for amyloid tracers) or `pons` or `pons2` (used for FDG).
45
+
-`PVC_PSF_TSV` is the TSV file containing the `psf_x`, `psf_y` and `psf_z` of the PSF for each PET image. It is expected to be generated by the user. More explanation is given in [PET Introduction](./PET_Introduction.md) page.
48
46
49
47
!!! info
50
48
Since the release of Clinica v0.3.8, the handling of PSF information has changed.
@@ -55,8 +53,9 @@ More explanation is given in [PET Introduction](../PET_Introduction) page.
55
53
56
54
Pipeline options:
57
55
58
-
-`-np`: This parameter specifies the number of threads to run in parallel.
59
-
We recommend using `your_number_of_cpu - 1`.
56
+
-`--reconstruction_method`: Select only images based on a [specific reconstruction method](./PET_Introduction.md#reconstruction-methods).
57
+
-`-np`: This parameter specifies the number of threads to run in parallel. We recommend using `your_number_of_cpu - 1`.
58
+
60
59
Please note that PETPVC is extremely demanding in terms of resources and may cause the pipeline to crash if many subjects happen to be partial volume corrected at the same time (Error : `Failed to allocate memory for image`).
61
60
To mitigate this issue, you can do the following:
62
61
@@ -108,7 +107,7 @@ surface at equal distance between the white matter/gray matter interface and the
108
107
TSV files summarizing the regional statistics on the labelled atlases (Desikan and Destrieux).
109
108
110
109
!!! note
111
-
The full list of output files from the `pet-surface` pipeline can be found in [The ClinicA Processed Structure (CAPS) specifications](../../CAPS/Specifications/#pet-surface-surface-based-processing-of-pet-images).
110
+
The full list of output files from the `pet-surface` pipeline can be found in [The ClinicA Processed Structure (CAPS) specifications](../CAPS/Specifications.md/#pet-surface---surface-based-processing-of-pet-images).
*<center><small>FDG PET SUVR projected onto the cortical surface (left hemisphere) for (from left to right) a cognitively normal subject (CN), a patient with Alzheimer’s disease (AD), a patient with semantic variant primary progressive aphasia (svPPA) and a patient with logopenic variant primary progressive aphasia (lvPPA).
@@ -148,11 +147,6 @@ The second row is the same signal for each subject, but warped to FsAverage afte
- You can use the [Clinica Google Group](https://groups.google.com/forum/#!forum/clinica-user) to ask for help!
154
-
- Report an issue on [GitHub](https://github.com/aramis-lab/clinica/issues).
155
-
156
150
## Appendix I: Diagram of the pipeline execution
157
151
158
152
<center>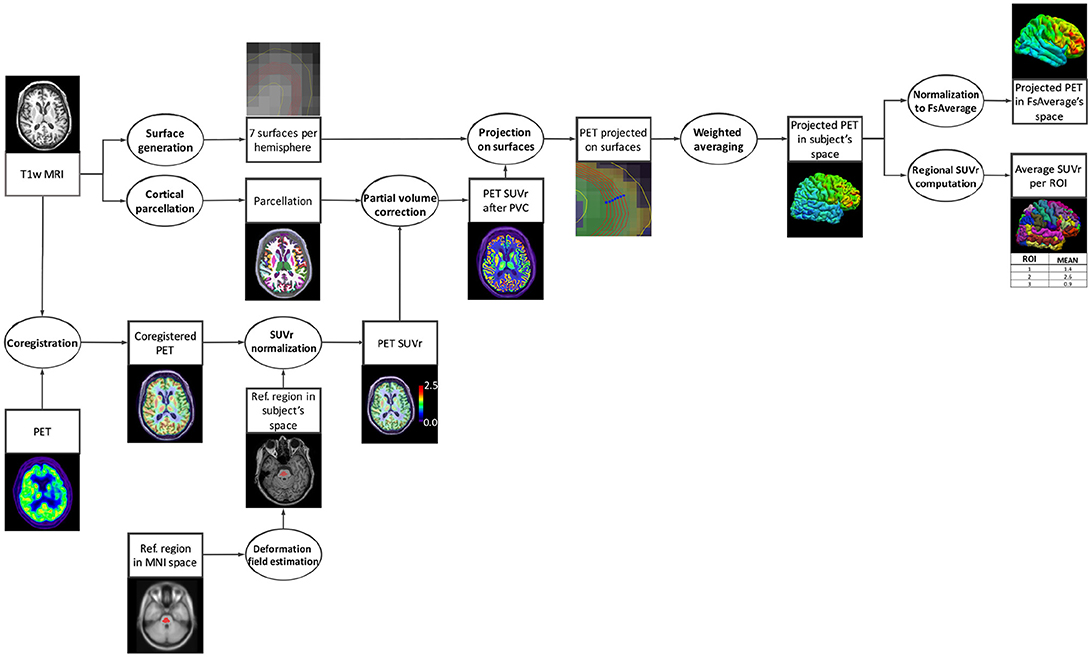</center>
Copy file name to clipboardExpand all lines: docs/Pipelines/PET_Volume.md
+15-16Lines changed: 15 additions & 16 deletions
Display the source diff
Display the rich diff
Original file line number
Diff line number
Diff line change
@@ -8,15 +8,15 @@ This pipeline performs several processing steps on [PET](../glossary.md#pet) dat
8
8
- inter-subject spatial normalization of the PET image into MNI space based on the DARTEL deformation model of SPM [[Ashburner, 2007](http://dx.doi.org/10.1016/j.neuroimage.2007.07.007)];
9
9
- intensity normalization using the average PET uptake in reference regions resulting in a standardized uptake value ratio ([SUVR](../glossary.md#suvr)) map;
10
10
- parcellation into anatomical regions based on an atlas and computation of average values within each region.
11
-
The list of available atlases can be found [here](../../Atlases).
11
+
The list of available atlases can be found [here](../Atlases.md).
12
12
13
13
!!! note "Clinica & BIDS specifications for PET modality"
14
14
Since Clinica `v0.6`, PET data following the official specifications in BIDS version 1.6.0 are now compatible with Clinica.
15
-
See [BIDS](../../BIDS) page for more information.
15
+
See [BIDS](../BIDS.md) page for more information.
16
16
17
17
## Prerequisite
18
18
19
-
You need to have performed the [`t1-volume`](../T1_Volume) pipeline on your T1-weighted MR images.
19
+
You need to have performed the [`t1-volume`](./T1_Volume.md) pipeline on your T1-weighted MR images.
-`BIDS_DIRECTORY` is the input folder containing the dataset in a [BIDS](../../BIDS) hierarchy.
39
-
-`CAPS_DIRECTORY` acts both as an input folder (where the results of the `t1-volume-*` pipeline are stored) and as the output folder containing the results in a [CAPS](../../CAPS/Introduction) hierarchy.
40
-
-`GROUP_LABEL` is the label of the group that is associated to the DARTEL template that you had created when running the [`t1-volume`](../T1_Volume) pipeline.
38
+
-`BIDS_DIRECTORY` is the input folder containing the dataset in a [BIDS](../BIDS.md) hierarchy.
39
+
-`CAPS_DIRECTORY` acts both as an input folder (where the results of the `t1-volume-*` pipeline are stored) and as the output folder containing the results in a [CAPS](../CAPS/Introduction.md) hierarchy.
40
+
-`GROUP_LABEL` is the label of the group that is associated to the DARTEL template that you had created when running the [`t1-volume`](./T1_Volume.md) pipeline.
41
41
-`ACQ_LABEL` is the label given to the PET acquisition, specifying the tracer used (`trc-<acq_label>`).
42
42
- The reference region is used to perform intensity normalization (i.e. dividing each voxel of the image by the average uptake in this region) resulting in a standardized uptake value ratio ([SUVR](../glossary.md#suvr)) map.
43
43
It can be `cerebellumPons` or `cerebellumPons2` (used for amyloid tracers) or `pons` or `pons2` (used for FDG).
44
44
45
45
Pipeline options:
46
46
47
-
-`--pvc_psf_tsv`: TSV file containing the `psf_x`, `psf_y` and `psf_z` of the PSF for each PET image.
48
-
More explanation is given in [PET Introduction](../PET_Introduction) page.
47
+
-`--pvc_psf_tsv`: TSV file containing the `psf_x`, `psf_y` and `psf_z` of the PSF for each PET image. More explanation is given in [PET Introduction](./PET_Introduction.md#partial-volume-correction-pvc) page.
49
48
-`--smooth`: a list of integers specifying the different isotropic full width at half maximum ([FWHM](../glossary.md#fwhm)) in millimeters to smooth the image. Default value is: 0, 8 (both without smoothing and with an isotropic smoothing of 8 mm)
49
+
-`--reconstruction_method`: Select only images based on a [specific reconstruction method](./PET_Introduction.md#reconstruction-methods).
50
+
51
+
!!! warning
52
+
It can happen that a [BIDS](../BIDS.md) dataset contains several [PET](../glossary.md#pet) scans for a given subject and session.
53
+
In this situation, these images will differ through at least one [BIDS](../BIDS.md) entity like the tracer or the reconstruction method.
54
+
When running the `pet-volume` pipeline, clinica will raise an error if more than one image matches the criteria provided through the command line.
55
+
To avoid that, it is important to specify values for these options such that a single image is selected per subject and session.
50
56
51
57
!!! info
52
58
Since the release of Clinica v0.3.8, the handling of PSF information in the TSV file has changed: `fwhm_x`, `fwhm_y`, `fwhm_z` columns have been replaced by `psf_x`, `psf_y`, `psf_z` and the `acq_label` column has been added.
@@ -61,9 +67,7 @@ More explanation is given in [PET Introduction](../PET_Introduction) page.
Results are stored in the following folder of the [CAPS hierarchy](../CAPS/Specifications.md#pet-volume---volume-based-processing-of-pet-images): `subjects/<participant_id>/<session_id>/pet/preprocessing`.
67
71
68
72
The main output files are:
69
73
@@ -108,8 +112,3 @@ The main output files are:
108
112
109
113
!!! tip
110
114
Easily access the papers cited on this page on [Zotero](https://www.zotero.org/groups/2240070/clinica_aramislab/items/collectionKey/INDXD9QQ).
111
-
112
-
## Support
113
-
114
-
- You can use the [Clinica Google Group](https://groups.google.com/forum/#!forum/clinica-user) to ask for help!
115
-
- Report an issue on [GitHub](https://github.com/aramis-lab/clinica/issues).
0 commit comments