diff --git a/protoBuilds/00222e-dd/README.json b/protoBuilds/00222e-dd/README.json
index dcafb0136d..e97da8a2b9 100644
--- a/protoBuilds/00222e-dd/README.json
+++ b/protoBuilds/00222e-dd/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sterile Workflows": [
"Cell Culture"
@@ -10,7 +10,7 @@
"internal": "00222e-dd",
"labware": "\nThermo Scientific Nunc 96 Well Plate 2000 \u00b5L #278743\nThermofisher 96 Well Plate 300 \u00b5L #167008\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sterile Workflows\n\t* Cell Culture\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a drug dosing assay in triplicate for up to 3 cell lines and 3 replicate plates.\n\n\n",
diff --git a/protoBuilds/00222e-seed/README.json b/protoBuilds/00222e-seed/README.json
index 20a9c0f03f..5055277c49 100644
--- a/protoBuilds/00222e-seed/README.json
+++ b/protoBuilds/00222e-seed/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sterile Workflows": [
"Seeding"
@@ -10,7 +10,7 @@
"internal": "00222e-seed",
"labware": "\nThermofisher 96 Well Plate 300 \u00b5L #167008\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sterile Workflows\n\t* Seeding\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a seeding assay in triplicate for up to 3 cell lines and 3 replicate plates.\n\n\n",
diff --git a/protoBuilds/00222e/README.json b/protoBuilds/00222e/README.json
index 1d29467575..b1aa57b81c 100644
--- a/protoBuilds/00222e/README.json
+++ b/protoBuilds/00222e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Serial Dilution and Screening"
@@ -10,7 +10,7 @@
"internal": "00222e",
"labware": "\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 96 Tip Rack 20 \u00b5L\nOpentrons 10 Tube Rack with Falcon 4x50 mL, 6x15 mL Conical\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Serial Dilution and Screening\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol adds a serially diluted stock solution to 12 plasma filled vials. The serial dilution can be toggled on and off in the variables. This protocol also predicts diluent and plasma height based on known labware dimensions. The initial liquid level needs to be specified for this to be accurate.\n\n\n",
diff --git a/protoBuilds/002af3/README.json b/protoBuilds/002af3/README.json
index f90e804938..51b5537242 100644
--- a/protoBuilds/002af3/README.json
+++ b/protoBuilds/002af3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol builds a 384 well plate with from up to 14 columns from up to 2 96 well plates. It is a pythonization of this protocol, which only could not run a variable number of columns. This protocol is used for building qPCR plates.\n\n\n\n8-Channel Electronic Pipette (GEN2) P20\nOpentrons 20uL Filter Tips\nHard-Shell 384-Well PCR Plates, thin wall, skirted, clear/white #HSP3805\nNEST 96 Deepwell Plate 2mL\n",
"internal": "002af3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol builds a 384 well plate with from up to 14 columns from up to 2 96 well plates. It is a pythonization of [this protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2020-07-17/uo03r2t/Covid-19%2012%20columns%20.json), which only could not run a variable number of columns. This protocol is used for building qPCR plates.\n\n---\n\n\n* [8-Channel Electronic Pipette (GEN2) P20](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 20uL Filter Tips](https://shop.opentrons.com/products/opentrons-20ul-filter-tips)\n* [Hard-Shell 384-Well PCR Plates, thin wall, skirted, clear/white #HSP3805](https://www.bio-rad.com/en-us/sku/hsp3805-hard-shell-384-well-pcr-plates-thin-wall-skirted-clear-white?ID=hsp3805)\n* [NEST 96 Deepwell Plate 2mL](http://www.cell-nest.com/page94?product_id=101&_l=en)\n\n",
"internal": "002af3\n",
diff --git a/protoBuilds/0045f1/README.json b/protoBuilds/0045f1/README.json
index bf7d470a6a..691c2d2bfb 100644
--- a/protoBuilds/0045f1/README.json
+++ b/protoBuilds/0045f1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "0045f1",
"labware": "\nOpentrons 10ul filter tips\nOpentrons 4-in-1 tube rack with Eppendorf 1.5mL safelock snapcap\nCustom slide plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "This protocol spots up to 6 slides with antibody. Each slide has 8 wells, to which the protocol will spot with 8 dots for wells 2-7. The user has the ability to manipulate the spacing between the dots, as well as the volume of the dispenses. The user can also specify the number of slides spotted. Please see below for the order in which slides are spotted.\n\nExplanation of complex parameters below:\n* `Number of tubes`: Specify the number of tubes this protocol will run.\n* `Number of Slides`: Specify the number of slides for this run (1-6).\n* `Spot Spacing`: Specify the spacing between dots on the slide wells.\n* `Spot Volume`: Specify the volume of each dot.\n* `Aspiration Flow Rate`: Specify the aspiration rate. A value of 1 is the default rate, a value of 1.2 is a 20% increase of the default rate, a value of 0.5 is half of the default rate.\n* `Dispense Flow Rate`: Specify the dispense rate. A value of 1 is the default rate, a value of 1.2 is a 20% increase of the default rate, a value of 0.5 is half of the default rate.\n* `P20 Single-Channel Mount`: Specify which mount (left or right) to host the P20 single-channel pipette.\n\n\n\n\n---\n\n\n",
diff --git a/protoBuilds/007992/README.json b/protoBuilds/007992/README.json
index 5126f3848f..919b0aec22 100644
--- a/protoBuilds/007992/README.json
+++ b/protoBuilds/007992/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Viral RNA"
@@ -8,7 +8,7 @@
"description": "This protocol automates a 3-wash RNA isolation using magnetic beads and a magnetic module GEN2. Purified elutions are transferred to a 384 well plate mounted on a temperature module GEN2.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons P300 GEN2 Multi-Channel Pipette\nOpentrons P20 GEN2 Single-Channel Pipette\nOpentrons 20uL and 200\u00b5L Filter Tips\nUSA Scientific 12-Well Reservoir, 22ml\nUSA Scientific 96-Deepwell Plate, 2.4mL #1896-2000\nCole Parmer 384-Well Plate, 50uL\n\n",
"internal": "007992",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Viral RNA\n\n",
"description": "This protocol automates a 3-wash RNA isolation using magnetic beads and a magnetic module GEN2. Purified elutions are transferred to a 384 well plate mounted on a temperature module GEN2.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons P300 GEN2 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons P20 GEN2 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 20uL and 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [USA Scientific 12-Well Reservoir, 22ml](https://www.usascientific.com/12-channel-automation-reservoir/p/1061-8150)\n* [USA Scientific 96-Deepwell Plate, 2.4mL #1896-2000](https://www.usascientific.com/plateone-96-deep-well-2ml/p/PlateOne-96-Deep-Well-2mL)\n* Cole Parmer 384-Well Plate, 50uL\n\n---\n\n",
"internal": "007992\n",
diff --git a/protoBuilds/00a214-protocol-1/README.json b/protoBuilds/00a214-protocol-1/README.json
index 398157777b..fbf325d6d7 100644
--- a/protoBuilds/00a214-protocol-1/README.json
+++ b/protoBuilds/00a214-protocol-1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Seeding Plate"
@@ -8,7 +8,7 @@
"description": "This protocol automates seeding plates with mammalian cells and is part of a five protocol suite. The five protocols are:\n\nProtocol 1: Seeding Plates with Mammalian Cells\nProtocol 2: DAPI Staining\nProtocol 3: Fix and DAPI Stain\nProtocol 4: Media Exchange\nProtocol 5: Primary Staining\n\nThe protocol begins with an empty reservoir for liquid waste, a 12-channel reservoir that contains what will be added to the plates, tips, eppendorf plate(s), and the P300-Multi attached to the left mount. Using up to 8 plates, the protocol will iterate through each plate and remove 50\u00b5L before adding 140\u00b5L from the reservoir. Each plate should have a designated well in the 12-well trough (i.e., slot 1 --> plate 1, slot 2 --> plate 2, etc.)\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons p300 Multi-Channel Pipette, GEN2\nOpentrons Tips\nNEST 12-Well Reservoir, 15mL\nNEST 1-Well Reservoir, 195mL\nEppendorf Plate\nReagents\n\n\n\nSlot 1: Opentrons Tips, note: protocol saves tip state and will prompt user to replace tips when new tips are needed.\n\nSlot 2: Eppendorf Plate (Plate 1)\n\nSlot 3: Eppendorf Plate (Plate 2)\n\nSlot 4: Eppendorf Plate (Plate 3)\n\nSlot 5: Eppendorf Plate (Plate 4)\n\nSlot 6: Eppendorf Plate (Plate 5)\n\nSlot 7: Eppendorf Plate (Plate 6)\n\nSlot 8: Eppendorf Plate (Plate 7)\n\nSlot 9: Eppendorf Plate (Plate 8)\n\nSlot 10: NEST 12-Well Reservoir, 15mL\n\nSlot 11: NEST 1-Well Reservoir, 195mL, for liquid waste\n\n\nUsing the customizations field (below), set up your protocol.\n Number of Plates: Specify the number of plates (1-8) to use.\n Aspiration/Dispense Speed: Specify the aspiration/dispense speed of the pipette (in \u00b5L/sec). Note: default rate is 43.46\u00b5L/sec.\n Aspiration Height: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when aspirating.\n Dispense Height: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when dispensing.\n\n",
"internal": "00a214-protocol-1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Seeding Plate\n\n\n",
"description": "This protocol automates seeding plates with mammalian cells and is part of a five protocol suite. The five protocols are:\n\nProtocol 1: [Seeding Plates with Mammalian Cells](https://develop.protocols.opentrons.com/protocol/00a214-protocol-1)\nProtocol 2: [DAPI Staining](https://develop.protocols.opentrons.com/protocol/00a214-protocol-2)\nProtocol 3: [Fix and DAPI Stain](https://develop.protocols.opentrons.com/protocol/00a214-protocol-3)\nProtocol 4: [Media Exchange](https://develop.protocols.opentrons.com/protocol/00a214-protocol-4)\nProtocol 5: [Primary Staining](https://develop.protocols.opentrons.com/protocol/00a214-protocol-5)\n\nThe protocol begins with an empty reservoir for liquid waste, a 12-channel reservoir that contains what will be added to the plates, tips, eppendorf plate(s), and the P300-Multi attached to the left mount. Using up to 8 plates, the protocol will iterate through each plate and remove 50\u00b5L before adding 140\u00b5L from the reservoir. Each plate should have a designated well in the 12-well trough (i.e., slot 1 --> plate 1, slot 2 --> plate 2, etc.)\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons p300 Multi-Channel Pipette, GEN2](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* Eppendorf Plate\n* Reagents\n\n\n\n---\n\n\nSlot 1: [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips), note: protocol saves tip state and will prompt user to replace tips when new tips are needed.\n\nSlot 2: Eppendorf Plate (Plate 1)\n\nSlot 3: Eppendorf Plate (Plate 2)\n\nSlot 4: Eppendorf Plate (Plate 3)\n\nSlot 5: Eppendorf Plate (Plate 4)\n\nSlot 6: Eppendorf Plate (Plate 5)\n\nSlot 7: Eppendorf Plate (Plate 6)\n\nSlot 8: Eppendorf Plate (Plate 7)\n\nSlot 9: Eppendorf Plate (Plate 8)\n\nSlot 10: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n\nSlot 11: [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml), for liquid waste\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Plates**: Specify the number of plates (1-8) to use.\n* **Aspiration/Dispense Speed**: Specify the aspiration/dispense speed of the pipette (in \u00b5L/sec). Note: default rate is 43.46\u00b5L/sec.\n* **Aspiration Height**: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when aspirating.\n* **Dispense Height**: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when dispensing.\n\n\n\n",
"internal": "00a214-protocol-1\n",
diff --git a/protoBuilds/00a214-protocol-2/README.json b/protoBuilds/00a214-protocol-2/README.json
index 5fe01c3630..421d0b0eba 100644
--- a/protoBuilds/00a214-protocol-2/README.json
+++ b/protoBuilds/00a214-protocol-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Staining"
@@ -8,7 +8,7 @@
"description": "This protocol automates DAPI staining and is part of a five protocol suite. The five protocols are:\n\nProtocol 1: Seeding Plates with Mammalian Cells\nProtocol 2: DAPI Staining\nProtocol 3: Fix and DAPI Stain\nProtocol 4: Media Exchange\nProtocol 5: Primary Staining\n\nThe protocol begins with an empty reservoir for liquid waste, a 12-channel reservoir that contains what will be added to the plates, tips, eppendorf plate(s), and the P300-Multi attached to the left mount. Using up to 4 plates, the protocol will iterate through each plate, adding reagents to the plate and removing liquid.\n\nThe following steps are performed to each plate:\nRemove 120\u00b5L of media\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 100\u00b5L of 2% PFA\nPause\nRemove 100\u00b5L of 2% PFA\nAdd 140\u00b5L of PBS+DAPI\nRemove 140\u00b5L of PBS+DAPI\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 140\u00b5L of PBS\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons p300 Multi-Channel Pipette, GEN2\nOpentrons Tips\nNEST 12-Well Reservoir, 15mL\nNEST 1-Well Reservoir, 195mL\nEppendorf Plate\nReagents\n\n\n\nSlot 1: Opentrons Tips, note: protocol saves tip state and will prompt user to replace tips when new tips are needed.\n\nSlot 2: Eppendorf Plate (Plate 1)\n\nSlot 3: Eppendorf Plate (Plate 2)\n\nSlot 5: Eppendorf Plate (Plate 3)\n\nSlot 6: Eppendorf Plate (Plate 4)\n\nSlot 7: NEST 12-Well Reservoir, 15mL\n\nColumns 1-4: PBS\nColumns 5-8: PBS\nColumns 9-12: 2% PFA\n\nSlot 10: NEST 12-Well Reservoir, 15mL\n\nColumns 1-4: PBS+DAPI\nColumns 5-8: PBS\nColumns 9-12: PBS\n\nSlot 11: NEST 1-Well Reservoir, 195mL, for liquid waste\n\nNote: When filling the 12-well reservoirs with reagents, each column corresponds to a plate (there are 4 columns allocated per reagent). For example, if running this protocol with two (2) plates, columns 1, 2, 5, and 6 would be filled with PBS in slot 7; columns 9 and 10 would be filled with 2% PFA; etc.\n\nUsing the customizations field (below), set up your protocol.\n Number of Plates: Specify the number of plates (1-4) to use.\n Aspiration/Dispense Speed: Specify the aspiration/dispense speed of the pipette (in \u00b5L/sec). Note: default rate is 43.46\u00b5L/sec.\n Aspiration Height: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when aspirating.\n Dispense Height: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when dispensing.\n\n",
"internal": "00a214-protocol-2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Staining\n\n\n",
"description": "This protocol automates DAPI staining and is part of a five protocol suite. The five protocols are:\n\nProtocol 1: [Seeding Plates with Mammalian Cells](https://develop.protocols.opentrons.com/protocol/00a214-protocol-1)\nProtocol 2: [DAPI Staining](https://develop.protocols.opentrons.com/protocol/00a214-protocol-2)\nProtocol 3: [Fix and DAPI Stain](https://develop.protocols.opentrons.com/protocol/00a214-protocol-3)\nProtocol 4: [Media Exchange](https://develop.protocols.opentrons.com/protocol/00a214-protocol-4)\nProtocol 5: [Primary Staining](https://develop.protocols.opentrons.com/protocol/00a214-protocol-5)\n\nThe protocol begins with an empty reservoir for liquid waste, a 12-channel reservoir that contains what will be added to the plates, tips, eppendorf plate(s), and the P300-Multi attached to the left mount. Using up to 4 plates, the protocol will iterate through each plate, adding reagents to the plate and removing liquid.\n\nThe following steps are performed to each plate:\nRemove 120\u00b5L of media\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 100\u00b5L of 2% PFA\nPause\nRemove 100\u00b5L of 2% PFA\nAdd 140\u00b5L of PBS+DAPI\nRemove 140\u00b5L of PBS+DAPI\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 140\u00b5L of PBS\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons p300 Multi-Channel Pipette, GEN2](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* Eppendorf Plate\n* Reagents\n\n\n\n---\n\n\nSlot 1: [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips), note: protocol saves tip state and will prompt user to replace tips when new tips are needed.\n\nSlot 2: Eppendorf Plate (Plate 1)\n\nSlot 3: Eppendorf Plate (Plate 2)\n\nSlot 5: Eppendorf Plate (Plate 3)\n\nSlot 6: Eppendorf Plate (Plate 4)\n\nSlot 7: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n\nColumns 1-4: PBS\nColumns 5-8: PBS\nColumns 9-12: 2% PFA\n\nSlot 10: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n\nColumns 1-4: PBS+DAPI\nColumns 5-8: PBS\nColumns 9-12: PBS\n\nSlot 11: [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml), for liquid waste\n\n**Note**: When filling the 12-well reservoirs with reagents, each column corresponds to a plate (there are 4 columns allocated per reagent). For example, if running this protocol with two (2) plates, columns 1, 2, 5, and 6 would be filled with PBS in slot 7; columns 9 and 10 would be filled with 2% PFA; etc.\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Plates**: Specify the number of plates (1-4) to use.\n* **Aspiration/Dispense Speed**: Specify the aspiration/dispense speed of the pipette (in \u00b5L/sec). Note: default rate is 43.46\u00b5L/sec.\n* **Aspiration Height**: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when aspirating.\n* **Dispense Height**: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when dispensing.\n\n\n\n",
"internal": "00a214-protocol-2\n",
diff --git a/protoBuilds/00a214-protocol-3/README.json b/protoBuilds/00a214-protocol-3/README.json
index f1f468b011..e485a19c53 100644
--- a/protoBuilds/00a214-protocol-3/README.json
+++ b/protoBuilds/00a214-protocol-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Staining"
@@ -8,7 +8,7 @@
"description": "This protocol automates DAPI staining and is part of a five protocol suite. The five protocols are:\n\nProtocol 1: Seeding Plates with Mammalian Cells\nProtocol 2: DAPI Staining\nProtocol 3: Fix and DAPI Stain\nProtocol 4: Media Exchange\nProtocol 5: Primary Staining\n\nThe protocol begins with an empty reservoir for liquid waste, a 12-channel reservoir that contains what will be added to the plates, tips, eppendorf plate(s), and the P300-Multi attached to the left mount. Using up to 4 plates, the protocol will iterate through each plate, adding reagents to the plate and removing liquid.\n\nThe following steps are performed to each plate:\nRemove 120\u00b5L of media\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 100\u00b5L of 2% PFA\nPause\nRemove 100\u00b5L of 2% PFA\nAdd 140\u00b5L of PBS+DAPI\nRemove 140\u00b5L of PBS+DAPI\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 140\u00b5L of PBS\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons p300 Multi-Channel Pipette, GEN2\nOpentrons Tips\nNEST 12-Well Reservoir, 15mL\nNEST 1-Well Reservoir, 195mL\nEppendorf Plate\nReagents\n\n\n\nSlot 1: Opentrons Tips, note: protocol saves tip state and will prompt user to replace tips when new tips are needed.\n\nSlot 2: Eppendorf Plate (Plate 1)\n\nSlot 3: Eppendorf Plate (Plate 2)\n\nSlot 5: Eppendorf Plate (Plate 3)\n\nSlot 6: Eppendorf Plate (Plate 4)\n\nSlot 7: NEST 12-Well Reservoir, 15mL\n\nColumns 1-4: PBS\nColumns 5-8: PBS\nColumns 9-12: 2% PFA\n\nSlot 10: NEST 12-Well Reservoir, 15mL\n\nColumns 1-4: PBS+DAPI\nColumns 5-8: PBS\nColumns 9-12: PBS\n\nSlot 11: NEST 1-Well Reservoir, 195mL, for liquid waste\n\nNote: When filling the 12-well reservoirs with reagents, each column corresponds to a plate (there are 4 columns allocated per reagent). For example, if running this protocol with two (2) plates, columns 1, 2, 5, and 6 would be filled with PBS in slot 7; columns 9 and 10 would be filled with 2% PFA; etc.\n\nUsing the customizations field (below), set up your protocol.\n Number of Plates: Specify the number of plates (1-4) to use.\n Aspiration/Dispense Speed: Specify the aspiration/dispense speed of the pipette (in \u00b5L/sec). Note: default rate is 43.46\u00b5L/sec.\n Aspiration Height: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when aspirating.\n Dispense Height: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when dispensing.\n\n",
"internal": "00a214-protocol-3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Staining\n\n\n",
"description": "This protocol automates DAPI staining and is part of a five protocol suite. The five protocols are:\n\nProtocol 1: [Seeding Plates with Mammalian Cells](https://develop.protocols.opentrons.com/protocol/00a214-protocol-1)\nProtocol 2: [DAPI Staining](https://develop.protocols.opentrons.com/protocol/00a214-protocol-2)\nProtocol 3: [Fix and DAPI Stain](https://develop.protocols.opentrons.com/protocol/00a214-protocol-3)\nProtocol 4: [Media Exchange](https://develop.protocols.opentrons.com/protocol/00a214-protocol-4)\nProtocol 5: [Primary Staining](https://develop.protocols.opentrons.com/protocol/00a214-protocol-5)\n\nThe protocol begins with an empty reservoir for liquid waste, a 12-channel reservoir that contains what will be added to the plates, tips, eppendorf plate(s), and the P300-Multi attached to the left mount. Using up to 4 plates, the protocol will iterate through each plate, adding reagents to the plate and removing liquid.\n\nThe following steps are performed to each plate:\nRemove 120\u00b5L of media\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 100\u00b5L of 2% PFA\nPause\nRemove 100\u00b5L of 2% PFA\nAdd 140\u00b5L of PBS+DAPI\nRemove 140\u00b5L of PBS+DAPI\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 140\u00b5L of PBS\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons p300 Multi-Channel Pipette, GEN2](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* Eppendorf Plate\n* Reagents\n\n\n\n---\n\n\nSlot 1: [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips), note: protocol saves tip state and will prompt user to replace tips when new tips are needed.\n\nSlot 2: Eppendorf Plate (Plate 1)\n\nSlot 3: Eppendorf Plate (Plate 2)\n\nSlot 5: Eppendorf Plate (Plate 3)\n\nSlot 6: Eppendorf Plate (Plate 4)\n\nSlot 7: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n\nColumns 1-4: PBS\nColumns 5-8: PBS\nColumns 9-12: 2% PFA\n\nSlot 10: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n\nColumns 1-4: PBS+DAPI\nColumns 5-8: PBS\nColumns 9-12: PBS\n\nSlot 11: [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml), for liquid waste\n\n**Note**: When filling the 12-well reservoirs with reagents, each column corresponds to a plate (there are 4 columns allocated per reagent). For example, if running this protocol with two (2) plates, columns 1, 2, 5, and 6 would be filled with PBS in slot 7; columns 9 and 10 would be filled with 2% PFA; etc.\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Plates**: Specify the number of plates (1-4) to use.\n* **Aspiration/Dispense Speed**: Specify the aspiration/dispense speed of the pipette (in \u00b5L/sec). Note: default rate is 43.46\u00b5L/sec.\n* **Aspiration Height**: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when aspirating.\n* **Dispense Height**: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when dispensing.\n\n\n\n",
"internal": "00a214-protocol-3\n",
diff --git a/protoBuilds/00a214-protocol-4/README.json b/protoBuilds/00a214-protocol-4/README.json
index 1aa5cd5200..d93e5137a0 100644
--- a/protoBuilds/00a214-protocol-4/README.json
+++ b/protoBuilds/00a214-protocol-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Staining"
@@ -8,7 +8,7 @@
"description": "This protocol automates Media Exchange and is part of a five protocol suite. The five protocols are:\n\nProtocol 1: Seeding Plates with Mammalian Cells\nProtocol 2: DAPI Staining\nProtocol 3: Fix and DAPI Stain\nProtocol 4: Media Exchange\nProtocol 5: Primary Staining\n\nThe protocol begins with an empty reservoir for liquid waste, a 12-channel reservoir that contains what will be added to the plates, tips, eppendorf plate(s), and the P300-Multi attached to the left mount. Using up to 4 plates, the protocol will iterate through each plate, adding reagents to the plate and removing liquid.\n\nThe following steps are performed to each plate:\nRemove 120\u00b5L of media\nAdd 140\u00b5L of differentiation media\nIf necessary (can select 'yes' or 'no' under Second Exchange)\nRemove 140\u00b5L of differentiation media\nAdd 140\u00b5L of differentiation media\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons p300 Multi-Channel Pipette, GEN2\nOpentrons Tips\nNEST 12-Well Reservoir, 15mL\nNEST 1-Well Reservoir, 195mL\nEppendorf Plate\nReagents\n\n\n\nSlot 1: Opentrons Tips, note: protocol saves tip state and will prompt user to replace tips when new tips are needed.\n\nSlot 2: Eppendorf Plate (Plate 1)\n\nSlot 3: Eppendorf Plate (Plate 2)\n\nSlot 4: Eppendorf Plate (Plate 3)\n\nSlot 5: Eppendorf Plate (Plate 4)\n\nSlot 5: Eppendorf Plate (Plate 5)\n\nSlot 6: Eppendorf Plate (Plate 6)\n\nSlot 10: NEST 12-Well Reservoir, 15mL\n\nColumns 1-6: Differentiation Media (for 1st exchange)\nColumns 7-22: Differentiation Media (for 2nd exchange)\n\nSlot 11: NEST 1-Well Reservoir, 195mL, for liquid waste\n\nNote: When filling the 12-well reservoirs with reagents, each column corresponds to a plate (there are 6 columns allocated per reagent). For example, if running this protocol with two (2) plates, columns 1 and 2 would be filled with differentiation media. If also doing the second exchange, then columns 7 and 8 would also need to be filled.\n\nUsing the customizations field (below), set up your protocol.\n Number of Plates: Specify the number of plates (1-4) to use.\n Aspiration/Dispense Speed: Specify the aspiration/dispense speed of the pipette (in \u00b5L/sec). Note: default rate is 43.46\u00b5L/sec.\n Aspiration Height: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when aspirating.\n Dispense Height: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when dispensing.\n* Second Exchange: Select whether or not to perform a second exchange of differentiation media.\n\n",
"internal": "00a214-protocol-4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Staining\n\n\n",
"description": "This protocol automates Media Exchange and is part of a five protocol suite. The five protocols are:\n\nProtocol 1: [Seeding Plates with Mammalian Cells](https://develop.protocols.opentrons.com/protocol/00a214-protocol-1)\nProtocol 2: [DAPI Staining](https://develop.protocols.opentrons.com/protocol/00a214-protocol-2)\nProtocol 3: [Fix and DAPI Stain](https://develop.protocols.opentrons.com/protocol/00a214-protocol-3)\nProtocol 4: [Media Exchange](https://develop.protocols.opentrons.com/protocol/00a214-protocol-4)\nProtocol 5: [Primary Staining](https://develop.protocols.opentrons.com/protocol/00a214-protocol-5)\n\nThe protocol begins with an empty reservoir for liquid waste, a 12-channel reservoir that contains what will be added to the plates, tips, eppendorf plate(s), and the P300-Multi attached to the left mount. Using up to 4 plates, the protocol will iterate through each plate, adding reagents to the plate and removing liquid.\n\nThe following steps are performed to each plate:\nRemove 120\u00b5L of media\nAdd 140\u00b5L of differentiation media\nIf necessary (can select 'yes' or 'no' under **Second Exchange**)\nRemove 140\u00b5L of differentiation media\nAdd 140\u00b5L of differentiation media\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons p300 Multi-Channel Pipette, GEN2](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* Eppendorf Plate\n* Reagents\n\n\n\n---\n\n\nSlot 1: [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips), note: protocol saves tip state and will prompt user to replace tips when new tips are needed.\n\nSlot 2: Eppendorf Plate (Plate 1)\n\nSlot 3: Eppendorf Plate (Plate 2)\n\nSlot 4: Eppendorf Plate (Plate 3)\n\nSlot 5: Eppendorf Plate (Plate 4)\n\nSlot 5: Eppendorf Plate (Plate 5)\n\nSlot 6: Eppendorf Plate (Plate 6)\n\nSlot 10: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n\nColumns 1-6: Differentiation Media (for 1st exchange)\nColumns 7-22: Differentiation Media (for 2nd exchange)\n\nSlot 11: [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml), for liquid waste\n\n**Note**: When filling the 12-well reservoirs with reagents, each column corresponds to a plate (there are 6 columns allocated per reagent). For example, if running this protocol with two (2) plates, columns 1 and 2 would be filled with differentiation media. If also doing the second exchange, then columns 7 and 8 would also need to be filled.\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Plates**: Specify the number of plates (1-4) to use.\n* **Aspiration/Dispense Speed**: Specify the aspiration/dispense speed of the pipette (in \u00b5L/sec). Note: default rate is 43.46\u00b5L/sec.\n* **Aspiration Height**: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when aspirating.\n* **Dispense Height**: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when dispensing.\n* **Second Exchange**: Select whether or not to perform a second exchange of differentiation media.\n\n\n\n",
"internal": "00a214-protocol-4\n",
diff --git a/protoBuilds/00a214-protocol-5/README.json b/protoBuilds/00a214-protocol-5/README.json
index ee5d8a080b..10d561962d 100644
--- a/protoBuilds/00a214-protocol-5/README.json
+++ b/protoBuilds/00a214-protocol-5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Staining"
@@ -8,7 +8,7 @@
"description": "This protocol automates primary staining and is part of a five protocol suite. The five protocols are:\n\nProtocol 1: Seeding Plates with Mammalian Cells\nProtocol 2: DAPI Staining\nProtocol 3: Fix and DAPI Stain\nProtocol 4: Media Exchange\nProtocol 5: Primary Staining\n\nThe protocol begins with an empty reservoir for liquid waste, a 12-channel reservoir that contains what will be added to the plates, tips, eppendorf plate(s), and the P300-Multi attached to the left mount. Using up to 4 plates, the protocol will iterate through each plate, adding reagents to the plate and removing liquid.\n\nThe following steps are performed to each plate:\nRemove 120\u00b5L of media\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 100\u00b5L of 2% PFA\nPause\nRemove 100\u00b5L of 2% PFA\nAdd 140\u00b5L of PBST\nPause\nRemove 140\u00b5L of PBST\nAdd 140\u00b5L of Blocking Buffer\nPause\nRemove 140\u00b5L of Blocking Buffer\nAdd 140\u00b5L of Primary Antibody\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons p300 Multi-Channel Pipette, GEN2\nOpentrons Tips\nNEST 12-Well Reservoir, 15mL\nNEST 1-Well Reservoir, 195mL\nEppendorf Plate\nReagents\n\n\n\nSlot 1: Opentrons Tips\n\nSlot 2: Eppendorf Plate (Plate 1)\n\nSlot 3: Eppendorf Plate (Plate 2)\n\nSlot 4: Opentrons Tips, note: protocol saves tip state and will prompt user to replace tips when new tips are needed.\n\nSlot 5: Eppendorf Plate (Plate 3)\n\nSlot 6: Eppendorf Plate (Plate 4)\n\nSlot 7: NEST 12-Well Reservoir, 15mL\n\nColumns 1-4: PBS\nColumns 5-8: PBS\nColumns 9-12: 2% PFA\n\nSlot 10: NEST 12-Well Reservoir, 15mL\n\nColumns 1-4: PBST\nColumns 5-8: Blocking Buffer\nColumns 9-12: Primary Antibody\n\nSlot 11: NEST 1-Well Reservoir, 195mL, for liquid waste\n\nNote: When filling the 12-well reservoirs with reagents, each column corresponds to a plate (there are 4 columns allocated per reagent). For example, if running this protocol with two (2) plates, columns 1, 2, 5, and 6 would be filled with PBS in slot 7; columns 9 and 10 would be filled with 2% PFA; etc.\n\nUsing the customizations field (below), set up your protocol.\n Number of Plates: Specify the number of plates (1-4) to use.\n Aspiration/Dispense Speed: Specify the aspiration/dispense speed of the pipette (in \u00b5L/sec). Note: default rate is 43.46\u00b5L/sec.\n Aspiration Height: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when aspirating.\n Dispense Height: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when dispensing.\n\n",
"internal": "00a214-protocol-5",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Staining\n\n\n",
"description": "This protocol automates primary staining and is part of a five protocol suite. The five protocols are:\n\nProtocol 1: [Seeding Plates with Mammalian Cells](https://develop.protocols.opentrons.com/protocol/00a214-protocol-1)\nProtocol 2: [DAPI Staining](https://develop.protocols.opentrons.com/protocol/00a214-protocol-2)\nProtocol 3: [Fix and DAPI Stain](https://develop.protocols.opentrons.com/protocol/00a214-protocol-3)\nProtocol 4: [Media Exchange](https://develop.protocols.opentrons.com/protocol/00a214-protocol-4)\nProtocol 5: [Primary Staining](https://develop.protocols.opentrons.com/protocol/00a214-protocol-5)\n\nThe protocol begins with an empty reservoir for liquid waste, a 12-channel reservoir that contains what will be added to the plates, tips, eppendorf plate(s), and the P300-Multi attached to the left mount. Using up to 4 plates, the protocol will iterate through each plate, adding reagents to the plate and removing liquid.\n\nThe following steps are performed to each plate:\nRemove 120\u00b5L of media\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 140\u00b5L of PBS\nRemove 140\u00b5L of PBS\nAdd 100\u00b5L of 2% PFA\nPause\nRemove 100\u00b5L of 2% PFA\nAdd 140\u00b5L of PBST\nPause\nRemove 140\u00b5L of PBST\nAdd 140\u00b5L of Blocking Buffer\nPause\nRemove 140\u00b5L of Blocking Buffer\nAdd 140\u00b5L of Primary Antibody\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons p300 Multi-Channel Pipette, GEN2](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* Eppendorf Plate\n* Reagents\n\n\n\n---\n\n\nSlot 1: [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 2: Eppendorf Plate (Plate 1)\n\nSlot 3: Eppendorf Plate (Plate 2)\n\nSlot 4: [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips), note: protocol saves tip state and will prompt user to replace tips when new tips are needed.\n\nSlot 5: Eppendorf Plate (Plate 3)\n\nSlot 6: Eppendorf Plate (Plate 4)\n\nSlot 7: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n\nColumns 1-4: PBS\nColumns 5-8: PBS\nColumns 9-12: 2% PFA\n\nSlot 10: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n\nColumns 1-4: PBST\nColumns 5-8: Blocking Buffer\nColumns 9-12: Primary Antibody\n\nSlot 11: [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml), for liquid waste\n\n**Note**: When filling the 12-well reservoirs with reagents, each column corresponds to a plate (there are 4 columns allocated per reagent). For example, if running this protocol with two (2) plates, columns 1, 2, 5, and 6 would be filled with PBS in slot 7; columns 9 and 10 would be filled with 2% PFA; etc.\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Plates**: Specify the number of plates (1-4) to use.\n* **Aspiration/Dispense Speed**: Specify the aspiration/dispense speed of the pipette (in \u00b5L/sec). Note: default rate is 43.46\u00b5L/sec.\n* **Aspiration Height**: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when aspirating.\n* **Dispense Height**: Specify how high (in mm) from the bottom of the well the pipette should be in the plate when dispensing.\n\n\n\n",
"internal": "00a214-protocol-5\n",
diff --git a/protoBuilds/00a577/README.json b/protoBuilds/00a577/README.json
index 548473c45b..acb4096e84 100644
--- a/protoBuilds/00a577/README.json
+++ b/protoBuilds/00a577/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Purification": [
"MP Biomedicals magGENic Plant DNA Kit"
@@ -10,7 +10,7 @@
"internal": "00a577",
"labware": "\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\nNEST 2 mL 96-Well Deep Well Plate, V Bottom #999-00103\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nNEST 1 Well Reservoir 195 mL #360103\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Purification\n\t* MP Biomedicals magGENic Plant DNA Kit\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs nucleic acid purification on up to 24 samples according to the [MP Biomedicals magGenic\u00a0Plant DNA Kit](https://www.mpbio.com/us/maggenic-plant-dna-kit).\n\n\n",
diff --git a/protoBuilds/00a6e5/README.json b/protoBuilds/00a6e5/README.json
index c2713274a1..252ee45f89 100644
--- a/protoBuilds/00a6e5/README.json
+++ b/protoBuilds/00a6e5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "00a6e5",
"labware": "\nOpentrons 4-in-1 tube rack\nOpentrons 1000ul tips\nOpentrons 300ul Tips\nNEST 12-Well Reservoir, 15mL\nAgilent 1 Well Reservoir, 290mL\nCustom 96 well plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n",
"description": "This protocol preps a 96 well plate for an LCMS extraction. The 96 well plate is loaded with negative urine samples. Reagents include enzyme, buffer, water, and spike INSTD. All transfers in the 96 well plate are by column. No mix steps are included and a 30 minute pause is included after water is added to the plate.\n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify the number of samples (1-96) for this protocol.\n* `P300 Mount`: Specify which mount (left or right) to host the P300 single-channel pipette.\n* `Aspiration height`: Specify the aspiration height for this run. A value of 1mm is the default from the bottom of the well. \n* `P1000 Mount`: Specify which mount (left or right) to host the P1000 single-channel pipette.\n\n\n\n---\n\n",
diff --git a/protoBuilds/00bbd7/README.json b/protoBuilds/00bbd7/README.json
index 828184a5e1..2c47929645 100644
--- a/protoBuilds/00bbd7/README.json
+++ b/protoBuilds/00bbd7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "00bbd7",
"labware": "\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nOpentrons 1000uL Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "Samples should be placed in tube racks by row (A1, A2, etc.) and by slot order 4, 5, 6, 7, 8, 9, 10.\n\n\n---\n\n",
"description": "This protocol preps a 96 well plate with Covid-19 saliva samples in up to 7 tube racks. Delays are inserted after aspiration to ensure the full volume of sample is achieved, as well as giving the user the ability to manipulate aspiration and dispense speeds.\n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify the number of samples for this run.\n* `Delay After Aspiration (seconds)`: Specify the number of seconds after aspirating Covid sample to allow time for the full volume to be achieved.\n* `Aspiration/Dispense Speed`: Specify the speed at which to aspirate/dispense Covid sample. A value of 1 is the default speed, a value of 0.5 is half of the default speed, etc.\n* `P1000 Single GEN2 Mount`: Specify whether the P1000 single channel pipette is on the left or right mount.\n\n\n---\n\n\n",
diff --git a/protoBuilds/00c517-pt2/README.json b/protoBuilds/00c517-pt2/README.json
index 5fb3a5722c..7fe4a25898 100644
--- a/protoBuilds/00c517-pt2/README.json
+++ b/protoBuilds/00c517-pt2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "00c517-pt2",
"labware": "\nGreiner_UV-star 96 Well Plate 392 \u00b5L #655801\n3DPrint 96 Tube Rack with Nippon 0.2 mL #U0014\nCostar 96 Well Plate 330 \u00b5L #Corning - 3779\nAgilent 96 Well Plate 500 \u00b5L #5042-1385\nAgilent 96 Well Plate 1400 \u00b5L #204355-100\nOpentrons 10 Tube Rack with Falcon 4x50 mL, 6x15 mL Conical\nOpentrons 96 Tip Rack 20 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol fills sds (slot 4) and sample (slot 2) according to a user specified csv.\n\n\n",
diff --git a/protoBuilds/00c517/README.json b/protoBuilds/00c517/README.json
index 929de616eb..2cd35bc122 100644
--- a/protoBuilds/00c517/README.json
+++ b/protoBuilds/00c517/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "00c517",
"labware": "\nGreiner_UV-star 96 Well Plate 392 \u00b5L\n3DPrint 96 Tube Rack with Nippon 0.2 mL #U0014\nCostar 96 Well Plate 330 \u00b5L #Corning - 3779\nAgilent 96 Well Plate 500 \u00b5L #5042-1385\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\nOpentrons 10 Tube Rack with Falcon 4x50 mL, 6x15 mL Conical\nOpentrons 96 Tip Rack 20 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\nAgilent 1 Well Reservoir 290 mL #201252-100\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n\n",
"description": "This protocol is part 1 of a series for the VIB UGENT protocol. [This protocol performs steps 1-19 of this detailed protocol description.](https://s3.amazonaws.com/pf-user-files-01/u-4256/uploads/2023-01-31/ok13urr/VIB-UGENT_Customized_protocol_S-trap_steps.xlsx). The protocol will automatically pause if it runs out of tips. \n\n\n",
diff --git a/protoBuilds/00f7b1_part10/README.json b/protoBuilds/00f7b1_part10/README.json
index e5e4bdcb99..62da847cac 100644
--- a/protoBuilds/00f7b1_part10/README.json
+++ b/protoBuilds/00f7b1_part10/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext\u00ae Ultra\u2122 II Directional RNA Library Prep"
@@ -10,7 +10,7 @@
"internal": "00f7b1_part10",
"labware": "\nArmadillo 96 Well Plate 200 \u00b5L PCR\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nNEST 1 Well Reservoir 195 mL #360103\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\u00ae Ultra\u2122 II Directional RNA Library Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol is part 10 of a 10 part series. Please look at deck map and liquid legend below. Note: the TE buffer can accommodate 3 columns of sample per column of wash. Each ethanol column can accommodate 4 columns of sample. If running more than one column of reagent, the volumes should be split equally among all columns.\n\n\n",
diff --git a/protoBuilds/00f7b1_part2/README.json b/protoBuilds/00f7b1_part2/README.json
index ed9effd82e..07fe74725c 100644
--- a/protoBuilds/00f7b1_part2/README.json
+++ b/protoBuilds/00f7b1_part2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext\u00ae Ultra\u2122 II Directional RNA Library Prep"
@@ -10,7 +10,7 @@
"internal": "00f7b1_part2",
"labware": "\nArmadillo 96 Well Plate 200 \u00b5L PCR\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nNEST 1 Well Reservoir 195 mL #360103\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\u00ae Ultra\u2122 II Directional RNA Library Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol is part 2 of a 10 part series. Please look at deck map and liquid legend below. Note: bead strip tubes can accommodate 2 columns of sample by volume (i.e. 6 columns of beads for a full plate of samples). The RNA binding buffer can accommodate 3 columns of sample per column of wash. There only needs to be one column of strand primer mix per run for all sample numbers. An overage of at least 10% should be used each run.\n\n\n",
diff --git a/protoBuilds/00f7b1_part3/README.json b/protoBuilds/00f7b1_part3/README.json
index 277b1af994..40032ba512 100644
--- a/protoBuilds/00f7b1_part3/README.json
+++ b/protoBuilds/00f7b1_part3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext\u00ae Ultra\u2122 II Directional RNA Library Prep"
@@ -10,7 +10,7 @@
"internal": "00f7b1_part3",
"labware": "\nThermo Fisher 96 Well Plate 200 \u00b5L #AB-3396\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nNEST 1 Well Reservoir 195 mL #360103\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\u00ae Ultra\u2122 II Directional RNA Library Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol is part 3 of a 10 part series. Please look at deck map and liquid legend below. This protocol adds the required reagents for the cDNA first strand synthesis\n\n\n",
diff --git a/protoBuilds/00f7b1_part4/README.json b/protoBuilds/00f7b1_part4/README.json
index d3ee1cb27a..1c363cd59b 100644
--- a/protoBuilds/00f7b1_part4/README.json
+++ b/protoBuilds/00f7b1_part4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext\u00ae Ultra\u2122 II Directional RNA Library Prep"
@@ -10,7 +10,7 @@
"internal": "00f7b1_part3",
"labware": "\nThermo Fisher 96 Well Plate 200 \u00b5L #AB-3396\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nNEST 1 Well Reservoir 195 mL #360103\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\u00ae Ultra\u2122 II Directional RNA Library Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol is part 4 of a 10 part series. Please look at deck map and liquid legend below. This protocol adds the required reagents for the cDNA second strand synthesis\n\n\n",
diff --git a/protoBuilds/00f7b1_part5/README.json b/protoBuilds/00f7b1_part5/README.json
index cb6ab958c0..beb74715ef 100644
--- a/protoBuilds/00f7b1_part5/README.json
+++ b/protoBuilds/00f7b1_part5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext\u00ae Ultra\u2122 II Directional RNA Library Prep"
@@ -10,7 +10,7 @@
"internal": "00f7b1_part5",
"labware": "\nArmadillo 96 Well Plate 200 \u00b5L PCR\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nNEST 1 Well Reservoir 195 mL #360103\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\u00ae Ultra\u2122 II Directional RNA Library Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol is part 5 of a 10 part series. Please look at deck map and liquid legend below. Note: the TE buffer can accommodate 3 columns of sample per column of wash. Each ethanol column can accommodate 4 columns of sample. If running more than one column of reagent, the volumes should be split equally among all columns.\n\n\n",
diff --git a/protoBuilds/00f7b1_part6/README.json b/protoBuilds/00f7b1_part6/README.json
index 0e8bbc17fc..fdf1b8ea73 100644
--- a/protoBuilds/00f7b1_part6/README.json
+++ b/protoBuilds/00f7b1_part6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext\u00ae Ultra\u2122 II Directional RNA Library Prep"
@@ -10,7 +10,7 @@
"internal": "00f7b1_part6",
"labware": "\nThermo Fisher 96 Well Plate 200 \u00b5L #AB-3396\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nNEST 1 Well Reservoir 195 mL #360103\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\u00ae Ultra\u2122 II Directional RNA Library Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs the end prep for cDNA in the NEBNext Ultra II Directional RNA Library Prep Kit.\nReaction buffer and enzyme mix are added to samples before mixing. The sample plate is then moved to\nan off-deck thermocycler.\n\n\n",
diff --git a/protoBuilds/00f7b1_part7/README.json b/protoBuilds/00f7b1_part7/README.json
index 5018564f9c..9fc21ea652 100644
--- a/protoBuilds/00f7b1_part7/README.json
+++ b/protoBuilds/00f7b1_part7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II Directional RNA Library Prep Kit for Illumina"
@@ -10,7 +10,7 @@
"internal": "00f7b1_part7",
"labware": "\nThermo Fisher 96 Well Plate 200 \u00b5L #AB-3396\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nNEST 1 Well Reservoir 195 mL #360103\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext Ultra II Directional RNA Library Prep Kit for Illumina\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol adds the necessary reagents for the adapter ligation in the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina.\n\n\n",
diff --git a/protoBuilds/00f7b1_part8/README.json b/protoBuilds/00f7b1_part8/README.json
index 43921e862c..dc493d8b78 100644
--- a/protoBuilds/00f7b1_part8/README.json
+++ b/protoBuilds/00f7b1_part8/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext\u00ae Ultra\u2122 II Directional RNA Library Prep"
@@ -10,7 +10,7 @@
"internal": "00f7b1_part10",
"labware": "\nArmadillo 96 Well Plate 200 \u00b5L PCR\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nNEST 1 Well Reservoir 195 mL #360103\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\u00ae Ultra\u2122 II Directional RNA Library Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol is part 8 of a 10 part series. Please look at deck map and liquid legend below. Note: the TE buffer can accommodate 3 columns of sample per column of wash. Each ethanol column can accommodate 4 columns of sample. If running more than one column of reagent, the volumes should be split equally among all columns.\n\n\n",
diff --git a/protoBuilds/00f7b1_part9/README.json b/protoBuilds/00f7b1_part9/README.json
index a4e2cfdd13..81bc03279a 100644
--- a/protoBuilds/00f7b1_part9/README.json
+++ b/protoBuilds/00f7b1_part9/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II Directional RNA Library Prep Kit for Illumina"
@@ -10,7 +10,7 @@
"internal": "00f7b1_part9",
"labware": "\nThermo Fisher 96 Well Plate 200 \u00b5L #AB-3396\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nNEST 1 Well Reservoir 195 mL #360103\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext Ultra II Directional RNA Library Prep Kit for Illumina\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol adds barcode primers and master mix to samples to prepare for PCR enrichment as outlined in the NEBNext Ultra II Directional RNA Library Prep Kit\n\n\n",
diff --git a/protoBuilds/010526/README.json b/protoBuilds/010526/README.json
index 6f42350525..fcbea05157 100644
--- a/protoBuilds/010526/README.json
+++ b/protoBuilds/010526/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Restriction Digest"
@@ -10,7 +10,7 @@
"internal": "010526",
"labware": "\n4-in-1 Tube Rack Set\nAluminum Block Set\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOpentrons Pipette Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Restriction Digest\n\n",
"deck-setup": "\n\n\n**Note: If you set `Samples Labware Type` to `1x 96 Well PCR Plate` only a 96 Well PCR plate should be placed in Slot 2.**\n\n",
"description": "This protocol performs restriction digests on a multitude of samples using various [restriction enzymes](https://www.neb.com/products/restriction-endonucleases/restriction-endonucleases?gclid=EAIaIQobChMInsuBgInC8QIVeXxvBB06IAdAEAAYASAAEgIWHvD_BwE) and then performs an incubation on the thermocycler module for heat inactivation. The sample data is inserted using a CSV file which contains the necessary data to load enzymes, transfer reagents and location details. An example of the CSV file can be downloaded [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/010526/Table1.csv).\n\nExplanation of complex parameters below:\n* `P20 Single Channel GEN2 Mount Position`: The position your pipette is mounted on the OT-2 (Left or Right).\n* `P300 Single Channel GEN2 Mount Position`: The position your pipette is mounted on the OT-2 (Left or Right).\n* `Samples Labware Type`: The type of labware used to hold your samples. You have the option to use 2x 24-well tuberacks (Slot 2 and 4) OR 1x 96-well NEST 100 uL PCR Plate (Slot 2).\n* `Input CSV File`: The input CSV file containing your sample data. You can download an example CSV file [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/010526/Table1.csv) or use the following formatting:\n```\nUID,ng/ul,800ng (ul),Water (ul),Enzyme,Location\n1301-1,328,2,33,EcoRI,A1\n1301-2,250,3,32,EcoRI,A2\n1301-3,180,4,31,EcoRI,A3\n1303-1,145,6,29,EcoRI,A4\n1303-2,169,5,30,EcoRI,A5\n1303-3,162,5,30,EcoRI,A6\n1362-1,150,5,30,BamHI; HindIII,A7\n1362-2,156,5,30,BamHI; HindIII,A8\n1362-3,231,3,32,BamHI; HindIII,A9\n1368-1,67,12,23,BamHI; HindIII,A10\n1368-2,183,4,31,BamHI; HindIII,A11\n1368-3,231,3,32,BamHI; HindIII,A12\n```\n* `Temperature Module Hold Temperature (C)`: The temperature the temperature module should reach and hold to keep reagents cool. \n* `Reaction Volume (uL)`: Total reaction volume for all samples in microliters (uL).\n* `Enzyme Volume (uL)`: The enzyme volume used for individual enzyme transfers to the sample wells in microliters (uL).\n* `Enzyme Transfer Aspirate Flow Rate (uL/s)`: The aspirate flow rate for the enzyme transfer step in microliters/second (uL/s).\n* `Enzyme Transfer Dispense Flow Rate (uL/s)`: The dispense flow rate for the enzyme transfer step in microliters/second (uL/s).\n* `Digest Duration (s)`: The total duration in seconds for the incubation at 37\u00b0C.\n* `Heat Kill Temperature (\u00b0C)`: The temperature for heat inactivation (default: 65\u00b0C).\n* `Heat Kill Duration (s)`: The duration in seconds for heat kill (default: 1200).\n\n---\n\n",
diff --git a/protoBuilds/010d6c/README.json b/protoBuilds/010d6c/README.json
index 6168dd181e..fba64df10a 100644
--- a/protoBuilds/010d6c/README.json
+++ b/protoBuilds/010d6c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "010d6c",
"labware": "\nCorning 12 Reservoir 2000 \u00b5L\nNunc 96 Well Plate 400 \u00b5L\nOpentrons 15 Tube Rack with eppendorf 5 mL\nPyramid 96 Well Plate 2000 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 24 Tube Rack with NEST 2 mL Snapcap\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a Ribrogreen assay - for detailed protocol steps, please see below. There is the option to perform duplicate/triplicate plating. The csv for sample input should include `source slot, source well, destination well` in the header.\n\n\n",
diff --git a/protoBuilds/0175a4/README.json b/protoBuilds/0175a4/README.json
index 95cd7c5fbb..01a34caf0b 100644
--- a/protoBuilds/0175a4/README.json
+++ b/protoBuilds/0175a4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Pooling"
@@ -8,7 +8,7 @@
"description": "This protocol uses two p1000-single channel (GEN2) pipettes to pool 500 ul aliquots of up to 96 patient samples to create up to 32 pools (when pool_size is specified in the parameters below as 3 samples combined) or up to 48 pools (when pool_size is specified in the parameters below as 2 samples combined) using an Opentrons OT-2 robot. This is a pythonized version (and modified to make use of two pipettes) of a Protocol Designer protocol. Sample racks are in deck slots 1, 3, 4, 6, 7, 9, and 10 and pool racks are in deck slots 2, 5, and 8 unless otherwise specified in the parameters below. 3:1 pools are kept apart on the deck to minimize risk of cross-contamination by leaving row \"B\" empty as much as possible. When more than 45 pools are created, column 5 of the last sample rack is used to locate the 46th, 47th and 48th pools.\n\n\n\nOpentrons P1000-single channel electronic pipettes (GEN2)\nOpentrons 1000ul Filter Tips\nOpentrons 15 Tube Rack with Falcon 15 mL Conical\n",
"internal": "0175a4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Pooling\n\n",
"description": "This protocol uses two p1000-single channel (GEN2) pipettes to pool 500 ul aliquots of up to 96 patient samples to create up to 32 pools (when pool_size is specified in the parameters below as 3 samples combined) or up to 48 pools (when pool_size is specified in the parameters below as 2 samples combined) using an Opentrons OT-2 robot. This is a pythonized version (and modified to make use of two pipettes) of a [Protocol Designer protocol](https://designer.opentrons.com/). Sample racks are in deck slots 1, 3, 4, 6, 7, 9, and 10 and pool racks are in deck slots 2, 5, and 8 unless otherwise specified in the parameters below. 3:1 pools are kept apart on the deck to minimize risk of cross-contamination by leaving row \"B\" empty as much as possible. When more than 45 pools are created, column 5 of the last sample rack is used to locate the 46th, 47th and 48th pools.\n\n---\n\n\n* [Opentrons P1000-single channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=5984549142557)\n* [Opentrons 1000ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips)\n* [Opentrons 15 Tube Rack with Falcon 15 mL Conical](https://labware.opentrons.com/opentrons_15_tuberack_falcon_15ml_conical?category=tubeRack)\n\n\n",
"internal": "0175a4\n",
diff --git a/protoBuilds/017860/README.json b/protoBuilds/017860/README.json
index 97616a5ef2..ae66d2bd02 100644
--- a/protoBuilds/017860/README.json
+++ b/protoBuilds/017860/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"CSV Transfer"
@@ -10,7 +10,7 @@
"internal": "017860",
"labware": "\nOpentrons 20 uL tipracks\n300 uL tipracks\nBrandTech 20 uL tipracks\nBrandTech 300 uL tipracks\nNunc 400 \u00b5L 96 well plate\nOpentrons tuberacks\nOpentrons tubes\nFalcon tubes\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* CSV Transfer\n\n",
"deck-setup": "\nSlots:\n1. Nunc 96 well plate 1\n2. Nunc 96 well plate 2 (optional)\n3. Nunc 96 well plate 3 (optional)\n4. Nunc 96 well plate 4 (optional)\n5. Nunc 96 well plate 5 (optional)\n6. Nunc 96 well plate 6 (optional)\n7. Tuberack with 15 mL tubes antibiotic tuberack (#2)\n8. Tuberack 4x50 mL/6x15 mL tubes - Media and antibiotics (#3)\n9. Tuberack with 15 mL tubes antibiotic tuberack (#1)\n10. 20 \u00b5l tiprack\n11. 300 \u00b5L tiprack\n\n\n",
"description": "This is an OT-2 compatible protocol that first transfers media and then up to 35 different antibiotics plus a barcoding dye to the wells of one to six 96 well plates based on an input CSV file. If there are more entries once the plate(s) have been filled the user is asked to replace the plates with fresh ones, as well as used tips, and the process continues until all rows of the input file have been processed.\n\nEach line in the csv corresponds to the transfers to a well on the plates. The plates are ordered sequentially from slot 1 to slot 6 meaning that the first 96 rows of the csv (excluding the header) correspond to the 96 wells on plate 1, and so on.\n\nExplanation of protocol parameters:\n* `input .csv file`: Here you should upload a .csv file formatted in the following way, being sure to include the header line: The first column is an identifier and is discarded by the protocol (but must be included in order for the file to be processed correctly), the 2nd column is the transfer volume of M9 media that is stored in 3 50 mL tubes on the tuberack on slot 8. The remaining columns up to n-1 are antibiotics stored on 15 mL tuberack 1 and 2 (see below). The nth column is the volume of barcoding dye.\n```\n,M9,antibiotic 1, antibiotic 2, ... , antibiotic n, barcoding dye\n99999,50,0,0,...,0,50\n100,90,10,0,...,0,0\n```\n* `20 uL pipette tips`: Brand of pipette tips, either Opentrons or BrandTech 20 uL tips\n* `300 uL pipette tips`: Brand of pipette tips, either Opentrons or BrandTech 300 uL tips\n* `Number of plates`: The number of Nunc 96 well plates on the deck per run, may range from one to six\n* `Height offset for dispensing antibiotics into target wells [mm]`: Offset for dispensing antibiotics into wells in mm (above the original level which was the calculated liquid level in the well minus 0.4 mm).\n\n---\n\n",
diff --git a/protoBuilds/019a7d/README.json b/protoBuilds/019a7d/README.json
index 988bbdca06..4d33a0b574 100644
--- a/protoBuilds/019a7d/README.json
+++ b/protoBuilds/019a7d/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Gibson Prep"
@@ -10,7 +10,7 @@
"internal": "019a7d",
"labware": "\n1x Azenta Life Sciences 12 Well Reservoir, 21 ml\n2x Azenta Life Sciences 96 Well Plate, 200 ul\nAluminum Block for Generic Screw Cap Tubes, 1.5 ml\n2x Opentrons 20ul Filter Tip Racks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Gibson Prep\n\n",
"deck-setup": "* Deck Layout, temp modules should initially be set to 4 C in both slot 1 and 4\n\n\n---\n\n",
"description": "This is part 1 of a 2 part protocol for prepping then loading nucleic acid onto agar plates. It requires both a multichannel and single channel 20ul pipette.\nPart two can be found [here](https://protocols.opentrons.com/protocol/019a7d_part_2)\n\n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify a number of samples. Can be any number from 1 to 96\n\n---\n\n",
diff --git a/protoBuilds/019a7d_part_2/README.json b/protoBuilds/019a7d_part_2/README.json
index d1df5c4139..a31965a9a6 100644
--- a/protoBuilds/019a7d_part_2/README.json
+++ b/protoBuilds/019a7d_part_2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Gibson Prep"
@@ -10,7 +10,7 @@
"internal": "019a7d",
"labware": "\n2x Azenta Life Sciences 96 Well Plate, 200 ul\n1x Greiner 96 Deep Well Plate, 2 ml\n5x Thermo Fisher Omni Nunc Tray\n1x Opentrons 20ul Filter Tip Racks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Gibson Prep\n\n",
"deck-setup": "* Deck Layout, temp modules should initially be set to 4 C in both slot 1 and 4\n\n\n",
"description": "This is part 2 of a 2 part protocol for prepping then loading nucleic acid onto agar plates. It requires both a multichannel and single channel 20ul pipette.\nPart one can be found [here](https://protocols.opentrons.com/protocol/019a7d)\n\nThe basic outline follows:\nThe assembly plate liquid is added to a Mix and Go plate. A heat shock can be performed, bringing the Mix and Go plate to 42 C for 40 seconds then back to the holding temperature of 4 C. After 30 minutes, agar plates are loaded with the resultant mixture as specified in the parameters. If desired, this Mix and Go plate's liquid can be added to a deep well plate and mixed with liquid culture.\n\nTips are reused for the same sample repeatedly, i.e. sample A1 on the Mix and Go will use A1 in the tip rack.\n\n\nExplanation of complex parameters below:\n* `Number of Samples`: Number of samples in Mix and Go plate. Can be any number between 1 and 96\n* `24 or 96 Distributions per Plate`: Choose to use up to four agar plates with 24 samples each and/or one agar plate with up to 96 samples. Distribution of samples follows a common layout for either 24 well plates or 96 well plates on the agar\n* `Heat Shock`: Choose yes or no to determine whether a heat shock will occur during the protocol. This heat shock takes place after adding to the Mix and Go plate. It raises the temperature to 42 C for 40 seconds before returning to 4 C\n* `Volume to Distribute on Agar`: Choose a liquid volume to add to the agar plate(s). This can theoretically be many volumes above 1 ul but caution should be exercised so as not to co-mingle samples on the agar by overloading\n* `Add to Deep Well Plate`: Choose yes or no to add to deep well plate. Yes results in a specified amount of liquid being added to the deep well plate 1 cm above the well bottom and mixed\n* `Volume to Add to Deep Well Plate`: Specifies volume to add to the deep well plate if yes is chosen under \"Add to Deep Well Plate\"\n\n---\n\n",
diff --git a/protoBuilds/01a6b9/README.json b/protoBuilds/01a6b9/README.json
index fb3c8262a5..649be7bd7b 100644
--- a/protoBuilds/01a6b9/README.json
+++ b/protoBuilds/01a6b9/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sterile Workflows": [
"Cell Culture"
@@ -9,7 +9,7 @@
"internal": "01a6b9",
"labware": "\nPhenoPlate 96 Well Plate 425 \u00b5L #6055302\nPhenoPlate 384 Well Plate 145 \u00b5L #6057302\nOpentrons 96 Filter Tip Rack 1000 \u00b5L\nNEST 1 Well Reservoir 195 mL #360103\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sterile Workflows\n\t* Cell Culture\n\n\n",
"description": "This protocol performs a custom media removal and re-addition for cell culture plates, in 24-, 96-, or 384-well format. The user can select which pipette they would like to use. Pipette compatibility with labware selection is automatically determined.\n\n\n",
"internal": "01a6b9\n",
diff --git a/protoBuilds/01b4a2/README.json b/protoBuilds/01b4a2/README.json
index e8c1a9cefe..f4ef95281c 100644
--- a/protoBuilds/01b4a2/README.json
+++ b/protoBuilds/01b4a2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "01b4a2",
"labware": "\nCRO Vial Holder\nMicronic Tube Rack\nOpentrons 6 Tube Rack 50mL\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 96 Tip Rack 1000 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol preps up to two 96 well plates with reagent and sample. The protocol takes a csv as input to determine which wells in the middle rack (see below) will be populated. The csv must include the header, with the following order in the header: `vial_id, parent_well, dmso_volume, daughter_column`. The protocol pauses midway through after the samples are placed and mixed in the middle plates for the user to inspect before resuming.\n\n\n",
diff --git a/protoBuilds/022548-2/README.json b/protoBuilds/022548-2/README.json
index 2bdb728e4e..d980f2e54d 100644
--- a/protoBuilds/022548-2/README.json
+++ b/protoBuilds/022548-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"DNA Extraction"
@@ -10,7 +10,7 @@
"internal": "022548-2",
"labware": "\nNEST 12-Well Reservoirs, 15 mL\nPRL tuberack for 32 15 mL tubes\nThermo Fisher Kingfisher 96 deepwell plate 2 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* DNA Extraction\n\n",
"deck-setup": "* Load 300 ul tip racks in order 6-3. Load tube racks in order from top down. Load samples in order down the column in each tube rack. Tube racks should be filled fully before proceeding to next tube rack. For example, 33 samples total would mean 32 in the top most tube rack and 1 sample in the middle tube rack.\n\n\n\n\n",
"description": "This protocol transfers samples from tubes in up to 3 PRL tuberacks to a destination plate followed by resuspending a bead/binding buffer mastermix before transfering it to the samples on the plate.\n\nLinks:\n* [Part 1: Master Mix Assembly](./022548)\n* [Part 2: Master Mix Distribution and Sample Transfer](./022548-2)\n\n\nExplanation of parameters below:\n* `Number of samples in tuberack 1 slot 10-11`: How many samples to transfer from rack 1: 1 to 32\n* `Number of samples in tuberack 2 slot 7-8`: 0 to 32\n* `Number of samples in tuberack 3 slot 4-5`: 0 to 32\n* `Master mix wells location`: Informs the protocol which wells of the reservoir contain mastermix, starting from A1 on the very left. For example 1-4 to specify wells A1 to A4. This parameter may also be a single number instead of a range\n* `Mastermix max volume per well (mL)`: Informs the protocol what the maximal volume of mastermix is in each reservoir well, default is 9.54 mL\n* `Mastermix mixing rate multiplier`: Controls the flow rate of mixing, 1.0 means standard flow rate, less is slower and more is faster.\n* `mastermix aspiration flow rate multiplier`: Controls the rate of aspiration of mastermix from the reservoir wells\n* `mastermix dispension flow rate multiplier`: Controls the rate of dispension of mastermix into wells on the target plate\n* `Mount for 300 uL single channel pipette`: Left or right\n* `Mount for 300 uL multi channel pipette`: Left or right\n* `Pause after mixing the mastermix for vortex/resuspension?`: Optional pause after (re-)mixing the mastermix where the reservoir may be taken out for manual resuspension such as vortexing, nutation or shaking if resuspension by pipetting is deemed insufficient.\n\n---\n\n",
diff --git a/protoBuilds/022548/README.json b/protoBuilds/022548/README.json
index 41759ca34b..20d4ae8520 100644
--- a/protoBuilds/022548/README.json
+++ b/protoBuilds/022548/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"DNA Extraction"
@@ -10,7 +10,7 @@
"internal": "022548-1",
"labware": "\nNEST 12-Well Reservoirs, 15 mL\nOpentrons tuberacks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* DNA Extraction\n\n",
"deck-setup": "\n\n",
"description": "This protocol create a mastermix of binding buffer and bead mix. For large volumes the deck should use a NEST 12 well reservoir for the bead mix, for smaller number of samples a tuberack may be used as source for bead mix.\n\nDesignate the wells used for binding buffer and bead mix and the mastermix is created starting in the well immediately after, and uses as many wells as required for creating mastermix for the indicated number of samples. The protocol creates a dead volume in addition to the 'active' volume of mastermix (1 mL for reservoirs) - this is to avoid the pipette aspirating air during mastermix dispension in part 2 of the protocol.\n\nThe protocol will create a maximum volume of 9.54 mL per well, of which 1/3rd is dead volume.\n\nThe protocol is written to account for the changing liquid level of bead source tubes so that the pipette will not plunge too far into tube solutions.\n\nLinks:\n* [Part 1: Master Mix Assembly](./022548)\n* [Part 2: Master Mix Distribution and Sample Transfer](./022548-2)\n\nExplanation of parameters below:\n* `Create mastermix for how many numbers of samples`: How many samples to create mastermix for.\n* `Binding buffer wells`: Designates which wells contain binding buffer, you may specify a range such as 1-4, or a single number if it is a single well. These number corresponds to the wells of the reservoir starting at the leftmost well.\n* `Bead mix well(s)`: Which well(s) contain bead mix. Just like the previous parameter it can be a number or a range, e.g. 5, or 5-6\n* `Volume of binding buffer per source well (mL)`: How many milliliters of binding buffer is contained in each source reservoir well.\n* `Volume of bead mix per source well (mL)`: How many milliliters of bead mix is contained in the source tube/reservoir well(s)\n* `P300 single channel pipette mount`: Left or right mount\n* `P300 multi channel pipette mount`: Left or right mount\n* `Mastermix tuberack (Optional, only for small volumes of mastermix)`: Specify what kind of tuberack contains your bead mix tube. If this parameter is set to none the bead source will be a NEST 12 well reservoir instead (default).\n* `Mastermix mixing rate multiplier`: The multiplier scales the flow rate of mixing aspirations and dispenses, e.g. 1.0 is the standard mixing rate, 0.5 would be half etc.\n* `Binding buffer aspiration flow rate multiplier`: Controls the flow rate of aspiration when aspirating binding buffer from a source well\n* `Binding buffer dispensing flow rate multiplier`: Controls the flow rate of dispension when dispensing binding buffer in a mastermix target well\n* `Bead solution aspiration flow rate multiplier`: Controls the flow rate of aspiration when aspirating from a bread mix source well\n* `Bead solution dispensing flow rate multiplier`: Controls the flow rate of dispension when dispensing bead mix in a mastermix target well\n* `How many times do you want to mix the mastermix?`: Indicates the number of times you want to mix the mastermix solution after the binding buffer and bead mix have been added.\n* `Mastermix mixing rate multiplier`: flow-rate modifier affecting the rate of aspiration and dispensing for mixing the mastermix after the addition of both components.\n* `Offsets from the edges of the reservoir wells for bead mix dispenses (mm)`: Offset defining how close the pipette may get to the edge of the reservoir well when dispensing bead mix into the mastermix wells (which is dispensed in three different locations inside of the well)\n* `Verbose protocol output?`: Indicates to the protocol whether it should output extra information about what it is doing.\n\n---\n\n",
diff --git a/protoBuilds/022a99/README.json b/protoBuilds/022a99/README.json
index fab3e69c4d..1b69a62082 100644
--- a/protoBuilds/022a99/README.json
+++ b/protoBuilds/022a99/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Aliquoting": [],
"Sample Prep": []
@@ -8,7 +8,7 @@
"internal": "022a99",
"labware": "\nAkura 384 Well Plate 50 \u00b5L #CS-09-005-02\nCorning 384 Well Plate 90 \u00b5L #3830BC\nOpentrons 96 Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "\n[Opentrons](https://opentrons.com/)\n\n",
+ "author": "\n[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "\n- Sample Prep\n - Aliquoting\n\n",
"description": "\nThis protocol transfers spheroids from a standard U-bottom 384-well plate to a specialized Akura 384-well plate. Samples are transferred to their corresponding wells. Tips are automatically changed after 16 columns to reflect new cell type. Flow rates and well positions are optimized to accurately transfer spheroid avoiding shearing.\n\n",
"internal": "\n022a99\n",
diff --git a/protoBuilds/029f5b/README.json b/protoBuilds/029f5b/README.json
index 4ca0e945b4..6dc011b894 100644
--- a/protoBuilds/029f5b/README.json
+++ b/protoBuilds/029f5b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"SAMPLE PREP": [
"Custom Sample Transfer"
@@ -10,7 +10,7 @@
"internal": "029f5b",
"labware": "\nAxygen 96well minitube system corning 96 wellplate 1320\u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\n[Opentrons Nest 1 well Reservoir 290 mL]\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* SAMPLE PREP\n\t* Custom Sample Transfer\n\n\n",
"deck-setup": "\n\n\n",
"description": "With this Protocol you can add lysis buffer, chloroform, and wash with Isopropanol to samples. This protocol is set up to do 384 samples across 4 different plates. \n\n\n",
diff --git a/protoBuilds/02b308/README.json b/protoBuilds/02b308/README.json
index 9c09369ccf..662ea44b4f 100644
--- a/protoBuilds/02b308/README.json
+++ b/protoBuilds/02b308/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"qPCR Prep"
@@ -10,7 +10,7 @@
"internal": "02b308",
"labware": "\nApplied Biosystems 384 Well Plate 40 \u00b5L #4309849\nEppendorf 96 Deepwell 1000uL #951032603\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* qPCR Prep\n\n\n",
"deck-setup": "\nFor transfer scheme, please view [this document](https://s3.amazonaws.com/pf-user-files-01/u-4256/uploads/2023-01-25/1p03tt5/Scheme.pdf).\n\n\n",
"description": "This protocol performs a custom PCR prep for up to 6 384-well qPCR plates.\n\n\n",
diff --git a/protoBuilds/02pnzp/README.json b/protoBuilds/02pnzp/README.json
index e6e5a016fd..6e1b48d9de 100644
--- a/protoBuilds/02pnzp/README.json
+++ b/protoBuilds/02pnzp/README.json
@@ -1,5 +1,5 @@
{
- "author": "Michael Fichtner",
+ "author": "Michael Fichtner\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"DNA/RNA": [
"DNA/RNA"
@@ -10,7 +10,7 @@
"internal": "02pnzp",
"labware": "\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 24 Well Aluminum Block with NEST 1.5 mL Screwcap\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\nOpentrons 96 Well Aluminum Block with NEST Well Plate 100 \u00b5L\nOpentrons 10 Tube Rack with Falcon 4x50 mL, 6x15 mL Conical\n",
"markdown": {
- "author": "Michael Fichtner\n\n\n",
+ "author": "Michael Fichtner\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* DNA/RNA\n\t* DNA/RNA\n\n\n",
"deck-setup": "[deck](https://drive.google.com/open?id=1E1W92HHveoQJEblfzXOynKiVUMofAmA0)\n\n\n",
"description": "This protocol will prepare and run a standard Endpoint-PCR with the opentrons Cycler Module (only tested with Gen1). It aims to provide sufficient flexibility in terms of sample and replicate numbers to avoid the need of multiple similar protocols.\n\nKey benefits of the protocol are:\n* The master mixes will be prepared in 50 ml falcons and all reagents and samples can be provided in 1.5 ml tubes.\n* Thus, no manual pipetting steps are involved.\n* Up to 12 different template samples can be used with up to 3 different master mixes.\n* Only the p300 and p20 pipettes are needed.\n* Needed volumes of the different reagents are calculated and shown in run tab.\n\nNotes:\n* A no-template control (NTC) will be automatically added for each master mix.\n* The first well of every master mix will always be in row A. If the sample number + NTC is not a multiple of 8, the remaining wells of the last column will be left empty to have an easier plate layout.\n* (e.g. You have 2 master mixes, 4 samples and 1 replicate each. The last well of master mix 1 will be E1 (NTC control). Master mix 2 will now be added to the wells A2 - E2, skipping the remaining wells of column 1. This is meant to make it less confusing down the road.)\n\n\n",
@@ -21,7 +21,8 @@
"pipettes": "* [Opentrons P20 Single Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/single-channel-electronic-pipette-p20/)\n* [Opentrons P300 Single Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/single-channel-electronic-pipette-p20/)\n\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit \"Run\".\n\n\n",
"protocol-steps": "1. Open the protocol and define the number of samples, replicates and master mixes. (Starting at line 140 of the script)\n2. Open the protocol in the opentrons app. The exact volumes of the needed reagents are shown in the run tab.\n3. (optional) The last required tip position of both boxes is shown at the end of the run tab.\n4. Load the robot according to the deck layout. If less than 12 samples are run, the respective slots can be left empty. Same is true for the primer tubes and empty master mix tubes, if less than 3 master mixes are used.\n5. Start run.\n6. The robot will start to cool the cycler block and the temperature module to 4\u00b0C. Thus, it might take a while to start pipetting.\n7. The robot will then prepare the master mixes and distribute it to the PCR plate.\n8. Samples will be added to the plate.\n9. All wells will be mixed.\n10. The robot will run the pre-defined PCR.\n11. Once finished, the cycler block will be cooled down to 4\u00b0C\n12. (optional) If selected in the protocol, the robot will sort the remaining tips in the tip boxes to the front.\n13. The PCR cycler will remain closed until opened manually.\n14. Neither the temperature module nor the cycler module will be deactived to ensure all temperature sensitive reagents remain at 4\u00b0C until removed by the operator.\n\n\n\n",
- "reagent-setup": "[reagents](https://drive.google.com/open?id=1vt3BBPGD7XKCplorlWHnOjGihgPvOTiU, https://drive.google.com/open?id=1Fx_SsQSPlXiwKEYsWtjFRVgN-vlHM7ci, https://drive.google.com/open?id=1ERF1CRX-2yoKkuyTm3ytyLM361joa1q7)\n\n\n"
+ "reagent-setup": "[reagents](https://drive.google.com/open?id=1vt3BBPGD7XKCplorlWHnOjGihgPvOTiU, https://drive.google.com/open?id=1Fx_SsQSPlXiwKEYsWtjFRVgN-vlHM7ci, https://drive.google.com/open?id=1ERF1CRX-2yoKkuyTm3ytyLM361joa1q7)\n\n\n",
+ "title": "Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library."
},
"modules": [
"Opentrons Thermocycler Module",
@@ -31,5 +32,6 @@
"pipettes": "\nOpentrons P20 Single Channel Electronic Pipette (GEN2)\nOpentrons P300 Single Channel Electronic Pipette (GEN2)\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit \"Run\".\n",
"protocol-steps": "\nOpen the protocol and define the number of samples, replicates and master mixes. (Starting at line 140 of the script)\nOpen the protocol in the opentrons app. The exact volumes of the needed reagents are shown in the run tab.\n(optional) The last required tip position of both boxes is shown at the end of the run tab.\nLoad the robot according to the deck layout. If less than 12 samples are run, the respective slots can be left empty. Same is true for the primer tubes and empty master mix tubes, if less than 3 master mixes are used.\nStart run.\nThe robot will start to cool the cycler block and the temperature module to 4\u00b0C. Thus, it might take a while to start pipetting.\nThe robot will then prepare the master mixes and distribute it to the PCR plate.\nSamples will be added to the plate.\nAll wells will be mixed.\nThe robot will run the pre-defined PCR.\nOnce finished, the cycler block will be cooled down to 4\u00b0C\n(optional) If selected in the protocol, the robot will sort the remaining tips in the tip boxes to the front.\nThe PCR cycler will remain closed until opened manually.\nNeither the temperature module nor the cycler module will be deactived to ensure all temperature sensitive reagents remain at 4\u00b0C until removed by the operator.\n",
- "reagent-setup": "reagents"
+ "reagent-setup": "reagents",
+ "title": "Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library."
}
\ No newline at end of file
diff --git a/protoBuilds/030dd8-amplify/README.json b/protoBuilds/030dd8-amplify/README.json
index 9f6f1969fa..567c75235c 100644
--- a/protoBuilds/030dd8-amplify/README.json
+++ b/protoBuilds/030dd8-amplify/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Broad Category": [
"Specific Category"
@@ -10,7 +10,7 @@
"internal": "030dd8-amplify",
"labware": "\nQuantgene 96 Aluminum Block 200 \u00b5L\nAgilent 96 Well Plate 200 \u00b5L #5042-8502\nAgilent With Nonskirted 96 Well Plate 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Broad Category\n\t* Specific Category\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol does stuff!\n\n\n",
diff --git a/protoBuilds/030dd8-amplify2/README.json b/protoBuilds/030dd8-amplify2/README.json
index 8c972989dc..f4729ba3fb 100644
--- a/protoBuilds/030dd8-amplify2/README.json
+++ b/protoBuilds/030dd8-amplify2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Broad Category": [
"Specific Category"
@@ -10,7 +10,7 @@
"internal": "030dd8-amplify2",
"labware": "\nQuantgene 96 Aluminum Block 200 \u00b5L\nAgilent 96 Well Plate 200 \u00b5L #5042-8502\nAgilent With Nonskirted 96 Well Plate 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Broad Category\n\t* Specific Category\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol does stuff!\n\n\n",
diff --git a/protoBuilds/030dd8-anneal/README.json b/protoBuilds/030dd8-anneal/README.json
index 27dcd3c301..2d1cacf22f 100644
--- a/protoBuilds/030dd8-anneal/README.json
+++ b/protoBuilds/030dd8-anneal/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Broad Category": [
"Specific Category"
@@ -10,7 +10,7 @@
"internal": "030dd8-ngs-anneal",
"labware": "\nQuantgene 96 Aluminum Block 200 \u00b5L\nAgilent 96 Well Plate 200 \u00b5L #5042-8502\nAgilent With Nonskirted 96 Well Plate 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Broad Category\n\t* Specific Category\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol does stuff!\n\n\n",
diff --git a/protoBuilds/030dd8-cleanup/README.json b/protoBuilds/030dd8-cleanup/README.json
index 40eae6bb72..42e7bdb4ff 100644
--- a/protoBuilds/030dd8-cleanup/README.json
+++ b/protoBuilds/030dd8-cleanup/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Broad Category": [
"Specific Category"
@@ -10,7 +10,7 @@
"internal": "030dd8-cleanup",
"labware": "\nQuantgene 96 Aluminum Block 200 \u00b5L\nAgilent 96 Well Plate 200 \u00b5L #5042-8502\nAgilent With Nonskirted 96 Well Plate 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Broad Category\n\t* Specific Category\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol does stuff!\n\n\n",
diff --git a/protoBuilds/030dd8-synthesize/README.json b/protoBuilds/030dd8-synthesize/README.json
index 75a9468c71..6a92d12db2 100644
--- a/protoBuilds/030dd8-synthesize/README.json
+++ b/protoBuilds/030dd8-synthesize/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Broad Category": [
"Specific Category"
@@ -10,7 +10,7 @@
"internal": "030dd8-synthesize",
"labware": "\nQuantgene 96 Aluminum Block 200 \u00b5L\nAgilent 96 Well Plate 200 \u00b5L #5042-8502\nAgilent With Nonskirted 96 Well Plate 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Broad Category\n\t* Specific Category\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol does stuff!\n\n\n",
diff --git a/protoBuilds/030dd8-tagment/README.json b/protoBuilds/030dd8-tagment/README.json
index 92f8eb9b87..7bf72584d1 100644
--- a/protoBuilds/030dd8-tagment/README.json
+++ b/protoBuilds/030dd8-tagment/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Broad Category": [
"Specific Category"
@@ -10,7 +10,7 @@
"internal": "030dd8-tagment",
"labware": "\nQuantgene 96 Aluminum Block 200 \u00b5L\nAgilent 96 Well Plate 200 \u00b5L #5042-8502\nAgilent With Nonskirted 96 Well Plate 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Broad Category\n\t* Specific Category\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol does stuff!\n\n\n",
diff --git a/protoBuilds/030dd8/README.json b/protoBuilds/030dd8/README.json
index e50e9ca956..bb9ad6fc48 100644
--- a/protoBuilds/030dd8/README.json
+++ b/protoBuilds/030dd8/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Broad Category": [
"Specific Category"
@@ -10,7 +10,7 @@
"internal": "030dd8",
"labware": "\nNEST 96 Deepwell Plate 2mL #503001\nNEST 1 Well Reservoir 195 mL #360103\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Broad Category\n\t* Specific Category\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol does stuff!\n\n\n",
diff --git a/protoBuilds/036016/README.json b/protoBuilds/036016/README.json
index 8cc7522a80..b0da2463df 100644
--- a/protoBuilds/036016/README.json
+++ b/protoBuilds/036016/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "036016",
"labware": "\nApplied Biosystems (ThermoFisher/Life) 4309849 With Barcode 384 Well Plate 30 \u00b5L \nOpentrons 96 Tip Rack 20 \u00b5L\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nCorning 96 Well Plate 360 \u00b5L Flat #3650\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol preps (4) 384 well plates with a source plate on the Thermocycler module. For a detailed description of protocol steps, please see below. Gantry speed slowed to retard dripping.\n\n\n",
diff --git a/protoBuilds/039ecb/README.json b/protoBuilds/039ecb/README.json
index ce467aad75..3024e635e7 100644
--- a/protoBuilds/039ecb/README.json
+++ b/protoBuilds/039ecb/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext\u00ae Ultra\u2122 II FS DNA Library Prep Kit for Illumina"
@@ -10,7 +10,7 @@
"internal": "039ecb",
"labware": "\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nOpentrons 24 Well Aluminum Block with NEST 1.5 mL Snapcap\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\u00ae Ultra\u2122 II FS DNA Library Prep Kit for Illumina\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol forms an automated NGS library prep for the [NEBNext\u00ae Ultra\u2122 II FS DNA Library Prep Kit for Illumina](https://www.neb.com/-/media/nebus/files/manuals/manuale6177-e7805.pdf?rev=60c9f0ba3c944a708539331d8a71dfbc&hash=B86DFE0923F3F6A7420D1C5A48BB62C5).\n\n\n",
diff --git a/protoBuilds/03f9cd/README.json b/protoBuilds/03f9cd/README.json
index b5b02ac069..3c61dc2512 100644
--- a/protoBuilds/03f9cd/README.json
+++ b/protoBuilds/03f9cd/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "03f9cd",
"labware": "\nV&P Scientific 48 Well Plate 2000 \u00b5L #VP 416-ALB-48\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 96 Tip Rack 1000 \u00b5L\nOpentrons 6 Tube Rack with Falcon 50 mL Conical\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs custom emulsions via two uploaded csv files - one for the aqueous solutions (slot 10) and another for the oil solutions (slot 11). Touch tips and delays are instilled after aspirations, and blowouts are instilled after dispensing from the top of the well. One liquid solution is dispensed at a time into as many rows that are provided in the csv. If a value of 0 is passed in the csv, the pipette will skip that well. For volumes less than 300ul, the P300 single channel pipette is used, otherwise for volumes 300-1000ul, the P1000 pipette is used. All aspirations and dispenses are in order of row for all racks and plates.\n\n\n\n* The csv should be formatted like so. Note that it should include the header, as well as a second row describing the initial volume in each tube in milliliters to ensure liquid height tracking, and avoid submerging the pipette plunger in solution. The aqueous and oil csv files do not necessarily need to have the same number of columns (liquid number), but they should have the same number of rows (up to 144 rows).\n\n",
diff --git a/protoBuilds/041adf/README.json b/protoBuilds/041adf/README.json
index cabf94cd4a..39100c89d4 100644
--- a/protoBuilds/041adf/README.json
+++ b/protoBuilds/041adf/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Screening"
@@ -10,7 +10,7 @@
"internal": "041adf",
"labware": "\nROAR_printed_ 48 Well Plate 1500 \u00b5L\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons 96 Tip Rack 20 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Screening\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs reaction library screening on up to 60 samples in the center of a 96-well plate on the Opentrons Heater-Shaker Module.\n\n\n",
diff --git a/protoBuilds/04318b/README.json b/protoBuilds/04318b/README.json
index 7548272425..42ee201edc 100644
--- a/protoBuilds/04318b/README.json
+++ b/protoBuilds/04318b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Serial Dilution"
@@ -10,7 +10,7 @@
"internal": "04318b",
"labware": "\nNEST 12 Well Reservoir 15 mL #360102\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Serial Dilution\n\n\n",
"deck-setup": "\n\n\n\n",
"description": "This protocol serially diluted compound stock with DMSO. There is an optional predilution step in this protocol. For detailed protocol steps, please see below. Slots 6, 7, 8, 9 are nest deep well plates, and slots 2, 4, 5, 10, 11 are NEST PCR plates.\n\n\n",
diff --git a/protoBuilds/0451f3/README.json b/protoBuilds/0451f3/README.json
index 5dec88d085..a48cb6557b 100644
--- a/protoBuilds/0451f3/README.json
+++ b/protoBuilds/0451f3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Purification & Extraction": [
"Zymo Zyppy\u2122-96 Plasmid MagBead Miniprep"
@@ -10,7 +10,7 @@
"internal": "0451f3",
"labware": "\nNEST 96 Deepwell Plate 2mL #503001\nBio-Rad 96 Well Plate 200 \u00b5L PCR #hsp9601\nNEST 1 Well Reservoir 195 mL #360103\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Purification & Extraction\n\t* Zymo Zyppy\u2122-96 Plasmid MagBead Miniprep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a semi-automated miniprep following the [Zymo Zyppy\u2122-96 Plasmid MagBead Miniprep](https://www.zymoresearch.com/products/zyppy-96-plasmid-magbead-kit#:~:text=The%20Zyppy%2D96%20Plasmid%20MagBead,coli.) protocol for up to 96 samples. The user is prompted to manually refill tipracks as necessary if running a high-throughput experiment (> 80 samples).\n\n",
diff --git a/protoBuilds/04552f/README.json b/protoBuilds/04552f/README.json
index 7ff3ae694d..185409b8b9 100644
--- a/protoBuilds/04552f/README.json
+++ b/protoBuilds/04552f/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -10,7 +10,7 @@
"internal": "04552f",
"labware": "\nCorning 384 Well Plate 112 \u00b5L Flat #3640\nOpentrons 96 Tip Rack 20 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 6 Tube Rack with Falcon 50 mL Conical\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a custom cherrypicking protocol for seeding up to 2 source plates to up to 2 destination plates. The input .csv file should be formatted as follows:\n\n```\nOriginal plate position in OT-2 workspace,Original position,Picking volume (ul),Destination plate position,Destination position,Seeding volume,Trashing volume,Cell medium original plate,Cell medium destination plate\n1.1,B1,20,2.1,B1,20,0,40,40\n1.1,B5,30,2.1,B2,20,10,30,40\n1.1,C16,50,2.2,B1,30,20,10,30\n1.2,B6,10,2.2,B2,10,0,50,50\n1.2,D4,20,2.2,B3,10,10,40,50\n1.2,E5,30,2.2,B4,20,10,30,40\n...\n```\n\nYou can also download a template file from [this link](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/04552f/ex.csv).\n\nTo reflect the changes of your input .csv file, perform the following: paste the content of your .csv file as a multi-line string on line 17 of your download protocol, setting the variable `INPUT_CSV`.\n\n",
diff --git a/protoBuilds/0479ad-cp/README.json b/protoBuilds/0479ad-cp/README.json
index 063a456df9..6d7f003cb4 100644
--- a/protoBuilds/0479ad-cp/README.json
+++ b/protoBuilds/0479ad-cp/README.json
@@ -1,5 +1,5 @@
{
- "author": "Breakthrough Genomics",
+ "author": "Breakthrough Genomics\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "0479ad-cp",
"labware": "\nthermofisher 96 well pcr plate semi-skirted 200 uL + plastic adapter\nthermofisher 96-well PCR Plate semi-skirted 200 uL + aluminum block\nUSA Scientific 12 Well Reservoir 22 mL #1061-8150\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Breakthrough Genomics](https://opentrons.com/)\n\n\n",
+ "author": "[Breakthrough Genomics](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs normalization on up to 96 samples in PCR plates using Opentrons single-channel pipettes. The `Volumes CSV` download parameter should be formatted as the following:\n\n```\nsample,vol_dna,vol_water\nA1,23.4,11.6\nB1,4.9,21.1\nC1,21.6,4.4\nD1,15.1,10.9\nE1,19.1,6.9\nF1,18.6,7.4\nG1,10.7,15.3\nH1,21.9,4.1\nA2,19.1,6.9\nB2,0.4,25.6\nC2,24.0,2.0\nD2,6.1,19.9\nE2,10.7,15.3\nF2,24.2,1.8\nG2,6.1,19.9\nH2,3.0,23.0\n...\n```\n\n\n",
diff --git a/protoBuilds/0479ad-part-2/README.json b/protoBuilds/0479ad-part-2/README.json
index 604ea9156b..d741565f85 100644
--- a/protoBuilds/0479ad-part-2/README.json
+++ b/protoBuilds/0479ad-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II RNA Library Prep Kit for Illumina"
@@ -10,7 +10,7 @@
"internal": "0479ad-part-2",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons Temperature Module] (https://shop.opentrons.com/modules/)\n[Opentrons Magnetic Module] (https://shop.opentrons.com/modules/)\nThermoFisher 96 Well PCR Plate 300 uL Semi-Skirted\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext Ultra II RNA Library Prep Kit for Illumina\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module with 96-Well Aluminum Block holding Reagent Plate (nest_96_wellplate_100ul_pcr_full_skirt) \n\n\n\n**Slot 4**: Opentrons Temperature Module with 96-Well Aluminum Block holding Input Plate (ThermoFisher 96 Well PCR Plate 300 uL Semi-Skirted) \n\n\n\n**Slot 9**: Opentrons Magnetic Module (empty) \n**Slot 7**: Opentrons 20 uL filter tips \n**Slot 6**: Opentrons 200 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses multi-channel P20 and P300 pipettes to perform library prep steps on 8-48 input total RNA samples according to the attached NEB user guide. This is part-2 of a six part process (first strand and second strand cDNA synthesis). Reagents are combined into two mixtures (mix1 is composed of water + first strand enzyme mix, mix2 is composed of second strand reaction buffer + second strand enzyme mix + water) to reduce the number of transfer steps. A user-specified parameter is available to specify the number of samples in the current run.\n\n\n---\n\n\n",
diff --git a/protoBuilds/0479ad-part-3/README.json b/protoBuilds/0479ad-part-3/README.json
index c1e5aa416b..8c4e20f9ac 100644
--- a/protoBuilds/0479ad-part-3/README.json
+++ b/protoBuilds/0479ad-part-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II RNA Library Prep Kit for Illumina"
@@ -10,7 +10,7 @@
"internal": "0479ad-part-3",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons Temperature Module] (https://shop.opentrons.com/modules/)\n[Opentrons Magnetic Module] (https://shop.opentrons.com/modules/)\n[USA Scientific 12 Well Reservoir 22 mL] (https://shop.opentrons.com)\nThermoFisher 96 Well PCR Plate 300 uL Semi-Skirted\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext Ultra II RNA Library Prep Kit for Illumina\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module with 96-Well Aluminum Block holding Output Plate (ThermoFisher 96 Well PCR Plate 300 uL Semi-Skirted) \n**Slot 2**: Reagent Reservoir for NEBNext Sample Purification Beads, 0.1X TE, 80 Percent EtOH, Liquid Waste (usascientific_12_reservoir_22ml) \n\n\n\n**Slot 4**: Opentrons Temperature Module empty \n**Slot 9**: Opentrons Magnetic Module (ThermoFisher 96 Well PCR Plate 300 uL Semi-Skirted on Custom Plastic Adapter containing 80 uL Double-Stranded cDNA Samples) \n\n\n\n**Slots 6,7,8,10,11**: Opentrons 200 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses multi-channel P20 and P300 pipettes to perform library prep steps on 8-48 input total RNA samples according to the attached NEB user guide. This is part-3 of a six part process (bead purification of double-stranded cDNA). User-determined parameters are available to specify the number of samples and the magnet engage height and time.\n\n\n---\n\n\n",
diff --git a/protoBuilds/0479ad-part-4/README.json b/protoBuilds/0479ad-part-4/README.json
index c5d94552ca..052ed32f4a 100644
--- a/protoBuilds/0479ad-part-4/README.json
+++ b/protoBuilds/0479ad-part-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II RNA Library Prep Kit for Illumina"
@@ -10,7 +10,7 @@
"internal": "0479ad-part-4",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons Temperature Module] (https://shop.opentrons.com/modules/)\n[Opentrons Magnetic Module] (https://shop.opentrons.com/modules/)\nThermoFisher 96 Well PCR Plate 300 uL Semi-Skirted\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext Ultra II RNA Library Prep Kit for Illumina\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module with 96-Well Aluminum Block holding cDNA Plate (ThermoFisher 96 Well PCR Plate 300 uL Semi-Skirted) \n\n\n\n**Slot 4**: Opentrons Temperature Module with 96-Well Aluminum Block holding Reagent Plate (nest_96_wellplate_100ul_pcr_full_skirt) \n\n\n\n**Slot 9**: Opentrons Magnetic Module (empty) \n**Slots 5,7**: Opentrons 20 uL filter tips \n**Slot 6**: Opentrons 200 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses multi-channel P20 and P300 pipettes to perform library prep steps on 8-48 input total RNA samples according to the attached NEB user guide. This is part-4 of a six part process (Part-4 - End Prep and Adapter Ligation). End prep reaction buffer + end prep enzyme mix are combined into a single mix to reduce the number of transfer steps. A user-specified parameter is available to specify the number of samples in the current run.\n\n\n---\n\n\n",
diff --git a/protoBuilds/0479ad-part-5/README.json b/protoBuilds/0479ad-part-5/README.json
index 990c439fd2..04d4834602 100644
--- a/protoBuilds/0479ad-part-5/README.json
+++ b/protoBuilds/0479ad-part-5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II RNA Library Prep Kit for Illumina"
@@ -10,7 +10,7 @@
"internal": "0479ad-part-5",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons Temperature Module] (https://shop.opentrons.com/modules/)\n[Opentrons Magnetic Module] (https://shop.opentrons.com/modules/)\n[USA Scientific 12 Well Reservoir 22 mL] (https://shop.opentrons.com)\nThermoFisher 96 Well PCR Plate 300 uL Semi-Skirted\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext Ultra II RNA Library Prep Kit for Illumina\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module with 96-Well Aluminum Block holding Output Plate (ThermoFisher 96 Well PCR Plate 300 uL Semi-Skirted) \n**Slot 2**: Reagent Reservoir for NEBNext Sample Purification Beads, 0.1X TE, 80 Percent EtOH, Liquid Waste (usascientific_12_reservoir_22ml) \n\n\n\n**Slot 4**: Opentrons Temperature Module empty \n**Slot 9**: Opentrons Magnetic Module (ThermoFisher 96 Well PCR Plate 300 uL Semi-Skirted on Custom Plastic Adapter containing 96 uL Adapter-Ligated cDNA Samples) \n\n\n\n**Slot 8**: Opentrons 20 uL filter tips \n**Slots 6,7,10**: Opentrons 200 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses multi-channel P20 and P300 pipettes to perform library prep steps on 8-48 input total RNA samples according to the attached NEB user guide. This is part-5 of a six part process (bead purification of the ligation reaction). User-determined parameters are available to specify the number of samples and the magnet engage height and time.\n\n\n---\n\n\n",
diff --git a/protoBuilds/0479ad/README.json b/protoBuilds/0479ad/README.json
index 0879bcb82a..8a03a82ad1 100644
--- a/protoBuilds/0479ad/README.json
+++ b/protoBuilds/0479ad/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II RNA Library Prep Kit for Illumina"
@@ -10,7 +10,7 @@
"internal": "0479ad",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons Temperature Module] (https://shop.opentrons.com/modules/)\n[Opentrons Magnetic Module] (https://shop.opentrons.com/modules/)\n[Nest 96 Well PCR Plates 100 uL, USA Scientific 12 Well Reservoir 22 mL] (https://shop.opentrons.com)\nThermoFisher 96 Well PCR Plate 300 uL Semi-Skirted\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext Ultra II RNA Library Prep Kit for Illumina\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module with 96-Well Aluminum Block holding Reagent Plate (nest_96_wellplate_100ul_pcr_full_skirt) \n\n\n\n**Slot 2**: Reagent Reservoir for 0.1X TE, Wash Buffer, Liquid Waste (usascientific_12_reservoir_22ml) \n\n\n\n**Slot 4**: Opentrons Temperature Module with 96-Well Aluminum Block holding Output Plate (ThermoFisher 96 Well PCR Plate 300 uL Semi-Skirted) \n**Slot 9**: Opentrons Magnetic Module (ThermoFisher 96 Well PCR Plate 300 uL Semi-Skirted on Custom Plastic Adapter containing 50 uL Total RNA Samples + 50 uL Oligo-dT Beads) \n\n\n\n**Slot 8**: Opentrons 20 uL filter tips \n**Slots 6,7,10,11**: Opentrons 200 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses multi-channel P20 and P300 pipettes to perform library prep steps on 8-48 input total RNA samples according to the attached NEB user guide (reagents were combined in a few instances to reduce the number of transfer steps required). User-determined parameters are available to specify the number of samples and the magnet engage height and time.\n\n\n---\n\n\n",
diff --git a/protoBuilds/04bec0/README.json b/protoBuilds/04bec0/README.json
index 933caf6891..d786987689 100644
--- a/protoBuilds/04bec0/README.json
+++ b/protoBuilds/04bec0/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "04bec0",
"labware": "\nBeckman Fed Batch Plate #Beckman Coulter\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\nOpentrons 6 Tube Rack with Falcon 50 mL Conical\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 1000 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a custom normalization and LDS addition across 3 sets of 1.5ml tuberacks using P300 and P1000 single-channel pipettes. The normalization volumes are automatically calculated.\n\nThe input .csv file specifying the biomasses of all sample input should be formatted as follows ***including first row for column names and first column for row names***. Note that wells that should be ignored should contain a `0` value:\n\n```\n,1,2,3,4,5,6,7,8\nA,0,0,0,0,0,0,0,0\nB,0,0,0,0,0,0,0,0\nC,80,60.72,82.79,207.23,370.24,265.78,270.34,342.88\nD,70,56.65,99.74,249.64,340.56,285.08,294,366.09\nE,117.17,162.28,88.7,135.69,322.46,321,285.73,375.51\nF,130.01,172.35,93.48,136.36,326.26,285.79,316.29,344.37\n```\n\nYou can also download a template from [this link](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/04bec0/ex.csv).\n\n\n",
diff --git a/protoBuilds/04eeb1-part-2/README.json b/protoBuilds/04eeb1-part-2/README.json
index 23d73ee20f..9058d9dee3 100644
--- a/protoBuilds/04eeb1-part-2/README.json
+++ b/protoBuilds/04eeb1-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina COVIDSeq Test"
@@ -10,7 +10,7 @@
"internal": "04eeb1-part-2",
"labware": "\nOpentrons 20 uL Filter Tips\nBio-Rad Hard-Shell 96 Well Plate 200 \u00b5L PCR\nNEST 2 mL 96-Well Deep Well Plate\nNEST 12-Well Reservoirs, 15 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina COVIDSeq Test\n\n",
"deck-setup": "\n\n**Note**: Master Mix should be added in Column 2 (A2) of any reservoir type.\n\n---\n\n",
"description": "The Illumina COVIDSeq Test is a high-throughput, next-generation sequencing test that is used fo detecting SARS-CoV-2 in patient samples. This protocol is the second part of a seven part protocol that is run on the OT-2 for this kit.\n\n* Part 1: [Anneal RNA](https://protocols.opentrons.com/protocol/04eeb1)\n* Part 2: [Synthesize First Strand cDNA](https://protocols.opentrons.com/protocol/04eeb1-part-2)\n* Part 3: [Amplify cDNA](https://protocols.opentrons.com/protocol/04eeb1-part-3)\n* Part 4: [Tagment PCR Amplicons](https://protocols.opentrons.com/protocol/04eeb1-part-4)\n* Part 5: [Post Tagmentation Clean Up](https://protocols.opentrons.com/protocol/04eeb1-part-5)\n* Part 6: [Amplify Tagmented Amplicons](https://protocols.opentrons.com/protocol/04eeb1-part-6)\n* Part 7: [Pool and Clean Up Libraries](https://protocols.opentrons.com/protocol/04eeb1-part-7)\n\nExplanation of complex parameters below:\n* `P20 Multichannel GEN2 Pipette Mount`: Choose the mount position of your P20 Multichannel pipette, either left or right.\n* `Reservoir Labware Type`: Choose the type of reservoir that will be used for holding the master mix. **Note: Only the Bio-Rad Hard-Shell 96 Well Plate 200 \u00b5L plate will resume after Plate 1 and prompt for a refill of the master mix.**\n* `Plate 1 Columns`: Choose which columns master mix should be added on plate 1. Separate column numbers with a comman (Ex: 1,2,3,4).\n* `Plate 2 Columns`: Choose which columns master mix should be added on plate 2. Separate column numbers with a comman (Ex: 1,2,3,4).\n* `Temperature (C)`: Choose the temperature the temperature module should be set at in the beginning of the protocol.\n\n---\n\n",
diff --git a/protoBuilds/04eeb1-part-3/README.json b/protoBuilds/04eeb1-part-3/README.json
index c1f606be54..c94bc7690d 100644
--- a/protoBuilds/04eeb1-part-3/README.json
+++ b/protoBuilds/04eeb1-part-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina COVIDSeq Test"
@@ -10,7 +10,7 @@
"internal": "04eeb1-part-3",
"labware": "\nOpentrons 200 uL Filter Tips\nBio-Rad Hard-Shell 96 Well Plate 200 \u00b5L PCR\nNEST 12-Well Reservoirs, 15 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina COVIDSeq Test\n\n",
"deck-setup": "\n\n---\n\n",
"description": "The Illumina COVIDSeq Test is a high-throughput, next-generation sequencing test that is used fo detecting SARS-CoV-2 in patient samples. This protocol is the third part of a seven part protocol that is run on the OT-2 for this kit.\n\n* Part 1: [Anneal RNA](https://protocols.opentrons.com/protocol/04eeb1)\n* Part 2: [Synthesize First Strand cDNA](https://protocols.opentrons.com/protocol/04eeb1-part-2)\n* Part 3: [Amplify cDNA](https://protocols.opentrons.com/protocol/04eeb1-part-3)\n* Part 4: [Tagment PCR Amplicons](https://protocols.opentrons.com/protocol/04eeb1-part-4)\n* Part 5: [Post Tagmentation Clean Up](https://protocols.opentrons.com/protocol/04eeb1-part-5)\n* Part 6: [Amplify Tagmented Amplicons](https://protocols.opentrons.com/protocol/04eeb1-part-6)\n* Part 7: [Pool and Clean Up Libraries](https://protocols.opentrons.com/protocol/04eeb1-part-7)\n\nExplanation of complex parameters below:\n* `P300 Multichannel GEN2 Pipette Mount`: Choose the mount position of your P300 Multichannel pipette, either left or right.\n* `Plate 1 Columns`: Choose which columns master mix should be added on plate 1. Separate column numbers with a comman (Ex: 1,2,3,4).\n* `Plate 2 Columns`: Choose which columns master mix should be added on plate 2. Separate column numbers with a comman (Ex: 1,2,3,4).\n* `Temperature (C)`: Choose the temperature the temperature module should be set at in the beginning of the protocol.\n* `Plate 1 Master Mix Column`: Choose the column on the 12-channel reservoir for the master mix for Plate 1.\n* `Plate 2 Master Mix Column`: Choose the column on the 12-channel reservoir for the master mix for Plate 2.\n\n---\n\n",
diff --git a/protoBuilds/04eeb1-part-4/README.json b/protoBuilds/04eeb1-part-4/README.json
index 6b5f3fcf83..b550633b3f 100644
--- a/protoBuilds/04eeb1-part-4/README.json
+++ b/protoBuilds/04eeb1-part-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina COVIDSeq Test"
@@ -10,7 +10,7 @@
"internal": "04eeb1-part-4",
"labware": "\nOpentrons 200 uL Filter Tips\nBio-Rad Hard-Shell 96 Well Plate 200 \u00b5L PCR\nNEST 12-Well Reservoirs, 15 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina COVIDSeq Test\n\n",
"deck-setup": "\n\n---\n\n",
"description": "The Illumina COVIDSeq Test is a high-throughput, next-generation sequencing test that is used fo detecting SARS-CoV-2 in patient samples. This protocol is the fourth part of a seven-part protocol that is run on the OT-2 for this kit.\n\n* Part 1: [Anneal RNA](https://protocols.opentrons.com/protocol/04eeb1)\n* Part 2: [Synthesize First Strand cDNA](https://protocols.opentrons.com/protocol/04eeb1-part-2)\n* Part 3: [Amplify cDNA](https://protocols.opentrons.com/protocol/04eeb1-part-3)\n* Part 4: [Tagment PCR Amplicons](https://protocols.opentrons.com/protocol/04eeb1-part-4)\n* Part 5: [Post Tagmentation Clean Up](https://protocols.opentrons.com/protocol/04eeb1-part-5)\n* Part 6: [Amplify Tagmented Amplicons](https://protocols.opentrons.com/protocol/04eeb1-part-6)\n* Part 7: [Pool and Clean Up Libraries](https://protocols.opentrons.com/protocol/04eeb1-part-7)\n\nExplanation of complex parameters below:\n* `P300 Multichannel GEN2 Pipette Mount`: Choose the mount position of your P20 Multichannel pipette, either left or right.\n* `Plate 1 Columns`: Choose which columns master mix should be added on plate 1. Separate column numbers with a comman (Ex: 1,2,3,4).\n* `Plate 2 Columns`: Choose which columns master mix should be added on plate 2. Separate column numbers with a comman (Ex: 1,2,3,4).\n* `Temperature (C)`: Choose the temperature the temperature module should be set at in the beginning of the protocol.\n* `Master Mix Column`: Choose the column on the 12-channel reservoir for the master mix for Plate 1.\n\n---\n\n",
diff --git a/protoBuilds/04eeb1-part-5/README.json b/protoBuilds/04eeb1-part-5/README.json
index 56dcac8793..45305b0b24 100644
--- a/protoBuilds/04eeb1-part-5/README.json
+++ b/protoBuilds/04eeb1-part-5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina COVIDSeq Test"
@@ -10,7 +10,7 @@
"internal": "04eeb1-part-5",
"labware": "\nOpentrons 200 uL Filter Tips\nBio-Rad Hard-Shell 96 Well Plate 200 \u00b5L PCR\nNEST 12-Well Reservoirs, 15 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina COVIDSeq Test\n\n",
"deck-setup": "\n\n**Note**: Wash Buffer should be added in Column 2 (A2).\n\n---\n\n",
"description": "The Illumina COVIDSeq Test is a high-throughput, next-generation sequencing test that is used fo detecting SARS-CoV-2 in patient samples. This protocol is the fifth part of a seven-part protocol that is run on the OT-2 for this kit.\n\n* Part 1: [Anneal RNA](https://protocols.opentrons.com/protocol/04eeb1)\n* Part 2: [Synthesize First Strand cDNA](https://protocols.opentrons.com/protocol/04eeb1-part-2)\n* Part 3: [Amplify cDNA](https://protocols.opentrons.com/protocol/04eeb1-part-3)\n* Part 4: [Tagment PCR Amplicons](https://protocols.opentrons.com/protocol/04eeb1-part-4)\n* Part 5: [Post Tagmentation Clean Up](https://protocols.opentrons.com/protocol/04eeb1-part-5)\n* Part 6: [Amplify Tagmented Amplicons](https://protocols.opentrons.com/protocol/04eeb1-part-6)\n* Part 7: [Pool and Clean Up Libraries](https://protocols.opentrons.com/protocol/04eeb1-part-7)\n\nExplanation of complex parameters below:\n* `P300 Multichannel GEN2 Pipette Mount`: Choose the mount position of your P300 Multichannel pipette, either left or right.\n* `Plate 1 Columns`: Choose which columns master mix should be added on plate 1. Separate column numbers with a comman (Ex: 1,2,3,4).\n* `Temperature (C)`: Choose the temperature the temperature module should be set at in the beginning of the protocol.\n* `Wash Buffer Column`: Choose the column on the 12-channel reservoir for the master mix for Plate 1.\n\n---\n\n",
diff --git a/protoBuilds/04eeb1-part-6/README.json b/protoBuilds/04eeb1-part-6/README.json
index 3f56aec5a9..0179b235a4 100644
--- a/protoBuilds/04eeb1-part-6/README.json
+++ b/protoBuilds/04eeb1-part-6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina COVIDSeq Test"
@@ -10,7 +10,7 @@
"internal": "04eeb1-part-6",
"labware": "\nOpentrons 200 uL Filter Tips\nBio-Rad Hard-Shell 96 Well Plate 200 \u00b5L PCR\nNEST 12-Well Reservoirs, 15 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina COVIDSeq Test\n\n",
"deck-setup": "\n\n**Note**: AMP Master Mix should be added in Column 3 (A3).\n\n---\n\n",
"description": "The Illumina COVIDSeq Test is a high-throughput, next-generation sequencing test that is used fo detecting SARS-CoV-2 in patient samples. This protocol is the sixth part of a seven-part protocol that is run on the OT-2 for this kit.\n\n* Part 1: [Anneal RNA](https://protocols.opentrons.com/protocol/04eeb1)\n* Part 2: [Synthesize First Strand cDNA](https://protocols.opentrons.com/protocol/04eeb1-part-2)\n* Part 3: [Amplify cDNA](https://protocols.opentrons.com/protocol/04eeb1-part-3)\n* Part 4: [Tagment PCR Amplicons](https://protocols.opentrons.com/protocol/04eeb1-part-4)\n* Part 5: [Post Tagmentation Clean Up](https://protocols.opentrons.com/protocol/04eeb1-part-5)\n* Part 6: [Amplify Tagmented Amplicons](https://protocols.opentrons.com/protocol/04eeb1-part-6)\n* Part 7: [Pool and Clean Up Libraries](https://protocols.opentrons.com/protocol/04eeb1-part-7)\n\nExplanation of complex parameters below:\n* `P300 Multichannel GEN2 Pipette Mount`: Choose the mount position of your P300 Multichannel pipette, either left or right.\n* `Plate 1 Columns`: Choose which columns master mix should be added on plate 1. Separate column numbers with a comman (Ex: 1,2,3,4).\n* `Temperature (C)`: Choose the temperature the temperature module should be set at in the beginning of the protocol.\n* `Master Mix Column`: Choose the column on the 12-channel reservoir for the master mix for Plate 1.\n---\n\n",
diff --git a/protoBuilds/04eeb1-part-7/README.json b/protoBuilds/04eeb1-part-7/README.json
index fdb7ec03bf..4a91f69409 100644
--- a/protoBuilds/04eeb1-part-7/README.json
+++ b/protoBuilds/04eeb1-part-7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina COVIDSeq Test"
@@ -10,7 +10,7 @@
"internal": "04eeb1-part-7",
"labware": "\nOpentrons 200 uL Filter Tips\nBio-Rad Hard-Shell 96 Well Plate 200 \u00b5L PCR\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina COVIDSeq Test\n\n",
"deck-setup": "\n\n**Note**: Add PCR strip on Column 1 of 96 Well Aluminum block in Slot 9. Add 1.7 mL tube to A1 of 24 Well Aluminum block on slot 6.\n\n---\n\n",
"description": "The Illumina COVIDSeq Test is a high-throughput, next-generation sequencing test that is used fo detecting SARS-CoV-2 in patient samples. This protocol is the seventh part of a seven-part protocol that is run on the OT-2 for this kit.\n\n* Part 1: [Anneal RNA](https://protocols.opentrons.com/protocol/04eeb1)\n* Part 2: [Synthesize First Strand cDNA](https://protocols.opentrons.com/protocol/04eeb1-part-2)\n* Part 3: [Amplify cDNA](https://protocols.opentrons.com/protocol/04eeb1-part-3)\n* Part 4: [Tagment PCR Amplicons](https://protocols.opentrons.com/protocol/04eeb1-part-4)\n* Part 5: [Post Tagmentation Clean Up](https://protocols.opentrons.com/protocol/04eeb1-part-5)\n* Part 6: [Amplify Tagmented Amplicons](https://protocols.opentrons.com/protocol/04eeb1-part-6)\n* Part 7: [Pool and Clean Up Libraries](https://protocols.opentrons.com/protocol/04eeb1-part-7)\n\nExplanation of complex parameters below:\n* `P300 Multichannel GEN2 Pipette Mount`: Choose the mount position of your P300 Multichannel pipette, either left or right.\n* `P20 Multichannel GEN2 Pipette Mount`: Choose the mount position of your P20 Multichannel pipette, either left or right.\n\n---\n\n",
diff --git a/protoBuilds/04eeb1/README.json b/protoBuilds/04eeb1/README.json
index 1d6d94d49b..dd4536dba8 100644
--- a/protoBuilds/04eeb1/README.json
+++ b/protoBuilds/04eeb1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina COVIDSeq Test"
@@ -10,7 +10,7 @@
"internal": "04eeb1",
"labware": "\nOpentrons 20 uL Filter Tips\nBio-Rad Hard-Shell 96 Well Plate 200 \u00b5L PCR\nNEST 2 mL 96-Well Deep Well Plate\nNEST 12-Well Reservoirs, 15 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina COVIDSeq Test\n\n",
"deck-setup": "\n\n**Note**: Master Mix should be added in Column 1 (A1) of any reservoir type.\n\n---\n\n",
"description": "The Illumina COVIDSeq Test is a high-throughput, next-generation sequencing test that is used fo detecting SARS-CoV-2 in patient samples. This protocol is the first part of a seven part protocol that is run on the OT-2 for this kit.\n\n* Part 1: [Anneal RNA](https://protocols.opentrons.com/protocol/04eeb1)\n* Part 2: [Synthesize First Strand cDNA](https://protocols.opentrons.com/protocol/04eeb1-part-2)\n* Part 3: [Amplify cDNA](https://protocols.opentrons.com/protocol/04eeb1-part-3)\n* Part 4: [Tagment PCR Amplicons](https://protocols.opentrons.com/protocol/04eeb1-part-4)\n* Part 5: [Post Tagmentation Clean Up](https://protocols.opentrons.com/protocol/04eeb1-part-5)\n* Part 6: [Amplify Tagmented Amplicons](https://protocols.opentrons.com/protocol/04eeb1-part-6)\n* Part 7: [Pool and Clean Up Libraries](https://protocols.opentrons.com/protocol/04eeb1-part-7)\n\nExplanation of complex parameters below:\n* `P20 Multichannel GEN2 Pipette Mount`: Choose the mount position of your P20 Multichannel pipette, either left or right.\n* `Reservoir Labware Type`: Choose the type of reservoir that will be used for holding the master mix. **Note: Only the Bio-Rad Hard-Shell 96 Well Plate 200 \u00b5L plate will resume after Plate 1 and prompt for a refill of the master mix.**\n* `Plate 1 Columns`: Choose which columns master mix should be added on plate 1. Separate column numbers with a comman (Ex: 1,2,3,4).\n* `Plate 2 Columns`: Choose which columns master mix should be added on plate 2. Separate column numbers with a comman (Ex: 1,2,3,4).\n* `Temperature (C)`: Choose the temperature the temperature module should be set at in the beginning of the protocol.\n\n---\n\n",
diff --git a/protoBuilds/04f310/README.json b/protoBuilds/04f310/README.json
index 65721b6c1c..ce614faeb0 100644
--- a/protoBuilds/04f310/README.json
+++ b/protoBuilds/04f310/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "04f310",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons 10-Tube Rack] (https://shop.opentrons.com/4-in-1-tube-rack-set/)\n[BioRad 200 uL PCR Plate] (https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate)\n[NEST 96 Deep Well Plate 2 mL] (https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate)\n[Agilent 290 mL Reservoir] (https://labware.opentrons.com/agilent_1_reservoir_290ml?category=reservoir)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons 10-Tube Rack with Falcon 50 mL Conical Tube (containing 40 mL water) \n**Slot 5**: BioRad 200 uL PCR Plate (output plate) \n**Slot 8**: BioRad 200 uL PCR Plate (intermediate dilution plate) \n**Slot 9**: Agilent 290 mL Reservoir (filled with 10 percent diluted bleach) \n**Slot 11**: NEST 96 Deep Well Plate 2 mL (containing input DNA samples >= 50 uL) \n**Slot 7**: Opentrons 20 uL filter tips \n**Slot 10**: Opentrons 200 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses single-channel P20 and P300 pipettes to normalize DNA concentrations of up to 96 input samples based on user input (input DNA sample concentration, target concentration, target volume) provided by csv file upload (see example file below). Intermediate dilutions are prepared as needed. A downloadable output file is generated to record calculated fold dilution, volume of sample and water transferred, if a sample was processed (or skipped due to insufficient input DNA concentration), and if an intermediate dilution was required.\n\nLinks:\n* [example input csv](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/04f310/normalization_example_input.csv)\n\n\n\nValues for CSV file input:\n* sample_id\n* dna_conc_initial (ng/ul)\n* dna_conc_target (ng/ul) 5-40 ng/uL\n* volume_target (ul) 30-100 uL\n\n\n\nAdditional Calculated Values in CSV file output:\n* fold dilution\n* sample_transfer (ul)\n* water_transfer (ul)\n* processed (0 - sample was skipped, 1 - sample was processed)\n* intermediate_dilution (0 - intermediate dilution not required, 1 - intermediate dilution required)\n\n---\n\n\n",
diff --git a/protoBuilds/04f4e7/README.json b/protoBuilds/04f4e7/README.json
index 50d0c10f66..ba7367a299 100644
--- a/protoBuilds/04f4e7/README.json
+++ b/protoBuilds/04f4e7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Purification"
@@ -10,7 +10,7 @@
"internal": "04f4e7",
"labware": "\nNunc\u2122 96-Well Polypropylene DeepWell\u2122 Storage Plates 278743\nBio-Rad 96 Well Plate 200 \u00b5L PCR\nNest 1 well Reservoir 195mL\nNest 12 well Reservoir 15mL\nOpentrons 300ul tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Nucleic Acid Purification\n\n",
"deck-setup": "* If the deck layout of a particular protocol is more or less static, it is often helpful to attach a preview of the deck layout, most descriptively generated with Labware Creator. Example:\n\n\n",
"description": "This protocol performs an RNA extraction on a user-specified number of samples. Beads are pre-mixed and added to sample with 5 sequential incubation-mix periods. Excess liquid is picked off the top of the wells and dispensed into waste, and the remaining liquid + beads are added to the magnetic plate. After two PBS washes, elution and neutralization buffer are added to the samples. Samples are then moved to the elution plate, with beads in the mag plate collected for later use. \n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify the number of samples to be processed this run.\n* `Slow Aspiration Rate (Step 7)`: Specify the aspiration rate for Step 7 in the protocol. A value of 1 is the default flow rate, a value of 0.5 would be half of the default flow rate, and so on.\n* `Aspiration Height to Remove Top Liquid (Step 7)`: Specify the aspiration height to aspirate 3 X 300ul from as specified in Step 7 in the protocol.\n* `Aspiration Height to Remove Supernatant`: Specify the aspiration height to remove supernatant from.\n* `Incubation Time After Engaging Magnet`: Specify the incubation time after engaging the magnet (in minutes).\n* `Length From Side of Well Opposite Magnetically Engaged Beads`: Specify the distance from the side of the well opposite magnetically engaged beads to aspirate from. A value of 2 is 2mm from the side of the well opposite magnetically engaged beads.\n* `P300 Multi-Channel Mount`: Specify which mount (left or right) for the P300 multi-channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/04fed3/README.json b/protoBuilds/04fed3/README.json
index 6ea2665c89..96b43b57dc 100644
--- a/protoBuilds/04fed3/README.json
+++ b/protoBuilds/04fed3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -9,7 +9,7 @@
"description": "This protocol uses a single tip on the p300 multi-channel pipette to distribute custom volumes of buffer (150-300 uL) from a 195 mL reservoir to specified wells of a 96-well destination plate followed by transfer of custom volumes of sample (2-15 uL) from up to three 24-tube racks to specified wells in the destination plate (volumes, rack locations and well locations are specified in a csv file uploaded at the time of protocol download).\nLinks:\n* example input csv",
"internal": "04fed3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Normalization\n\n",
"deck-setup": "\n\n* Opentrons 300ul tips (Deck Slot 11)\n* Opentrons 20ul tips (Deck Slots 10)\n* 96-well destination plate genesee25221_96_wellplate_300ul (Deck Slot 6)\n* 24-tube Opentrons racks with 1.5 mL snap cap tubes opentrons_24_tuberack_eppendorf_1.5ml_safelock_snapcap (Deck Slots 1,4,7)\n* Reservoir nest_1_reservoir_195ml (Deck Slot 5)\n\n\n\n",
"description": "\nThis protocol uses a single tip on the p300 multi-channel pipette to distribute custom volumes of buffer (150-300 uL) from a 195 mL reservoir to specified wells of a 96-well destination plate followed by transfer of custom volumes of sample (2-15 uL) from up to three 24-tube racks to specified wells in the destination plate (volumes, rack locations and well locations are specified in a csv file uploaded at the time of protocol download).\n\nLinks:\n* [example input csv](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/04fed3/example.csv)\n\n",
diff --git a/protoBuilds/051557/README.json b/protoBuilds/051557/README.json
index 57498ea1a5..36362d8c2d 100644
--- a/protoBuilds/051557/README.json
+++ b/protoBuilds/051557/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Standard Curve"
@@ -9,7 +9,7 @@
"internal": "051557",
"labware": "\nAbgene 96 Well Plate 2200 \u00b5L #AB-0932\nEppendorf 7 Reservoir 100000 \u00b5L #960051017\nEppendorf 7 Reservoir 30000 \u00b5L #960051009\nOpentrons 24 Well Aluminum Block with NEST 1.5 mL Snapcap\nOpentrons 96 Tip Rack 20 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Standard Curve\n\n\n",
"description": "This performs sample and standard preparation for lipid quantification.\n\n\n",
"internal": "051557\n",
diff --git a/protoBuilds/0523c2/README.json b/protoBuilds/0523c2/README.json
index c342b2aedc..beb9d58163 100644
--- a/protoBuilds/0523c2/README.json
+++ b/protoBuilds/0523c2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "0523c2",
"labware": "\nEppendorf Twin.tec 96 Well Plate 150 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 24 Well Aluminum Block with NEST 1.5 mL Screwcap\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a custom PCR preparation for up to 32 samples, including mastermix creation and sample barcoding.\n\n\n",
diff --git a/protoBuilds/052d03/README.json b/protoBuilds/052d03/README.json
index 1c41c47a64..4eb678edc6 100644
--- a/protoBuilds/052d03/README.json
+++ b/protoBuilds/052d03/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR"
@@ -9,7 +9,7 @@
"internal": "052d03",
"labware": "\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\nOpentrons 96 Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR\n\n\n",
"description": "This protocol performs a custom dilution and PCR protocol on the Opentrons Thermocycler GEN2 Module. The number of samples will be automatically calculated from the input cherrypicking .csv list, which should be formatted as the following example **excluding the positive control**:\n\n```\nA4,tc\nB4,tc\nC1,dil\nD1,dil\nB1,dil\nH12,dil\n\n```\n\nNote that the first value of each line should specify the well location, and the second value should specify whether to pick from the thermocycler plate or the dilution plate.\n\n\n",
"internal": "052d03\n",
diff --git a/protoBuilds/0530d8-pt2/README.json b/protoBuilds/0530d8-pt2/README.json
index 98ab2b0193..bcafc1ca1a 100644
--- a/protoBuilds/0530d8-pt2/README.json
+++ b/protoBuilds/0530d8-pt2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"DNA Extraction"
@@ -10,7 +10,7 @@
"internal": "0530d8-pt2",
"labware": "\nAbgene 96 deep well plate\nOpentrons 200ul Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* DNA Extraction\n\n",
"deck-setup": "\n\n",
"description": "This protocol performs a DNA extraction with the samples gathered from part 1 of the protocol. For specific protocol steps, please see below. A field for aspiration height when transferring the samples from the heater shaker to the magnetic module allows the user to ensure beads are not picked up, and and aspiration rate of 10% of the default is instilled to ensure so. Note: for reagent 4 in columns 4 & 5, reagent 5 in columns 6 & 7, and reagent 6 in columns 8 & 9, split the total calculated reagent volume EQUALLY between the pair of columns, as the protocol will circle between the pair of columns as the source.\n\n\n\n---\n\n",
diff --git a/protoBuilds/0530d8/README.json b/protoBuilds/0530d8/README.json
index 34708ea5ab..f9c3e9332b 100644
--- a/protoBuilds/0530d8/README.json
+++ b/protoBuilds/0530d8/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"DNA Extraction"
@@ -10,7 +10,7 @@
"internal": "0530d8",
"labware": "\nAbgene 96 deep well plate\nOpentrons 200ul Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* DNA Extraction\n\n",
"deck-setup": "\n\n",
"description": "This protocol takes up to 3 csv files per source plate to dispense into a 96 well plate on the heater shaker module. Samples can be in any order in the source plate as long as they are specified in the csv, but will always be loaded into the final plate by column. A maximum number of 90 samples can be loaded into the 96 well plate on the heater shaker, leaving room for the 6 controls (and subsequently 90 samples between all 3 csv files). Note: for reagent 4 in columns 4 & 5, reagent 5 in columns 6 & 7, and reagent 6 in columns 8 & 9, split the total calculated reagent volume EQUALLY between the pair of columns, as the protocol will circle between the pair of columns as the source.\n\n* The first row of all 3 csv files should be populated with the header, with the relevant parsing information to start in row 2 of each csv file. The header should consist of the following in the following order: ```barcode, rack_num, sample_rack_row, sample_rack_col,plate_row, plate_col, dna_extract_row, dna_extract_col```. All relevant information for the protocol is just taken from the ```dna_extract_row, dna_extract_col``` columns. The number of csv files must equal the number of source plates selected below.\n\n\n---\n\n",
diff --git a/protoBuilds/053386-pt2/README.json b/protoBuilds/053386-pt2/README.json
index e52899f2fb..a6ed39cf1c 100644
--- a/protoBuilds/053386-pt2/README.json
+++ b/protoBuilds/053386-pt2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "053386-pt2",
"labware": "\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "For detailed protocol steps, please see below. The trash will be used as a waste reservoir for supernatant removal. This protocol is for full plates (96 samples) exclusively. Well 12 of the reservoir will be used for pooled samples. \n\n\n",
diff --git a/protoBuilds/053386-pt3/README.json b/protoBuilds/053386-pt3/README.json
index b46b857cdd..342e86631d 100644
--- a/protoBuilds/053386-pt3/README.json
+++ b/protoBuilds/053386-pt3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "053386-pt3",
"labware": "\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "For detailed protocol steps, please see below. The trash will be used as a waste reservoir for supernatant removal. This protocol is for full plates (96 samples) exclusively. Well 11 of the reservoir will be used for pooled samples.\n\n\n",
diff --git a/protoBuilds/053386/README.json b/protoBuilds/053386/README.json
index 048922227f..2b9677ffd9 100644
--- a/protoBuilds/053386/README.json
+++ b/protoBuilds/053386/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "053386",
"labware": "\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "For detailed protocol steps, please see below. The trash will be used as a waste reservoir for supernatant removal. This protocol is for full plates (96 samples) exclusively. The protocol will pause to prompt the user to replace tips when needed, and pause for certain centrifuging steps.\n\n\n",
diff --git a/protoBuilds/0556be-bmda/README.json b/protoBuilds/0556be-bmda/README.json
index da548b713c..b2d84382db 100644
--- a/protoBuilds/0556be-bmda/README.json
+++ b/protoBuilds/0556be-bmda/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -9,7 +9,7 @@
"description": "This protocol performs various transfers of 12 different components to create a master mix. The master mix is then distributed in 32 PCR tube strips.\n\n\n\nP300 single-channel GEN2 electronic pipette\nOpentrons Aluminum Blocks\n\n\n",
"internal": "0556be-bmda",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"deck-setup": "* 200uL Filter Tips (Slot 1)\n* Temperature Module + 24 Well Aluminum Block (Slot 7)\n* 96 Well Aluminum Block with PCR Strips (Slot 8)\n\n",
"description": "This protocol performs various transfers of 12 different components to create a master mix. The master mix is then distributed in 32 PCR tube strips.\n\n---\n\n\n\n* [P300 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons Aluminum Blocks](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set?_ga=2.44012454.2010707504.1610321113-1181961818.1604785212)\n\n---\n\n\n",
diff --git a/protoBuilds/0556be-covid-mm-qc-v3/README.json b/protoBuilds/0556be-covid-mm-qc-v3/README.json
index a8cf1fe919..5e39e9b692 100644
--- a/protoBuilds/0556be-covid-mm-qc-v3/README.json
+++ b/protoBuilds/0556be-covid-mm-qc-v3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -9,7 +9,7 @@
"description": "This protocol performs the transfer of master mix into 24 tubes in an aluminum block on top of the temperature module.\n\n\n\nP1000 single-channel GEN2 electronic pipette\nOpentrons Aluminum Blocks\n\n\n",
"internal": "0556be-covid-mm-qc-v3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "* 1000 uL Filter Tips (Slot 1)\n* Opentrons 10 Tube Rack (Slot 4)\n* Temperature Module + 24 Well Aluminum Block (Slot 10)\n\n\n",
"description": "This protocol performs the transfer of master mix into 24 tubes in an aluminum block on top of the temperature module.\n\n---\n\n\n\n* [P1000 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons Aluminum Blocks](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set?_ga=2.44012454.2010707504.1610321113-1181961818.1604785212)\n\n---\n\n\n",
diff --git a/protoBuilds/0556be-covid-mm-qc/README.json b/protoBuilds/0556be-covid-mm-qc/README.json
index 9780613f53..5b54609efe 100644
--- a/protoBuilds/0556be-covid-mm-qc/README.json
+++ b/protoBuilds/0556be-covid-mm-qc/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -9,7 +9,7 @@
"description": "This protocol performs the transfer of 2 components into multiple 2 mL tubes in the first step. In the second step, the protocol transfers reagent from the first tube to 9 select wells in the PCR tube strips. In the final step, it transfers Component 3 into the first 3 wells of the second strip.\nThis protocol combines v2 and v3 of the COVID MM-QC Protocol. The volumes of Component 1 and Component 2 can be changed in the parameters below.\n\n\n\nP300 single-channel GEN2 electronic pipette\nOpentrons Aluminum Blocks\n\n\n",
"internal": "0556be-covid-mm-qc",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "* Opentrons 10 Tube Rack (Slot 8)\n* Temperature Module + 24 Well Aluminum Block (Slot 9)\n* 24 Well Aluminum Block (Slot 5)\n* 96 Well Aluminum Block with PCR Strips (Slot 6)\n* 200 uL Filter Tips (Slot 4)\n* 20 uL Filter Tips (Slot 1)\n\n",
"description": "This protocol performs the transfer of 2 components into multiple 2 mL tubes in the first step. In the second step, the protocol transfers reagent from the first tube to 9 select wells in the PCR tube strips. In the final step, it transfers Component 3 into the first 3 wells of the second strip.\n\nThis protocol combines v2 and v3 of the COVID MM-QC Protocol. The volumes of Component 1 and Component 2 can be changed in the parameters below.\n\n---\n\n\n\n* [P300 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons Aluminum Blocks](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set?_ga=2.44012454.2010707504.1610321113-1181961818.1604785212)\n\n---\n\n\n",
diff --git a/protoBuilds/0556be-covid-ntc/README.json b/protoBuilds/0556be-covid-ntc/README.json
index eea955a862..4b2aad4549 100644
--- a/protoBuilds/0556be-covid-ntc/README.json
+++ b/protoBuilds/0556be-covid-ntc/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -9,7 +9,7 @@
"description": "This protocol performs the transfer of nuclease-free water into multiple 2 mL tubes.\n\n\n\nP300 single-channel GEN2 electronic pipette\nOpentrons Aluminum Blocks\n\n\n",
"internal": "0556be-covid-ntc",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "* Opentrons 10 Tube Rack (Slot 10)\n* 24 Well Aluminum Block (Slot 11)\n* 1000uL Filter Tips (Slot 1)\n\n",
"description": "This protocol performs the transfer of nuclease-free water into multiple 2 mL tubes.\n\n---\n\n\n\n* [P300 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons Aluminum Blocks](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set?_ga=2.44012454.2010707504.1610321113-1181961818.1604785212)\n\n---\n\n\n",
diff --git a/protoBuilds/055b94/README.json b/protoBuilds/055b94/README.json
index 0e728bd601..3ab6458007 100644
--- a/protoBuilds/055b94/README.json
+++ b/protoBuilds/055b94/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "055b94",
"labware": "\nKingFisher 96 Well Plate 200 \u00b5L #97002540\nThermoFisher Microamp 96 Aluminum Block 200 \u00b5L #4346906\nMicroamp 384 Well Plate 40 \u00b5L #4483273\ncustom 24 Tube Rack with Custom 2 mL\nKingFisher 96 Deep Well Plate #95040450\nCustom 96 Well Plate 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"deck-setup": "\n* yellow on 96-wellplates: sample\n* blue on tuberack: prepared mastermix\n* purple on strips: mastermix strips for plating (loaded empty)\n\n",
"description": "This protocol provides a custom qPCR prep on the OT-2 for up to 4x source 96-well plates into 1x destination 384-wellplate. Mastermix is prepared manually and transferred to each reaction well on the OT-2.\n\nEach protocol run generates an output .csv file that is compatible with Quantstudio instruments. To retrieve the file, launch a Jupyter Notebook server on your robot by following [these instructions](https://support.opentrons.com/s/article/Uploading-files-through-Jupyter-Notebook). The file will be named after the 9000 plate barcode scan, followed by \".csv.\"\n\n",
diff --git a/protoBuilds/055f76/README.json b/protoBuilds/055f76/README.json
index 377da0d4aa..6ef8ec787f 100644
--- a/protoBuilds/055f76/README.json
+++ b/protoBuilds/055f76/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "055f76",
"labware": "\nKingFisher 96 Well Plate 200 \u00b5L #97002540\nNEST 96 Deepwell Plate 2mL #503001\nUSA Scientific 12 Well Reservoir 22 mL #1061-8150\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 1000 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a custom reagent plate filling for the [MagMax Core Protocol](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fmanuals%2FMAN0015944_MagMAXCORE_NA_Kit_UG.pdf).\n\n\n",
diff --git a/protoBuilds/056543/README.json b/protoBuilds/056543/README.json
index 075598af8e..6e77ce98dc 100644
--- a/protoBuilds/056543/README.json
+++ b/protoBuilds/056543/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Compound Plating"
@@ -10,7 +10,7 @@
"internal": "056543",
"labware": "\nBio-Rad 96 Well Plate 200 \u00b5L PCR #hsp9601\nCorning 384 Well Plate 112 \u00b5L Flat #3640\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\nOpentrons 96 Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Compound Plating\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a custom compound plating protocol as specified by a user-input .csv file. You can download an example .csv template [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/056543/ex.csv)!\n\nThe same tip is used for each source, and the user is prompted to refill the deck state if all destination plates have been processed.\n\n\n",
diff --git a/protoBuilds/056f47/README.json b/protoBuilds/056f47/README.json
index 89d9787d5b..59bbee1071 100644
--- a/protoBuilds/056f47/README.json
+++ b/protoBuilds/056f47/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Serial Dilution"
@@ -10,7 +10,7 @@
"internal": "056f47",
"labware": "\nNEST 12 Well Reservoir 15 mL #360102\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Serial Dilution\n\n\n",
"deck-setup": "\n\n\n\n",
"description": "This protocol serially diluted compound stock with DMSO. For detailed protocol steps, please see below. Note: if n < 15, use NEST PCR plates, and if n > 15, use deep well plates. \n\n\n",
diff --git a/protoBuilds/058124/README.json b/protoBuilds/058124/README.json
index e209e0bbfb..57d3f11e24 100644
--- a/protoBuilds/058124/README.json
+++ b/protoBuilds/058124/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Serial Dilution"
@@ -10,7 +10,7 @@
"internal": "058124",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons 24-Tube Racks] (https://shop.opentrons.com/4-in-1-tube-rack-set/)\nNest 1.5 mL Snap Cap Tubes\n[Nest 96 Deep Well Plates 2 mL] (https://shop.opentrons.com/nest-2-ml-96-well-deep-well-plate-v-bottom/)\n[Axygen 90 mL Reservoir] (https://labware.opentrons.com/axygen_1_reservoir_90ml/)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Serial Dilution\n\n",
"deck-setup": "\n\n\n**Slots 1,4**: Opentrons 24-Tube Rack with 1.5 mL Snap Cap Tubes (up to 48 Protein Samples) \n**Slots 2,3,5,6**: Nest 96 Deep Well 2 mL Plates (up to 4 Dilution Plates) \n**Slot 10**: Opentrons 20 uL filter tips \n**Slot 11**: Opentrons 200 uL filter tips \n\n\n\n\n---\n\n",
"description": "This protocol uses a multi-channel P300 and single-channel P20 pipette to perform custom serial dilution of up to 48 input protein samples. There are six dilution schemes (defined below) which differ in diluent volume, sample volume, number of mix repeats, fold dilution and number of dilution points. An input csv file assigns a dilution scheme and a sample number to each sample (sample numbers 1-48 in column-wise order spanning two 24-tube racks). Samples using the same dilution scheme are assigned (up to 8 at a time) to a block of four consecutive dilution plate columns. The p300 will pick up a number of tips corresponding to the current number of samples (up to 8) and fill two to four dilution plate columns (depending on the assigned dilution scheme) with dilution buffer. The p20 will add sample to the filled wells of the first column. The p300 will then perform serial dilution steps (mixing and transfer to the next column). User-specified parameters are available to indicate the labware to be used for the diluent reservoir, and the necessary amount of deadvolume (small amount of inaccessible liquid in the bottom of the reservoir) for that reservoir. A downloadable output file (see instructions below) will be generated which indicates the dilution plate destination well for each input sample.\n\nLinks:\n* [example input csv](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/058124/input_csv.csv)\n\n\n\n\n\n---\n\n\n",
diff --git a/protoBuilds/059126/README.json b/protoBuilds/059126/README.json
index 0283514b99..d7c8beb6f1 100644
--- a/protoBuilds/059126/README.json
+++ b/protoBuilds/059126/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Reagent Addition"
@@ -10,7 +10,7 @@
"internal": "059126",
"labware": "\nGreiner 96 Well Plate 340uL\nGreiner 384 Well Plate 50uL\nCytive 24 Well Plate 2000uL used as Vial Holder\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Reagent Addition\n\n",
"deck-setup": "* Starting Deck Setup for 2 Reagent Vials:\n\n\n---\n\n",
"description": "This is a specialized reagent addition to add reagent from vials in a Cytiva well plate to a 96 well plate which are then transferred to a 384 well plate. The vials and transfer volumes are specified via a CSV file. The tips are picked up in a non-standard manner with a single tip picked up from the bottom of a specified column continuing up and to the left with each new tip pick-up. E.g. Column 11 is specified, first tip picked up will be H11, then G11, F11,.....H10, G10, etc. These single tips are used to add from the vials to the 96 well plate. The columns of the 96 well plate will then have a specified amount of each column added to the 384 well plate, reusing tips as much as possible. Transfer volumes from the vials to the 96 well plate, transfer volumes from the 96 well plate to the 384 well plate, and which vials will be used is specified by uploading a CSV. The specific format can be found in this [example CSV located here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/059126/example_trans.csv)\n\nExplanation of complex parameters below:\n* `Number of Columns in 384 Well Plate`: How many columns will be in the 384 well plate starting from the left side\n* `Starting Tip Column (1-12)`: Which tip will be picked up first. Number corresponds to column from left to right. Bottom tip will be picked up first in each column.\n* `P20 Multi GEN2 Mount`: Which mount the P20 multi-channel is connected to. The P300 will be connected to the opposite mount.\n* `Transfer .CSV File`: Upload the CSV file with vial list and transfer volumes.\n\n---\n\n",
diff --git a/protoBuilds/05f673/README.json b/protoBuilds/05f673/README.json
index 4a8ce2ce23..3c66a9d4f1 100644
--- a/protoBuilds/05f673/README.json
+++ b/protoBuilds/05f673/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "05f673",
"labware": "Corning 24-Well Plate, 3.4mL, Flat or Corning 96-Well Plate, 360\u00b5L, Flat\nAgilent 1-Well Reservoir, 290mL\nOpentrons 1000uL Tips\nOpentrons 300uL Tips, optional",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n\n",
"deck-setup": "Source plates containing samples can be placed in deck slots 1, 2, 3, and 5 (and should be referred to as 1-4 in the CSV). Destination plates (where buffer and cells will be added) can be placed in deck slots 6, 8, 9, and 11 (and should be referred to as 1-4 in the CSV). Tips should be placed in deck slot 7.\n\n---\n\n",
"description": "This protocol allows for a robust cherrypicking and normalization in one step.\n\nUsing a [P1000 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette), buffer will be transferred to all wells specified in the **Normalization CSV** (see below), before transferring samples from up to four source plates to four destination plates (specific plate and well location specifed in **Normalization CSV**).\n\nThe .csv file should be formatted as shown in the example below, **including headers**:\n\n```\nSource Plate, Source Well, Dest Plate, Dest Well, Buffer Volume (in ul), Culture Volume (in ul)\n2,A1,1,A1,1591,1870\n2,B1,2,B1,1629,808\n```\n\n\n**Update**: This protocol has been updated per user request and now allows for the option to use 96-well plates and (if using 96-well plates), prefill the plates with a [P300 8-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette).\nExplanation of complex parameters below:\n* **P1000 (GEN2) Mount**: Specify whether the P1000 single channel pipette is on the left or right mount.\n* **Normalization CSV**: CSV containing transfer information for normalization (as outlined above).\n* **Labware Type**: Select between Corning 24-Well Plate and Corning 96-Well Plate.\n* **Prefill Volume**: Specify the volume (in \u00b5L) to prefill the 96-Well plates.\n\n\n---\n\n\n",
diff --git a/protoBuilds/05fb0f/README.json b/protoBuilds/05fb0f/README.json
index a8e9348ed3..43c65750c5 100644
--- a/protoBuilds/05fb0f/README.json
+++ b/protoBuilds/05fb0f/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Assay"
@@ -10,7 +10,7 @@
"internal": "05fb0f",
"labware": "\nAxygen 4 Reservoir 70 \u00b5L #RES-MW4-HP\nFrameStar PCR 96 Well Plate 200 \u00b5L #4ti-0960\nThermo 1 Reservoir 300 \u00b5L #1200-1301\nTecan 96 Well Plate 1300 \u00b5L\nEppendorf Deepwell 96 Well Plate 1200 \u00b5L #0030501241\nOpentrons 96 Tip Rack 20 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Assay\n\n\n",
"deck-setup": "\n\n* Note: All volumes shown for 96 samples.\n\n\n",
"description": "This protocol performs a custom GSH assay on up to 96 samples. The user is automatically prompted to replace tipracks if necessary mid-protocol.\n\n\n",
diff --git a/protoBuilds/060c4f/README.json b/protoBuilds/060c4f/README.json
index f38202187a..808dc47220 100644
--- a/protoBuilds/060c4f/README.json
+++ b/protoBuilds/060c4f/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol automates the distribution of samples from a 96-well plate (on the Temperature Module) to a 384-well plate.\n\nGiven the mapping differences between a 96-well plate and a 384-well plate, the user can specify to transfer to odd rows (A/C/E...) or even rows (B/D/F...).\n\nThis protocol uses a custom labware definition for the Greiner Bio-One 384-well plate. When downloading the protocol, the labware definition (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: Using labware in your protocols\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P300 Multi-Channel Pipette\nOpentrons 300\u00b5L Tips\nOpentrons Temperature Module\n96-Well Aluminum Block\n96-Well Plate\nGreiner Bio-One 384-well plate\n\n\n\nSlot 1: Greiner Bio-One 384-well plate\nSlot 2: Opentrons 300\u00b5L Tips\nSlot 4: Opentrons Temperature Module with 96-Well Aluminum Block & 96-Well Plate containing samples in columns 1-8\n\n\nUsing the customizations field (below), set up your protocol.\n Destination Rows: Specify which rows the multi-channel pipette will dispense in.\n P300-Multi Mount: Select which mount (left or right) the P300-Multi is attached to.",
"internal": "060c4f",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol automates the distribution of samples from a 96-well plate (on the Temperature Module) to a 384-well plate.\n\nGiven the mapping differences between a 96-well plate and a 384-well plate, the user can specify to transfer to *odd* rows (A/C/E...) or *even* rows (B/D/F...).\n\nThis protocol uses a custom labware definition for the [Greiner Bio-One 384-well plate](https://shop.gbo.com/en/row/products/bioscience/microplates/384-well-microplates/384-deep-well-small-volume-polypropylene-microplate/784201.html). When downloading the protocol, the labware definition (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: [Using labware in your protocols](https://support.opentrons.com/en/articles/3136506-using-labware-in-your-protocols)\n\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [96-Well Aluminum Block](https://shop.opentrons.com/collections/verified-labware/products/aluminum-block-set)\n* [96-Well Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Greiner Bio-One 384-well plate](https://shop.gbo.com/en/row/products/bioscience/microplates/384-well-microplates/384-deep-well-small-volume-polypropylene-microplate/784201.html)\n\n\n\n---\n\n\nSlot 1: [Greiner Bio-One 384-well plate](https://shop.gbo.com/en/row/products/bioscience/microplates/384-well-microplates/384-deep-well-small-volume-polypropylene-microplate/784201.html)\n\nSlot 2: [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 4: [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [96-Well Aluminum Block](https://shop.opentrons.com/collections/verified-labware/products/aluminum-block-set) & [96-Well Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) containing samples in columns 1-8\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Destination Rows**: Specify which rows the multi-channel pipette will dispense in.\n* **P300-Multi Mount**: Select which mount (left or right) the P300-Multi is attached to.\n\n\n\n",
"internal": "060c4f\n",
diff --git a/protoBuilds/063611-part-2/README.json b/protoBuilds/063611-part-2/README.json
index 2f91adfa85..5b16538be3 100644
--- a/protoBuilds/063611-part-2/README.json
+++ b/protoBuilds/063611-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -9,7 +9,7 @@
"description": "This protocol uses a p300 multi-channel pipette to run a separate, preparative process to put 300 ul tips into a custom arrangement required for use of a multi-channel pipette with only four tips attached on alternating nozzles for use with a custom labware rack which contains 6 columns of 4 tubes. The starting deck arrangement requires either 2, 4 or 6 tip boxes. Half of those tip boxes must be completely full while the other half must be completely empty.\nLinks:\n Custom Cap Filling\n Custom Tip Rack Formatting\nThis protocol was developed as a separate preparative process to set up a custom arrangement of 300 ul tips needed to use a custom rack (with 6 columns of 4 wells each) in the attached Custom Cap Filling protocol.",
"internal": "063611-part-2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Plate Filling\n\n",
"deck-setup": "\n\n* Opentrons p300 tips opentrons_96_tiprack_300ul (these boxes should be full) (Deck Slots 7, 8, 9)\n* Opentrons p300 tips opentrons_96_tiprack_300ul (these boxes are initially empty) (Deck Slots 4, 5, 6)\n\n",
"description": "\nThis protocol uses a p300 multi-channel pipette to run a separate, preparative process to put 300 ul tips into a custom arrangement required for use of a multi-channel pipette with only four tips attached on alternating nozzles for use with a custom labware rack which contains 6 columns of 4 tubes. The starting deck arrangement requires either 2, 4 or 6 tip boxes. Half of those tip boxes must be completely full while the other half must be completely empty.\n\nLinks:\n* [Custom Cap Filling](https://protocols.opentrons.com/protocol/063611)\n* [Custom Tip Rack Formatting](https://protocols.opentrons.com/protocol/063611-part-2)\n\nThis protocol was developed as a separate preparative process to set up a custom arrangement of 300 ul tips needed to use a custom rack (with 6 columns of 4 wells each) in the attached Custom Cap Filling protocol.\n\n",
diff --git a/protoBuilds/063611/README.json b/protoBuilds/063611/README.json
index 38dc1fa87b..29d0205f5a 100644
--- a/protoBuilds/063611/README.json
+++ b/protoBuilds/063611/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -9,7 +9,7 @@
"description": "This protocol uses a p300 multi-channel pipette to top-dispense 250 ul of liquid from a reservoir into as many as 96 caps housed in up to four custom racks (each rack has 6 columns of 4 caps each). The 300 ul tips are required to be in a custom arrangement (tiprack rows B, D, F, H are empty to permit a mode of non-standard, multi-channel pipetting with 4 tips mounted on alternating nozzles) to support use of custom racks having 6 columns of 4 wells each. The custom tip arrangement can be set up manually, with a separate preparative OT-2 protocol (part-2) attached on this page (recommended), or as a first step at the start of the main protocol. \nLinks:\n Custom Cap Filling\n Custom Tip Arranging\nThis protocol was developed to transfer 250 ul of liquid from a reservoir into as many as 96 caps arranged in columns of 4 in up to 4 custom racks. Tip change after every rack is optional. Tip tracking to keep track of the starting tip from run to run is optional. Preparation of the custom tip arrangement at the start of this main protocol is optional (recommendation is to handle tip arrangement as a separate process by using protocol Custom Tip Arranging attached above as part 2).",
"internal": "063611",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Plate Filling\n\n",
"deck-setup": "\n\n* Opentrons p300 tips in a custom arrangement (Deck Slots 10, 11)\n* Custom Racks (4 columns of 6) (Deck Slots 4,1,5,2)\n* Reservoir nest_1_reservoir_195ml (Deck Slot 3)\n\n",
"description": "\nThis protocol uses a p300 multi-channel pipette to top-dispense 250 ul of liquid from a reservoir into as many as 96 caps housed in up to four custom racks (each rack has 6 columns of 4 caps each). The 300 ul tips are required to be in a custom arrangement (tiprack rows B, D, F, H are empty to permit a mode of non-standard, multi-channel pipetting with 4 tips mounted on alternating nozzles) to support use of custom racks having 6 columns of 4 wells each. The custom tip arrangement can be set up manually, with a separate preparative OT-2 protocol (part-2) attached on this page (recommended), or as a first step at the start of the main protocol. \n\nLinks:\n* [Custom Cap Filling](https://protocols.opentrons.com/protocol/063611)\n* [Custom Tip Arranging](https://protocols.opentrons.com/protocol/063611-part-2)\n\nThis protocol was developed to transfer 250 ul of liquid from a reservoir into as many as 96 caps arranged in columns of 4 in up to 4 custom racks. Tip change after every rack is optional. Tip tracking to keep track of the starting tip from run to run is optional. Preparation of the custom tip arrangement at the start of this main protocol is optional (recommendation is to handle tip arrangement as a separate process by using protocol Custom Tip Arranging attached above as part 2).\n\n",
diff --git a/protoBuilds/06ab09/README.json b/protoBuilds/06ab09/README.json
index 03c61698f7..10f8a70ebe 100644
--- a/protoBuilds/06ab09/README.json
+++ b/protoBuilds/06ab09/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol is designed to transfer 10\u00b5L of sample DNA from the Micronics Tubes to the wells of an Axygen 96-Well Plate using the P10-Multi.\nUsing the customizations fields, below set up your protocol.\n* P10 Multi Mount: Select which mount (left or right) the P10 Multi is attached to.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons P10 Multi-Channel Pipette\nOpentrons 10\u00b5L Tips\nAxygen 96-Well Plate (PCR-96-FS-C)\nMicronics 96-Tubes in Plate (MP52757S-Y20)\nSamples\n\n\n\nSlot 2: Axygen 96-Well Plate (PCR-96-FS-C)\nSlot 3: Micronics 96-Tubes in Plate (MP52757S-Y20)\nSlot 5: Opentrons 10\u00b5L Tips",
"internal": "06ab09",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol is designed to transfer 10\u00b5L of sample DNA from the Micronics Tubes to the wells of an Axygen 96-Well Plate using the P10-Multi.\n\n\n\n**Using the customizations fields, below set up your protocol.**\n* **P10 Multi Mount**: Select which mount (left or right) the P10 Multi is attached to.\n\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P10 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* Axygen 96-Well Plate (PCR-96-FS-C)\n* Micronics 96-Tubes in Plate (MP52757S-Y20)\n* Samples\n\n\n---\n\n\n\nSlot 2: Axygen 96-Well Plate (PCR-96-FS-C)\n\nSlot 3: Micronics 96-Tubes in Plate (MP52757S-Y20)\n\nSlot 5: [Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n\n\n",
"internal": "06ab09\n",
diff --git a/protoBuilds/06da40/README.json b/protoBuilds/06da40/README.json
index 49564915b0..9a18bc3f46 100644
--- a/protoBuilds/06da40/README.json
+++ b/protoBuilds/06da40/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization & PCR Prep"
@@ -10,7 +10,7 @@
"internal": "06da40",
"labware": "\nNEST 96-Well PCR Plate 100\u00b5L\nOpentrons 24 Tube Rack with Eppendorf 1.5mL Safe-Lock Snapcap\nOpentrons Tip Rack, 20\u00b5L\nOpentrons Tip Rack, 300\u00b5L\nSemi-skirted Plate on Aluminum Block\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization & PCR Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol prepares plates for ddPCR with a normalization step beforehand.\n\nBased on a user-supplied CSV, the protocol begins by performing a normalization from the plate in deck slot 4 to the plate in deck slot 5 (allowing one to preserve their original samples).\n\nAfter the normalization step, there is an optional reaction mix creation step. Once reaction mix is ready, each well of the destination plate(s) receive 18\u00b5L of reaction mix.\n\nThe protocol finishes by adding 4\u00b5L of each sample to a well containing reaction mix, per replicate. For example, if 3 replicates are selected, 4\u00b5L of the normalized sample in A1 would be dispensed in A1, B1, and C1 of the destination plate, followed by 4\u00b5L of normalized sample from A2 to D1, E1, F1, etc.\n\n**Explanation of complex parameters below:**\n* **.CSV Input File**: Upload CSV file. Should include a header row and the first three rows: Well (A1), Sample Volume (\u00b5L), Dilutent Volume (\u00b5L)\n* **Automate Reaction Mix Creation**: Select whether to automate the creation of Reaction Mix.\n* **Number of Replicates**: Specify the number of replicates\n\n",
diff --git a/protoBuilds/06e5b6/README.json b/protoBuilds/06e5b6/README.json
index e51c0ce4d6..dab43a10c7 100644
--- a/protoBuilds/06e5b6/README.json
+++ b/protoBuilds/06e5b6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Crystallization"
@@ -10,7 +10,7 @@
"internal": "06e5b6",
"labware": "\nEppendorf 96 Well Plate 500 \u00b5L #0030501136\nGreiner Bio-One CRYSTALQUICK LP 96 Well Plate 140ul #609171\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 24 Well Aluminum Block with NEST 2 mL Snapcap\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Crystallization\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs protein crystallization on 96-samples.\n\n\n",
diff --git a/protoBuilds/072463/README.json b/protoBuilds/072463/README.json
index 95757da2b7..003762d63d 100644
--- a/protoBuilds/072463/README.json
+++ b/protoBuilds/072463/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "072463",
"labware": "\nRRL 1 Well Plate 180000 \u00b5L #v 1.0\nRRL Custom 40 Well Plate 1500 \u00b5L #v 1.0\nZinsser 96 Well Plate 1898 \u00b5L\nHellma 96 Well Plate 300ul\nUSA Scientific 96 Deep Well Plate 2.4 mL #1896-2000\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 96 Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs the Simplified Fe Quantification Assay. If in Test Mode, the protocol will skip over all incubations and temperature changes. The protocol will automatically pause if tips have to be replaced, prompting the user. Pauses are included according to the protocol.\n\n\n",
diff --git a/protoBuilds/075bee/README.json b/protoBuilds/075bee/README.json
index 457fc596da..db586ef729 100644
--- a/protoBuilds/075bee/README.json
+++ b/protoBuilds/075bee/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "075bee",
"labware": "\nTWD_Tradewinds 24 Tube Rack with 4mL_Scintillation_Vials 4 mL\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 15 Tube Rack with Falcon 15 mL Conical\nOpentrons 10 Tube Rack with Falcon 4x50 mL, 6x15 mL Conical\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol preps up to (4) 24 tube custom tube racks with unique volumes of reagents 1-18 coming from a 15mL tube rack, as well as a 50mL tube rack. CSV's for destination custom racks should all include the following header `Rack, Well, Label, DEAEMA (mL), AEMAm (mL), DIPAEMA (mL), DMAEMA (mL), MPC (mL), PEGMA500 (mL), HEMA (mL), PEGMA300 (mL), PEGMA950 (mL), GlyMA (mL), PHPMA (mL), Initiator (mL), CTA (mL), Initiator 2 (mL), CTA 2 (mL), Solv (mL), Solv2, Solv3`. Headers may include whatever nomenclature is desired, but must have the same number of columns. The initial volumes CSV informs the initial volume of all 15 tubes in slots 2 and 5, and includes the following as the header `Slot (2 or 5), Tube Position (A1, A2, etc.), Initial Volume (mL)`. Note: the initial volume CSV should include 18 rows not including the header - 15 for the 15 tube rack, and 3 for the 50mL rack. \n\n\n",
diff --git a/protoBuilds/076f67/README.json b/protoBuilds/076f67/README.json
index 76dc672ecc..4fa1e8cd8d 100644
--- a/protoBuilds/076f67/README.json
+++ b/protoBuilds/076f67/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "076f67",
"labware": "\nFalcon 96 Well Plate 200 \u00b5L\nCorning 96 Well Plate 360 \u00b5L Flat #3650\nAgilent 1 Well Reservoir 290 mL #201252-100\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n\n",
"description": "This protocol performs the SARS-COV2 (VSV) Neutralization Assay on the OT-2. The user may select how many source-destination plate pairs (1-3) for this protocol, as well as select labware. The protocol will pause if the tips are depleted, prompting the user to refill tips. \n\n\n",
diff --git a/protoBuilds/079db2/README.json b/protoBuilds/079db2/README.json
index 7eadf19be9..b16c7047cb 100644
--- a/protoBuilds/079db2/README.json
+++ b/protoBuilds/079db2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "079db2",
"labware": "\nCustom 24 Tube Rack\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol reformats tubes from up to 4 custom tube racks to a 96 deepwell plate. The user has the option to dispense 25ul or 50ul into the deepwell plate. The sample input starts at A11 of the deepwell plate, moving by row.\n\n\n",
diff --git a/protoBuilds/07c65a/README.json b/protoBuilds/07c65a/README.json
index 60b2648a20..c06834e1ac 100644
--- a/protoBuilds/07c65a/README.json
+++ b/protoBuilds/07c65a/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "07c65a",
"labware": "\nArmadillo 96 Well Plate 200 \u00b5L PCR Full Skirt #AB2396\nOpentrons 24 Well Aluminum Block with NEST 2 mL Snapcap\nOpentrons 96 Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol dilutes sample with water at 4C, mixing with the transfer volume for 3 repetitions upon dispensing. The multi-channel pipette will be used as a single for this protocol.\n\n* `CSV Sample Format`: Below, you should upload a .csv file formatted in the following way, being sure to include the header line:\n```\nDestination Well, Transfer Volume\nA1, 5\nB1, 2\netc.\n```\n\n",
diff --git a/protoBuilds/07caf7/README.json b/protoBuilds/07caf7/README.json
index ff2e16a6ba..7758e34c3e 100644
--- a/protoBuilds/07caf7/README.json
+++ b/protoBuilds/07caf7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Mass Spec"
@@ -10,7 +10,7 @@
"internal": "07caf7",
"labware": "\nAxygen 96 Well PCR Microplate 200uL #PCR-96-FS-C\nGreiner Bio-One 96 Well Plate 2000 \u00b5L #780285\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\nNEST 96 Deepwell Plate 2mL #503001\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Mass Spec\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a custom mass spec sample prep.\n\n\n",
diff --git a/protoBuilds/08207f/README.json b/protoBuilds/08207f/README.json
index 9a901a70b4..7ac1d38966 100644
--- a/protoBuilds/08207f/README.json
+++ b/protoBuilds/08207f/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Normalization": [
"Normalization and Pooling"
@@ -10,7 +10,7 @@
"internal": "08207f",
"labware": "\nEppendorf Twin.tec 96 Well Plate 150 \u00b5L #0030129512\nEppendorf Twin.Tec 96 on Aluminum Block 150 \u00b5L\nStarlab 96-Well PCR Plate, Skirted 200 \u00b5L #E1403-5200\nStarlab 96-Well PCR Plate on Aluminum Block 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 10 Tube Rack with Falcon 4x50 mL, 6x15 mL Conical\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Normalization\n\t* Normalization and Pooling\n\n\n",
"deck-setup": "\n\n\n\n",
"description": "This is a normalization protocol that transfers diluent buffer from a 50 mL tube into an empty 96-well PCR plate, then transfers sample from a 96-well PCR plate on the temperature module into the plate with the diluent.\n\n\n",
diff --git a/protoBuilds/0845ab/README.json b/protoBuilds/0845ab/README.json
index 5860134120..7d926383bb 100644
--- a/protoBuilds/0845ab/README.json
+++ b/protoBuilds/0845ab/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "0845ab",
"labware": "\nTMTpro18 PCR strip plate #72.985.002\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nOpentrons 24 Tube Rack with Eppendorf 2 mL Safe-Lock Snapcap\nOpentrons 96 Tip Rack 300 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Tip Rack 20 \u00b5L\nNEST 1 Well Reservoir 195 mL #360103\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocols performs a blead cleanup on the OT-2 with overnight digestion step for 16, 32, or 48 samples. For detailed description of requirements and steps, please see below.\n\n",
diff --git a/protoBuilds/0849c2/README.json b/protoBuilds/0849c2/README.json
index 3e6595dc2d..1619d680c8 100644
--- a/protoBuilds/0849c2/README.json
+++ b/protoBuilds/0849c2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Extraction"
@@ -10,7 +10,7 @@
"internal": "0849c2",
"labware": "\nBio-Rad 96 Well Plate 350 \u00b5L #HSS9601\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Nucleic Acid Extraction\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a custom nucleic acid purification on up to 96 samples using the Opentrons Magnetic Module GEN2 and [Beckman Coulter Ampure XP Reagent for PCR Purification](https://www.beckman.com/reagents/genomic/cleanup-and-size-selection/pcr). The user is prompted to refill tipracks when necessary.\n\n\n",
diff --git a/protoBuilds/08878b/README.json b/protoBuilds/08878b/README.json
index 7c723e3dc4..f3194adf6a 100644
--- a/protoBuilds/08878b/README.json
+++ b/protoBuilds/08878b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol utilizes the P300 (GEN2) and P1000 pipettes to transfer and mix reagents between their original container and destination plate.\nThe protocol begins by making four (4) dilutions of reagent with heparin or sample. These dilutions are then added to a PCR plate on the temperature module and other reagents are added.\nNote about Temperature Module: The temperature of the Temperature Module can now be set in the Opentrons App. To expedite a heating or cooling step in your protocol, you should set your Temperature Module to the desired temperature in the Opentrons App, prior to running your protocol.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP300 Single GEN2 Pipette\nP1000 Single Pipette\nOpentrons 300\u00b5L Tiprack\nOpentrons 1000\u00b5L Tiprack\nOpentrons Temperature Module with Aluminum Block Set\nBio-Rad 96-Well Plate 200\u00b5L PCR\nOpentrons 4-in-1 Tube Rack Set (2mL and 15mL+50mL tops)\nTubes (2mL, 15mL, and 50mL) for Tube Rack\n\n\n\nFor this protocol, be sure that the pipettes (P300 Single GEN2 and P1000 Single) are attached.\nUsing the customization fields below, set up your protocol.\n P300 Single GEN2 Mount: Specify which mount the P300 is on (left or right).\n P1000 Single Mount: Specify which mount the P1000 is on (left or right.\n* Incubation Time (in minutes): Specify how many minutes to have have the robot wait (incubate) after adding each reagent to the samples on the temperature module.\nSlot 4: Opentrons Temperature Module with 96-Well Aluminum Block (from set) and Bio-Rad Plate, clean and empty\nSlot 5: 15mL+50mL Tube Rack\n * A1: Reagent 1 (15mL Tube)\n * A2: Reagent 2 (15mL Tube)\n * A3: Reagent 4 (50mL Tube)\n * B1: Reagent 3 (15mL Tube)\n * B2: 20% Acetic Acid (15mL Tube)\nSlot 6: 2mL Tube Rack with 2mL tubes in A1-A5 and B1-B5\n * A1: Heparin Standard\n * B1: Sample\n * Remaining tubes should be empty to begin\nSlot 5: 12 Column Reservoir\n * Column 1: Proeinase K\n * Column 3: Lysis Solution\n * Column 5: Magnetic Beads\n * Column 7: Elution\nSlot 8: Opentrons 300\u00b5L Tiprack\nSlot 9: Opentrons 1000\u00b5L Tiprack\nSlot 11: Opentrons 300\u00b5L Tiprack",
"internal": "08878b",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol utilizes the P300 (GEN2) and P1000 pipettes to transfer and mix reagents between their original container and destination plate.\n\nThe protocol begins by making four (4) dilutions of reagent with heparin or sample. These dilutions are then added to a PCR plate on the temperature module and other reagents are added.\n\n**Note about Temperature Module**: The temperature of the Temperature Module can now be set in the Opentrons App. To expedite a heating or cooling step in your protocol, you should set your Temperature Module to the desired temperature in the Opentrons App, prior to running your protocol.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P300 Single GEN2 Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P1000 Single Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 1000\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [Aluminum Block Set](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set)\n* [Bio-Rad 96-Well Plate 200\u00b5L PCR](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) (2mL and 15mL+50mL tops)\n* Tubes (2mL, 15mL, and 50mL) for Tube Rack\n\n\n\n---\n\n\nFor this protocol, be sure that the pipettes (P300 Single GEN2 and P1000 Single) are attached.\n\nUsing the customization fields below, set up your protocol.\n* P300 Single GEN2 Mount: Specify which mount the P300 is on (left or right).\n* P1000 Single Mount: Specify which mount the P1000 is on (left or right.\n* Incubation Time (in minutes): Specify how many minutes to have have the robot wait (incubate) after adding each reagent to the samples on the temperature module.\n\nSlot 4: [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [96-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) (from set) and [Bio-Rad Plate](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate), clean and empty\n\nSlot 5: [15mL+50mL Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n\t* A1: Reagent 1 (15mL Tube)\n\t* A2: Reagent 2 (15mL Tube)\n\t* A3: Reagent 4 (50mL Tube)\n\t* B1: Reagent 3 (15mL Tube)\n\t* B2: 20% Acetic Acid (15mL Tube)\n\nSlot 6: [2mL Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 2mL tubes in A1-A5 and B1-B5\n\t* A1: Heparin Standard\n\t* B1: Sample\n\t* Remaining tubes should be empty to begin\n\nSlot 5: [12 Column Reservoir](https://www.agilent.com/store/en_US/LCat-SubCat1ECS_112089/Reservoirs)\n\t* Column 1: Proeinase K\n\t* Column 3: Lysis Solution\n\t* Column 5: Magnetic Beads\n\t* Column 7: Elution\n\nSlot 8: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 9: [Opentrons 1000\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n\nSlot 11: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n",
"internal": "08878b\n",
diff --git a/protoBuilds/089108/README.json b/protoBuilds/089108/README.json
index 4b56e34651..04b55887a5 100644
--- a/protoBuilds/089108/README.json
+++ b/protoBuilds/089108/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NUCLEIC ACID EXTRACTION & PURIFICATION": [
"Nucleic Acid Purification"
@@ -10,7 +10,7 @@
"internal": "089108",
"labware": "\nNEST 1 Well Reservoir\nOpentrons 300ul Tips\nNEST 1 Well Reservoir\nUSA Scientific 96 Deepwell Plate 2.4mL\nCustom 96 plates\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NUCLEIC ACID EXTRACTION & PURIFICATION\n\t* Nucleic Acid Purification\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This protocol performs the [96-well Plasmid ezFilter Miniprep Kit](https://www.bioland-sci.com/index.php?main_page=product_info&products_id=137&zenid=6d2c9844db2a12e37e573e104fedbba5). For a detailed list of protocol steps, please see below.\n\nExplanation of complex parameters below:\n* `Number of plates`: Specify whether using 1 (slot 2 alone) or two (slots 2 and 5) plates for this run.\n* `Number of Samples Plate 1 & 2`: Select the number of samples per plate. If the number of plates variable = 1 above, then the number of samples for plate 2 will be ignored.\n* `P300 Mount`: Specify which mount (left or right) to host the P300 Multi-Channel Pipette\n\n---\n\n",
diff --git a/protoBuilds/08aeaa/README.json b/protoBuilds/08aeaa/README.json
index c841ab170f..0da3184f4c 100644
--- a/protoBuilds/08aeaa/README.json
+++ b/protoBuilds/08aeaa/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -8,7 +8,7 @@
"description": "\nThis protocol performs the normalization of samples by adjusting the concentration in Micronic 96 rack. Once normalized it will transfer nuclease-free water to the corresponding wells in a PCR plate and then transfer samples to the corresponding wells in the PCR plate.\nIn this protocol, the ThermoFisher Nunc 96 Well Plate 2mL is used as the reservoir for water. You can define the volume of water placed in each well at the Water Reservoir Volume per Well parameter below. By default this value is 1500 uL. The robot will assume that each well it needs has a volume of 1500 uL and will keep track of how much water it has used per well before moving onto a new well. While, it keeps track of the volume per well, it will also adjust the height of how low the pipette moves to aspirate.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 App\nOpentrons Single-Channel Pipettes and corresponding Tips\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nCSV Format\nYour file must be saved as a comma separated value (.csv) file type. Your CSV must contain values corresponding to volumes in microliters (\u03bcL). Below is an example of the format for the CSV.\nPlate position,ng/ul,Volume,ng TOT,ul Final Dil,ul Water\nA1,19.9,15,298.7,19.9,4.9\nB1,22.4,15,335.5,22.4,7.4\nC1,22.2,15,332.9,22.2,7.2",
"internal": "08aeaa",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"description": "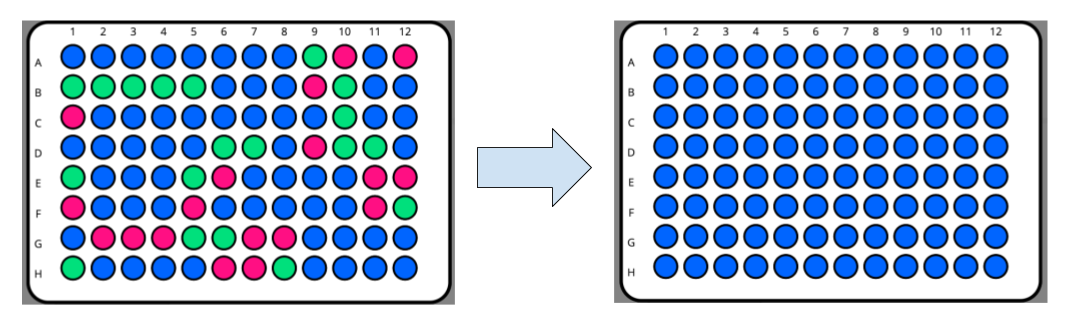\n\nThis protocol performs the normalization of samples by adjusting the concentration in Micronic 96 rack. Once normalized it will transfer nuclease-free water to the corresponding wells in a PCR plate and then transfer samples to the corresponding wells in the PCR plate.\n\nIn this protocol, the ThermoFisher Nunc 96 Well Plate 2mL is used as the reservoir for water. You can define the volume of water placed in each well at the `Water Reservoir Volume per Well` parameter below. By default this value is 1500 uL. The robot will assume that each well it needs has a volume of 1500 uL and will keep track of how much water it has used per well before moving onto a new well. While, it keeps track of the volume per well, it will also adjust the height of how low the pipette moves to aspirate.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 App](https://opentrons.com/ot-app/)\n* [Opentrons Single-Channel Pipettes](https://shop.opentrons.com/collections/ot-2-pipettes) and corresponding [Tips](https://shop.opentrons.com/collections/opentrons-tips)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n---\n\n\n**CSV Format**\n\nYour file must be saved as a comma separated value (.csv) file type. Your CSV must contain values corresponding to volumes in microliters (\u03bcL). Below is an example of the format for the CSV.\n\n\n```\nPlate position,ng/ul,Volume,ng TOT,ul Final Dil,ul Water\nA1,19.9,15,298.7,19.9,4.9\nB1,22.4,15,335.5,22.4,7.4\nC1,22.2,15,332.9,22.2,7.2\n```\n\n",
"internal": "08aeaa\n",
diff --git a/protoBuilds/08bbaf/README.json b/protoBuilds/08bbaf/README.json
index dc40f5185b..f2c4d64fd8 100644
--- a/protoBuilds/08bbaf/README.json
+++ b/protoBuilds/08bbaf/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Standard Curve"
@@ -9,7 +9,7 @@
"internal": "08bbaf",
"labware": "\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons 24 Well Aluminum Block with NEST 1.5 mL Snapcap\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Standard Curve\n\n\n",
"description": "This protocol performs up to 8 11-point standard curve preparations in blood media from up to 8 different stocks.\n\n\n",
"internal": "08bbaf\n",
diff --git a/protoBuilds/08e3eb/README.json b/protoBuilds/08e3eb/README.json
index 675d9901ec..4924665d35 100644
--- a/protoBuilds/08e3eb/README.json
+++ b/protoBuilds/08e3eb/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol automates the distribution of assays from a 48-well reservoir to 9, 96-well plates.\n\nUsing the customizable parameters below, you can specify which column to start picking up tips from and how many \"runs\" to perform. During each run, all 9 plates will be filled with assay from the reservoir. At the conclusion of filling the 9 plates, the OT-2 will flash, pause, and prompt the user to replace the plates on the deck.\n\nThis protocol uses custom labware definitions for the 48-well reservoir (Agilent) and 96-well plate (MicroAmp Endura). When downloading the protocol, the labware definitions (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: Using labware in your protocols\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P50 Multi-Channel Pipette\nOpentrons 50/300\u00b5L Tips\nAgilent 48-Well Reservoir\nMicroAmp Endura 96-Well Plate\nReagents\n\n\n\nSlot 1: MicroAmp Endura 96-Well Plate\nSlot 2: MicroAmp Endura 96-Well Plate\nSlot 3: MicroAmp Endura 96-Well Plate\nSlot 4: MicroAmp Endura 96-Well Plate\nSlot 5: MicroAmp Endura 96-Well Plate\nSlot 6: MicroAmp Endura 96-Well Plate\nSlot 7: MicroAmp Endura 96-Well Plate\nSlot 8: MicroAmp Endura 96-Well Plate\nSlot 9: MicroAmp Endura 96-Well Plate\nSlot 10: Opentrons 50/300\u00b5L Tips\nSlot 11: Agilent 48-Well Reservoir with Reagents\n\n\nUsing the customizations field (below), set up your protocol.\n Starting Tip Column: Specify which column of tips to begin drawing from. Over the course of the protocol, the multi-channel pipette will pick up tips from\n P300-Multi Mount: Select which mount (left or right) the P300-Multi is attached to.",
"internal": "08e3eb",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol automates the distribution of assays from a 48-well reservoir to 9, 96-well plates.\n\nUsing the customizable parameters below, you can specify which column to start picking up tips from and how many *\"runs\"* to perform. During each *run*, all 9 plates will be filled with assay from the reservoir. At the conclusion of filling the 9 plates, the OT-2 will flash, pause, and prompt the user to replace the plates on the deck.\n\nThis protocol uses custom labware definitions for the 48-well reservoir (Agilent) and 96-well plate (MicroAmp Endura). When downloading the protocol, the labware definitions (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: [Using labware in your protocols](https://support.opentrons.com/en/articles/3136506-using-labware-in-your-protocols)\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P50 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* Agilent 48-Well Reservoir\n* MicroAmp Endura 96-Well Plate\n* Reagents\n\n\n\n---\n\n\nSlot 1: MicroAmp Endura 96-Well Plate\n\nSlot 2: MicroAmp Endura 96-Well Plate\n\nSlot 3: MicroAmp Endura 96-Well Plate\n\nSlot 4: MicroAmp Endura 96-Well Plate\n\nSlot 5: MicroAmp Endura 96-Well Plate\n\nSlot 6: MicroAmp Endura 96-Well Plate\n\nSlot 7: MicroAmp Endura 96-Well Plate\n\nSlot 8: MicroAmp Endura 96-Well Plate\n\nSlot 9: MicroAmp Endura 96-Well Plate\n\nSlot 10: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 11: Agilent 48-Well Reservoir with Reagents\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Starting Tip Column**: Specify which column of tips to begin drawing from. Over the course of the protocol, the multi-channel pipette will pick up tips from\n* **P300-Multi Mount**: Select which mount (left or right) the P300-Multi is attached to.\n\n\n\n",
"internal": "08e3eb\n",
diff --git a/protoBuilds/08fd01-pt2/README.json b/protoBuilds/08fd01-pt2/README.json
index beb279e42e..27acd17fed 100644
--- a/protoBuilds/08fd01-pt2/README.json
+++ b/protoBuilds/08fd01-pt2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "08fd01-pt2",
"labware": "\nNest 12 Well Reservoir\nOpentrons 20ul Filter tips\nCustom 384 well plate\n96 Index Plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "This protocol is Stage 2 of a 3 part PCR Prep protocol. Parts 1 and 3 can be found below. For detailed protocol details, please reference the \"Protocol Steps\" section.\n\n[Stage 1 Protocol](https://protocols.opentrons.com/protocol/08fd01)\n[Stage 3 Protocol](https://protocols.opentrons.com/protocol/08fd01-pt3)\n\n---\n\n",
diff --git a/protoBuilds/08fd01-pt3/README.json b/protoBuilds/08fd01-pt3/README.json
index ecdbf162a1..fc6273d28f 100644
--- a/protoBuilds/08fd01-pt3/README.json
+++ b/protoBuilds/08fd01-pt3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "08fd01-pt3",
"labware": "\nOpentrons 20ul Filter tips\nCustom 384 well plate(link to labware on shop.opentrons.com when applicable)\nNest 96 Wellplate 100ul PCR Full Skirt\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "This protocol is Stage 3 of a 3 part PCR Prep protocol. Parts 1 and 2 can be found below. For detailed protocol details, please reference the \"Protocol Steps\" section.\n\n[Stage 1 Protocol](https://protocols.opentrons.com/protocol/08fd01)\n[Stage 2 Protocol](https://protocols.opentrons.com/protocol/08fd01-pt2)\n\n---\n\n",
diff --git a/protoBuilds/08fd01/README.json b/protoBuilds/08fd01/README.json
index 59c3190857..7328e4b123 100644
--- a/protoBuilds/08fd01/README.json
+++ b/protoBuilds/08fd01/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "08fd01",
"labware": "\nNest 12 Well Reservoir](https://shop.opentrons.com/consumables/\nOpentrons 20ul Filter tips\nCustom 384 well plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "This protocol is Stage 1 of a 3 part PCR Prep protocol. Parts 2 and 3 can be found below. For detailed protocol details, please reference the \"Protocol Steps\" section. This protocol pools 4 plates one at a time into a fresh 384 well plate with water. Each PCR plate is pooled in order A1, A2, B1, B2 to A1 pool, then A3, A4, B3, B4 to A3 pool, etc. The next plate will pool all of the same source wells, but the destination wells of the pool will start at B1, skipping every other well. PCR plate 3 will start pooling at B1, and PCR plate wills start pooling at B2.\n\n[Stage 2 Protocol](https://protocols.opentrons.com/protocol/08fd01-pt2)\n[Stage 3 Protocol](https://protocols.opentrons.com/protocol/08fd01-pt3)\n\n---\n\n",
diff --git a/protoBuilds/0909e6/README.json b/protoBuilds/0909e6/README.json
index fc8aebd602..0ab3e64517 100644
--- a/protoBuilds/0909e6/README.json
+++ b/protoBuilds/0909e6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sterile Workflows": [
"Cell Culture"
@@ -10,7 +10,7 @@
"internal": "0909e6",
"labware": "\nOpentrons 6 Tube Rack with Falcon 50 mL Conical\nUSA Scientific 96 Deep Well Plate 2.4 mL #1896-2000\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 1000 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sterile Workflows\n\t* Cell Culture\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a custom 12-factor cell culture assay in a 96-deepwell plate. Please use [this example .csv template](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/0909e6/ex4.csv) to format your worklist! When uploading your file below, please ensure it is saved in the format `.csv`.\n\n\n",
diff --git a/protoBuilds/090c2e/README.json b/protoBuilds/090c2e/README.json
index e8489bb742..0aaa9e6b39 100644
--- a/protoBuilds/090c2e/README.json
+++ b/protoBuilds/090c2e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Purification": [],
"Omega Bio-Tek Mag-Bind Environmental DNA 96 Kit": []
@@ -9,7 +9,7 @@
"internal": "090c2e",
"labware": "\nQIAGEN 96 Tube Rack 1.2mL #19560\nNEST 12 Well Reservoir 15 mL #360102\nNEST 1 Well Reservoir 195 mL #360103\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\n",
"markdown": {
- "author": "\n[Opentrons](https://opentrons.com/)\n\n",
+ "author": "\n[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "\n- Nucleic Acid Purification\n - Omega Bio-Tek Mag-Bind Environmental DNA 96 Kit\n\n",
"deck-setup": "\n\n\n",
"description": "\nThis protocol performs a nucleic acid purification according to the [Omega Bio-Tek Mag-Bind Environmental DNA 96 Kit](https://s3.amazonaws.com/pf-user-files-01/u-4256/uploads/2023-03-14/is13ts3/QMF27.0248.M5645%20v9.0%202.pdf).\n\n",
diff --git a/protoBuilds/0944e4/README.json b/protoBuilds/0944e4/README.json
index 977b48de11..6d3689566e 100644
--- a/protoBuilds/0944e4/README.json
+++ b/protoBuilds/0944e4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Ampure XP"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom NGS library cleanup using Ampure XP beads in a variable ratio with samples. The samples are mounted on an Opentrons magnetic module, and the final elute is transferred to a fresh PCR plate.\n\n\n\nOpentrons Magnetic Module GEN2\nUSA Scientific 12-channel reservoir 22ml #1061-8150\nEppendorf twin.tec 96-well PCR plate #0030129512\nP20 multi-channel GEN2 electronic pipette\nP300 multi-channel electronic pipette\nOpentrons 20ul filter tiprack\nOpentrons 200ul filter tiprack\n\n\n\n12-channel reservoir (slot 7)\n channel 1: Ampure XP beads\n channel 2: 80% ethanol\n channel 3: EB buffer\n channels 11-12: liquid waste (loaded empty)",
"internal": "0944e4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Ampure XP\n\n",
"description": "This protocol performs a custom NGS library cleanup using Ampure XP beads in a variable ratio with samples. The samples are mounted on an Opentrons magnetic module, and the final elute is transferred to a fresh PCR plate.\n\n---\n\n\n* [Opentrons Magnetic Module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [USA Scientific 12-channel reservoir 22ml #1061-8150](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n* [Eppendorf twin.tec 96-well PCR plate #0030129512](https://www.fishersci.com/shop/products/eppendorf-twin-tec-96-lobind-pcr-plates-skirted-clear/e0030129512)\n* [P20 multi-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [P300 multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 20ul filter tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-filter-tip)\n* [Opentrons 200ul filter tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n\n---\n\n\n12-channel reservoir (slot 7)\n* channel 1: Ampure XP beads\n* channel 2: 80% ethanol\n* channel 3: EB buffer\n* channels 11-12: liquid waste (loaded empty)\n\n",
"internal": "0944e4\n",
diff --git a/protoBuilds/0961d2-part1/README.json b/protoBuilds/0961d2-part1/README.json
index 54849e3218..0eca1c134e 100644
--- a/protoBuilds/0961d2-part1/README.json
+++ b/protoBuilds/0961d2-part1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"plexWell LP384"
@@ -8,7 +8,7 @@
"description": "This protocol is part 1 of a 4-part protocol designed to accomplish the Sample-Barcoding Module of the plexWell 384 Low Pass Library Preparation.\nPart 1\nPart 2\nPart 3\nPart 4\nThis protocol accomplishes Step 1 (Sample-Barcoding, SB, Reaction Set-up). During this protocol, 6\u00b5L of input DNA in each well of two 96-well plates is transferred to the SBP96 plate containing specific reagents. 5\u00b5L of Coding Buffer (3X) is then added to each well of the SBP96 plate.\n\n\n\nplexWell LP 384 Kit\nSBP96 Plate\nCoding Buffer (3X)\nOpentrons P10 Multi Channel Pipette\nBio-Rad 96-Well Plate, 200\u00b5L PCR, containing samples\nNEST 12-Well Reservoir, 15mL\nOpentrons 10\u00b5L Filter Tips\n\n\n\nSlot 1: Opentrons 10\u00b5L Filter Tips\nSlot 2: Bio-Rad Plate, containing samples (Source 1)\nSlot 3: SBP96 Plate (Destination 1)\nSlot 4: Opentrons 10\u00b5L Filter Tips\nSlot 5: Bio-Rad Plate, containing samples (Source 2)\nSlot 6: SBP96 Plate (Destination 2)\nSlot 7: Opentrons 10\u00b5L Filter Tips\nSlot 8: Empty\nSlot 9: Nest 12-Well Reservoir\n* Column 1: Coding Buffer (3X)\nSlot 10: Opentrons 10\u00b5L Filter Tips",
"internal": "0961d2-part1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* plexWell LP384\n\n\n",
"description": "This protocol is part 1 of a 4-part protocol designed to accomplish the Sample-Barcoding Module of the [plexWell 384 Low Pass Library Preparation](https://seqwell.com/products/plexwell-lp-384/).\n\n\n[Part 1](https://protocols.opentrons.com/protocol/0961d2-part1)\n[Part 2](https://protocols.opentrons.com/protocol/0961d2-part2)\n[Part 3](https://protocols.opentrons.com/protocol/0961d2-part3)\n[Part 4](https://protocols.opentrons.com/protocol/0961d2-part4)\n\n\nThis protocol accomplishes Step 1 (Sample-Barcoding, SB, Reaction Set-up). During this protocol, 6\u00b5L of input DNA in each well of two 96-well plates is transferred to the SBP96 plate containing specific reagents. 5\u00b5L of Coding Buffer (3X) is then added to each well of the SBP96 plate.\n\n---\n\n\n* [plexWell LP 384 Kit](https://seqwell.com/products/plexwell-lp-384/)\n- SBP96 Plate\n- Coding Buffer (3X)\n* [Opentrons P10 Multi Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Bio-Rad 96-Well Plate, 200\u00b5L PCR](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate), containing samples\n* [NEST 12-Well Reservoir, 15mL](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* [Opentrons 10\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-filter-tip)\n\n\n---\n\n\nSlot 1: Opentrons 10\u00b5L Filter Tips\n\nSlot 2: Bio-Rad Plate, containing samples (Source 1)\n\nSlot 3: SBP96 Plate (Destination 1)\n\nSlot 4: Opentrons 10\u00b5L Filter Tips\n\nSlot 5: Bio-Rad Plate, containing samples (Source 2)\n\nSlot 6: SBP96 Plate (Destination 2)\n\nSlot 7: Opentrons 10\u00b5L Filter Tips\n\nSlot 8: *Empty*\n\nSlot 9: Nest 12-Well Reservoir\n* Column 1: Coding Buffer (3X)\n\nSlot 10: Opentrons 10\u00b5L Filter Tips\n\n\n",
"internal": "0961d2-part1\n",
diff --git a/protoBuilds/0961d2-part2/README.json b/protoBuilds/0961d2-part2/README.json
index 13bcdb8d5e..63f4b52cb6 100644
--- a/protoBuilds/0961d2-part2/README.json
+++ b/protoBuilds/0961d2-part2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"plexWell LP384"
@@ -8,7 +8,7 @@
"description": "This protocol is part 2 of a 4-part protocol designed to accomplish the Sample-Barcoding Module of the plexWell 384 Low Pass Library Preparation.\nPart 1\nPart 2\nPart 3\nPart 4\nThis protocol accomplishes Step 2 (SB Reaction Stop). During this protocol, 7.5\u00b5L of X Solution is added to each well of a 96-Well Plate (SBP96), containing samples from Part 1. With this protocol, you are given the option to use 1-4 plates for this step, for higher throughputs. The SBP96 plates should be loaded in the column containing slots 2, 5, 8, and 11 (sequentially). For each plate, a tip rack should be placed in the adjoining slot (slots 1, 4, 7, and 10).\n\n\n\nplexWell LP 384 Kit\nSBP96 Plate, containing samples from Part 1\nX Solution\nOpentrons P10 Multi Channel Pipette\nNEST 12-Well Reservoir, 15mL\nOpentrons 10\u00b5L Filter Tips\n\n\n\nSlot 1: Opentrons 10\u00b5L Filter Tips\nSlot 2: SBP96 Plate, containing samples\nSlot 3: Empty\nSlot 4: Opentrons 10\u00b5L Filter Tips (optional)\nSlot 5: SBP96 Plate, containing samples (optional)\nSlot 6: Empty\nSlot 7: Opentrons 10\u00b5L Filter Tips (optional)\nSlot 8: SBP96 Plate, containing samples (optional)\nSlot 9: Nest 12-Well Reservoir\n* Column 2: X Solution\nSlot 10: Opentrons 10\u00b5L Filter Tips (optional)\nSlot 11: SBP96 Plate, containing samples (optional)",
"internal": "0961d2-part2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* plexWell LP384\n\n\n",
"description": "This protocol is part 2 of a 4-part protocol designed to accomplish the Sample-Barcoding Module of the [plexWell 384 Low Pass Library Preparation](https://seqwell.com/products/plexwell-lp-384/).\n\n\n[Part 1](https://protocols.opentrons.com/protocol/0961d2-part1)\n[Part 2](https://protocols.opentrons.com/protocol/0961d2-part2)\n[Part 3](https://protocols.opentrons.com/protocol/0961d2-part3)\n[Part 4](https://protocols.opentrons.com/protocol/0961d2-part4)\n\n\nThis protocol accomplishes Step 2 (SB Reaction Stop). During this protocol, 7.5\u00b5L of X Solution is added to each well of a 96-Well Plate (SBP96), containing samples from [Part 1](https://protocols.opentrons.com/protocol/0961d2-part1). With this protocol, you are given the option to use 1-4 plates for this step, for higher throughputs. The SBP96 plates should be loaded in the column containing slots 2, 5, 8, and 11 (sequentially). For each plate, a tip rack should be placed in the adjoining slot (slots 1, 4, 7, and 10).\n\n---\n\n\n* [plexWell LP 384 Kit](https://seqwell.com/products/plexwell-lp-384/)\n- SBP96 Plate, containing samples from [Part 1](https://protocols.opentrons.com/protocol/0961d2-part1)\n- X Solution\n* [Opentrons P10 Multi Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [NEST 12-Well Reservoir, 15mL](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* [Opentrons 10\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-filter-tip)\n\n\n---\n\n\nSlot 1: Opentrons 10\u00b5L Filter Tips\n\nSlot 2: SBP96 Plate, containing samples\n\nSlot 3: *Empty*\n\nSlot 4: Opentrons 10\u00b5L Filter Tips (optional)\n\nSlot 5: SBP96 Plate, containing samples (optional)\n\nSlot 6: *Empty*\n\nSlot 7: Opentrons 10\u00b5L Filter Tips (optional)\n\nSlot 8: SBP96 Plate, containing samples (optional)\n\nSlot 9: Nest 12-Well Reservoir\n* Column 2: X Solution\n\nSlot 10: Opentrons 10\u00b5L Filter Tips (optional)\n\nSlot 11: SBP96 Plate, containing samples (optional)\n\n\n",
"internal": "0961d2-part2\n",
diff --git a/protoBuilds/0961d2-part3/README.json b/protoBuilds/0961d2-part3/README.json
index 2fb4cca956..888b2636ec 100644
--- a/protoBuilds/0961d2-part3/README.json
+++ b/protoBuilds/0961d2-part3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"plexWell LP384"
@@ -8,7 +8,7 @@
"description": "This protocol is part 3 of a 4-part protocol designed to accomplish the Sample-Barcoding Module of the plexWell 384 Low Pass Library Preparation.\nPart 1\nPart 2\nPart 3\nPart 4\nThis protocol performs the first half of Step 3 (SB Pooling). During this protocol, 1-4 SBP96 plates from Part 2 can be loaded on to the deck of the OT-2. Every six columns from the SBP96 plates are pooled into one column of a clean, 96-well plate (columns 1-6 of SBP96 plate 1 are pooled into column 1 of clean plate; columns 7-12 of SBP96 plate 1 are pooled into column 2 of clean plate; columns 1-6 of SBP96 plate 2 are pooled into column 3 of clean plate, etc).\n\n\n\nplexWell LP 384 Kit\nSBP96 Plate, containing samples from Part 2\nOpentrons P10 Multi Channel Pipette\nBio-Rad 96-Well Plate, 200\u00b5L PCR, clean and empty\nOpentrons 10\u00b5L Filter Tips\n\n\n\nSlot 1: Opentrons 10\u00b5L Filter Tips\nSlot 2: SBP96 Plate, containing samples\nSlot 3: Bio-Rad Plate, clean and empty\nSlot 4: Opentrons 10\u00b5L Filter Tips (optional)\nSlot 5: SBP96 Plate, containing samples (optional)\nSlot 6: Empty\nSlot 7: Opentrons 10\u00b5L Filter Tips (optional)\nSlot 8: SBP96 Plate, containing samples (optional)\nSlot 9: Empty\nSlot 10: Opentrons 10\u00b5L Filter Tips (optional)\nSlot 11: SBP96 Plate, containing samples (optional)",
"internal": "0961d2-part3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* plexWell LP384\n\n\n",
"description": "This protocol is part 3 of a 4-part protocol designed to accomplish the Sample-Barcoding Module of the [plexWell 384 Low Pass Library Preparation](https://seqwell.com/products/plexwell-lp-384/).\n\n\n[Part 1](https://protocols.opentrons.com/protocol/0961d2-part1)\n[Part 2](https://protocols.opentrons.com/protocol/0961d2-part2)\n[Part 3](https://protocols.opentrons.com/protocol/0961d2-part3)\n[Part 4](https://protocols.opentrons.com/protocol/0961d2-part4)\n\n\nThis protocol performs the first half of Step 3 (SB Pooling). During this protocol, 1-4 SBP96 plates from [Part 2](https://protocols.opentrons.com/protocol/0961d2-part2) can be loaded on to the deck of the OT-2. Every six columns from the SBP96 plates are pooled into one column of a clean, 96-well plate (columns 1-6 of SBP96 plate 1 are pooled into column 1 of clean plate; columns 7-12 of SBP96 plate 1 are pooled into column 2 of clean plate; columns 1-6 of SBP96 plate 2 are pooled into column 3 of clean plate, etc).\n\n---\n\n\n* [plexWell LP 384 Kit](https://seqwell.com/products/plexwell-lp-384/)\n- SBP96 Plate, containing samples from [Part 2](https://protocols.opentrons.com/protocol/0961d2-part2)\n* [Opentrons P10 Multi Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Bio-Rad 96-Well Plate, 200\u00b5L PCR](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate), clean and empty\n* [Opentrons 10\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-filter-tip)\n\n\n---\n\n\nSlot 1: Opentrons 10\u00b5L Filter Tips\n\nSlot 2: SBP96 Plate, containing samples\n\nSlot 3: Bio-Rad Plate, clean and empty\n\nSlot 4: Opentrons 10\u00b5L Filter Tips (optional)\n\nSlot 5: SBP96 Plate, containing samples (optional)\n\nSlot 6: *Empty*\n\nSlot 7: Opentrons 10\u00b5L Filter Tips (optional)\n\nSlot 8: SBP96 Plate, containing samples (optional)\n\nSlot 9: *Empty*\n\nSlot 10: Opentrons 10\u00b5L Filter Tips (optional)\n\nSlot 11: SBP96 Plate, containing samples (optional)\n\n\n",
"internal": "0961d2-part3\n",
diff --git a/protoBuilds/0961d2-part4/README.json b/protoBuilds/0961d2-part4/README.json
index a0a51c4dd7..2372ab4d6f 100644
--- a/protoBuilds/0961d2-part4/README.json
+++ b/protoBuilds/0961d2-part4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"plexWell LP384"
@@ -8,7 +8,7 @@
"description": "This protocol is part 4 of a 4-part protocol designed to accomplish the Sample-Barcoding Module of the plexWell 384 Low Pass Library Preparation.\nPart 1\nPart 2\nPart 3\nPart 4\nThis protocol performs the second half of Step 3 (SB Pooling). During this protocol, each column of the destination plate in Part 3 is pooled into a 2mL DNA LoBind tube. The user has the option to pool up to 12 columns (the entire plate) into individual tubes for each column.\n\n\n\nOpentrons P300 Single Channel Pipette (GEN2)\nBio-Rad 96-Well Plate, 200\u00b5L PCR, containing samples from Part 3\nOpentrons 200\u00b5L Filter Tips\nOpentrons 24 Tube Rack\n2mL DNA LoBind Tubes\n\n\n\nSlot 1: Opentrons 200\u00b5L Filter Tips\nSlot 2: Empty\nSlot 3: Bio-Rad Plate, containing samples\nSlot 4: Empty\nSlot 5: Empty\nSlot 6: Tube Rack with 2mL Tubes",
"internal": "0961d2-part4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* plexWell LP384\n\n\n",
"description": "This protocol is part 4 of a 4-part protocol designed to accomplish the Sample-Barcoding Module of the [plexWell 384 Low Pass Library Preparation](https://seqwell.com/products/plexwell-lp-384/).\n\n\n[Part 1](https://protocols.opentrons.com/protocol/0961d2-part1)\n[Part 2](https://protocols.opentrons.com/protocol/0961d2-part2)\n[Part 3](https://protocols.opentrons.com/protocol/0961d2-part3)\n[Part 4](https://protocols.opentrons.com/protocol/0961d2-part4)\n\n\nThis protocol performs the second half of Step 3 (SB Pooling). During this protocol, each column of the destination plate in [Part 3](https://protocols.opentrons.com/protocol/0961d2-part3) is pooled into a 2mL DNA LoBind tube. The user has the option to pool up to 12 columns (the entire plate) into individual tubes for each column.\n\n---\n\n\n* [Opentrons P300 Single Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [Bio-Rad 96-Well Plate, 200\u00b5L PCR](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate), containing samples from [Part 3](https://protocols.opentrons.com/protocol/0961d2-part3)\n* [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 24 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [2mL DNA LoBind Tubes](https://labware.opentrons.com/opentrons_24_tuberack_eppendorf_2ml_safelock_snapcap?category=tubeRack)\n\n\n---\n\n\nSlot 1: Opentrons 200\u00b5L Filter Tips\n\nSlot 2: *Empty*\n\nSlot 3: Bio-Rad Plate, containing samples\n\nSlot 4: *Empty*\n\nSlot 5: *Empty*\n\nSlot 6: Tube Rack with 2mL Tubes\n\n\n",
"internal": "0961d2-part4\n",
diff --git a/protoBuilds/0966ae/README.json b/protoBuilds/0966ae/README.json
index 18fdb80a4b..494718dea0 100644
--- a/protoBuilds/0966ae/README.json
+++ b/protoBuilds/0966ae/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sterile Workflows": [
"Cell Culture"
@@ -9,7 +9,7 @@
"internal": "0966ae",
"labware": "\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 24 Tube Rack with Eppendorf 2 mL Safe-Lock Snapcap\nAnanda 384 Well Plate 100 \u00b5L Flat\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sterile Workflows\n\t* Cell Culture\n\n\n",
"description": "This protocol performs a custom sterile 384-wellplate prep on up to 9 plates.\n\n\n",
"internal": "0966ae\n",
diff --git a/protoBuilds/09cdbe/README.json b/protoBuilds/09cdbe/README.json
index 3b8fc1f186..2e18d0f6da 100644
--- a/protoBuilds/09cdbe/README.json
+++ b/protoBuilds/09cdbe/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Consolidation"
@@ -8,7 +8,7 @@
"description": "This protocol performs a sample consolidation from .csv file. The file should be organized in the following way including header line:\nPlate Position,Source Well ID,Destination Tube ID,Volume,,\n1,A1,A1,3.0,,\n1,B1,A1,4.0,,\n1,C1,B1,5.0,,\n1,D1,A1,3.0,,\n...\nEmpty lines are ignored. You can download a template file here.\nThe destination tube for all sources will be a specified location (A1-D6) of the custom tuberack in slot 9.\n\n\n\nAbgene 96-Well Microplates 330\u00b5l (ThermoFisher #AB0796)\nAxygen\u2122 96-well PCR Microplates, barcoded 200\u00b5l #14-222-327\nAbgene\u2122 96 Well 2.2mL Polypropylene Deepwell Storage Plate\nOpentrons P20 GEN2 single-channel electronic pipette\nOpentrons 20\u00b5l tipracks\nVWR 5ml tubes seated in Opentrons 3x5 tuberack\n\n\n\n",
"internal": "09cdbe",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Consolidation\n\n",
"description": "This protocol performs a sample consolidation from `.csv` file. The file should be organized in the following way **including header line**:\n\n```\nPlate Position,Source Well ID,Destination Tube ID,Volume,,\n1,A1,A1,3.0,,\n1,B1,A1,4.0,,\n1,C1,B1,5.0,,\n1,D1,A1,3.0,,\n...\n```\n\nEmpty lines are ignored. You can download a template file [here](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2020-03-07/b303m2c/Verogen-CP-Example-File.xlsx).\n\nThe destination tube for all sources will be a specified location (A1-D6) of the custom tuberack in slot 9.\n\n---\n\n\n* [Abgene 96-Well Microplates 330\u00b5l (ThermoFisher #AB0796)](https://www.thermofisher.com/order/catalog/product/AB0796#/AB0796)\n* [Axygen\u2122 96-well PCR Microplates, barcoded 200\u00b5l #14-222-327](https://www.fishersci.com/shop/products/axygen-96-well-full-skirted-pcr-microplates-clear-sterilized/14222327)\n* [Abgene\u2122 96 Well 2.2mL Polypropylene Deepwell Storage Plate](https://www.thermofisher.com/order/catalog/product/AB0661#/AB0661)\n* [Opentrons P20 GEN2 single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 20\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* VWR 5ml tubes seated in [Opentrons 3x5 tuberack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n\n---\n\n\n\n\n",
"internal": "09cdbe\n",
diff --git a/protoBuilds/0a23c6/README.json b/protoBuilds/0a23c6/README.json
index dc71682d60..8d740b02bf 100644
--- a/protoBuilds/0a23c6/README.json
+++ b/protoBuilds/0a23c6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"qPCR setup"
@@ -8,7 +8,7 @@
"description": "This protocol automates the setup of a qPCR reaction using the Opentrons robots. It scales with quantity of samples, but has a practical lower limit of 24 samples. This is a pythonized version of a Protocol Designer protocol.\n\n\n\nOpentrons P300-single channel electronic pipettes (GEN2)\nOpentrons P20-multi channel electronic pipettes (GEN2)\nOpentrons 20ul Filter Tips\nOpentrons 200ul Filter Tips\nOpentrons Aluminum Block Set\n",
"internal": "0a23c6",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* qPCR setup\n\n",
"description": "This protocol automates the setup of a qPCR reaction using the Opentrons robots. It scales with quantity of samples, but has a practical lower limit of 24 samples. This is a pythonized version of a [Protocol Designer protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2020-09-21/tm23shu/Full%20PCR%20Plate%20with%20Master%20Mix%20.json).\n\n---\n\n\n* [Opentrons P300-single channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=5984549109789)\n* [Opentrons P20-multi channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 20ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons Aluminum Block Set](https://shop.opentrons.com/products/aluminum-block-set)\n\n",
"internal": "0a23c6\n",
diff --git a/protoBuilds/0a25f2/README.json b/protoBuilds/0a25f2/README.json
index b87d947857..88816130ff 100644
--- a/protoBuilds/0a25f2/README.json
+++ b/protoBuilds/0a25f2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Tube Filling"
@@ -8,7 +8,7 @@
"description": "This protocol dispenses a variable amount of oil from a source container in the usual location of the Opentrons trash bin to up to 300 capsules seated in 3 10x10 racks on the deck. The racks are filled in order of top left, then bottom left, then bottom right, depending on the number of samples. Within each rack, transfers are carried out down each column, then across each row. A single tip is used for the entire process\n\n\n\n3x Custom 10x10 capsule racks in custom Opentrons adapter\nOpentrons 4-in-1 tuberack set with 4x6 insert holding 2ml Eppendorf snapcap tubes\nOpentrons P1000-single channel electronic pipettes (GEN2)\nOpentrons 1000ul tiprack\n",
"internal": "0a25f2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Tube Filling\n\n",
"description": "This protocol dispenses a variable amount of oil from a source container in the usual location of the Opentrons trash bin to up to 300 capsules seated in 3 10x10 racks on the deck. The racks are filled in order of top left, then bottom left, then bottom right, depending on the number of samples. Within each rack, transfers are carried out down each column, then across each row. A single tip is used for the entire process\n\n---\n\n\n* 3x Custom 10x10 capsule racks in custom Opentrons adapter\n* [Opentrons 4-in-1 tuberack set](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 4x6 insert holding [2ml Eppendorf snapcap tubes](https://online-shop.eppendorf.us/US-en/Laboratory-Consumables-44512/Tubes-44515/Eppendorf-Safe-Lock-Tubes-PF-8863.html)\n* [Opentrons P1000-single channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 1000ul tiprack](https://shop.opentrons.com/collections/opentrons-tips)\n\n",
"internal": "0a25f2\n",
diff --git a/protoBuilds/0a33e8/README.json b/protoBuilds/0a33e8/README.json
index bce09ae2ab..75925deb94 100644
--- a/protoBuilds/0a33e8/README.json
+++ b/protoBuilds/0a33e8/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "0a33e8",
"labware": "\nThermofisher 96 well plate\nThermofisher 384 well plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This protocol preps a 384 well plate with 5ul from (4) 96 deep well plates as the source. Deepwell plates should be placed in order of slot 4, 1, 5, 2 as sample number increases, with tips placed in order of slot 10, 7, 11, and 8. Samples in Plate 1 on slot 4 are completely transferred before moving onto Plate 2 on slot 1, so on and so forth. Plate 1 (slot 4) is dispensed into A1-A23 of the 384 plate, Plate 2 (slot 1) -> B1-B23 of the 384 plate, Plate 3 (slot 5) -> A2-A24 of the 384 plate, Plate 4 (slot 2) -> B2-B24 of the 384 plate. Negative and positive controls should be pre-loaded into O24 and P24 of the 384 well plate, and thus G12 and H12 of the Plate 4 on slot 2 should be empty.\n\n\nExplanation of complex parameters below:\n* `Number of Samples (1-382)`: Specify number of samples for this run. Negative and positive controls are always in O24 and P24 of the 384 plate, and so samples should not be placed in G12 and H12 of plate 4 on slot 2.\n* `P20 Mount`: Specify which mount (left or right) to host the P20 multi-channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/0a9b58/README.json b/protoBuilds/0a9b58/README.json
index e25dad8f8a..ad243cd119 100644
--- a/protoBuilds/0a9b58/README.json
+++ b/protoBuilds/0a9b58/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "0a9b58",
"labware": "\nAgilent 1 Well Reservoir 290 mL #201252-100\nBio-Rad 96 Well Plate 200 \u00b5L PCR #hsp9601\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n* This is the deck setup for the normalization protocol. For the pooling protocol, the reservoir is not needed in slot 1, and the tip rack in slot 5 is not needed.\n\n\n\n",
"description": "This protocol performs a pooling or normalization protocol depending on the user's selection. Please format the csv to the examples below (two header rows before the transfer information). The normalization csv should include \"normalization\", and the pooling csv should include the word \"pooling\" in the top row. Volumes must be over 1.0ul for all transfers. Volumes should be \"0\" if they are to be skipped for a well.\n\n\n* [Normalization CSV Example](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/0a9b58/normalization.png)\n* [Pooling CSV Example](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/0a9b58/pooling.png)\n\n\n\n\n",
diff --git a/protoBuilds/0aa51a/README.json b/protoBuilds/0aa51a/README.json
index 071658abdc..1a1146d9c2 100644
--- a/protoBuilds/0aa51a/README.json
+++ b/protoBuilds/0aa51a/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"Complete PCR Workflow"
@@ -10,7 +10,7 @@
"internal": "0aa51a",
"labware": "\nBio-Rad 96 Well Plate 200 \u00b5L PCR #hsp9601\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* Complete PCR Workflow\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a PCR plating and PCR reaction on the Opentrons Thermocycler Module. The worklist input for where to transfer oligos should be specified in the following format, **including header line**:\n\n```\nPipettor,Slot 5 well (Oligo source plate),Slot 10 well (Destination plate),Volume uL\nP20 single channel,A6,A1,10\nP20 single channel,C1,A1,10\nP20 single channel,A1,B1,10\nP20 single channel,A7,B1,2.5\nP20 single channel,A8,B1,2.5\nP20 single channel,A9,B1,2.5\n...\n```\n\nYou can download a template .csv file [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/0aa51a/ex.csv).\n\n\n",
diff --git a/protoBuilds/0add76/README.json b/protoBuilds/0add76/README.json
index c3d07a1236..59bb93ae42 100644
--- a/protoBuilds/0add76/README.json
+++ b/protoBuilds/0add76/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol utilizes a CSV (.csv) file to dictate transfers of two reagents (DNA Stock and Water) to a VWR 96-well PCR plate. Simply upload the properly formatted CSV (examples below), set parameters, then download your protocol for use with the OT-2.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\n[P10 Single Pipette]\nP300 Single Pipette\nGEB 96 Tip Rack 10 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\nVWR Universal Pipet Tips 10ul, XL, Low Retention\nVWR Universal Pipet Tips 300ul, Low Retention\nVWR Flat 96 Well PCR Plate 200ul\nVWR Microplate 96 Square 2ml\nEK Scientific Reservoir Without Lid 290 mL\n\n\n\nFor this protocol, be sure that the pipettes (P10 and P50) are attached.\nUsing the customization fields below, set up your protocol.\n Transfer CSV: Upload your properly formatted (see below) CSV.\n P10 Single Mount: Specify which mount the P10 is on (left or right).\n* P300 Single Mount: Specify which mount the P50 is on (left or right).\nNote about CSV\nThe CSV should be formatted like so:\nSamples | DNA Stock Volume (\u00b5L) | H20 Stock Volume (\u00b5L) | Well\nThe first row (A1, B1, C1, D1) can contain headers (like above) or simply have the desired information. All of the following rows should just have the necessary information.\nLabware Setup\nSlot 6: EK Scientific Reservoir Without Lid 290 mL\nSlot 8: VWR Flat 96 Well PCR Plate 200ul\nSlot 9: Opentrons 96 Tip Rack 300 \u00b5L\nSlot 10: VWR Square Microplate 96 Square 2ml\nSlot 11: GEB 96 Tip Rack 10 \u00b5L",
"internal": "0add76",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "This protocol utilizes a CSV (.csv) file to dictate transfers of two reagents (DNA Stock and Water) to a VWR 96-well PCR plate. Simply upload the properly formatted CSV (examples below), set parameters, then download your protocol for use with the OT-2.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P10 Single Pipette]\n* [P300 Single Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [GEB 96 Tip Rack 10 \u00b5L](https://labware.opentrons.com/geb_96_tiprack_10ul?category=tipRack)\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://labware.opentrons.com/opentrons_96_tiprack_300ul?category=tipRack)\n* [VWR Universal Pipet Tips 10ul, XL, Low Retention](https://us.vwr.com/store/catalog/product.jsp?catalog_number=76323-388)\n* [VWR Universal Pipet Tips 300ul, Low Retention](https://us.vwr.com/store/catalog/product.jsp?catalog_number=76322-148)\n* [VWR Flat 96 Well PCR Plate 200ul](https://us.vwr.com/store/catalog/product.jsp?catalog_number=82006-636)\n* [VWR Microplate 96 Square 2ml](https://us.vwr.com/store/product?keyword=75870-792)\n* [EK Scientific Reservoir Without Lid 290 mL](https://us.vwr.com/store/catalog/product.jsp?catalog_number=89049-028)\n\n\n\n---\n\n\nFor this protocol, be sure that the pipettes (P10 and P50) are attached.\n\nUsing the customization fields below, set up your protocol.\n* Transfer CSV: Upload your properly formatted (see below) CSV.\n* P10 Single Mount: Specify which mount the P10 is on (left or right).\n* P300 Single Mount: Specify which mount the P50 is on (left or right).\n\n**Note about CSV**\n\nThe CSV should be formatted like so:\n\n`Samples` | `DNA Stock Volume (\u00b5L)` | `H20 Stock Volume (\u00b5L)` | `Well`\n\nThe first row (A1, B1, C1, D1) can contain headers (like above) or simply have the desired information. All of the following rows should just have the necessary information.\n\n**Labware Setup**\n\nSlot 6: EK Scientific Reservoir Without Lid 290 mL\n\nSlot 8: VWR Flat 96 Well PCR Plate 200ul\n\nSlot 9: Opentrons 96 Tip Rack 300 \u00b5L\n\nSlot 10: VWR Square Microplate 96 Square 2ml\n\nSlot 11: GEB 96 Tip Rack 10 \u00b5L\n\n",
"internal": "0add76\n",
diff --git a/protoBuilds/0aee8a/README.json b/protoBuilds/0aee8a/README.json
index cae88612f6..ea0b5f393a 100644
--- a/protoBuilds/0aee8a/README.json
+++ b/protoBuilds/0aee8a/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Serial Dilution"
@@ -10,7 +10,7 @@
"internal": "0aee8a",
"labware": "\nOpentrons 200ul Filter Tips \nOpentrons 20ul Filter Tips\nNEST 12-Well Reservoirs 15mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Serial Dilution\n\n\n",
"deck-setup": "\nVolume Definitions:\n* VOL1= DMSO transfer volume for vertical plates (post dilution volume) \n* VOL2= DMSO transfer volume for horizontal plates (post dilution volume)\n* VOL3= Compound C predilution volume DMSO (optional)\n* VOL4= Compound D predilution volume DMSO (optional)\n* VOL5= Compound A predilution volume DMSO (optional)\n* VOL6= Compound B predilution volume DMSO (optional)\n* VOL7= Compound C predilution volume compound (optional)\n* VOL8= Compound D predilution volume compound (optional)\n* VOL9= Compound A predilution volume compound (optional)\n* VOL10= Compound B predilution volume compound (optional)\n* VOL11= Compound C/D initial volume \n* VOL12= Compound C/D Serial dilution volume \n* VOL13= Compound A/B initial volume\n* VOL14= Compound A/B serial dilution volume\n* VOL15= Final plate volume for integra transfer \n\n",
"description": "This protocol outlines serial dilution of compound stock. There is an optional predilution step for each compounds involved. For detailed protocol steps, please see below. Labware setup consists of NEST 12 well 15mL reservoir in slot 1, Nest 96 well 100 uL plate in slot 2,4,5,7,8,10 & 11 as well as P20 filter tips in slot 3 and P200 filter tips in slot 6.\n\n\n",
diff --git a/protoBuilds/0b83b7/README.json b/protoBuilds/0b83b7/README.json
index 400b3e6f0d..5bf413aa1a 100644
--- a/protoBuilds/0b83b7/README.json
+++ b/protoBuilds/0b83b7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Cleanup"
@@ -9,7 +9,7 @@
"internal": "0b83b7",
"labware": "\nNEST 0.2 mL 96-Well PCR Plate, Full Skirt\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Cleanup\n\n\n",
"description": "This protocol is equipped to perform any of 4x cleanups for the Paragon NGS prep protocol.\n\n\n",
"internal": "0b83b7\n",
diff --git a/protoBuilds/0b97ae-protocol-2A/README.json b/protoBuilds/0b97ae-protocol-2A/README.json
index a7386210d9..dd4fe00eb2 100644
--- a/protoBuilds/0b97ae-protocol-2A/README.json
+++ b/protoBuilds/0b97ae-protocol-2A/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS LIBRARY PREP": [
"QIASeq FastSelect"
@@ -10,7 +10,7 @@
"internal": "0b97ae-protocol-2A",
"labware": "\nPerkin Elmer 12 Reservoir 21000 \u00b5L\nApplied Biosystems Enduraplate 96 Aluminum Block 220 \u00b5L\nOpentrons 96 Tip Rack 20 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 96 Well Aluminum Block with Bio-Rad Well Plate 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS LIBRARY PREP\n\t* QIASeq FastSelect\n\n\n",
"deck-setup": "\n\n\n",
"description": "This is Part 2 to the QIAseq FastSelect Protocol. This protocol is used to perform the addition of samples with mastermix into a plate.\n\nLinks:\n* [Part 1: Sample Normalization](http://protocols.opentrons.com/protocol/0b97ae)\n* [Part 2: QIAseq FastSelect Fragmentation](http://protocols.opentrons.com/protocol/0b97ae-protocol-2A)\n\n",
diff --git a/protoBuilds/0b97ae-protocol-2B/README.json b/protoBuilds/0b97ae-protocol-2B/README.json
index bb98bb0974..f34c9b4e46 100644
--- a/protoBuilds/0b97ae-protocol-2B/README.json
+++ b/protoBuilds/0b97ae-protocol-2B/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS LIBRARY PREP": [
"QIASeq FastSelect"
@@ -10,7 +10,7 @@
"internal": "0b97ae-protocol-2B",
"labware": "\nPerkin Elmer 12 Reservoir 21000 \u00b5L\nApplied Biosystems Enduraplate 96 Aluminum Block 220 \u00b5L\nOpentrons 96 Tip Rack 20 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 96 Well Aluminum Block with Bio-Rad Well Plate 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS LIBRARY PREP\n\t* QIASeq FastSelect\n\n\n",
"deck-setup": "\n\n\n",
"description": "This is Part 2 to the QIAseq FastSelect 5s, 16s, 23s Protocol. This protocol is used to perform the addition of samples with mastermix into a plate.\nPart 1 to this protocol is the normalization of samples.\nPart 3 to this protocol is the Extraction and Clean-Up steps.\n\nLinks:\n* [Part 1: Sample Normalization](http://protocols.opentrons.com/protocol/0b97ae)\n* [Part 2: QIAseq FastSelect 5s, 16s, 23s Fragmentation](http://protocols.opentrons.com/protocol/0b97ae-protocol-2B)\n* [Part 3: QIAseq FastSelect 5s, 16s, 23s Extraction](http://protocols.opentrons.com/protocol/0b97ae-protocol-3B)\n\n\n\n",
diff --git a/protoBuilds/0b97ae-protocol-3B/README.json b/protoBuilds/0b97ae-protocol-3B/README.json
index 58e81b6c9d..7845f17b7b 100644
--- a/protoBuilds/0b97ae-protocol-3B/README.json
+++ b/protoBuilds/0b97ae-protocol-3B/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS LIBRARY PREP": [
"QIASeq FastSelect"
@@ -10,7 +10,7 @@
"internal": "0b97ae-protocol-3B",
"labware": "\nPerkin Elmer 12 Reservoir 21000 \u00b5L\nApplied Biosystems Enduraplate 96 Aluminum Block 220 \u00b5L\nBio-Rad 96 Well Plate 200 \u00b5L PCR #hsp9601\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 96 Well Aluminum Block with Bio-Rad Well Plate 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS LIBRARY PREP\n\t* QIASeq FastSelect\n\n\n",
"deck-setup": "\nWater Reservoir (slot 2):\n\tColumn 1: Nuclease Free Water\n\tColumn 2: Binding Buffer\n\tColumn 3 & 4: Ethanol\n\tColumn 10, 11 & 12: Empty for Supernatent Removal\nDiluted RNA Plate (slot 7 Temperature Module):\n\tRNA Samples Starting Plate\nReagent Plate (Slot 10):\n\tColumn 1: MasterMix\n\tColumn 2: Magentic Beads\n\n",
"description": "This is Part 3 to the QIAseq FastSelect 5s, 16s, 23s Protocol. This protocol is used to perform the addition of samples with mastermix into a plate.\nPart 1 to this protocol is the normalization of samples.\nPart 2 to this protocol is the Fragementation.\n\nLinks:\n* [Part 1: Sample Normalization](http://protocols.opentrons.com/protocol/0b97ae)\n* [Part 2: QIAseq FastSelect 5s, 16s, 23s Fragmentation](http://protocols.opentrons.com/protocol/0b97ae-protocol-2B)\n* [Part 3: QIAseq FastSelect 5s, 16s, 23s Extraction](http://protocols.opentrons.com/protocol/0b97ae-protocol-3B)\n\n\n",
diff --git a/protoBuilds/0b97ae/README.json b/protoBuilds/0b97ae/README.json
index 118af11ddf..537e3618fb 100644
--- a/protoBuilds/0b97ae/README.json
+++ b/protoBuilds/0b97ae/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"SAMPLE PREP": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "0b97ae",
"labware": "\nPerkin Elmer 12 Reservoir 21000 \u00b5L\nApplied Biosystems Enduraplate 96 Aluminum Block 220 \u00b5L\nOpentrons 96 Tip Rack 20 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 96 Well Aluminum Block with Bio-Rad Well Plate 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* SAMPLE PREP\n\t* Normalization\n\n\n",
"deck-setup": "\n\n\n",
"description": "With this Protocol you can Normalize RNA samples using RNA Concentration, Goal Concentration, Total Volume. This Normalization is the normalization protocol to preceed the QIAseq FastSelect and QIAsec FastSelect 5s , 16S and 23S. \n\n\n",
diff --git a/protoBuilds/0b98cc/README.json b/protoBuilds/0b98cc/README.json
index afbdfa31a3..6bc78266f4 100644
--- a/protoBuilds/0b98cc/README.json
+++ b/protoBuilds/0b98cc/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Serial Dilution"
@@ -8,7 +8,7 @@
"description": "This protocol performs viral titration (serial dilution) of viral samples. It adds media to the dilution plates and then begins the serial dilution process of up to 12 columns worth of samples. It will dilute the samples in one of four dilution plates and then add it to the analysis dish in a specific order (skipping 3 columns for every dilution).\n\n\n\nOpentrons 200uL Filter Tips\nCorning 96 Well Round Bottom 1mL\nCorning 96-well Flat Clear Bottom Black Polystyrene TC-treated Microplate\nNEST 1 Well 195mL Reservoir\nP300 Multichannel GEN2\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nNote: The Dilution Plates utilize the Corning 96 Well Round Bottom 1mL and the Analysis Dishes utilize the Corning 96-well Flat Clear Bottom Black Polystyrene TC-treated Microplate.\n\nLabware Type\n Dilution Plate: Corning 96 Well Round Bottom 1mL\n Analysis Dish: Corning 96-well Flat Clear Bottom Black Polystyrene TC-treated Microplate\n Samples: NEST 96 Well Plate 100uL PCR Full Skirt\n Media Reservoir: NEST 1 Well 195mL Reservoir\nProtocol Steps\n1. Transfer appropriate volumes of media from the reservoir to each dilution plate on a per column basis using the multichannel pipette.\n2. Sample from the first column (A1) is transferred to A1 of Dilution Plate 1 where it is mixed and then transffered to A1 on analysis dish 1.\n3. Diluted sample is transferred from A1 of Dilution Plate 1 to A1 of Dilution Plate 2. It is thoroughly mixed and then transferred to A4 on Analysis Dish 1.\n4. Diluted sample is transferred from A1 of Dilution Plate 2 to A1 of Dilution Plate 3. It is thoroughly mixed and then transferred to A7 on Analysis Dish 1.\n5. Diluted sample is transferred from A1 of Dilution Plate 3 to A1 of Dilution Plate 4. It is thoroughly mixed and then transferred to A10 on Analysis Dish 1.\n6. This completes one set of samples. This cycle will repeat for up to 12 columns worth of samples across four analysis dishes.",
"internal": "0b98cc",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Serial Dilution\n\n",
"description": "This protocol performs viral titration (serial dilution) of viral samples. It adds media to the dilution plates and then begins the serial dilution process of up to 12 columns worth of samples. It will dilute the samples in one of four dilution plates and then add it to the analysis dish in a specific order (skipping 3 columns for every dilution).\n\n---\n\n\n* [Opentrons 200uL Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Corning 96 Well Round Bottom 1mL](https://ecatalog.corning.com/life-sciences/b2c/US/en/Genomics-&-Molecular-Biology/Automation-Consumables/Deep-Well-Plate/Corning%C2%AE-96-well-Polypropylene-Storage-Blocks/p/3958)\n* [Corning 96-well Flat Clear Bottom Black Polystyrene TC-treated Microplate](https://ecatalog.corning.com/life-sciences/b2c/US/en/Microplates/Assay-Microplates/96-Well-Microplates/Corning%C2%AE-96-well-Black-Clear-and-White-Clear-Bottom-Polystyrene-Microplates/p/3603)\n* [NEST 1 Well 195mL Reservoir](https://shop.opentrons.com/collections/reservoirs/products/nest-1-well-reservoir-195-ml)\n* [P300 Multichannel GEN2](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n\n---\n\n\n**Note:** The Dilution Plates utilize the Corning 96 Well Round Bottom 1mL and the Analysis Dishes utilize the Corning 96-well Flat Clear Bottom Black Polystyrene TC-treated Microplate.\n\n\n**Labware Type**\n* Dilution Plate: Corning 96 Well Round Bottom 1mL\n* Analysis Dish: Corning 96-well Flat Clear Bottom Black Polystyrene TC-treated Microplate\n* Samples: NEST 96 Well Plate 100uL PCR Full Skirt\n* Media Reservoir: NEST 1 Well 195mL Reservoir\n\n**Protocol Steps**\n1. Transfer appropriate volumes of media from the reservoir to each dilution plate on a per column basis using the multichannel pipette.\n2. Sample from the first column (A1) is transferred to A1 of Dilution Plate 1 where it is mixed and then transffered to A1 on analysis dish 1.\n3. Diluted sample is transferred from A1 of Dilution Plate 1 to A1 of Dilution Plate 2. It is thoroughly mixed and then transferred to A4 on Analysis Dish 1.\n4. Diluted sample is transferred from A1 of Dilution Plate 2 to A1 of Dilution Plate 3. It is thoroughly mixed and then transferred to A7 on Analysis Dish 1.\n5. Diluted sample is transferred from A1 of Dilution Plate 3 to A1 of Dilution Plate 4. It is thoroughly mixed and then transferred to A10 on Analysis Dish 1.\n6. This completes one set of samples. This cycle will repeat for up to 12 columns worth of samples across four analysis dishes.\n\n",
"internal": "0b98cc",
diff --git a/protoBuilds/0ba998-2/README.json b/protoBuilds/0ba998-2/README.json
index 96caecb288..509dd10ba1 100644
--- a/protoBuilds/0ba998-2/README.json
+++ b/protoBuilds/0ba998-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina DNA Prep"
@@ -10,7 +10,7 @@
"internal": "0ba998",
"labware": "\nNEST 1 Well Reservoir 195 mL #360103\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina DNA Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs part 2/4 of the [Illumina DNA Prep](https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/illumina_prep/illumina-dna-prep-reference-guide-1000000025416-09.pdf) protocol: Post Tagmentation Cleanup. The user is prompted to move plates and perform manual centrifugation, sealing, and thermocycling steps when necessary.\n\nLinks:\n\n1. [Tagment Genomic DNA](./0ba998)\n2. [Post Tagmentation Cleanup](./0ba998-2)\n3. [Amplify Tagmented DNA](./0ba998-3)\n4. [Clean Up Libraries](./0ba998-4)\n\n\n",
diff --git a/protoBuilds/0ba998-3/README.json b/protoBuilds/0ba998-3/README.json
index 93c660875b..3af817888d 100644
--- a/protoBuilds/0ba998-3/README.json
+++ b/protoBuilds/0ba998-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina DNA Prep"
@@ -10,7 +10,7 @@
"internal": "0ba998",
"labware": "\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina DNA Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs part 3/4 of the [Illumina DNA Prep](https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/illumina_prep/illumina-dna-prep-reference-guide-1000000025416-09.pdf) protocol: Amplify Tagmented DNA. The user is prompted to move plates and perform manual centrifugation, sealing, and thermocycling steps when necessary.\n\nLinks:\n\n1. [Tagment Genomic DNA](./0ba998)\n2. [Post Tagmentation Cleanup](./0ba998-2)\n3. [Amplify Tagmented DNA](./0ba998-3)\n4. [Clean Up Libraries](./0ba998-4)\n\n\n",
diff --git a/protoBuilds/0ba998-4/README.json b/protoBuilds/0ba998-4/README.json
index ac34db0267..85422610a5 100644
--- a/protoBuilds/0ba998-4/README.json
+++ b/protoBuilds/0ba998-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina DNA Prep"
@@ -10,7 +10,7 @@
"internal": "0ba998",
"labware": "\nNEST 1 Well Reservoir 195 mL #360103\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina DNA Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs part 4/4 of the [Illumina DNA Prep](https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/illumina_prep/illumina-dna-prep-reference-guide-1000000025416-09.pdf) protocol: Clean Up Libraries. The user is prompted to move plates and perform manual centrifugation, sealing, and thermocycling steps when necessary.\n\nLinks:\n\n1. [Tagment Genomic DNA](./0ba998)\n2. [Post Tagmentation Cleanup](./0ba998-2)\n3. [Amplify Tagmented DNA](./0ba998-3)\n4. [Clean Up Libraries](./0ba998-4)\n\n\n",
diff --git a/protoBuilds/0ba998/README.json b/protoBuilds/0ba998/README.json
index 2484be7517..f1e8217431 100644
--- a/protoBuilds/0ba998/README.json
+++ b/protoBuilds/0ba998/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina DNA Prep"
@@ -10,7 +10,7 @@
"internal": "0ba998",
"labware": "\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina DNA Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs part 1/4 of the [Illumina DNA Prep](https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/illumina_prep/illumina-dna-prep-reference-guide-1000000025416-09.pdf) protocol: Tagment Genomic DNA. The user is prompted to move plates and perform manual centrifugation, sealing, and thermocycling steps when necessary.\n\nLinks:\n\n1. [Tagment Genomic DNA](./0ba998)\n2. [Post Tagmentation Cleanup](./0ba998-2)\n3. [Amplify Tagmented DNA](./0ba998-3)\n4. [Clean Up Libraries](./0ba998-4)\n\n\n",
diff --git a/protoBuilds/0bd707/README.json b/protoBuilds/0bd707/README.json
index ce9d60f238..a55c231752 100644
--- a/protoBuilds/0bd707/README.json
+++ b/protoBuilds/0bd707/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Aliquoting"
@@ -8,7 +8,7 @@
"description": "This protocol is a modified version of this sample aliquoting protocol.\n\nWith this version of the protocol, the user can specify the number of plates they would like to fill (up to 6). When run, the robot will transfer the specified volume two times (2x) to each tube, in each tube rack, and change tip after each liquid transfer.\n\nPlease note that this protocol uses a custom labware definition for the tube rack (definition included with download). For more information regarding custom labware definition usage on the OT-2, please see this article: Using labware in your protocols.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.17.0 or later)\nNEST 1 Well Reservoir 195 mL\nOpentrons P1000 GEN2 single-channel electronic pipette\nOpentrons 96 Tip Rack 1000 \u00b5L\n24-Tube Tube Rack plus Tubes\n\n\n\nTube Racks\nLoad sequentially deck slots, in this order: 2, 3, 5, 6, 7, 8, 9\n\nTip Racks\nOne tip rack is needed for every two tube racks. The tip racks should be loaded in deck slots: 1, 4\n\nReagent Reservoirs\nOne reagent reservoir should be used for every three tube racks. The reservoirs should be loaded in deck slots: 11 (will be used first), 10",
"internal": "0bd707",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Aliquoting\n\n",
"description": "This protocol is a modified version of this [sample aliquoting protocol](https://develop.protocols.opentrons.com/protocol/53134e).\n\nWith this version of the protocol, the user can specify the number of plates they would like to fill (up to 6). When run, the robot will transfer the specified volume two times (2x) to each tube, in each tube rack, and change tip after each liquid transfer.\n\nPlease note that this protocol uses a custom labware definition for the tube rack (definition included with download). For more information regarding custom labware definition usage on the OT-2, please see this article: [Using labware in your protocols](https://support.opentrons.com/en/articles/3136506-using-labware-in-your-protocols).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.17.0 or later)](https://opentrons.com/ot-app/)\n* [NEST 1 Well Reservoir 195 mL](https://labware.opentrons.com/nest_1_reservoir_195ml)\n* [Opentrons P1000 GEN2 single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette?variant=5984549142557)\n* [Opentrons 96 Tip Rack 1000 \u00b5L](https://labware.opentrons.com/opentrons_96_tiprack_1000ul)\n* 24-Tube Tube Rack plus Tubes\n\n---\n\n\n**Tube Racks**\nLoad sequentially deck slots, in this order: *2, 3, 5, 6, 7, 8, 9*\n\n**Tip Racks**\nOne tip rack is needed for every two tube racks. The tip racks should be loaded in deck slots: *1, 4*\n\n**Reagent Reservoirs**\nOne reagent reservoir should be used for every three tube racks. The reservoirs should be loaded in deck slots: *11 (will be used first), 10*\n\n",
"internal": "0bd707\n",
diff --git a/protoBuilds/0be1bf/README.json b/protoBuilds/0be1bf/README.json
index 592461ead2..cdfb3910c2 100644
--- a/protoBuilds/0be1bf/README.json
+++ b/protoBuilds/0be1bf/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Aliquoting"
@@ -10,7 +10,7 @@
"internal": "0be1bf",
"labware": "\nOpentrons 15 Tube Rack with Eppendorf 5 mL\nOpentrons 96 Tip Rack 20 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 24 Tube Rack with Eppendorf 2 mL Safe-Lock Snapcap\nNEST 12 Well Reservoir 15 mL #360102\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Aliquoting\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs custom IVT aliquoting for up to 24 reactions in 2ml Eppendorf tubes. For liquid layout, please reference the liquid setup section of your protocol in the Opentrons App.\n\n\n",
diff --git a/protoBuilds/0bf4f4-pt2/README.json b/protoBuilds/0bf4f4-pt2/README.json
index 28abd03720..07ebe6e95c 100644
--- a/protoBuilds/0bf4f4-pt2/README.json
+++ b/protoBuilds/0bf4f4-pt2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina DNA Prep"
@@ -10,7 +10,7 @@
"internal": "0bf4f4-pt2",
"labware": "\nBio-RAD 96 well plate 200ul\nNest 12 Well Reservoir 15mL\nOpentrons 20ul Filter Tip Rack\nOpentrons 200ul Filter Tip Rack\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina DNA Prep\n\n",
"deck-setup": "\n* Deck setup with a full plate of samples. Tip rack on slot 11 will be used to park tips in ethanol wash steps.\n\n\n\n",
"description": "This protocol is part 2 of a 3 part series which preps a 96 well Bio-Rad 200ul plate in accordance with the [Ilumina DNA Prep Kit](https://emea.support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/illumina_prep/illumina-dna-prep-reference-guide-1000000025416-09.pdf).\n\n* [Part 1: Tagment DNA](https://protocols.opentrons.com/protocol/0bf4f4)\n* [Part 3: Cleanup libraries](https://protocols.opentrons.com/protocol/0bf4f4-pt3)\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples for this run.\n* `Index start column`: Specify which column to start aspirating from on the index plate.\n* `P300 tip start column on slot 11 (1-12)`: The number of columns of tips used on slot 11 is equal to the number of columns of samples. Slot 11 tip rack is used for tip parking on the two ethanol washes to save tips. Specify which column to start picking tips up from.\n* `Length from side`: Specify the length from the side of the well to aspirate from magnetically engaged beads. A value of 1 would be 1mm from the side of the well opposite beads. A value of 2.73 means the exact center of the well. This distance is also used when re-suspending beads, and mixing at bead location.\n* `Aspiration height`: Specify how many millimeters from the bottom of the well to aspirate from at the `Length from side` distance specified above.\n* `P20 Multi-Channel GEN2 Mount`: Specify which side (left or right) the P20 multi channel pipette is mounted.\n* `P300 Multi Channel GEN2 Mount`: Specify which side (left or right) the P300 multi channel pipette is mounted.\n\n\n",
diff --git a/protoBuilds/0bf4f4-pt3/README.json b/protoBuilds/0bf4f4-pt3/README.json
index a91cfb80d8..591a94e480 100644
--- a/protoBuilds/0bf4f4-pt3/README.json
+++ b/protoBuilds/0bf4f4-pt3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina DNA Prep"
@@ -10,7 +10,7 @@
"internal": "0bf4f4-pt3",
"labware": "\nBio-RAD 96 well plate 200ul\nNest 12 Well Reservoir 15mL\nOpentrons 20ul Filter Tip Rack\nOpentrons 200ul Filter Tip Rack\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina DNA Prep\n\n",
"deck-setup": "\n* Deck setup with a full plate of samples and pre-populated mag beads in Plate B. Note: if the protocol runs out of tips, it will pause and prompt the user to replace all tip racks. Tip boxes will be used in the order of the following slots for 200ul tips: Slot 7, 8, 9, 10. Tip rack on slot 11 will always be used for the ethanol wash step, as it will use parked tips, no matter how many samples. \n\n\n\n",
"description": "This protocol is part 3 of a 3 part series which preps a 96 well Bio-Rad 200ul plate in accordance with the [Ilumina DNA Prep Kit](https://emea.support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/illumina_prep/illumina-dna-prep-reference-guide-1000000025416-09.pdf).\n\n* [Part 1: Tagment DNA](https://protocols.opentrons.com/protocol/0bf4f4)\n* [Part 2: Cleanup libraries](https://protocols.opentrons.com/protocol/0bf4f4-pt2)\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples for this run.\n* `Plate A Start Column`: Specify which column to start on plate A.\n* `Plate B Start Column`: Specify which column to start on plate B.\n* `Plate C Start Column`: Specify which column to start on plate C.\n* `P300 tip start column on slot 11 (1-12)`: The number of columns of tips used on slot 11 is equal to the number of columns of samples. Slot 11 tip rack is used for tip parking on the two ethanol washes to save tips. Specify which column to start picking tips up from.\n* `Length from side`: Specify the length from the side of the well to aspirate from magnetically engaged beads. A value of 1 would be 1mm from the side of the well opposite beads. A value of 2.73 means the exact center of the well. This distance is also used when re-suspending beads, and mixing at bead location.\n* `Aspiration height`: Specify how many millimeters from the bottom of the well to aspirate from at the `Length from side` distance specified above.\n* `P20 Multi-Channel GEN2 Mount`: Specify which side (left or right) the P20 multi channel pipette is mounted.\n* `P300 Multi Channel GEN2 Mount`: Specify which side (left or right) the P300 multi channel pipette is mounted.\n\n\n",
diff --git a/protoBuilds/0bf4f4/README.json b/protoBuilds/0bf4f4/README.json
index c03c70d329..171410796f 100644
--- a/protoBuilds/0bf4f4/README.json
+++ b/protoBuilds/0bf4f4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina DNA Prep"
@@ -10,7 +10,7 @@
"internal": "0bf4f4",
"labware": "\nBio-RAD 96 well plate 200ul\nNest 12 Well Reservoir 15mL\nOpentrons 20ul Filter Tip Rack\nOpentrons 200ul Filter Tip Rack\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina DNA Prep\n\n",
"deck-setup": "\n* Deck setup with a full plate of samples. Mastermix is always kept in column 1 regardless of number of samples, as is water in the first column of the Nest 12 well reservoir.\n\n\n\n---\n\n",
"description": "This protocol is part 1 of a 3 part series which preps a 96 well Bio-Rad 200ul plate with diluted DNA and mastermix in accordance with the [Ilumina DNA Prep Kit](https://emea.support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/illumina_prep/illumina-dna-prep-reference-guide-1000000025416-09.pdf). DNA is diluted with water before mastermix is added. Find part 2 and part 3 of protocol below:\n\n* [Part 2: Post Tagmentation Cleanup](https://protocols.opentrons.com/protocol/0bf4f4-pt2)\n* [Part 3: Cleanup libraries](https://protocols.opentrons.com/protocol/0bf4f4-pt3)\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples for this run.\n* `P300 Tip Start Column on Slot 8 (1-12)`: One column of the 200ul tip rack will be used per run. Specify which column to pick up tips from for this run.\n* `P20 Multi-Channel GEN2 Mount`: Specify which side (left or right) the P20 multi channel pipette is mounted.\n* `P300 Multi Channel GEN2 Mount`: Specify which side (left or right) the P300 multi channel pipette is mounted.\n\n\n",
diff --git a/protoBuilds/0c24ca/README.json b/protoBuilds/0c24ca/README.json
index 2836d11a79..6d763f5cc0 100644
--- a/protoBuilds/0c24ca/README.json
+++ b/protoBuilds/0c24ca/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "0c24ca",
"labware": "\nThermofisher 96 Well Plate 1400 \u00b5L\nThermofisher 96 Well Plate 500 \u00b5L\nCorning 96 Well Plate 340 \u00b5L\nOpentrons 10 Tube Rack with Falcon 4x50 mL, 6x15 mL Conical\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol will transfer scattered sample tubes to a specified destination plate according to a csv. The csv header should read `sample well, sample slot, destination well`. Include the header in the csv and specify the volumes below. THe protocol will automatically pause if the user runs out of tips, rprompting the user to replace tips. A P20 or P300 pipette is specified depending on the volume selected by the user. Liquid height tracking is performed on the 50mL tube so that the pipette does not submerge.\n\n\n",
diff --git a/protoBuilds/0c3e45/README.json b/protoBuilds/0c3e45/README.json
index 40ee71d4d4..c6592239cd 100644
--- a/protoBuilds/0c3e45/README.json
+++ b/protoBuilds/0c3e45/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "0c3e45",
"labware": "\nNEST 96 Well Plate 200 \u00b5L Flat #701011\nNEST 1 Well Reservoir 195 mL #360103\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a basic plate filling protocol for 9 plates from one source reservoir. The same set of tips is used for the entire protocol, and tips are tracked across protocols.\n\nThe protocol is adapted to Python from [this Protocol Designer protocol](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/0c3e45/pd.json).\n\n\n",
diff --git a/protoBuilds/0ca318-pt2/README.json b/protoBuilds/0ca318-pt2/README.json
index db137b3d39..51c1e1028e 100644
--- a/protoBuilds/0ca318-pt2/README.json
+++ b/protoBuilds/0ca318-pt2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR"
@@ -10,7 +10,7 @@
"internal": "0ca318",
"labware": "\nAbgene\u2122 96 Well 0.8mL Thermo Scientific #AB0859\nNEST 12 Well Reservoir 15 mL #360102\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR\n\n\n",
"deck-setup": "\n\n\n",
"description": "The protocol begins with a bead-based cleanup. Note: each well for the ethanol in the reservoir is per 24 samples. That is, for 48 samples, load two wells of ethanol. For 48-72 samples, 3 wells of ethanol, so on and so forth. The ethanol should be distributed evenly among all wells (volume is the same in each well). The protocol will automatically pause when the tips are depleted, asking the user to resume the protocol.\n\nSince this protocol uses a custom plate on the magnetic module, please ensure that the magnetic height variable down below is set to the correct height - the user will have to adjust this number to what's best for their plate. Ideally, the top of the magnet is to come just below, but not touching the bottom of the plate when engaged. The reagent rack should be placed on the temperature module which will be at 4C to keep the mastermix cool.\n\nThe last 4 wells of the reservoir are used for trash.\n\n\n",
diff --git a/protoBuilds/0ca318/README.json b/protoBuilds/0ca318/README.json
index 5d44f2a942..f1a29f65ba 100644
--- a/protoBuilds/0ca318/README.json
+++ b/protoBuilds/0ca318/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR"
@@ -10,7 +10,7 @@
"internal": "0ca318",
"labware": "\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nOpentrons 24 Well Aluminum Block with NEST 1.5 mL Snapcap\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR\n\n\n",
"deck-setup": "\n\n\n\n\n\n",
"description": "This protocol begins with mastermix addition, then proceeds to do pcr. Note: mastermix is loaded on the temperature module on slot 4. One column of mastermix is for 24 (3 columns) of samples. That is, for 48 samples, you should load two columns of mastermix. For 48-72 samples, 3 columns of mastermix, so on and so forth. If there are multiple columns of mastermix, the volume should be split evenly between the number of columns you are running. For unfilled columns, mastermix should always be placed in column 6 of the plate on slot 3.\n\nIf you select tube racks for mastermix, each tube accomodates 24 samples in slot 3, and should be placed in the first row (maximum 4 tubes in first row for 96 samples). Mastermix volume should be distributed equally for all tubes in the first row. \n\nThe NEST 96 100ul plate should be placed on the 96 aluminum adaptor on the temperature module, which will be at 4C to keep the temperature cool.\n\nSee below for the two types of deck states before pcr. If you select the tube rack deck state, tubes should be placed by row in one tube rack before moving onto another, in the order of slots 4, 5, 1, 2. That is, for 27 samples, you should fill the entire first tube rack on slot 4, then the first 3 tubes in the first row of the tube rack in slot 5.\n\n\n\n",
diff --git a/protoBuilds/0cded6-amplify/README.json b/protoBuilds/0cded6-amplify/README.json
index d3cc4a2b49..f48c6ee08e 100644
--- a/protoBuilds/0cded6-amplify/README.json
+++ b/protoBuilds/0cded6-amplify/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina COVIDSeq"
@@ -10,7 +10,7 @@
"internal": "0cded6",
"labware": "\nQuantgene 96 Aluminum Block 200 \u00b5L\nAgilent 96 Well Plate 200 \u00b5L #5042-8502\nAgilent With Nonskirted 96 Well Plate 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina COVIDSeq\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs Amplify cDNA for the Illumina COVIDSeq protocol.\n\nLinks: \n* [1. Anneal RNA](./0cded6)\n* [2. Synthesize First Strand cDNA](./0cded6-synthesize)\n* [3. Amplify cDNA](./0cded6-amplify)\n* [4. Tagment PCR Amplicons](./0cded6-tagment)\n* [5. Post Tagmentation Clean Up](./0cded6-cleanup)\n* [6. Amplify Tagmented Amplicons](./0cded6-amplify2)\n\n\n",
diff --git a/protoBuilds/0cded6-amplify2/README.json b/protoBuilds/0cded6-amplify2/README.json
index b9ec3d54a4..9a1f4404d8 100644
--- a/protoBuilds/0cded6-amplify2/README.json
+++ b/protoBuilds/0cded6-amplify2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina COVIDSeq"
@@ -10,7 +10,7 @@
"internal": "0cded6",
"labware": "\nQuantgene 96 Aluminum Block 200 \u00b5L\nAgilent 96 Well Plate 200 \u00b5L #5042-8502\nAgilent With Nonskirted 96 Well Plate 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina COVIDSeq\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs Amplify Tagmented Amplicons for the Illumina COVIDSeq protocol.\n\nLinks: \n* [1. Anneal RNA](./0cded6)\n* [2. Synthesize First Strand cDNA](./0cded6-synthesize)\n* [3. Amplify cDNA](./0cded6-amplify)\n* [4. Tagment PCR Amplicons](./0cded6-tagment)\n* [5. Post Tagmentation Clean Up](./0cded6-cleanup)\n* [6. Amplify Tagmented Amplicons](./0cded6-amplify2)\n\n\n",
diff --git a/protoBuilds/0cded6-cleanup/README.json b/protoBuilds/0cded6-cleanup/README.json
index 984648c891..c9cc6eeec1 100644
--- a/protoBuilds/0cded6-cleanup/README.json
+++ b/protoBuilds/0cded6-cleanup/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina COVIDSeq"
@@ -10,7 +10,7 @@
"internal": "0cded6",
"labware": "\nQuantgene 96 Aluminum Block 200 \u00b5L\nAgilent 96 Well Plate 200 \u00b5L #5042-8502\nAgilent With Nonskirted 96 Well Plate 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina COVIDSeq\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs Post Tagmentation Cleanup for the Illumina COVIDSeq protocol.\n\nLinks: \n* [1. Anneal RNA](./0cded6)\n* [2. Synthesize First Strand cDNA](./0cded6-synthesize)\n* [3. Amplify cDNA](./0cded6-amplify)\n* [4. Tagment PCR Amplicons](./0cded6-tagment)\n* [5. Post Tagmentation Clean Up](./0cded6-cleanup)\n* [6. Amplify Tagmented Amplicons](./0cded6-amplify2)\n\n\n",
diff --git a/protoBuilds/0cded6-synthesize/README.json b/protoBuilds/0cded6-synthesize/README.json
index 9f61e1d1b7..15a2cf6146 100644
--- a/protoBuilds/0cded6-synthesize/README.json
+++ b/protoBuilds/0cded6-synthesize/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina COVIDSeq"
@@ -10,7 +10,7 @@
"internal": "0cded6",
"labware": "\nQuantgene 96 Aluminum Block 200 \u00b5L\nAgilent 96 Well Plate 200 \u00b5L #5042-8502\nAgilent With Nonskirted 96 Well Plate 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina COVIDSeq\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs Syntehsize First Strand cDNA for the Illumina COVIDSeq protocol.\n\nLinks: \n* [1. Anneal RNA](./0cded6)\n* [2. Synthesize First Strand cDNA](./0cded6-synthesize)\n* [3. Amplify cDNA](./0cded6-amplify)\n* [4. Tagment PCR Amplicons](./0cded6-tagment)\n* [5. Post Tagmentation Clean Up](./0cded6-cleanup)\n* [6. Amplify Tagmented Amplicons](./0cded6-amplify2)\n\n\n",
diff --git a/protoBuilds/0cded6-tagment/README.json b/protoBuilds/0cded6-tagment/README.json
index 4a29420354..b49ec14224 100644
--- a/protoBuilds/0cded6-tagment/README.json
+++ b/protoBuilds/0cded6-tagment/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina COVIDSeq"
@@ -10,7 +10,7 @@
"internal": "0cded6",
"labware": "\nQuantgene 96 Aluminum Block 200 \u00b5L\nAgilent 96 Well Plate 200 \u00b5L #5042-8502\nAgilent With Nonskirted 96 Well Plate 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina COVIDSeq\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs Tagment PCR Amplicons for the Illumina COVIDSeq protocol.\n\nLinks: \n* [1. Anneal RNA](./0cded6)\n* [2. Synthesize First Strand cDNA](./0cded6-synthesize)\n* [3. Amplify cDNA](./0cded6-amplify)\n* [4. Tagment PCR Amplicons](./0cded6-tagment)\n* [5. Post Tagmentation Clean Up](./0cded6-cleanup)\n* [6. Amplify Tagmented Amplicons](./0cded6-amplify2)\n\n\n",
diff --git a/protoBuilds/0cded6/README.json b/protoBuilds/0cded6/README.json
index ad6a5337e7..0e21aee801 100644
--- a/protoBuilds/0cded6/README.json
+++ b/protoBuilds/0cded6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina COVIDSeq"
@@ -10,7 +10,7 @@
"internal": "0cded6",
"labware": "\nQuantgene 96 Aluminum Block 200 \u00b5L\nAgilent 96 Well Plate 200 \u00b5L #5042-8502\nAgilent With Nonskirted 96 Well Plate 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina COVIDSeq\n\n\n",
"deck-setup": "\n\n\n",
"description": "\nThis protocol performs Anneal RNA for the Illumina COVIDSeq protocol.\n\nLinks: \n* [1. Anneal RNA](./0cded6)\n* [2. Synthesize First Strand cDNA](./0cded6-synthesize)\n* [3. Amplify cDNA](./0cded6-amplify)\n* [4. Tagment PCR Amplicons](./0cded6-tagment)\n* [5. Post Tagmentation Clean Up](./0cded6-cleanup)\n* [6. Amplify Tagmented Amplicons](./0cded6-amplify2)\n\n\n",
diff --git a/protoBuilds/0d2950/README.json b/protoBuilds/0d2950/README.json
index 467492a636..92705c3d9d 100644
--- a/protoBuilds/0d2950/README.json
+++ b/protoBuilds/0d2950/README.json
@@ -17,7 +17,7 @@
"internal": "0d2950\n",
"labware": "* [NEST 2 mL 96-Well Deep Well Plate, V Bottom](https://shop.opentrons.com/collections/lab-plates/products/nest-0-2-ml-96-well-deep-well-plate-v-bottom)\n* [Opentrons 1000uL Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* [Opentrons 4-in-1 tube rack](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[appliedbiosystems](https://www.thermofisher.com/content/dam/LifeTech/Documents/PDFs/clinical/taqpath-COVID-19-combo-kit-full-instructions-for-use.pdf)\n\n",
+ "partner": "[appliedbiosystems](https://www.thermofisher.com/content/dam/LifeTech/Documents/PDFs/clinical/taqpath-COVID-19-combo-kit-full-instructions-for-use.pdf)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P1000 GEN2 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Pick up tip.\n2. 200ul of sample is aspirated.\n3. 200ul of sample is dispensed into deepwell plate, by column.\n4. Drop tip.\n5. Repeat up to the specified number of steps.\n\n\n\n",
@@ -25,7 +25,7 @@
"title": "Extraction Prep for TaqPath Covid-19 Combo Kit"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "appliedbiosystems",
+ "partner": "appliedbiosystems\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP1000 GEN2 Single Channel Pipette\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nPick up tip.\n200ul of sample is aspirated.\n200ul of sample is dispensed into deepwell plate, by column.\nDrop tip.\nRepeat up to the specified number of steps.\n",
diff --git a/protoBuilds/0d868e-part2/README.json b/protoBuilds/0d868e-part2/README.json
index 0fd0f688cf..4f4f604a16 100644
--- a/protoBuilds/0d868e-part2/README.json
+++ b/protoBuilds/0d868e-part2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Generic"
@@ -10,7 +10,7 @@
"internal": "0d868e-part2",
"labware": "\nFroggabio 96 Well Plate 300 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 24 Well Aluminum Block with NEST 2 mL Snapcap\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Generic\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs sections 3 to 8 of the Ilumina TruSeq Stranded mRNA SOP (Synthesize First Strand cDNA to the end of Cleanup Ligated Fragments). Pauses are included in the protocol for user intervention. Pauses will include details from the SOP for the user to follow for manual steps - the user may see these details in the Opentrons App during the pause. In the event of tip depletion, the protocol will automatically pause prompting the user to refill tips. Magnetic engagement height should be determined and adjusted accordingly, since the plate on the magnetic module is not in the labware library. If reagents take more than one column in the reagent plate (beads for example), then the reagent volume in each well should be per sample, otherwise, reagent liquid volume should be per protocol if reagent takes one column.\n\n\n",
diff --git a/protoBuilds/0d868e/README.json b/protoBuilds/0d868e/README.json
index 32c39bdbbf..601134fbe8 100644
--- a/protoBuilds/0d868e/README.json
+++ b/protoBuilds/0d868e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Generic"
@@ -10,7 +10,7 @@
"internal": "0d868e",
"labware": "\nFroggabio 96 Well Plate 300 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Generic\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs sections 1 & 2 of the Ilumina TruSeq Stranded mRNA SOP (Purify mRNA to the end of Fragment mRNA). Pauses are included in the protocol for user intervention. Pauses will include details from the SOP for the user to follow for manual steps - the user may see these details in the Opentrons App during the pause. In the event of tip depletion, the protocol will automatically pause prompting the user to refill tips. Magnetic engagement height should be determined and adjusted accordingly, since the plate on the magnetic module is not in the labware library. If reagents take more than one column in the reagent plate (BWB for example), then the reagent volume in each well should be per sample, otherwise, reagent liquid volume should be per protocol if reagent takes one column.\n\n\n",
diff --git a/protoBuilds/0dda91/README.json b/protoBuilds/0dda91/README.json
index fcfb7c1b46..482ed5b59a 100644
--- a/protoBuilds/0dda91/README.json
+++ b/protoBuilds/0dda91/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Pooling"
@@ -10,7 +10,7 @@
"internal": "0dda91",
"labware": "\nOT-2 Filter Tips, 20\u00b5L\nOpentrons Tough 0.2 mL 96-Well PCR Plate, Full Skirt\n4-in-1 Tube Rack Set\nFisherbrand 2.0 mL Microcentrifuge Tubes with Locking Snap Cap\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Pooling\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "This protocol uses a .csv file to pool a calculated volume of each sample from a PCR plate into one or more 1.5 mL snapcap tube(s). \n\nExplanation of complex parameters below:\n* `input .csv file`: Here, you should upload a .csv file formatted in the following way, being sure to include the header line:\n```\n Sample Location,Qubits,Volume (ul) for ng: 70,Sample Destination,,,,,,,,,,,\nA1,3.5,20.0,A1,,,,,,,,,,,\nA2,15,4.7,A2,,,,,,,,,,,\n```\n\n---\n\n",
diff --git a/protoBuilds/0df680/README.json b/protoBuilds/0df680/README.json
index e6bddbc471..5b596c00a1 100644
--- a/protoBuilds/0df680/README.json
+++ b/protoBuilds/0df680/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -9,7 +9,7 @@
"internal": "0df680",
"labware": "\nOpentrons 96 Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n\n",
"description": "This protocol performs a custom cherrypicking for any number of source and destination plates using a P20 single-channel pipette. An output file mapping each sample ID, source plate ID, and destination plate ID is output for each run.\n\nThe input .csv file should be formatted as follows **including header line**\n\n```\n,clone ID,0ur plate ID,Source Labware,Source Slot,Source Well,Source Aspiration Height Above Bottom (in mm),Dest Labware,Dest Slot,Dest. Plate ID,Dest Well,Volume (in ul)\nn1,CL-1418591,1,biorad_96_wellplate_200ul_pcr,2,B06,1,biorad_96_wellplate_200ul_pcr,11,RAILPB03,A01,15\nn2,CL-1418640,1,biorad_96_wellplate_200ul_pcr,2,D12,1,biorad_96_wellplate_200ul_pcr,11,RAILPB03,A02,15\nn3,CL-1418621,1,biorad_96_wellplate_200ul_pcr,2,H09,1,biorad_96_wellplate_200ul_pcr,11,RAILPB03,A03,15\nn4,CL-1418715,2,biorad_96_wellplate_200ul_pcr,3,H09,1,biorad_96_wellplate_200ul_pcr,11,RAILPB03,A04,15\nn5,CL-1418738,3,biorad_96_wellplate_200ul_pcr,4,A01,1,biorad_96_wellplate_200ul_pcr,11,RAILPB03,A05,15\nn6,CL-1418818,3,biorad_96_wellplate_200ul_pcr,4,A11,1,biorad_96_wellplate_200ul_pcr,11,RAILPB03,A06,15\nn7,CL-1418744,3,biorad_96_wellplate_200ul_pcr,4,G01,1,biorad_96_wellplate_200ul_pcr,11,RAILPB03,A07,15\nn8,CL-1418866,4,biorad_96_wellplate_200ul_pcr,5,C05,1,biorad_96_wellplate_200ul_pcr,11,RAILPB03,A08,15\nn9,CL-1418976,5,biorad_96_wellplate_200ul_pcr,6,C07,1,biorad_96_wellplate_200ul_pcr,11,RAILPB03,A09,15\n...\n```\n\nYou can also access a template ***[here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/0df680/ex.csv)***.\n\n\n",
"internal": "0df680\n",
diff --git a/protoBuilds/0e39fc/README.json b/protoBuilds/0e39fc/README.json
index 478624d4e8..7c62cc1cc2 100644
--- a/protoBuilds/0e39fc/README.json
+++ b/protoBuilds/0e39fc/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "0e39fc",
"labware": "\nBio-Rad 96 Tube Rack with Applied Biosystems 0.2 mL #HSP9601\nBio-Rad 96 Well Plate 200 \u00b5L PCR #hsp9601\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs PCR prep on up to 384 samples. NTC with PCR mastermix will be plated in wells P17-19. cDNA IPC template will be plated in wells P20-22, and NTC with IPC mastermix will be plated in wells P23-24.\n\n\n",
diff --git a/protoBuilds/0e525c/README.json b/protoBuilds/0e525c/README.json
index 328a88a776..861e2cb28d 100644
--- a/protoBuilds/0e525c/README.json
+++ b/protoBuilds/0e525c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "0e525c",
"labware": "\nOT-2 Filter Tips, 200\u00b5L (999-00081)\nOT-2 Filter Tips, 20\u00b5L (999-00099)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n\n\n",
"description": "This protocol performs a custom ddPCR prep in semi-skirted PCR plates using an adapter to fit into the deck slots.\n\n---\n",
diff --git a/protoBuilds/0e696e/README.json b/protoBuilds/0e696e/README.json
index 8bbc445805..6ee8cf2b70 100644
--- a/protoBuilds/0e696e/README.json
+++ b/protoBuilds/0e696e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -10,7 +10,7 @@
"internal": "0e696e",
"labware": "\nAny verified labware found in our Labware Library\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"deck-setup": "* Example deck setup - tip racks loaded onto remining slots.\n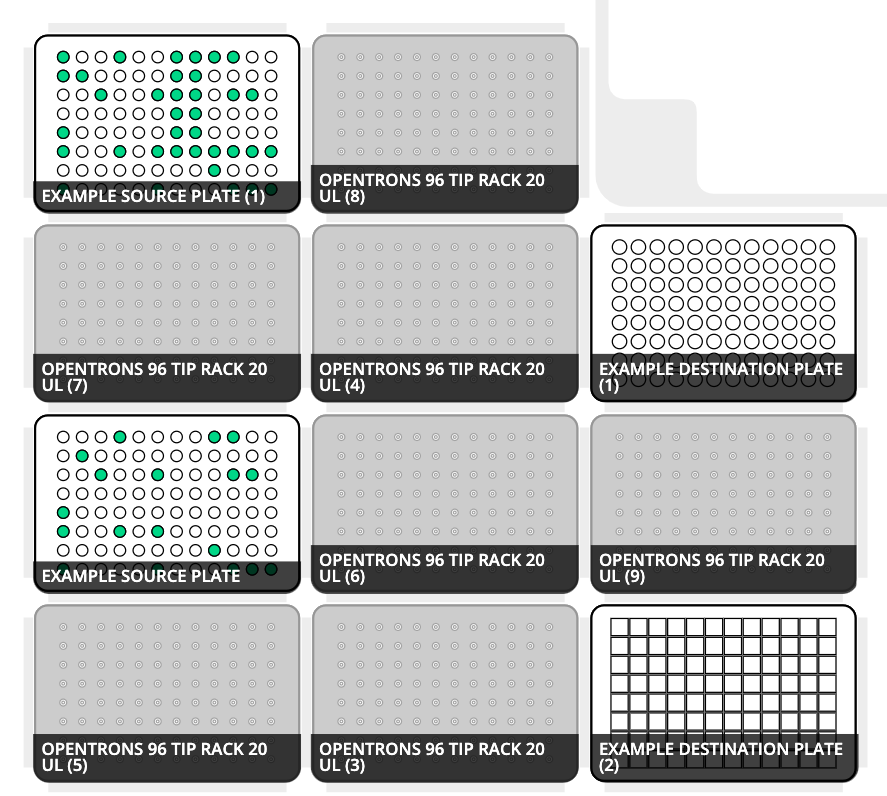\n\n---\n\n",
"description": "\nOur most robust cherrypicking protocol. Specify aspiration height, labware, pipette, as well as source and destination wells with this all inclusive cherrypicking protocol.\n\n\n\nExplanation of complex parameters below:\n\n* `input .csv file`: Here, you should upload a .csv file formatted in the [following way](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/0e696e/example.csv), making sure to include headers in your csv file. Refer to our [Labware Library](https://labware.opentrons.com/?category=wellPlate) to copy API names for labware to include in the `Source Labware` column of the .csv.\n* `Pipette Model`: Select which pipette you will use for this protocol.\n* `Pipette Mount`: Specify which mount your single-channel pipette is on (left or right)\n* `Tip Type`: Specify whether you want to use filter tips.\n* `Tip Usage Strategy`: Specify whether you'd like to use a new tip for each transfer, or keep the same tip throughout the protocol.\n\n\n---\n\n\n",
diff --git a/protoBuilds/0e7175-pt2/README.json b/protoBuilds/0e7175-pt2/README.json
index 60ecd6423e..c297eec8c0 100644
--- a/protoBuilds/0e7175-pt2/README.json
+++ b/protoBuilds/0e7175-pt2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "0e7175-pt2",
"labware": "\nNEST 12 Well Reservoir 195mL\nEppendorf Isopaks\nGreiner 96 well Chimney Bottom\nOpentrons 300ul Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\n\n---\n\n",
"description": "This protocol provides sample and reagents to 2 96 well plates according to the [Luminex MagPlex\u00ae Microspheres kit](https://www.luminexcorp.com/magplex-microspheres/#overview). Column 1 of the source isopak 1 is dispensed into rows A, C, E, G of the destination plate 1 in columns 1 and 2. Column 2 of the source isopak is dispensed into rows A, C, E, G of the destination plate 1 in columns 3 and 4, etc. Column 1 of the source isopak 2 is dispensed into rows B, D, F, H of the destination plate 1 in columns 1 and 2, etc. This is then repeated for destination plate 2. Tips are removed from rows B, D, F, H of the tipracks on slot 10 and 11 to allow for the multi-channel pipette to access the isopaks. The tiprack on slot 9 should be a full tip rack.\n\n\nExplanation of complex parameters below:\n* `Sample Volume`: Specify the sample volume to be replicated in microliters.\n* `Use middle two columns?`: Specify whether using the middle two columns in source isopaks. See below for deck map if selected. If selected, the protocol will pause after the first 96 plate is filled to prompt the user to replace the alternate tip rack on slot 11.\n* `Antibody Volume`: Specify the sample volume to be replicated in microliters.\n* `SA-PE Volume`: Specify the SA-PE volume to be replicated in microliters.\n* `P300 Mount`: Specify which mount (left or right) to host the P300 Multi-Channel Pipette.\n\n---\n\n",
diff --git a/protoBuilds/0e7175/README.json b/protoBuilds/0e7175/README.json
index c98044b9e6..866d2b278f 100644
--- a/protoBuilds/0e7175/README.json
+++ b/protoBuilds/0e7175/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "0e7175",
"labware": "\nEppendorf Isopaks\nGreiner 96 well Chimney Bottom\nOpentrons 300ul Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "This protocol replicates 6 or 7 isopaks from 1 or 2 source isopaks depending on the sample volume specified by the user according to the [Luminex MagPlex\u00ae Microspheres kit](https://www.luminexcorp.com/magplex-microspheres/#overview). Tips are removed from rows B, D, F, H of the tiprack to allow for the multi-channel pipette to access the isopaks.\n\n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify whether running 24 or 48 samples. If running 24 samples, one isopak will be replicated into 7 isopaks. If running 48 samples, isopak 1 on slot 10 will supply empty isopaks in slots 1, 2, and 3, and isopak 2 on slot 11 will supply empty isopaks in slot 4, 5, and 6. See below for diagrams.\n* `Use middle two columns?`: Specify whether using the middle two columns in source isopak(s). If this option is selected, the middle two columns of the source isopaks will be ignored, and the middle two tubes in the destination isopaks will also be ignored for dispensing.\n* `Sample Volume`: Specify the sample volume to be replicated in microliters.\n* `P300 Mount`: Specify which mount (left or right) to host the P300 Multi-Channel Pipette.\n\n---\n\n",
diff --git a/protoBuilds/0e8475/README.json b/protoBuilds/0e8475/README.json
index 9ca5139660..03f7726212 100644
--- a/protoBuilds/0e8475/README.json
+++ b/protoBuilds/0e8475/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina GUIDE-seq"
@@ -10,7 +10,7 @@
"internal": "0e8475",
"labware": "\nBio-Rad 96 Well Plate 200 \u00b5L PCR #hsp9601\nOpentrons 96 Well Aluminum Block with Bio-Rad Well Plate 200 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina GUIDE-seq\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a semi-automated protocol for the Illumina GUIDE-seq NGS Library Prep for up to 16 samples. The user is prompted to move the plate to and from a thermal cycler at various points throughout the protocol.\n\n\n",
diff --git a/protoBuilds/0e8e3/README.json b/protoBuilds/0e8e3/README.json
index ff1867ac68..c63401fc1d 100644
--- a/protoBuilds/0e8e3/README.json
+++ b/protoBuilds/0e8e3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -9,7 +9,7 @@
"internal": "0e8e3",
"labware": "\nThermoFisher Nunc\u2122 96-Well Microplates #249944\nBio-Rad 96 Well Plate 200 \u00b5L PCR #HSP9601\nNEST 12-Well Reservoirs, 15 mL\nOpentrons 20\u00b5l and 300\u00b5l 96 Tipracks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol performs a custom PCR prep across 6 96-well plates. It accommodates selection of P20 and P300 GEN2 multi- or single-channel pipettes.\n\n---\n\n",
"internal": "0e8e3\n",
diff --git a/protoBuilds/0eaeb5/README.json b/protoBuilds/0eaeb5/README.json
index 754451261b..208559aa63 100644
--- a/protoBuilds/0eaeb5/README.json
+++ b/protoBuilds/0eaeb5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS": [
"Paragon Genomics CleanPlex NGS Panel Kit"
@@ -10,7 +10,7 @@
"internal": "0eaeb5",
"labware": "\nBio-Rad 96 Well Plate 200 uL PCR\nNEST 96 Deepwell Plate 2mL\nNEST 12-Well Reservoir, 15 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS\n\t* Paragon Genomics CleanPlex NGS Panel Kit\n\n",
"deck-setup": "* `Slot 1`: Magnetic Module + Bio-Rad 96 Well Plate 200 uL PCR (mag_plate)\n* `Slot 2`: Bio-Rad 96 Well Plate 200 uL PCR (sample_plate) for calibration purposes. This labwared will be transferred among Slot 2, Slot 1, and external thermocycler after.\n* `Slot 3`: Temperature Module + Bio-Rad 96 Well Plate 200 uL PCR (reagent_plate)\n* `Slots 4/7/6`: Opentrons 96 Filter Tip Rack 200 uL\n* `Slots 9/8/5`: Opentrons 96 Filter Tip Rack 20 uL\n* `Slot 10`: NEST 12-Well Reservoir, 15 mL (reagent_reservoir)\n* `Slot 11`: NEST 12-Well Reservoir, 15 mL (waste_reservoir)\n\n\n\n",
"description": "This protocol performs the [CleanPlex\u00ae NGS Panel](https://www.paragongenomics.com/wp-content/uploads/2021/01/UG1001-08-CleanPlex-NGS-Panel-User-Guide-v2.pdf) for up to 96 samples.\n\nThe protocol is broken down into 6 main parts:\n* `1A`: Multiplex PCR (mPCR) reaction\n* `1B`: Post-mPCR Purification\n* `2A`: Digestion Reaction\n* `2B`: Post-Digestion Purification\n* `3A`: Second PCR Reaction\n* `3B`: Post-Second PCR Purification\n\nExplanation of complex parameters below:\n* Test parameters\n * `DRYRUN`: The default setting is `False` . If set to `True`, the protocol will be in dry run testing mode. The pipette will return the tip to the tiprack after each liquid transfer.\n\t* `TEST_MODE_BEADS`: The default setting is `False` . If set to `True`, the protocol will be in beads testing mode. The beads incubation and magnet engaging time will be in unit of seconds rather than minutes. Meanwhile, the pipette will return the tip to the tiprack after each liquid transfer.\n* Timer parameters\n * `timer_h`: Set up the time for hours\n\t* `timer_m`: Set up the time for minutes\n* Flow rate parameters\n * `water_rate`: Set up the flow rate of transferring water liquids, unit: folds of the default speed, 1.2 means 120 % of the default speed while 0.6 indicates 60% of the default speed\n\t* `buffer_mmx_primer_rate`: Set up the flow rate of transferring buffer/mastermix/primer liquids, unit: the same as above\n * `sample_rate`: Set up the flow rate of transferring sample-related liquids, unit: folds of the default speed\n\t* `beads_rate`: Set up the flow rate of transferring bead liquids, unit: the same as above\n\t* `ethanol`: Set up the flow rate of transferring ethanol liquids, unit: the same as above\n* beads mixing parameters\n * `beads_mix_reps`: The frequencies that robot will mix the beads\n\t* `beads_mix_rate`: The flow rate that the robot will mix the beads, the same as `Flow rate parameters`\n* Magnetic Module parameters\n * `magdeck_engage_height`: The height of magnet that will engage, in `mm`\n\t* `ethnaol_dis_zoffset`: The height of dispensing ethanol from the top of the well during ethanol washing, in `mm`\n\t* `incubation_time`: The time it takes for DNA binding before engaging the magnet, unit in `minutes` when `TEST_MODE_BEADS` is `False`\n * `beads_engaging_time`: The time it takes for beads binding after engaging the magnet, unit in `minutes` when `TEST_MODE_BEADS` is `False`\n\t* `airdry_time`: The time it takes for airdrying after removing the enthanol, unit in `minutes` when `TEST_MODE_BEADS` is `False`\n\t* `ethanol_wash_time`: The time it takes for ethanol washing after adding it in the well, unit in `minutes` when `TEST_MODE_BEADS` is `False`\n\n---\n\n",
diff --git a/protoBuilds/0edc68/README.json b/protoBuilds/0edc68/README.json
index 9cf521df56..12b9533fd7 100644
--- a/protoBuilds/0edc68/README.json
+++ b/protoBuilds/0edc68/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Purification"
@@ -10,7 +10,7 @@
"internal": "0edc68",
"labware": "\nNEST 96 Deepwell Plate 2mL #503001 on Deepwell Adapter\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 24 Well Aluminum Block with Generic 2 mL Screwcap\nOpentrons 96 Well Aluminum Block with NEST Well Plate 100 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Purification\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a custom VHH protein purification assay.\n\n\n",
diff --git a/protoBuilds/0ef96f/README.json b/protoBuilds/0ef96f/README.json
index 4d9a9f650b..a45a26c9d4 100644
--- a/protoBuilds/0ef96f/README.json
+++ b/protoBuilds/0ef96f/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom cherrypicking from a 4x6 Opentrons tuberack containing 2ml Eppendorf tubes to an IDT 96-deepwell plate, and back to new tubes on the tuberack. The information for this workflow should be input in .csv file formatted as shown in the following example (including headers):\nLabel,Pos,nM,FC,Rxn,Vol,h2O,Tube\nP_F3,A1,28.9,0.8,100,10,30,T1\nP_B3,A2,28.32,0.8,100,10,,T1\nP_FIP,A3,53.8,1.6,100,20,,T1\nP_BIP,A4,82,1.6,100,20,,T1\nP_LF,A5,32.5,0.4,100,5,,T1\nP_LB,A6,29.4,0.4,100,5,,T1\nP_F3,A7,28.9,0.8,100,10,35,T2\nP_B3,A8,28.32,0.8,100,10,,T2\nP_FIP,A9,53.8,1.6,100,20,,T2\nP_BIP,A10,82,1.6,100,20,,T2\nP_LF,A11,32.5,0.4,100,5,,T2\nP_LB,A12,29.4,0.4,100,5,,T2\n\n\n\nIDT 96-deepwell plate\nOpentrons 4-in-1 tuberack set with 4x6 insert holding 2ml Eppendorf snapcap tubes\nOpentrons P20- and P300-single channel electronic pipettes (GEN2)\nOpentrons 20 and 300ul tipracks\n\n\n\n4x6 tuberack with 2ml Eppendorf tubes (slot 4)\n",
"internal": "0ef96f",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"description": "This protocol performs a custom cherrypicking from a 4x6 Opentrons tuberack containing 2ml Eppendorf tubes to an IDT 96-deepwell plate, and back to new tubes on the tuberack. The information for this workflow should be input in `.csv` file formatted as shown in the following example (**including headers**):\n\n```\nLabel,Pos,nM,FC,Rxn,Vol,h2O,Tube\nP_F3,A1,28.9,0.8,100,10,30,T1\nP_B3,A2,28.32,0.8,100,10,,T1\nP_FIP,A3,53.8,1.6,100,20,,T1\nP_BIP,A4,82,1.6,100,20,,T1\nP_LF,A5,32.5,0.4,100,5,,T1\nP_LB,A6,29.4,0.4,100,5,,T1\nP_F3,A7,28.9,0.8,100,10,35,T2\nP_B3,A8,28.32,0.8,100,10,,T2\nP_FIP,A9,53.8,1.6,100,20,,T2\nP_BIP,A10,82,1.6,100,20,,T2\nP_LF,A11,32.5,0.4,100,5,,T2\nP_LB,A12,29.4,0.4,100,5,,T2\n```\n\n---\n\n\n* IDT 96-deepwell plate\n* [Opentrons 4-in-1 tuberack set](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 4x6 insert holding [2ml Eppendorf snapcap tubes](https://online-shop.eppendorf.us/US-en/Laboratory-Consumables-44512/Tubes-44515/Eppendorf-Safe-Lock-Tubes-PF-8863.html)\n* [Opentrons P20- and P300-single channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 20 and 300ul tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n\n---\n\n\n4x6 tuberack with 2ml Eppendorf tubes (slot 4)\n\n\n",
"internal": "0ef96f\n",
diff --git a/protoBuilds/0f3b60/README.json b/protoBuilds/0f3b60/README.json
index d350cc5b21..882e4209ff 100644
--- a/protoBuilds/0f3b60/README.json
+++ b/protoBuilds/0f3b60/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "0f3b60",
"labware": "\nNEST 96 Deepwell Plate 2mL #503001\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol produces a cleanup as part of the NEBNext kit. For detailed protocol steps, please see below. \n\n\n",
diff --git a/protoBuilds/0f409e/README.json b/protoBuilds/0f409e/README.json
index 7120494bba..827e2e0f9c 100644
--- a/protoBuilds/0f409e/README.json
+++ b/protoBuilds/0f409e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "0f409e",
"labware": "\nCorning 24 Well Plate 3.4 mL Flat #3337\nCorning 96 Well Plate 360 \u00b5L Flat #3650\nAgilent 1 Well Reservoir 290 mL #201252-100\nOpentrons 96 Filter Tip Rack 1000 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a custom plate filling protocol from a 24-well source plate into one quadrant of a 96-well plate.\n\n\n",
diff --git a/protoBuilds/0f4405/README.json b/protoBuilds/0f4405/README.json
index fa7491ba48..5a07c0a071 100644
--- a/protoBuilds/0f4405/README.json
+++ b/protoBuilds/0f4405/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext\u00ae Ultra\u2122 II DNA Library Prep Kit for Illumina\u00ae"
@@ -10,7 +10,7 @@
"internal": "0f4405",
"labware": "\nEppendorf twin.tec \u00ae PCR Plate 96 #0030128648\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nOpentrons 96-Well Aluminum Block\nOpentrons 20\u00b5L Filter Tips\nOpentrons 200\u00b5L Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\u00ae Ultra\u2122 II DNA Library Prep Kit for Illumina\u00ae\n\n",
"deck-setup": " \n\nreservoir (slot 9): \n* column 1: EtOH\n* column 2: elution buffer\n* column 12: waste (loaded empty)\n\nThermocycler plate (slot 7, 8, 10, 11): \n* columns 1-3: DNA samples\n\nMagnetic Module plate (slot 1): \n* columns 1-6: beads\n\nTemperature Module plate (slot 3):\n* column 1: mastermix 1\n* column 2: adaptor\n* column 3: mastermix 2\n* column 4: USER\n* columns 5-7: final PCR reagents\n\n---\n\n",
"description": "This protocol performs the [NEBNext\u00ae Ultra\u2122 II DNA Library Prep Kit for Illumina\u00ae](https://www.neb.com/products/e7645-nebnext-ultra-ii-dna-library-prep-kit-for-illumina#Protocols,%20Manuals%20&%20Usage). The protocol includes a fully automated End Repair PCR step on the Opentrons Thermocycler. Up to 24 samples can be processed in one protocol run without the need to replenish tipracks or change out labware.\n\n---\n\n",
diff --git a/protoBuilds/0f5985/README.json b/protoBuilds/0f5985/README.json
index 9bae0a7319..885b451288 100644
--- a/protoBuilds/0f5985/README.json
+++ b/protoBuilds/0f5985/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Blood Sample"
@@ -10,7 +10,7 @@
"internal": "0f5985",
"labware": "\nNEST 1 Well Reservoir 195 mL\nNEST 12 Well Reservoir 15 mL\nUSA Scientific PlateOne\u00ae Deep 96-Well 2 mL Polypropylene Plate (P/N 1896-2800)\nAbgene\u2122 96 Well 0.8mL Polypropylene Deepwell Storage Plate (Thermo AB-0859)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Blood Sample\n\n",
"deck-setup": "\n\n",
"description": "This protocol automates the GenFind V3 protocol from Beckman Coulter for [DNA isolation from whole blood or serum with magnetic particles](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2022-01-24/kz234fm/C36038AB.pdf). This protocol utilizes a P300-Multi channel pipette Gen2 (and optionally a p300-Single channel pipette Gen2) and gives the user the option to select a variable number of samples (Ideally a multiple of 8 for efficient use of reagents) and whether to dispense liquid waste in a 1-well reservoir in slot 6 or in the fixed trash container. The user can also choose whether they want to use Proteinase K and elution buffer from a tube-rack rather than the 12 well reservoir. \u00dfThis protocol is designed for holding the samples on the [USA Scientific PlateOne\u00ae Deep 96-Well 2 mL Polypropylene Plate (P/N 1896-2800)](https://www.usascientific.com/plateone-96-deep-well-2ml/p/PlateOne-96-Deep-Well-2mL) on a 2nd generation Magnetic module. The purified samples are transferred to a [Abgene\u2122 96 Well 0.8mL Polypropylene Deepwell Storage Plate (Thermo AB-0859)](https://www.thermofisher.com/order/catalog/product/AB0859) in the final step. The protocol uses the [NEST 12 Well Reservoir 15 mL](https://shop.opentrons.com/nest-12-well-reservoirs-15-ml/) and the [NEST 1 Well Reservoir 195 mL](https://shop.opentrons.com/nest-1-well-reservoirs-195-ml/) as sources of reagents. All reagents except the two wash buffers (Wash WBB and Wash WBC are stored either in the twelve well reservoir or the tube rack)\n\n\nThe protocol is broken down into 5 main parts:\n* Lysis buffer and proteinase K is added to the samples. The protocol is paused and the samples are incubated until the user is ready to continue. The pause allows the user to resuspend the magnetic particles in Bind BBB before adding the solution to the twelve well reservoir.\n* Bind BBB is added and the samples are incubated in order to bind DNA in the lysed solution.\n* The supernatant is discarded and the beads are washed two times with Wash WBB and two times with Wash WBC buffers.\n* Elution buffer is added to the samples which are then incubated and mixed two times.\n* Finally the supernatants containing DNA are transferred to the target plate.\n\nExplanation of parameters below:\n* `Number of samples`: How many samples to run, it should ideally be a multiple of 8 for efficient use of resources.\n* `Blood cells or serum?`: Choose what kind of samples to use for the protocol. This parameter will set the sample volume to 200 uL for blood cells, and 400 for serum samples. It will also set the elution volume to 200 uL for blood cell samples, and 40 uL for serum samples.\nThis setting will affect the volume requirements for all other reagents as well since they are all based on the starting volume.\n* `x offset for bead aspiration`: How many millimeters away from the center of the sample wells to aspirate bead supernatant from in the x (left/right) direction, the default is 1 mm.\n* `Magnet extension (mm)`: How far from the base of the sample plate to extend the magnets of the magnet deck (default is 4.7 mm)\n* `Proteinase K location`: Choose whether the proteinase K is located in tube 1 of the tube rack, or well 1 of the 12 well reservoir\n* `Elution buffer location`: Choose whether the elution buffer is located in tube 2 to 8 of the tube rack, or well 6 and 7 of the 12 well reservoir\n* `Liquid waste reservoir?`: If set to `yes` the protocol will use a NEST 1 well 195 mL reservoir as a target of liquid waste, otherwise it will empty liquid waste into the regular trash bin. \n\n---\n\n",
diff --git a/protoBuilds/0f5c31/README.json b/protoBuilds/0f5c31/README.json
index 4a2f164c2b..422c68271c 100644
--- a/protoBuilds/0f5c31/README.json
+++ b/protoBuilds/0f5c31/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteiomics": [
"TPP"
@@ -10,7 +10,7 @@
"internal": "0f5c31",
"labware": "\nPall 96 Well Plate 350 \u00b5L #8036\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nOpentrons 96 Well Aluminum Block with NEST Well Plate 100 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteiomics\n\t* TPP\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a custom Thermal Proteome Profiling (TPP) protocol using the Opentrons Thermocycler and Temperature Modules. The user is prompted as necessary to replace plates, reagents, and tips as needed throughout the protocol.\n\n\n",
diff --git a/protoBuilds/0f7910/README.json b/protoBuilds/0f7910/README.json
index 3bdce5e2b0..a73989994a 100644
--- a/protoBuilds/0f7910/README.json
+++ b/protoBuilds/0f7910/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "0f7910",
"labware": "\nCorning 384 Well Plate 112 \u00b5L Flat\nCorning 96 Well Plate 360 \u00b5L\nOpentrons 96 Tip Rack 20 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "Select the format for the source and destination plates. See diagram below. If a 384 plate is selected for destination plate, it should always be in slot 2. If a 96 plate is selected for destination, it should always be in slot 1. Source plates always start from slot 3 to 9, and tip racks are always the same. You can select which tip the protocol will start on in the fields below. If the global transfer volume is over 20ul, then the P300 will be used. If the transfer volume is 20 or less, the P20 will be used. A value of \"8\" for the starting position of the tip would mean to start H1 of the tip rack, and a value of 10 would mean to start at B2 of the tip rack, since it iterates down by column. The csv should be formatted as such in the header:\n\n```\nSource plate barcode, Source plate slot (3-9), Source well (A1, B1, etc.), Destination plate barcode, Destination well\n```\n\n\n",
diff --git a/protoBuilds/0fa015/README.json b/protoBuilds/0fa015/README.json
index 5be096f596..a0f6cd5b1b 100644
--- a/protoBuilds/0fa015/README.json
+++ b/protoBuilds/0fa015/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol fillsNEST 96-Well PCR Plate with QE using a GEN2 Multi-Channel Pipette.\n\nThe protocol can fill up to 9 plates and will dispense 10\u00b5L of QE into each well\n\nThis protocol is based largely on this protocol and will employ similar behaviors (touch tip after aspiration, blow out, and aspiration at the top of the well to prevent cross-contamination).\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons Multi-Channel Pipette, GEN2\nOpentrons Tips\nNEST 12-Well Reservoir\nNEST 96-Well PCR Plate\nReagents (QE)\n\n\n\nSlots 1 - 9: NEST 96-Well PCR Plate\nSlot 10: Opentrons Tips\nSlot 11: NEST 12-Well Reservoir\nA1: QE (for plates 1-5)\nA2: QE (for plates 6-9)\n\n\nUsing the customizations field (below), set up your protocol.\n Pipette Type: Select which GEN2 Multi-Channel Pipette (p300 or p20) will be used\n Pipette Mount: Select which mount (left or right) the Multi-Channel Pipette is attached to\n* Number of Plates (1-9): Specify the number of plates to fill",
"internal": "0fa015",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol fills[NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) with QE using a [GEN2 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes).\n\nThe protocol can fill up to 9 plates and will dispense 10\u00b5L of QE into each well\n\nThis protocol is based largely on [this protocol](https://develop.protocols.opentrons.com/protocol/17cb2d) and will employ similar behaviors (touch tip after aspiration, blow out, and aspiration at the top of the well to prevent cross-contamination).\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Multi-Channel Pipette, GEN2](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* Reagents (QE)\n\n\n---\n\n\nSlots 1 - 9: [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n\nSlot 10: [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\nSlot 11: [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\nA1: QE (for plates 1-5)\nA2: QE (for plates 6-9)\n\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Pipette Type**: Select which GEN2 Multi-Channel Pipette (p300 or p20) will be used\n* **Pipette Mount**: Select which mount (left or right) the Multi-Channel Pipette is attached to\n* **Number of Plates (1-9)**: Specify the number of plates to fill\n\n\n\n",
"internal": "0fa015\n",
diff --git a/protoBuilds/0fd3dd/README.json b/protoBuilds/0fd3dd/README.json
index fa41e62fac..ff9953789c 100644
--- a/protoBuilds/0fd3dd/README.json
+++ b/protoBuilds/0fd3dd/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PROTEINS & PROTEOMICS": [
"ELISA"
@@ -9,7 +9,7 @@
"internal": "0fd3dd",
"labware": "\nStarlab 12 Reservoir 22000 \u00b5L #E2110-1200\nStarlab 1 Reservoir 240000 \u00b5L #E2999-2496\nThermo Maxisorb #Maxisorb\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PROTEINS & PROTEOMICS\n\t* ELISA\n\n\n",
"description": "This protocol performs a custom sample plating and ELISA for up to 88 samples on up to 3 test (ELISA) plates.\n\n\n",
"internal": "0fd3dd\n",
diff --git a/protoBuilds/0fdc01/README.json b/protoBuilds/0fdc01/README.json
index 6d8da42ae4..6f3e35e66d 100644
--- a/protoBuilds/0fdc01/README.json
+++ b/protoBuilds/0fdc01/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -9,7 +9,7 @@
"internal": "0fdc01",
"labware": "\nNEST 12-Well Reservoirs, 15 mL\nOpentrons 300uL Tips\nOpentrons 20\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This is a PCR Prep protocol. The protocol begins by adding mastermix to the PCR plate and then adds in the samples. The user has prompted to pause at various points.\n\n",
"internal": "0fdc01\n",
diff --git a/protoBuilds/0ff0c3/README.json b/protoBuilds/0ff0c3/README.json
index 34617c96c9..a25b804d88 100644
--- a/protoBuilds/0ff0c3/README.json
+++ b/protoBuilds/0ff0c3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Purification"
@@ -10,7 +10,7 @@
"internal": "0ff0c3",
"labware": "\nOT-2 Filter Tips, 200\u00b5L\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nNEST 1-Well Reservoir, 195 mL\n\nOpentrons Tough 0.2 mL 96-Well PCR Plate, Full Skirt\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Nucleic Acid Purification\n\n",
"deck-setup": "\n\n\n\n---\n\n",
"description": "This is a Nucleaic Acid Purification Protocol for the OT-2 liquid handling system\n\nThe protocol is broken down into 3 main parts:\n* Pipette mixing of lysates, binding buffer and magnetic beads\n* Bead washing 3x using magnetic module\n* Final elution to PCR plate\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 6) to the elution PCR plate (slot 3). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\n---\n\n",
diff --git a/protoBuilds/1033g1/README.json b/protoBuilds/1033g1/README.json
index 05fb95bf40..abdb66cc74 100644
--- a/protoBuilds/1033g1/README.json
+++ b/protoBuilds/1033g1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Vasudha Nair",
+ "author": "Vasudha Nair\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Cell and Tissue Culture": [
"Cell and Tissue Culture"
@@ -10,7 +10,7 @@
"internal": "1033g1",
"labware": "\nCorning 96 Well Plate 360 \u00b5L Flat #3650\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 10 Tube Rack with Falcon 4x50 mL, 6x15 mL Conical\n",
"markdown": {
- "author": "Vasudha Nair\n\n\n",
+ "author": "Vasudha Nair\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Cell and Tissue Culture\n\t* Cell and Tissue Culture\n\n\n",
"deck-setup": "[deck](https://drive.google.com/open?id=1vvyaQvEp-MFuKYR9x25cYHy11sXaXxJm, https://drive.google.com/open?id=1FxNxAm_1GPeJA24OzmhPK7ofKM-ku2A3)\n\n\n",
"description": "This protocol can be used to measure the viability and cytotoxicity of two different cell types (suspension and adherent) that have been treated with respective drugs, using Cell Viability assay kit (Cell Titer Glo 2.0) and Cytotoxicity assay kit (CellTox Green) from Promega on the OT-2. This protocol is designed for the 96-well plate format and both assays can be processed in the same plate. In the case of K562 cells, the protocol is broken down into 2 main parts: a) Seeding/ Plating of K562 cells and bortezomib additions to the cells b) After 72 hours, completion of both the assays in the same plate i.e. sequential multiplexing of CellTox Green Cytotoxicity Assay and CellTiter Glo 2.0 Assay.\n\n\n",
diff --git a/protoBuilds/10bf60-station-B/README.json b/protoBuilds/10bf60-station-B/README.json
index 5d16904b87..2e50a9128a 100644
--- a/protoBuilds/10bf60-station-B/README.json
+++ b/protoBuilds/10bf60-station-B/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Covid Workstation": [
"RNA Extraction"
@@ -8,7 +8,7 @@
"description": "This is a flexible protocol accommodating a wide range of commercial RNA extraction workflows for COVID-19 sample processing. The protocol is broken down into 5 main parts:\n binding buffer addition to samples\n bead wash 2x using magnetic module\n* final elution to chilled PCR strips\nLysed samples should be loaded on the magnetic module in a NEST or USA Scientific 96-deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 4) to the elution PCR plate (temperature module, slot 1). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\nExplanation of complex parameters below:\n park tips: If set to yes (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to no, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n track tips across protocol runs: If set to yes, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to no, tips will be picked up from column 1 of the 1st tiprack.\n* flash: If set to yes, the robot rail lights will flash during any automatic pauses in the protocol. If set to no, the lights will not flash.\n\n \nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons magnetic module\nOpentrons temperature module\nNEST 12 Well Reservoir 15 mL or USA Scientific 12 Well Reservoir 22 mL\nNEST 1 Well Reservoir 195 mL\nBio-Rad 96 Well Plate 200 \u00b5L PCR\nNEST 96 Deepwell Plate 2mL or USA Scientific 96 Deep Well Plate 2.4 mL\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n\n\n\n\nReservoir 1: slot 5\nReservoir 2: slot 2 (12mL per channel for full 96-sample run)\n\n",
"internal": "10bf60",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Covid Workstation\n * RNA Extraction\n\n",
"description": "This is a flexible protocol accommodating a wide range of commercial RNA extraction workflows for COVID-19 sample processing. The protocol is broken down into 5 main parts:\n* binding buffer addition to samples\n* bead wash 2x using magnetic module\n* final elution to chilled PCR strips\n\nLysed samples should be loaded on the magnetic module in a NEST or USA Scientific 96-deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 4) to the elution PCR plate (temperature module, slot 1). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\nExplanation of complex parameters below:\n* `park tips`: If set to `yes` (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to `no`, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n* `flash`: If set to `yes`, the robot rail lights will flash during any automatic pauses in the protocol. If set to `no`, the lights will not flash.\n\n---\n\n \n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons magnetic module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons temperature module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [NEST 12 Well Reservoir 15 mL](https://labware.opentrons.com/nest_12_reservoir_15ml) or [USA Scientific 12 Well Reservoir 22 mL](https://labware.opentrons.com/usascientific_12_reservoir_22ml)\n* [NEST 1 Well Reservoir 195 mL](https://labware.opentrons.com/nest_1_reservoir_195ml)\n* [Bio-Rad 96 Well Plate 200 \u00b5L PCR](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr)\n* [NEST 96 Deepwell Plate 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep) or [USA Scientific 96 Deep Well Plate 2.4 mL](https://labware.opentrons.com/usascientific_96_wellplate_2.4ml_deep)\n* [Opentrons 96 Filter Tip Rack 200 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n\n---\n\n\n* Reservoir 1: slot 5\n* Reservoir 2: slot 2 (12mL per channel for full 96-sample run)\n\n\n",
"internal": "10bf60\n",
diff --git a/protoBuilds/10bf60-station-C/README.json b/protoBuilds/10bf60-station-C/README.json
index 13003511b2..c2db8c9fd5 100644
--- a/protoBuilds/10bf60-station-C/README.json
+++ b/protoBuilds/10bf60-station-C/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -9,7 +9,7 @@
"internal": "10bf60",
"labware": "\nApplied Biosystems MicroAmp 96 Aluminum Block 200 \u00b5L\nOpentrons 96 Tip Rack 20\u00b5l\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This is a flexible protocol accommodating a wide range of PCR setups. Mastermix is transferred from a 2ml snapcap tube to clean PCR plate using a P20 GEN2 single-channel pipette. Then, a P20 GEN2 multi-channel pipette transfers eluates from RNA extraction to the PCR plate containing mastermix. Samples are transferred to their corresponding positions from plate to plate (A1 to A1, B1, to B1, etc.)\n\n---\n\n",
"internal": "10bf60\n",
diff --git a/protoBuilds/111210-part-10/README.json b/protoBuilds/111210-part-10/README.json
index 172029ae64..de36c18f10 100644
--- a/protoBuilds/111210-part-10/README.json
+++ b/protoBuilds/111210-part-10/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung RNA Fusion UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "111210-part-10",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung RNA Fusion UMI Panel Kit\n\n",
"deck-setup": "* The example below illustrates the starting deck layout for Part 6 (Cleanup of Universal PCR with QIAact Beads).\n\n\n",
"description": "This protocol automates the tenth part of a ten part protocol for the [GeneRead QIAact Lung RNA Fusion UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/?catno=181936) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Cleanup of Target Enrichment PCR with QIAseq Beads part described in the [GeneRead QIAact Lung RNA Fusion UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=1a71d98a-c45c-44fa-b4af-874cd1d2b61f&lang=en).\n\n* Part 1: [GeneRead QIAact Lung RNA Fusion UMI](https://protocols.opentrons.com/protocol/111210)\n* Part 2: [Reverse transcription](https://protocols.opentrons.com/protocol/111210-part-2)\n* Part 3: [Second strand synthesis](https://protocols.opentrons.com/protocol/111210-part-3)\n* Part 4: [End repair / dA tailing](https://protocols.opentrons.com/protocol/111210-part-4)\n* Part 5: [Adaptor ligation](https://protocols.opentrons.com/protocol/111210-part-5)\n* Part 6: [Cleanup of Adapter-ligated DNA with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-6)\n* Part 7: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/111210-part-7)\n* Part 8: [Cleanup of Target Enrichment PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-8)\n* Part 9: [Universal PCR Amplification](https://protocols.opentrons.com/protocol/111210-part-9)\n* Part 10: [Cleanup of Universal PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-10)\n\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P300 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `Magnetic Module Engage Height (mm)`: The height the magnets should raise.\n\n---\n\n",
diff --git a/protoBuilds/111210-part-2/README.json b/protoBuilds/111210-part-2/README.json
index 39471ab764..9dcc5d79b5 100644
--- a/protoBuilds/111210-part-2/README.json
+++ b/protoBuilds/111210-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung RNA Fusion UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "111210-part-2",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung RNA Fusion UMI Panel Kit\n\n",
"deck-setup": "* The example below illustrates the deck layout when the samples are placed in 1.5 mL Screwcap tubes in an Opentrons Tube Rack.\n\n\n",
"description": "This protocol automates the second part of a ten part protocol for the [GeneRead QIAact Lung RNA Fusion UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/?catno=181936) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Reverse transcription part described in the [GeneRead QIAact Lung RNA Fusion UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=1a71d98a-c45c-44fa-b4af-874cd1d2b61f&lang=en).\n\n* Part 1: [GeneRead QIAact Lung RNA Fusion UMI](https://protocols.opentrons.com/protocol/111210)\n* Part 2: [Reverse transcription](https://protocols.opentrons.com/protocol/111210-part-2)\n* Part 3: [Second strand synthesis](https://protocols.opentrons.com/protocol/111210-part-3)\n* Part 4: [End repair / dA tailing](https://protocols.opentrons.com/protocol/111210-part-4)\n* Part 5: [Adaptor ligation](https://protocols.opentrons.com/protocol/111210-part-5)\n* Part 6: [Cleanup of Adapter-ligated DNA with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-6)\n* Part 7: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/111210-part-7)\n* Part 8: [Cleanup of Target Enrichment PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-8)\n* Part 9: [Universal PCR Amplification](https://protocols.opentrons.com/protocol/111210-part-9)\n* Part 10: [Cleanup of Universal PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-10)\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n\n---\n\n",
diff --git a/protoBuilds/111210-part-3/README.json b/protoBuilds/111210-part-3/README.json
index e40131d92f..5c879e73d6 100644
--- a/protoBuilds/111210-part-3/README.json
+++ b/protoBuilds/111210-part-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung RNA Fusion UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "111210-part-3",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung RNA Fusion UMI Panel Kit\n\n",
"deck-setup": "* The example below illustrates the deck layout when the samples are placed in 1.5 mL Screwcap tubes in an Opentrons Tube Rack.\n\n\n",
"description": "This protocol automates the third part of a ten part protocol for the [GeneRead QIAact Lung RNA Fusion UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/?catno=181936) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Second strand synthesis part described in the [GeneRead QIAact Lung RNA Fusion UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=1a71d98a-c45c-44fa-b4af-874cd1d2b61f&lang=en).\n\n* Part 1: [GeneRead QIAact Lung RNA Fusion UMI](https://protocols.opentrons.com/protocol/111210)\n* Part 2: [Reverse transcription](https://protocols.opentrons.com/protocol/111210-part-2)\n* Part 3: [Second strand synthesis](https://protocols.opentrons.com/protocol/111210-part-3)\n* Part 4: [End repair / dA tailing](https://protocols.opentrons.com/protocol/111210-part-4)\n* Part 5: [Adaptor ligation](https://protocols.opentrons.com/protocol/111210-part-5)\n* Part 6: [Cleanup of Adapter-ligated DNA with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-6)\n* Part 7: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/111210-part-7)\n* Part 8: [Cleanup of Target Enrichment PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-8)\n* Part 9: [Universal PCR Amplification](https://protocols.opentrons.com/protocol/111210-part-9)\n* Part 10: [Cleanup of Universal PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-10)\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n\n---\n\n",
diff --git a/protoBuilds/111210-part-4/README.json b/protoBuilds/111210-part-4/README.json
index c5f8488a12..8799dca72b 100644
--- a/protoBuilds/111210-part-4/README.json
+++ b/protoBuilds/111210-part-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung RNA Fusion UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "111210-part-4",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung RNA Fusion UMI Panel Kit\n\n",
"deck-setup": "\n\n",
"description": "This protocol automates the fourth part of a ten part protocol for the [GeneRead QIAact Lung RNA Fusion UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/?catno=181936) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the End repair / dA tailing part described in the [GeneRead QIAact Lung RNA Fusion UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=1a71d98a-c45c-44fa-b4af-874cd1d2b61f&lang=en).\n\n* Part 1: [GeneRead QIAact Lung RNA Fusion UMI](https://protocols.opentrons.com/protocol/111210)\n* Part 2: [Reverse transcription](https://protocols.opentrons.com/protocol/111210-part-2)\n* Part 3: [Second strand synthesis](https://protocols.opentrons.com/protocol/111210-part-3)\n* Part 4: [End repair / dA tailing](https://protocols.opentrons.com/protocol/111210-part-4)\n* Part 5: [Adaptor ligation](https://protocols.opentrons.com/protocol/111210-part-5)\n* Part 6: [Cleanup of Adapter-ligated DNA with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-6)\n* Part 7: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/111210-part-7)\n* Part 8: [Cleanup of Target Enrichment PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-8)\n* Part 9: [Universal PCR Amplification](https://protocols.opentrons.com/protocol/111210-part-9)\n* Part 10: [Cleanup of Universal PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-10)\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n\n---\n\n",
diff --git a/protoBuilds/111210-part-5/README.json b/protoBuilds/111210-part-5/README.json
index e3516772a0..d1e6984d0c 100644
--- a/protoBuilds/111210-part-5/README.json
+++ b/protoBuilds/111210-part-5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung RNA Fusion UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "111210-part-5",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung RNA Fusion UMI Panel Kit\n\n",
"deck-setup": "\n\n",
"description": "This protocol automates the fifth part of a ten part protocol for the [GeneRead QIAact Lung RNA Fusion UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/?catno=181936) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Adaptor ligation part described in the [GeneRead QIAact Lung RNA Fusion UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=1a71d98a-c45c-44fa-b4af-874cd1d2b61f&lang=en).\n\n* Part 1: [GeneRead QIAact Lung RNA Fusion UMI](https://protocols.opentrons.com/protocol/111210)\n* Part 2: [Reverse transcription](https://protocols.opentrons.com/protocol/111210-part-2)\n* Part 3: [Second strand synthesis](https://protocols.opentrons.com/protocol/111210-part-3)\n* Part 4: [End repair / dA tailing](https://protocols.opentrons.com/protocol/111210-part-4)\n* Part 5: [Adaptor ligation](https://protocols.opentrons.com/protocol/111210-part-5)\n* Part 6: [Cleanup of Adapter-ligated DNA with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-6)\n* Part 7: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/111210-part-7)\n* Part 8: [Cleanup of Target Enrichment PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-8)\n* Part 9: [Universal PCR Amplification](https://protocols.opentrons.com/protocol/111210-part-9)\n* Part 10: [Cleanup of Universal PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-10)\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `P300 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n\n---\n\n",
diff --git a/protoBuilds/111210-part-6/README.json b/protoBuilds/111210-part-6/README.json
index 96598c3a51..a1d996950e 100644
--- a/protoBuilds/111210-part-6/README.json
+++ b/protoBuilds/111210-part-6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung RNA Fusion UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "111210-part-6",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung RNA Fusion UMI Panel Kit\n\n",
"deck-setup": "\n\n",
"description": "This protocol automates the sixth part of a ten part protocol for the [GeneRead QIAact Lung RNA Fusion UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/?catno=181936) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Cleanup of Adapter-ligated DNA with QIAseq Beads part described in the [GeneRead QIAact Lung RNA Fusion UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=1a71d98a-c45c-44fa-b4af-874cd1d2b61f&lang=en).\n\n* Part 1: [GeneRead QIAact Lung RNA Fusion UMI](https://protocols.opentrons.com/protocol/111210)\n* Part 2: [Reverse transcription](https://protocols.opentrons.com/protocol/111210-part-2)\n* Part 3: [Second strand synthesis](https://protocols.opentrons.com/protocol/111210-part-3)\n* Part 4: [End repair / dA tailing](https://protocols.opentrons.com/protocol/111210-part-4)\n* Part 5: [Adaptor ligation](https://protocols.opentrons.com/protocol/111210-part-5)\n* Part 6: [Cleanup of Adapter-ligated DNA with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-6)\n* Part 7: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/111210-part-7)\n* Part 8: [Cleanup of Target Enrichment PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-8)\n* Part 9: [Universal PCR Amplification](https://protocols.opentrons.com/protocol/111210-part-9)\n* Part 10: [Cleanup of Universal PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-10)\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `P300 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `Magnetic Module Engage Height`: The height the magnets should raise.\n\n---\n\n",
diff --git a/protoBuilds/111210-part-7/README.json b/protoBuilds/111210-part-7/README.json
index 3e740f4712..198536bab6 100644
--- a/protoBuilds/111210-part-7/README.json
+++ b/protoBuilds/111210-part-7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung RNA Fusion UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "111210-part-7",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung RNA Fusion UMI Panel Kit\n\n",
"deck-setup": "* The example below illustrates the starting deck layout for Part 4 (Target Enrichment PCR).\n\n\n",
"description": "This protocol automates the seventh part of a ten part protocol for the [GeneRead QIAact Lung RNA Fusion UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/?catno=181936) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Target Enrichment PCR part described in the [GeneRead QIAact Lung RNA Fusion UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=1a71d98a-c45c-44fa-b4af-874cd1d2b61f&lang=en).\n\n* Part 1: [GeneRead QIAact Lung RNA Fusion UMI](https://protocols.opentrons.com/protocol/111210)\n* Part 2: [Reverse transcription](https://protocols.opentrons.com/protocol/111210-part-2)\n* Part 3: [Second strand synthesis](https://protocols.opentrons.com/protocol/111210-part-3)\n* Part 4: [End repair / dA tailing](https://protocols.opentrons.com/protocol/111210-part-4)\n* Part 5: [Adaptor ligation](https://protocols.opentrons.com/protocol/111210-part-5)\n* Part 6: [Cleanup of Adapter-ligated DNA with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-6)\n* Part 7: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/111210-part-7)\n* Part 8: [Cleanup of Target Enrichment PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-8)\n* Part 9: [Universal PCR Amplification](https://protocols.opentrons.com/protocol/111210-part-9)\n* Part 10: [Cleanup of Universal PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-10)\n\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n\n---\n\n",
diff --git a/protoBuilds/111210-part-8/README.json b/protoBuilds/111210-part-8/README.json
index d68f5df040..6f0a0434c1 100644
--- a/protoBuilds/111210-part-8/README.json
+++ b/protoBuilds/111210-part-8/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung RNA Fusion UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "111210-part-8",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung RNA Fusion UMI Panel Kit\n\n",
"deck-setup": "* The example below illustrates the starting deck layout for Part 5 (Cleanup of Target Enrichment PCR with QIAact Beads).\n\n\n",
"description": "This protocol automates the eigth part of a ten part protocol for the [GeneRead QIAact Lung RNA Fusion UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/?catno=181936) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Cleanup of Target Enrichment PCR with QIAseq Beads part described in the [GeneRead QIAact Lung RNA Fusion UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=1a71d98a-c45c-44fa-b4af-874cd1d2b61f&lang=en).\n\n* Part 1: [GeneRead QIAact Lung RNA Fusion UMI](https://protocols.opentrons.com/protocol/111210)\n* Part 2: [Reverse transcription](https://protocols.opentrons.com/protocol/111210-part-2)\n* Part 3: [Second strand synthesis](https://protocols.opentrons.com/protocol/111210-part-3)\n* Part 4: [End repair / dA tailing](https://protocols.opentrons.com/protocol/111210-part-4)\n* Part 5: [Adaptor ligation](https://protocols.opentrons.com/protocol/111210-part-5)\n* Part 6: [Cleanup of Adapter-ligated DNA with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-6)\n* Part 7: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/111210-part-7)\n* Part 8: [Cleanup of Target Enrichment PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-8)\n* Part 9: [Universal PCR Amplification](https://protocols.opentrons.com/protocol/111210-part-9)\n* Part 10: [Cleanup of Universal PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-10)\n\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P300 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `Magnetic Module Engage Height (mm)`: The height the magnets should raise.\n\n---\n\n",
diff --git a/protoBuilds/111210-part-9/README.json b/protoBuilds/111210-part-9/README.json
index 50b56aa467..66a4bc9799 100644
--- a/protoBuilds/111210-part-9/README.json
+++ b/protoBuilds/111210-part-9/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung DNA UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "111210-part-9",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung DNA UMI Panel Kit\n\n",
"deck-setup": "* The example below illustrates the starting deck layout for Part 6 (Universal PCR Amplification).\n\n\n",
"description": "This protocol automates the ninth part of a ten part protocol for the [GeneRead QIAact Lung RNA Fusion UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/?catno=181936) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Cleanup of Target Enrichment PCR with QIAseq Beads part described in the [GeneRead QIAact Lung RNA Fusion UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=1a71d98a-c45c-44fa-b4af-874cd1d2b61f&lang=en).\n\n* Part 1: [GeneRead QIAact Lung RNA Fusion UMI](https://protocols.opentrons.com/protocol/111210)\n* Part 2: [Reverse transcription](https://protocols.opentrons.com/protocol/111210-part-2)\n* Part 3: [Second strand synthesis](https://protocols.opentrons.com/protocol/111210-part-3)\n* Part 4: [End repair / dA tailing](https://protocols.opentrons.com/protocol/111210-part-4)\n* Part 5: [Adaptor ligation](https://protocols.opentrons.com/protocol/111210-part-5)\n* Part 6: [Cleanup of Adapter-ligated DNA with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-6)\n* Part 7: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/111210-part-7)\n* Part 8: [Cleanup of Target Enrichment PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-8)\n* Part 9: [Universal PCR Amplification](https://protocols.opentrons.com/protocol/111210-part-9)\n* Part 10: [Cleanup of Universal PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-10)\n\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P300 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n\n---\n\n",
diff --git a/protoBuilds/111210/README.json b/protoBuilds/111210/README.json
index 1f16e95009..15c4d90a53 100644
--- a/protoBuilds/111210/README.json
+++ b/protoBuilds/111210/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung RNA Fusion UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "111210",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung RNA Fusion UMI Panel Kit\n\n",
"deck-setup": "* The example below illustrates the deck layout when the samples are placed in 1.5 mL Screwcap tubes in an Opentrons Tube Rack.\n\n\n",
"description": "This protocol automates the first part of a ten part protocol for the [GeneRead QIAact Lung RNA Fusion UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/?catno=181936) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the First strand cDNA synthesis part described in the [GeneRead QIAact Lung RNA Fusion UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=1a71d98a-c45c-44fa-b4af-874cd1d2b61f&lang=en).\n\n* Part 1: [GeneRead QIAact Lung RNA Fusion UMI](https://protocols.opentrons.com/protocol/111210)\n* Part 2: [Reverse transcription](https://protocols.opentrons.com/protocol/111210-part-2)\n* Part 3: [Second strand synthesis](https://protocols.opentrons.com/protocol/111210-part-3)\n* Part 4: [End repair / dA tailing](https://protocols.opentrons.com/protocol/111210-part-4)\n* Part 5: [Adaptor ligation](https://protocols.opentrons.com/protocol/111210-part-5)\n* Part 6: [Cleanup of Adapter-ligated DNA with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-6)\n* Part 7: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/111210-part-7)\n* Part 8: [Cleanup of Target Enrichment PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-8)\n* Part 9: [Universal PCR Amplification](https://protocols.opentrons.com/protocol/111210-part-9)\n* Part 10: [Cleanup of Universal PCR with QIAseq Beads](https://protocols.opentrons.com/protocol/111210-part-10)\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `Samples Labware Type`: The starting samples can be placed in either 1.5 mL tubes on the Opentrons Tube Rack OR in a 96 Well Plate.\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n\n---\n\n",
diff --git a/protoBuilds/115d98/README.json b/protoBuilds/115d98/README.json
index bb2e40472a..fe9606f9f6 100644
--- a/protoBuilds/115d98/README.json
+++ b/protoBuilds/115d98/README.json
@@ -13,13 +13,13 @@
"description": "This flexible protocol automates the transferring of samples in 12mL tubes to dilution buffer, then to Visby device for rapid Covid testing.\n\n\n---\n\n**Protocol Steps**\n1. The protocol begins by transferring (up to 5) samples from row A in slot 2 to A1, A3, A5, C1 and C3 (in that order) in slot 5, containing dilution buffer.\n2. Once all samples are transferred to dilution buffer, *total volume of samples* will be added from slot 5 to Visby.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 4.0.0 or later)](https://opentrons.com/ot-app/)\n* [P1000 Single-Channel GEN2](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 1000\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips)\n* [Visby Device with Adapter](https://www.visbymedical.com/covid-19-test/)\n* Custom Tube Rack with 12mL Tubes\n\n---\n\n\n**Deck Layout**\n\n**Slot 1**: [Opentrons 1000\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips)\n\n**Slot 2**: Custom Tube Rack with 12mL Tubes, containing samples in row A\n\n**Slot 5**: Custom Tube Rack with 12mL Tubes, cotaining dilution buffer in A1, A3, A5, C1, and C3 (depending on the number of samples)\n\n**Slot 7**: [Visby 3](https://www.visbymedical.com/covid-19-test/)\n\n**Slot 8**: [Visby 4](https://www.visbymedical.com/covid-19-test/)\n\n**Slot 9**: [Visby 5](https://www.visbymedical.com/covid-19-test/)\n\n**Slot 10**: [Visby 1](https://www.visbymedical.com/covid-19-test/)\n\n**Slot 11**: [Visby 2](https://www.visbymedical.com/covid-19-test/)\n\n\nExplanation of adjustable parameters below:\n* `P1000 Single GEN2 Mount`: Select which mount (left, right) the pipette is attached to.\n* `Sample Volume (\u00b5L)`: Specify the volume (in \u00b5L) that should be transferred to the Visby. This volume will be transferred to the dilution buffer, before being transferred to the Visby.\n* `Number of Samples`: Select the number of samples to test (1-5).\n\n\n",
"internal": "115d98\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[BasisDx](https://www.basisdx.org/)\n\n",
+ "partner": "[BasisDx](https://www.basisdx.org/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"robot": "* [OT-2](https://opentrons.com/ot-2)\n\n",
"title": "Visby Test without Pooling p1000"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "BasisDx",
+ "partner": "BasisDx\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"robot": [
"OT-2"
diff --git a/protoBuilds/1185d3/README.json b/protoBuilds/1185d3/README.json
index 14efdf3a78..cb2df76c39 100644
--- a/protoBuilds/1185d3/README.json
+++ b/protoBuilds/1185d3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"MagMAX Nucleic Acid Isolation Kit"
@@ -8,7 +8,7 @@
"description": "This protocol automates the wash steps of the MagMAX Viral/Pathogen Nucleic Acid Isolation Kit. As inputs, it requires a deep well plate with sample beads, a single well trough of 80% ethanol, a single well trough of wash buffer, a single well trash, and an output 96 well plate.\nThere are 2 pauses during the protocol that require human intervention - to drain the liquid trash + to replace tip racks + to add elution buffer, and to add the deep well plate to a 65c incubator for 10 minutes.\n\n\n\nOpentrons P300-multi channel electronic pipettes (GEN2)\nOpentrons 200ul Filter Tips\nNEST 1 Well Reservoir 195 mL\nNEST 96 Deepwell Plate 2mL\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nMagMAX Viral/Pathogen Nucleic Acid Isolation Kit\n",
"internal": "1185d3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* MagMAX Nucleic Acid Isolation Kit\n\n",
"description": "This protocol automates the wash steps of the MagMAX Viral/Pathogen Nucleic Acid Isolation Kit. As inputs, it requires a deep well plate with sample beads, a single well trough of 80% ethanol, a single well trough of wash buffer, a single well trash, and an output 96 well plate.\n\nThere are 2 pauses during the protocol that require human intervention - to drain the liquid trash + to replace tip racks + to add elution buffer, and to add the deep well plate to a 65c incubator for 10 minutes.\n\n---\n\n\n* [Opentrons P300-multi channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [NEST 1 Well Reservoir 195 mL](http://www.cell-nest.com/page94?_l=en&product_id=102)\n* [NEST 96 Deepwell Plate 2mL](http://www.cell-nest.com/page94?product_id=101&_l=en)\n* [NEST 0.1 mL 96-Well PCR Plate, Full Skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [MagMAX Viral/Pathogen Nucleic Acid Isolation Kit](https://www.thermofisher.com/order/catalog/product/A42352?SID=srch-hj-A42352#/A42352?SID=srch-hj-A42352)\n\n",
"internal": "1185d3\n",
diff --git a/protoBuilds/118e8c/README.json b/protoBuilds/118e8c/README.json
index 422cc6f90d..c737df1832 100644
--- a/protoBuilds/118e8c/README.json
+++ b/protoBuilds/118e8c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "FluoGene HLA NX 96-Well or 384-Well Setup. This protocol performs the setup of two Fluogene 96-well trays or a single 384-well tray (columns 1-12) to perform a multiplexed TaqMan-based assay.\nLinks:\n* json protocol\nThis protocol is a translation of the attached json protocol to python with a couple additional features for faster completion time and tracking of reservoir columns and tips between protocol runs: A multi-channel p20 pipette is used to dispense 4 ul of water and 4 ul pcr mix to the last well of the first column of each tray by using only the rear-most channel of the pipette. Then the p300 single is used to mix the DNA dilution with the pcr mix and transfer it to the reagent reservoir. The p20 multi is then used to distribute 8 ul of this mixture to each of the remaining wells of the two 96-well trays or to columns 1-12 of a single 384-well tray.\nThe deck layout can be displayed by uploading the attached json protocol to the Opentrons Protocol Designer web page.",
"internal": "118e8c",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * PCR Prep\n\n",
"description": "FluoGene HLA NX 96-Well or 384-Well Setup. This protocol performs the setup of two Fluogene 96-well trays or a single 384-well tray (columns 1-12) to perform a multiplexed TaqMan-based assay.\n\nLinks:\n* [json protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-04-02/dc23h0c/FluoGene%20HLA%20NX%20Match-96%20ver.%2057%20with%207.5.json)\n\nThis protocol is a translation of the attached json protocol to python with a couple additional features for faster completion time and tracking of reservoir columns and tips between protocol runs: A multi-channel p20 pipette is used to dispense 4 ul of water and 4 ul pcr mix to the last well of the first column of each tray by using only the rear-most channel of the pipette. Then the p300 single is used to mix the DNA dilution with the pcr mix and transfer it to the reagent reservoir. The p20 multi is then used to distribute 8 ul of this mixture to each of the remaining wells of the two 96-well trays or to columns 1-12 of a single 384-well tray.\nThe deck layout can be displayed by uploading the attached json protocol to the [Opentrons Protocol Designer web page](https://opentrons.com/protocols/designer/).\n",
"internal": "118e8c\n",
diff --git a/protoBuilds/11bb6a-part-2/README.json b/protoBuilds/11bb6a-part-2/README.json
index 2d7a663a9b..1aba21f3a8 100644
--- a/protoBuilds/11bb6a-part-2/README.json
+++ b/protoBuilds/11bb6a-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext"
@@ -10,7 +10,7 @@
"internal": "11bb6a-part-2",
"labware": "\nOpentrons Filter Tips for the P20 and P300 (https://shop.opentrons.com)\nOpentrons Temperature Module (https://shop.opentrons.com/modules/)\nOpentrons Magnetic Module (https://shop.opentrons.com/modules/)\nSelected 96-well PCR plate ([see parameter dropdown list below] https://labware.opentrons.com/)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module empty \n**Slot 5**: Reagent Reservoir (Nest 12-well 15 mL reservoir) \n\n\n\n**Slot 6**: Ethanol (Agilent 290 mL reservoir) \n**Slot 8**: Liquid Waste (Agilent 290 mL reservoir) \n**Slot 9**: Magnetic Module with dropdown-selected 96-well PCR plate \n**Slot 10**: Opentrons 20 uL filter tips \n**Slots 2,3,4,7,11**: Opentrons 200 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses multi-channel P20 and P300 pipettes to perform bead-based sample cleanup steps on 48-96 input samples. User-determined parameters are available for selection of the labware for the 96-well PCR plate on the magnetic module, the sample volume, and the bead ratio.\n\n---\n\n\n",
diff --git a/protoBuilds/11bb6a-part-3/README.json b/protoBuilds/11bb6a-part-3/README.json
index ce0103d44c..0b352f5b11 100644
--- a/protoBuilds/11bb6a-part-3/README.json
+++ b/protoBuilds/11bb6a-part-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext"
@@ -10,7 +10,7 @@
"internal": "11bb6a-part-3",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons Temperature Module] (https://shop.opentrons.com/modules/)\n[Opentrons Magnetic Module] (https://shop.opentrons.com/modules/)\n[Selected 96-well PCR plate] ([see parameter dropdown list below] https://labware.opentrons.com/)\n[Selected aluminum block and tube combination] ([see parameter dropdown list below] https://labware.opentrons.com/)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module with dropdown-selected aluminum block and tube combination \n\n\n\n**Slot 3**: Library Prep Plate (dropdown-selected 96-well PCR plate) \n**Slot 6**: Reagent Plate (dropdown-selected 96-well PCR plate) \n**Slot 9**: Magnetic Module with dropdown-selected 96-well PCR plate \n**Slot 8**: Opentrons 200 uL filter tips \n**Slots 7, 10, 11**: Opentrons 20 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses multi-channel P20 and P300 pipettes to perform end prep steps on 48-96 input samples. User-determined parameters are available for selection of the labware for the library prep plate, the reagent plate, and the 96-well PCR plate on the magnetic module and the labware for reagent tubes on the temperature module.\n\nLinks:\n* [Part 1: Enzymatic Fragmentation and End Prep](http://protocols.opentrons.com/protocol/11bb6a)\n* [Part 2: Sample Cleanup for Covaris Samples](http://protocols.opentrons.com/protocol/11bb6a-part-2)\n* [Part 3: End Prep](http://protocols.opentrons.com/protocol/11bb6a-part-3)\n* [Part 4: Adapter Ligation](http://protocols.opentrons.com/protocol/11bb6a-part-4)\n* [Part 5: Size Selection](http://protocols.opentrons.com/protocol/11bb6a-part-5)\n* [Part 6: Clean Up](http://protocols.opentrons.com/protocol/11bb6a-part-6)\n* [Part 7: PCR Enrichment](http://protocols.opentrons.com/protocol/11bb6a-part-7)\n* [Part 8: Sample Clean Up](http://protocols.opentrons.com/protocol/11bb6a-part-8)\n* [Part 9: Fragment Analyzer](http://protocols.opentrons.com/protocol/11bb6a-part-9)\n* [Part 10: Pooling According to Concentration](http://protocols.opentrons.com/protocol/11bb6a-part-10)\n\n---\n\n\n",
diff --git a/protoBuilds/11bb6a-part-4/README.json b/protoBuilds/11bb6a-part-4/README.json
index 3b7e95ac14..e96e9942f0 100644
--- a/protoBuilds/11bb6a-part-4/README.json
+++ b/protoBuilds/11bb6a-part-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext"
@@ -10,7 +10,7 @@
"internal": "11bb6a-part-4",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons Temperature Module] (https://shop.opentrons.com/modules/)\n[Selected 96-well PCR plate] ([see parameter dropdown list below] https://labware.opentrons.com/)\n[Selected aluminum block and tube combination] ([see parameter dropdown list below] https://labware.opentrons.com/)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module with dropdown-selected aluminum block and tube combination \n\n\n\n**Slot 3**: Library Prep Plate (dropdown-selected 96-well PCR plate) \n**Slot 6**: Reagent Plate (dropdown-selected 96-well PCR plate) \n**Slot 9**: Magnetic Module empty \n**Slot 8**: Opentrons 200 uL filter tips \n**Slots 7, 10, 11**: Opentrons 20 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses multi-channel P20 and P300 pipettes to perform adapter ligation steps on 48-96 input samples. User-determined parameters are available for selection of the labware for the library prep plate, the reagent plate, and the labware for reagent tubes on the temperature module.\n\nLinks:\n* [Part 1: Enzymatic Fragmentation and End Prep](http://protocols.opentrons.com/protocol/11bb6a)\n* [Part 2: Sample Cleanup for Covaris Samples](http://protocols.opentrons.com/protocol/11bb6a-part-2)\n* [Part 3: End Prep](http://protocols.opentrons.com/protocol/11bb6a-part-3)\n* [Part 4: Adapter Ligation](http://protocols.opentrons.com/protocol/11bb6a-part-4)\n* [Part 5: Size Selection](http://protocols.opentrons.com/protocol/11bb6a-part-5)\n* [Part 6: Clean Up](http://protocols.opentrons.com/protocol/11bb6a-part-6)\n* [Part 7: PCR Enrichment](http://protocols.opentrons.com/protocol/11bb6a-part-7)\n* [Part 8: Sample Clean Up](http://protocols.opentrons.com/protocol/11bb6a-part-8)\n* [Part 9: Fragment Analyzer](http://protocols.opentrons.com/protocol/11bb6a-part-9)\n* [Part 10: Pooling According to Concentration](http://protocols.opentrons.com/protocol/11bb6a-part-10)\n\n---\n\n\n",
diff --git a/protoBuilds/11bb6a-part-5/README.json b/protoBuilds/11bb6a-part-5/README.json
index c2bbc36bef..60ba61bfd8 100644
--- a/protoBuilds/11bb6a-part-5/README.json
+++ b/protoBuilds/11bb6a-part-5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext"
@@ -10,7 +10,7 @@
"internal": "11bb6a-part-5",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons Temperature Module] (https://shop.opentrons.com/modules/)\n[Opentrons Magnetic Module] (https://shop.opentrons.com/modules/)\n[Selected 96-well PCR plate] ([see parameter dropdown list below] https://labware.opentrons.com/)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module empty \n**Slot 3**: Size Plate (dropdown-selected 96-well PCR plate) \n**Slot 5**: Reagent Reservoir (Nest 12-well 15 mL reservoir) \n\n\n\n**Slot 6**: Ethanol (Agilent 290 mL reservoir) \n**Slot 8**: Liquid Waste (Agilent 290 mL reservoir) \n**Slot 9**: Opentrons Magnetic Module with library prep plate (dropdown-selected 96-well PCR plate) \n**Slot 10**: Opentrons 20 uL filter tips \n**Slots 2,4,7,11**: Opentrons 200 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses multi-channel P20 and P300 pipettes to perform size selection steps on 48-96 input samples. User-determined parameters are available for selection of the labware for the library prep plate and the size plate.\n\nLinks:\n* [Part 1: Enzymatic Fragmentation and End Prep](http://protocols.opentrons.com/protocol/11bb6a)\n* [Part 2: Sample Cleanup for Covaris Samples](http://protocols.opentrons.com/protocol/11bb6a-part-2)\n* [Part 3: End Prep](http://protocols.opentrons.com/protocol/11bb6a-part-3)\n* [Part 4: Adapter Ligation](http://protocols.opentrons.com/protocol/11bb6a-part-4)\n* [Part 5: Size Selection](http://protocols.opentrons.com/protocol/11bb6a-part-5)\n* [Part 6: Clean Up](http://protocols.opentrons.com/protocol/11bb6a-part-6)\n* [Part 7: PCR Enrichment](http://protocols.opentrons.com/protocol/11bb6a-part-7)\n* [Part 8: Sample Clean Up](http://protocols.opentrons.com/protocol/11bb6a-part-8)\n* [Part 9: Fragment Analyzer](http://protocols.opentrons.com/protocol/11bb6a-part-9)\n* [Part 10: Pooling According to Concentration](http://protocols.opentrons.com/protocol/11bb6a-part-10)\n\n---\n\n\n",
diff --git a/protoBuilds/11bb6a-part-6/README.json b/protoBuilds/11bb6a-part-6/README.json
index d9384f4d55..d583f4bfbf 100644
--- a/protoBuilds/11bb6a-part-6/README.json
+++ b/protoBuilds/11bb6a-part-6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext"
@@ -10,7 +10,7 @@
"internal": "11bb6a-part-6",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons Temperature Module] (https://shop.opentrons.com/modules/)\n[Opentrons Magnetic Module] (https://shop.opentrons.com/modules/)\n[Selected 96-well PCR plate] ([see parameter dropdown list below] https://labware.opentrons.com/)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module empty \n**Slot 5**: Reagent Reservoir (Nest 12-well 15 mL reservoir) \n\n\n\n**Slot 6**: Ethanol (Agilent 290 mL reservoir) \n**Slot 8**: Liquid Waste (Agilent 290 mL reservoir) \n**Slot 9**: Opentrons Magnetic Module with library prep plate (dropdown-selected 96-well PCR plate) \n**Slot 10**: Opentrons 20 uL filter tips \n**Slots 2,4,7,11**: Opentrons 200 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses multi-channel P20 and P300 pipettes to perform bead cleanup steps on 48-96 input samples. User-determined parameters are available for selection of the labware for the library prep plate and the size plate.\n\nLinks:\n* [Part 1: Enzymatic Fragmentation and End Prep](http://protocols.opentrons.com/protocol/11bb6a)\n* [Part 2: Sample Cleanup for Covaris Samples](http://protocols.opentrons.com/protocol/11bb6a-part-2)\n* [Part 3: End Prep](http://protocols.opentrons.com/protocol/11bb6a-part-3)\n* [Part 4: Adapter Ligation](http://protocols.opentrons.com/protocol/11bb6a-part-4)\n* [Part 5: Size Selection](http://protocols.opentrons.com/protocol/11bb6a-part-5)\n* [Part 6: Clean Up](http://protocols.opentrons.com/protocol/11bb6a-part-6)\n* [Part 7: PCR Enrichment](http://protocols.opentrons.com/protocol/11bb6a-part-7)\n* [Part 8: Sample Clean Up](http://protocols.opentrons.com/protocol/11bb6a-part-8)\n* [Part 9: Fragment Analyzer](http://protocols.opentrons.com/protocol/11bb6a-part-9)\n* [Part 10: Pooling According to Concentration](http://protocols.opentrons.com/protocol/11bb6a-part-10)\n\n---\n\n\n",
diff --git a/protoBuilds/11bb6a-part-7/README.json b/protoBuilds/11bb6a-part-7/README.json
index 8e599b1d42..3f7b3d4535 100644
--- a/protoBuilds/11bb6a-part-7/README.json
+++ b/protoBuilds/11bb6a-part-7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext"
@@ -10,7 +10,7 @@
"internal": "11bb6a-part-7",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons Temperature Module] (https://shop.opentrons.com/modules/)\n[Opentrons Magnetic Module] (https://shop.opentrons.com/modules/)\n[Selected 96-well PCR plate] ([see parameter dropdown list below] https://labware.opentrons.com/)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module (dropdown-selected aluminum block and tube combination) \n\n\n\n**Slot 2**: i5 indexes (eppendorf_96_wellplate_200ul) \n**Slot 5**: i7 indexes (eppendorf_96_wellplate_200ul) \n**Slot 8**: PCR plate (dropdown-selected 96-well PCR plate) \n**Slot 9**: Opentrons Magnetic Module with library prep plate or size plate (dropdown-selected 96-well PCR plate) \n**Slots 10,3,6**: Opentrons 20 uL filter tips \n**Slot 11**: Opentrons 200 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses multi-channel P20 and P300 pipettes to perform PCR enrichment setup steps on 48-96 input samples. User-determined parameters are available for selection of the i7 primer plate row, the i5 primer plate column, sample count, and labware.\n\nLinks:\n* [Part 1: Enzymatic Fragmentation and End Prep](http://protocols.opentrons.com/protocol/11bb6a)\n* [Part 2: Sample Cleanup for Covaris Samples](http://protocols.opentrons.com/protocol/11bb6a-part-2)\n* [Part 3: End Prep](http://protocols.opentrons.com/protocol/11bb6a-part-3)\n* [Part 4: Adapter Ligation](http://protocols.opentrons.com/protocol/11bb6a-part-4)\n* [Part 5: Size Selection](http://protocols.opentrons.com/protocol/11bb6a-part-5)\n* [Part 6: Clean Up](http://protocols.opentrons.com/protocol/11bb6a-part-6)\n* [Part 7: PCR Enrichment](http://protocols.opentrons.com/protocol/11bb6a-part-7)\n* [Part 8: Sample Clean Up](http://protocols.opentrons.com/protocol/11bb6a-part-8)\n* [Part 9: Fragment Analyzer](http://protocols.opentrons.com/protocol/11bb6a-part-9)\n* [Part 10: Pooling According to Concentration](http://protocols.opentrons.com/protocol/11bb6a-part-10)\n\n---\n\n\n",
diff --git a/protoBuilds/11bb6a-part-8/README.json b/protoBuilds/11bb6a-part-8/README.json
index 284715001c..28f803b850 100644
--- a/protoBuilds/11bb6a-part-8/README.json
+++ b/protoBuilds/11bb6a-part-8/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext"
@@ -10,7 +10,7 @@
"internal": "11bb6a-part-8",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons Temperature Module] (https://shop.opentrons.com/modules/)\n[Opentrons Magnetic Module] (https://shop.opentrons.com/modules/)\n[Selected 96-well PCR plate] ([see parameter dropdown list below] https://labware.opentrons.com/)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module empty \n**Slot 3**: Output Plate (dropdown selected 96-well PCR plate) \n**Slot 5**: Reagent Reservoir (Nest 12-well 15 mL reservoir) \n\n\n\n**Slot 6**: Ethanol (Agilent 290 mL reservoir) \n**Slot 8**: Liquid Waste (Agilent 290 mL reservoir) \n**Slot 9**: Opentrons Magnetic Module with PCR plate (dropdown-selected 96-well PCR plate) \n**Slots 10,11**: Opentrons 20 uL filter tips \n**Slots 2,4,7**: Opentrons 200 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses multi-channel P20 and P300 pipettes to perform bead cleanup steps on 48-96 input samples. User-determined parameters are available for selection of the labware for the library prep plate and the output plate.\n\nLinks:\n* [Part 1: Enzymatic Fragmentation and End Prep](http://protocols.opentrons.com/protocol/11bb6a)\n* [Part 2: Sample Cleanup for Covaris Samples](http://protocols.opentrons.com/protocol/11bb6a-part-2)\n* [Part 3: End Prep](http://protocols.opentrons.com/protocol/11bb6a-part-3)\n* [Part 4: Adapter Ligation](http://protocols.opentrons.com/protocol/11bb6a-part-4)\n* [Part 5: Size Selection](http://protocols.opentrons.com/protocol/11bb6a-part-5)\n* [Part 6: Clean Up](http://protocols.opentrons.com/protocol/11bb6a-part-6)\n* [Part 7: PCR Enrichment](http://protocols.opentrons.com/protocol/11bb6a-part-7)\n* [Part 8: Sample Clean Up](http://protocols.opentrons.com/protocol/11bb6a-part-8)\n* [Part 9: Fragment Analyzer](http://protocols.opentrons.com/protocol/11bb6a-part-9)\n* [Part 10: Pooling According to Concentration](http://protocols.opentrons.com/protocol/11bb6a-part-10)\n\n---\n\n\n",
diff --git a/protoBuilds/11bb6a-part-9/README.json b/protoBuilds/11bb6a-part-9/README.json
index 16fd9f0e71..f1b695f534 100644
--- a/protoBuilds/11bb6a-part-9/README.json
+++ b/protoBuilds/11bb6a-part-9/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext"
@@ -10,7 +10,7 @@
"internal": "11bb6a-part-9",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons Temperature Module] (https://shop.opentrons.com/modules/)\n[Opentrons Magnetic Module] (https://shop.opentrons.com/modules/)\n[Selected 96-well PCR plate] ([see parameter dropdown list below] https://labware.opentrons.com/)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module empty \n**Slot 3**: Libraries Plate (dropdown selected 96-well PCR plate) \n**Slot 5**: Reagent Reservoir (Nest 12-well 15 mL reservoir) \n**Slot 6**: Fragment Analyzer Plate on Opentrons 96-Well Aluminum Block (biorad_96_fragment_analyzer_plate_aluminumblock_200ul) \n**Slot 9**: Opentrons Magnetic Module empty \n**Slot 10**: Opentrons 20 uL filter tips \n**Slot 7**: Opentrons 200 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses multi-channel P20 and P300 pipettes to perform fragment analyzer set up steps on 48-96 output libraries. User-determined parameters are available for selection of the labware for the libraries plate.\n\nLinks:\n* [Part 1: Enzymatic Fragmentation and End Prep](http://protocols.opentrons.com/protocol/11bb6a)\n* [Part 2: Sample Cleanup for Covaris Samples](http://protocols.opentrons.com/protocol/11bb6a-part-2)\n* [Part 3: End Prep](http://protocols.opentrons.com/protocol/11bb6a-part-3)\n* [Part 4: Adapter Ligation](http://protocols.opentrons.com/protocol/11bb6a-part-4)\n* [Part 5: Size Selection](http://protocols.opentrons.com/protocol/11bb6a-part-5)\n* [Part 6: Clean Up](http://protocols.opentrons.com/protocol/11bb6a-part-6)\n* [Part 7: PCR Enrichment](http://protocols.opentrons.com/protocol/11bb6a-part-7)\n* [Part 8: Sample Clean Up](http://protocols.opentrons.com/protocol/11bb6a-part-8)\n* [Part 9: Fragment Analyzer](http://protocols.opentrons.com/protocol/11bb6a-part-9)\n* [Part 10: Pooling According to Concentration](http://protocols.opentrons.com/protocol/11bb6a-part-10)\n\n---\n\n\n",
diff --git a/protoBuilds/11bb6a/README.json b/protoBuilds/11bb6a/README.json
index 9343bfd347..e7a1b1e036 100644
--- a/protoBuilds/11bb6a/README.json
+++ b/protoBuilds/11bb6a/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext"
@@ -10,7 +10,7 @@
"internal": "11bb6a",
"labware": "\nOpentrons Filter Tips for the P20 and P300 (https://shop.opentrons.com)\nOpentrons Temperature Module (https://shop.opentrons.com/modules/)\nSelected 96-well PCR plate ([see parameter dropdown list below] https://labware.opentrons.com/)\nSelected 24-well Aluminum Block and Tube Combination ([see parameter dropdown list below] https://labware.opentrons.com/)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module with dropdown-selected 24-well Aluminum Block and Tube combination \n\n\n\n**Slot 3**: Library Prep Plate (dropdown-selected 96-well PCR plate) \n**Slot 6**: Reagent Plate (dropdown-selected 96-well PCR plate) \n**Slot 10**: Opentrons 20 uL Filter Tips \n**Slot 11**: Opentrons 200 uL Filter Tips \n\n\n---\n\n",
"description": "This protocol uses multi-channel P20 and P300 pipettes to perform enzymatic fragmentation and end prep steps on 48-96 input samples. User-determined parameters are available for selection of the labware for the 96-well library prep and reagent plates and the aluminum block and tube combination holding reagents on the temperature module.\n\n---\n\n\n",
diff --git a/protoBuilds/11beaa/README.json b/protoBuilds/11beaa/README.json
index 173e7e2786..d1273149f8 100644
--- a/protoBuilds/11beaa/README.json
+++ b/protoBuilds/11beaa/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "11beaa",
"labware": "\nPerkinElmer Deepwell Pro Plate 96V 450\u00b5L (cat 6008299)\nBIO-RAD #HSP3865: Hard-Shell 384-Well PCR Plate, thin wall, skirted.\nBIO-RAD #HSP9601: HardShell 96Well PCR Plates, low profile, thin wall, skirted\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n",
"description": "This protocol preps a 384 well BIO-RAD plate for PCR. The protocol can be considered in 3 main parts:\n\n* cDNA is added to the SuperMix and mixed\n* Resulting solution (mastermix) is added to each well in the 384 well PCR plate\n* Primer is added to each well in the 384 well plate\n\nSince this protocol relies on the transfer of small amounts of liquid (in some cases 1ul transfers), aspiration and dispense flow rates are slowed, as well as incorporating blowout steps and proper aspiration/dispense heights to ensure proper distribution.\n\nExplanation of complex parameters below:\n\n* `Number of Samples`: Specify the number of primer pairs you will be running in this run (1-12, and wholly divisible by 2). A value of 2 will provide enough primer for 4 columns on the 384 well PCR plate, a value of 4 will accommodate 8 columns, and so forth.\n* `cDNA Column`: Specify which columns the cDNA will be hosted in Slot 4 of the deck. Select `0` for column 2 if only running one cDNA column.\n* `Aspiration height from well bottom`: Specify (in mm) the distance above the well bottom all aspirations steps will aspirate from.\n* `Dispense height from well bottom`: Specify (in mm) the distance above the well bottom all dispense steps will dispense from.\n* `Aspiration flow rate for P10-Multi (ul/sec)`: Specify the aspiration flow rate for the P10-Multi.\n* `Aspiration flow rate for P50-Multi (ul/sec)`: Specify the aspiration flow rate for the P50-Multi.\n* `Dispense flow rate for P10-Multi (ul/sec)`: Specify the dispense flow rate for the P10-Multi.\n* `Dispense flow rate for P50-Multi (ul/sec)`: Specify the dispense flow rate for the P50-Multi.\n* `Use Temperature Module`: If `Yes` is selected, the temperature module will be engaged over the course of the protocol with the user-specified temperature. A selection of `No` disengages the temperature module over the course of the protocol.\n* `Temperature Module Temperature`: Specify the temperature the temperature module will hold over the duration of the protocol if `Yes` is selected in the `Use Temperature Module` parameter.\n* `Small Volume Pipette`: Specify where a GEN1 or GEN2 pipette will be employed.\n* `Large Volume Pipette`: Specify where a GEN1 or GEN2 pipette will be employed.\n* `P10-Multi mount`: Specify which side the P10-Multi pipette will be mounted.\n* `P50-Multi mount`: Specify which side the P10-Multi pipette will be mounted.\n\n\n---\n\n",
diff --git a/protoBuilds/1211/README.json b/protoBuilds/1211/README.json
index d301a2b9c6..9c875ed61c 100644
--- a/protoBuilds/1211/README.json
+++ b/protoBuilds/1211/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -8,7 +8,7 @@
"description": "With this protocol, your robot can perform multiple well-to-well liquid transfers using a single-channel pipette by parsing through a user-defined .csv file.\nThe .csv file should be formatted as shown in the example below, including headers:\nSource Labware,Source Slot,Source Well,Source Aspiration Height Above Bottom (in mm),Dest Labware,Dest Slot,Dest Well,Volume (in ul)\nagilent_1_reservoir_290ml,1,A1,1,nest_96_wellplate_100ul_pcr_full_skirt,4,A11,1\nnest_12_reservoir_15ml,2,A1,1,nest_96_wellplate_2ml_deep,5,A5,3\nnest_1_reservoir_195ml,3,A1,1,nest_96_wellplate_2ml_deep,5,H12,7\nAll available empty slots will be filled with the necessary tipracks, and the user will be prompted to refill the tipracks if all are emptied in the middle of the protocol.",
"internal": "1211",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n\n",
"description": "With this protocol, your robot can perform multiple well-to-well liquid transfers using a single-channel pipette by parsing through a user-defined .csv file.\n\nThe .csv file should be formatted as shown in the example below, **including headers**:\n\n```\nSource Labware,Source Slot,Source Well,Source Aspiration Height Above Bottom (in mm),Dest Labware,Dest Slot,Dest Well,Volume (in ul)\nagilent_1_reservoir_290ml,1,A1,1,nest_96_wellplate_100ul_pcr_full_skirt,4,A11,1\nnest_12_reservoir_15ml,2,A1,1,nest_96_wellplate_2ml_deep,5,A5,3\nnest_1_reservoir_195ml,3,A1,1,nest_96_wellplate_2ml_deep,5,H12,7\n```\n\nAll available empty slots will be filled with the necessary tipracks, and the user will be prompted to refill the tipracks if all are emptied in the middle of the protocol.\n\n",
"internal": "1211\n",
diff --git a/protoBuilds/121d15-2-384/README.json b/protoBuilds/121d15-2-384/README.json
index 4a936c97c2..a1788515b4 100644
--- a/protoBuilds/121d15-2-384/README.json
+++ b/protoBuilds/121d15-2-384/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -10,7 +10,7 @@
"internal": "121d15",
"labware": "\nCustom 48-tuberack\nGreiner MASTERBLOCK 384 Well Plate 225 \u00b5L\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Manual Cleave](./121d15)\n
\n
\n* [Manual Cleave Elution (Off-deck Vacuum)](./121d15-4)\n
\n
\n* [Manual Cleave, ACN + Elution (Off-deck Vacuum)](./121d15-5)\n
\n
\n* [Manual Cleave, ACN + Elution (On-deck Vacuum)](./121d15-6)\n
\n
\n* [HPLC Picking](./121d15-3)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 500 \u00b5L)](./121d15-2-96-Greiner-500)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 1000 \u00b5L)](./121d15-2-96-Greiner-1000)\n
\n
\n* [Redo Replacement Picking (Irish Life Sciences 96 Well Plate 2200 \u00b5L)](./121d15-2-96-Irish-2200)\n
\n
\n* [Redo Replacement Picking (Greiner Masterblock 384 Well Plate 225 \u00b5L)](./121d15-2-384)\n
\n
\n* [Aliquoting - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-aliquot)\n
\n
\n* [Aliquoting - 15ml Tuberack to 2ml Tuberack](./121d15-7-2ml-15ml-aliquot)\n
\n
\n* [Pooling - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-pool)\n
\n
\n* [Pooling - 2ml Tuberack to 15ml Tuberack](./121d15-7-2ml-15ml-pool)\n
\n
\n\nThis protocol performs a custom Redo Replacement Picking protocol from a worklist. The worklist should be specified as follows:\n\n```\nNumber of Redo\n9\npos TB_RCK_1,pos MTP_1,disposal_vol,transfer_vol\nTUBE BAR,PLATE BAR\n1,1,200,200\n2,135,200,200\n3,2,150,200\n4,90,100,100\n5,84\n6,262\n7,242\n8,218\n9,219\n...\n```\n\n---\n\n",
diff --git a/protoBuilds/121d15-2-96-Greiner-1000/README.json b/protoBuilds/121d15-2-96-Greiner-1000/README.json
index d63f26c94f..318880e87f 100644
--- a/protoBuilds/121d15-2-96-Greiner-1000/README.json
+++ b/protoBuilds/121d15-2-96-Greiner-1000/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -10,7 +10,7 @@
"internal": "121d15",
"labware": "\nCustom 48-tuberack\nGreiner MASTERBLOCK 96 Well Plate 500 \u00b5L\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Manual Cleave](./121d15)\n
\n
\n* [Manual Cleave Elution (Off-deck Vacuum)](./121d15-4)\n
\n
\n* [Manual Cleave, ACN + Elution (Off-deck Vacuum)](./121d15-5)\n
\n
\n* [Manual Cleave, ACN + Elution (On-deck Vacuum)](./121d15-6)\n
\n
\n* [HPLC Picking](./121d15-3)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 500 \u00b5L)](./121d15-2-96-Greiner-500)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 1000 \u00b5L)](./121d15-2-96-Greiner-1000)\n
\n
\n* [Redo Replacement Picking (Irish Life Sciences 96 Well Plate 2200 \u00b5L)](./121d15-2-96-Irish-2200)\n
\n
\n* [Redo Replacement Picking (Greiner Masterblock 384 Well Plate 225 \u00b5L)](./121d15-2-384)\n
\n
\n* [Aliquoting - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-aliquot)\n
\n
\n* [Aliquoting - 15ml Tuberack to 2ml Tuberack](./121d15-7-2ml-15ml-aliquot)\n
\n
\n* [Pooling - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-pool)\n
\n
\n* [Pooling - 2ml Tuberack to 15ml Tuberack](./121d15-7-2ml-15ml-pool)\n
\n
\n\nThis protocol performs a custom Redo Replacement Picking protocol from a worklist. The worklist should be specified as follows:\n\n```\nNumber of Redo\n8\npos TB_RCK_1,pos MTP_1,disposal_vol,transfer_vol\nTUBE BAR,PLATE BAR\n1,1,200,200\n2,35,200,200\n3,2,150,200\n4,90,100,100\n5,84\n6,62\n7,42\n8,18\n...\n```\n\n---\n\n",
diff --git a/protoBuilds/121d15-2-96-Greiner-500/README.json b/protoBuilds/121d15-2-96-Greiner-500/README.json
index 1ef2295875..d77e062da8 100644
--- a/protoBuilds/121d15-2-96-Greiner-500/README.json
+++ b/protoBuilds/121d15-2-96-Greiner-500/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -10,7 +10,7 @@
"internal": "121d15",
"labware": "\nCustom 48-tuberack\nGreiner MASTERBLOCK 96 Well Plate 500 \u00b5L\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Manual Cleave](./121d15)\n
\n
\n* [Manual Cleave Elution (Off-deck Vacuum)](./121d15-4)\n
\n
\n* [Manual Cleave, ACN + Elution (Off-deck Vacuum)](./121d15-5)\n
\n
\n* [Manual Cleave, ACN + Elution (On-deck Vacuum)](./121d15-6)\n
\n
\n* [HPLC Picking](./121d15-3)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 500 \u00b5L)](./121d15-2-96-Greiner-500)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 1000 \u00b5L)](./121d15-2-96-Greiner-1000)\n
\n
\n* [Redo Replacement Picking (Irish Life Sciences 96 Well Plate 2200 \u00b5L)](./121d15-2-96-Irish-2200)\n
\n
\n* [Redo Replacement Picking (Greiner Masterblock 384 Well Plate 225 \u00b5L)](./121d15-2-384)\n
\n
\n* [Aliquoting - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-aliquot)\n
\n
\n* [Aliquoting - 15ml Tuberack to 2ml Tuberack](./121d15-7-2ml-15ml-aliquot)\n
\n
\n* [Pooling - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-pool)\n
\n
\n* [Pooling - 2ml Tuberack to 15ml Tuberack](./121d15-7-2ml-15ml-pool)\n
\n
\n\nThis protocol performs a custom Redo Replacement Picking protocol from a worklist. The worklist should be specified as follows:\n\n```\nNumber of Redo\n8\npos TB_RCK_1,pos MTP_1,disposal_vol,transfer_vol\nTUBE BAR,PLATE BAR\n1,1,200,200\n2,35,200,200\n3,2,150,200\n4,90,100,100\n5,84\n6,62\n7,42\n8,18\n...\n```\n\n---\n\n",
diff --git a/protoBuilds/121d15-2-96-Irish-2200/README.json b/protoBuilds/121d15-2-96-Irish-2200/README.json
index 83d7eada44..ed0fc8d3c9 100644
--- a/protoBuilds/121d15-2-96-Irish-2200/README.json
+++ b/protoBuilds/121d15-2-96-Irish-2200/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -10,7 +10,7 @@
"internal": "121d15",
"labware": "\nCustom 48-tuberack\nGreiner MASTERBLOCK 96 Well Plate 500 \u00b5L\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Manual Cleave](./121d15)\n
\n
\n* [Manual Cleave Elution (Off-deck Vacuum)](./121d15-4)\n
\n
\n* [Manual Cleave, ACN + Elution (Off-deck Vacuum)](./121d15-5)\n
\n
\n* [Manual Cleave, ACN + Elution (On-deck Vacuum)](./121d15-6)\n
\n
\n* [HPLC Picking](./121d15-3)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 500 \u00b5L)](./121d15-2-96-Greiner-500)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 1000 \u00b5L)](./121d15-2-96-Greiner-1000)\n
\n
\n* [Redo Replacement Picking (Irish Life Sciences 96 Well Plate 2200 \u00b5L)](./121d15-2-96-Irish-2200)\n
\n
\n* [Redo Replacement Picking (Greiner Masterblock 384 Well Plate 225 \u00b5L)](./121d15-2-384)\n
\n
\n* [Aliquoting - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-aliquot)\n
\n
\n* [Aliquoting - 15ml Tuberack to 2ml Tuberack](./121d15-7-2ml-15ml-aliquot)\n
\n
\n* [Pooling - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-pool)\n
\n
\n* [Pooling - 2ml Tuberack to 15ml Tuberack](./121d15-7-2ml-15ml-pool)\n
\n
\n\nThis protocol performs a custom Redo Replacement Picking protocol from a worklist. The worklist should be specified as follows:\n\n```\nNumber of Redo\n8\npos TB_RCK_1,pos MTP_1,disposal_vol,transfer_vol\nTUBE BAR,PLATE BAR\n1,1,200,200\n2,35,200,200\n3,2,150,200\n4,90,100,100\n5,84\n6,62\n7,42\n8,18\n...\n```\n\n---\n\n",
diff --git a/protoBuilds/121d15-3/README.json b/protoBuilds/121d15-3/README.json
index ee5bdeacc5..e07094f7e7 100644
--- a/protoBuilds/121d15-3/README.json
+++ b/protoBuilds/121d15-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -10,7 +10,7 @@
"internal": "121d15",
"labware": "\nIrish Life Sciences 2.2mL Deep Well Plate, V-Bottom #2.2S96-008V\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Manual Cleave](./121d15)\n
\n
\n* [Manual Cleave Elution (Off-deck Vacuum)](./121d15-4)\n
\n
\n* [Manual Cleave, ACN + Elution (Off-deck Vacuum)](./121d15-5)\n
\n
\n* [Manual Cleave, ACN + Elution (On-deck Vacuum)](./121d15-6)\n
\n
\n* [HPLC Picking](./121d15-3)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 500 \u00b5L)](./121d15-2-96-Greiner-500)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 1000 \u00b5L)](./121d15-2-96-Greiner-1000)\n
\n
\n* [Redo Replacement Picking (Irish Life Sciences 96 Well Plate 2200 \u00b5L)](./121d15-2-96-Irish-2200)\n
\n
\n* [Redo Replacement Picking (Greiner Masterblock 384 Well Plate 225 \u00b5L)](./121d15-2-384)\n
\n
\n* [Aliquoting - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-aliquot)\n
\n
\n* [Aliquoting - 15ml Tuberack to 2ml Tuberack](./121d15-7-2ml-15ml-aliquot)\n
\n
\n* [Pooling - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-pool)\n
\n
\n* [Pooling - 2ml Tuberack to 15ml Tuberack](./121d15-7-2ml-15ml-pool)\n
\n
\n\nThis protocol performs a custom HPLC Picking protocol from a worklist. The worklist should be specified as follows:\n\n```\nsource,dest,volume\n1,A1,300\n2,A1,300\n3,A1,300\n4,A1,300\n10,B1,300\n12,B1,300\n13,B1,300\n14,B1,\n```\n\n---\n\n",
diff --git a/protoBuilds/121d15-4/README.json b/protoBuilds/121d15-4/README.json
index 8189327008..c5604b08b0 100644
--- a/protoBuilds/121d15-4/README.json
+++ b/protoBuilds/121d15-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "121d15",
"labware": "\nCustom 96-tuberack\nNEST 1 Reservoir 195ml\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Manual Cleave](./121d15)\n
\n
\n* [Manual Cleave Elution (Off-deck Vacuum)](./121d15-4)\n
\n
\n* [Manual Cleave, ACN + Elution (Off-deck Vacuum)](./121d15-5)\n
\n
\n* [Manual Cleave, ACN + Elution (On-deck Vacuum)](./121d15-6)\n
\n
\n* [HPLC Picking](./121d15-3)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 500 \u00b5L)](./121d15-2-96-Greiner-500)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 1000 \u00b5L)](./121d15-2-96-Greiner-1000)\n
\n
\n* [Redo Replacement Picking (Irish Life Sciences 96 Well Plate 2200 \u00b5L)](./121d15-2-96-Irish-2200)\n
\n
\n* [Redo Replacement Picking (Greiner Masterblock 384 Well Plate 225 \u00b5L)](./121d15-2-384)\n
\n
\n* [Aliquoting - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-aliquot)\n
\n
\n* [Aliquoting - 15ml Tuberack to 2ml Tuberack](./121d15-7-2ml-15ml-aliquot)\n
\n
\n* [Pooling - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-pool)\n
\n
\n* [Pooling - 2ml Tuberack to 15ml Tuberack](./121d15-7-2ml-15ml-pool)\n
\n
\n\nThis protocol performs a custom plate filling elution for up to 4x 96-well plates from a single reagent reservoir. The user can input the number of rows and columns to be filled for each plate. Liquid handling parameters are automatically determined from the user's selection of which reagent will be added during the run.\n\n---\n\n",
diff --git a/protoBuilds/121d15-5/README.json b/protoBuilds/121d15-5/README.json
index b1be7dd1c9..ff480eca22 100644
--- a/protoBuilds/121d15-5/README.json
+++ b/protoBuilds/121d15-5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "121d15",
"labware": "\nCustom 96-tuberack\nNEST 1 Reservoir 195ml\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Manual Cleave](./121d15)\n
\n
\n* [Manual Cleave Elution (Off-deck Vacuum)](./121d15-4)\n
\n
\n* [Manual Cleave, ACN + Elution (Off-deck Vacuum)](./121d15-5)\n
\n
\n* [Manual Cleave, ACN + Elution (On-deck Vacuum)](./121d15-6)\n
\n
\n* [HPLC Picking](./121d15-3)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 500 \u00b5L)](./121d15-2-96-Greiner-500)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 1000 \u00b5L)](./121d15-2-96-Greiner-1000)\n
\n
\n* [Redo Replacement Picking (Irish Life Sciences 96 Well Plate 2200 \u00b5L)](./121d15-2-96-Irish-2200)\n
\n
\n* [Redo Replacement Picking (Greiner Masterblock 384 Well Plate 225 \u00b5L)](./121d15-2-384)\n
\n
\n* [Aliquoting - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-aliquot)\n
\n
\n* [Aliquoting - 15ml Tuberack to 2ml Tuberack](./121d15-7-2ml-15ml-aliquot)\n
\n
\n* [Pooling - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-pool)\n
\n
\n* [Pooling - 2ml Tuberack to 15ml Tuberack](./121d15-7-2ml-15ml-pool)\n
\n
\n\nThis protocol performs a custom plate filling and elution for up to 4x 96-well plates from a single reagent reservoir. The user can input the number of rows and columns to be filled for each plate. Liquid handling parameters are automatically determined from the user's selection of which reagent will be added during the run.\n\n---\n\n",
diff --git a/protoBuilds/121d15-6/README.json b/protoBuilds/121d15-6/README.json
index d0ecd9c61d..9794014d8c 100644
--- a/protoBuilds/121d15-6/README.json
+++ b/protoBuilds/121d15-6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "121d15",
"labware": "\nCustom 96-tuberack\nNEST 1 Reservoir 195ml\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Manual Cleave](./121d15)\n
\n
\n* [Manual Cleave Elution (Off-deck Vacuum)](./121d15-4)\n
\n
\n* [Manual Cleave, ACN + Elution (Off-deck Vacuum)](./121d15-5)\n
\n
\n* [Manual Cleave, ACN + Elution (On-deck Vacuum)](./121d15-6)\n
\n
\n* [HPLC Picking](./121d15-3)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 500 \u00b5L)](./121d15-2-96-Greiner-500)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 1000 \u00b5L)](./121d15-2-96-Greiner-1000)\n
\n
\n* [Redo Replacement Picking (Irish Life Sciences 96 Well Plate 2200 \u00b5L)](./121d15-2-96-Irish-2200)\n
\n
\n* [Redo Replacement Picking (Greiner Masterblock 384 Well Plate 225 \u00b5L)](./121d15-2-384)\n
\n
\n* [Aliquoting - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-aliquot)\n
\n
\n* [Aliquoting - 15ml Tuberack to 2ml Tuberack](./121d15-7-2ml-15ml-aliquot)\n
\n
\n* [Pooling - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-pool)\n
\n
\n* [Pooling - 2ml Tuberack to 15ml Tuberack](./121d15-7-2ml-15ml-pool)\n
\n
\n\nThis protocol performs a custom plate filling and elution for up to 4x 96-well plates from a single reagent reservoir. The user can input the number of rows and columns to be filled for each plate. Liquid handling parameters are automatically determined from the user's selection of which reagent will be added during the run.\n\n---\n\n",
diff --git a/protoBuilds/121d15-7-2ml-15ml-aliquot/README.json b/protoBuilds/121d15-7-2ml-15ml-aliquot/README.json
index 12924aea67..3585d2fbc1 100644
--- a/protoBuilds/121d15-7-2ml-15ml-aliquot/README.json
+++ b/protoBuilds/121d15-7-2ml-15ml-aliquot/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Pooling"
@@ -10,7 +10,7 @@
"internal": "121d15",
"labware": "\nCustom 96-tuberacks\nOpentrons 4-in-1 Tube Rack Set with 3x5 15ml Tube Insert\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Pooling\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Manual Cleave](./121d15)\n
\n
\n* [Manual Cleave Elution (Off-deck Vacuum)](./121d15-4)\n
\n
\n* [Manual Cleave, ACN + Elution (Off-deck Vacuum)](./121d15-5)\n
\n
\n* [Manual Cleave, ACN + Elution (On-deck Vacuum)](./121d15-6)\n
\n
\n* [HPLC Picking](./121d15-3)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 500 \u00b5L)](./121d15-2-96-Greiner-500)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 1000 \u00b5L)](./121d15-2-96-Greiner-1000)\n
\n
\n* [Redo Replacement Picking (Irish Life Sciences 96 Well Plate 2200 \u00b5L)](./121d15-2-96-Irish-2200)\n
\n
\n* [Redo Replacement Picking (Greiner Masterblock 384 Well Plate 225 \u00b5L)](./121d15-2-384)\n
\n
\n* [Aliquoting - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-aliquot)\n
\n
\n* [Aliquoting - 15ml Tuberack to 2ml Tuberack](./121d15-7-2ml-15ml-aliquot)\n
\n
\n* [Pooling - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-pool)\n
\n
\n* [Pooling - 2ml Tuberack to 15ml Tuberack](./121d15-7-2ml-15ml-pool)\n
\n
\n\nThis protocol performs a custom pooling protocol from a worklist. The worklist should be specified as follows:\n\n```\nNumber\n8\npos TB_RCK_1,pos MTP_1,transfer_vol\nTUBE BAR 1,TUBE BAR 1\n1,1,200\n2,35,200\n3,2,150\n4,90,100\n5,84\n6,62\n7,42\n8,18\n...\n```\n\n---\n\n",
diff --git a/protoBuilds/121d15-7-2ml-15ml-pool/README.json b/protoBuilds/121d15-7-2ml-15ml-pool/README.json
index 12924aea67..3585d2fbc1 100644
--- a/protoBuilds/121d15-7-2ml-15ml-pool/README.json
+++ b/protoBuilds/121d15-7-2ml-15ml-pool/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Pooling"
@@ -10,7 +10,7 @@
"internal": "121d15",
"labware": "\nCustom 96-tuberacks\nOpentrons 4-in-1 Tube Rack Set with 3x5 15ml Tube Insert\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Pooling\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Manual Cleave](./121d15)\n
\n
\n* [Manual Cleave Elution (Off-deck Vacuum)](./121d15-4)\n
\n
\n* [Manual Cleave, ACN + Elution (Off-deck Vacuum)](./121d15-5)\n
\n
\n* [Manual Cleave, ACN + Elution (On-deck Vacuum)](./121d15-6)\n
\n
\n* [HPLC Picking](./121d15-3)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 500 \u00b5L)](./121d15-2-96-Greiner-500)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 1000 \u00b5L)](./121d15-2-96-Greiner-1000)\n
\n
\n* [Redo Replacement Picking (Irish Life Sciences 96 Well Plate 2200 \u00b5L)](./121d15-2-96-Irish-2200)\n
\n
\n* [Redo Replacement Picking (Greiner Masterblock 384 Well Plate 225 \u00b5L)](./121d15-2-384)\n
\n
\n* [Aliquoting - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-aliquot)\n
\n
\n* [Aliquoting - 15ml Tuberack to 2ml Tuberack](./121d15-7-2ml-15ml-aliquot)\n
\n
\n* [Pooling - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-pool)\n
\n
\n* [Pooling - 2ml Tuberack to 15ml Tuberack](./121d15-7-2ml-15ml-pool)\n
\n
\n\nThis protocol performs a custom pooling protocol from a worklist. The worklist should be specified as follows:\n\n```\nNumber\n8\npos TB_RCK_1,pos MTP_1,transfer_vol\nTUBE BAR 1,TUBE BAR 1\n1,1,200\n2,35,200\n3,2,150\n4,90,100\n5,84\n6,62\n7,42\n8,18\n...\n```\n\n---\n\n",
diff --git a/protoBuilds/121d15-7-2ml-2ml-aliquot/README.json b/protoBuilds/121d15-7-2ml-2ml-aliquot/README.json
index 396804910a..d8b2697907 100644
--- a/protoBuilds/121d15-7-2ml-2ml-aliquot/README.json
+++ b/protoBuilds/121d15-7-2ml-2ml-aliquot/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Aliquoting"
@@ -10,7 +10,7 @@
"internal": "121d15",
"labware": "\nCustom 96-tuberacks\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Aliquoting\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Manual Cleave](./121d15)\n
\n
\n* [Manual Cleave Elution (Off-deck Vacuum)](./121d15-4)\n
\n
\n* [Manual Cleave, ACN + Elution (Off-deck Vacuum)](./121d15-5)\n
\n
\n* [Manual Cleave, ACN + Elution (On-deck Vacuum)](./121d15-6)\n
\n
\n* [HPLC Picking](./121d15-3)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 500 \u00b5L)](./121d15-2-96-Greiner-500)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 1000 \u00b5L)](./121d15-2-96-Greiner-1000)\n
\n
\n* [Redo Replacement Picking (Irish Life Sciences 96 Well Plate 2200 \u00b5L)](./121d15-2-96-Irish-2200)\n
\n
\n* [Redo Replacement Picking (Greiner Masterblock 384 Well Plate 225 \u00b5L)](./121d15-2-384)\n
\n
\n* [Aliquoting - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-aliquot)\n
\n
\n* [Aliquoting - 15ml Tuberack to 2ml Tuberack](./121d15-7-2ml-15ml-aliquot)\n
\n
\n* [Pooling - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-pool)\n
\n
\n* [Pooling - 2ml Tuberack to 15ml Tuberack](./121d15-7-2ml-15ml-pool)\n
\n
\n\nThis protocol performs a custom aliquoting protocol from a worklist. The worklist should be specified as follows:\n\n```\nNumber\n8\npos TB_RCK_1,pos MTP_1,transfer_vol\nTUBE BAR 1,TUBE BAR 1\n1,1,200\n2,35,200\n3,2,150\n4,90,100\n5,84\n6,62\n7,42\n8,18\n...\n```\n\n---\n\n",
diff --git a/protoBuilds/121d15-7-2ml-2ml-pool/README.json b/protoBuilds/121d15-7-2ml-2ml-pool/README.json
index 83f704de92..2faafccca0 100644
--- a/protoBuilds/121d15-7-2ml-2ml-pool/README.json
+++ b/protoBuilds/121d15-7-2ml-2ml-pool/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Pooling"
@@ -10,7 +10,7 @@
"internal": "121d15",
"labware": "\nCustom 96-tuberacks\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Pooling\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Manual Cleave](./121d15)\n
\n
\n* [Manual Cleave Elution (Off-deck Vacuum)](./121d15-4)\n
\n
\n* [Manual Cleave, ACN + Elution (Off-deck Vacuum)](./121d15-5)\n
\n
\n* [Manual Cleave, ACN + Elution (On-deck Vacuum)](./121d15-6)\n
\n
\n* [HPLC Picking](./121d15-3)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 500 \u00b5L)](./121d15-2-96-Greiner-500)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 1000 \u00b5L)](./121d15-2-96-Greiner-1000)\n
\n
\n* [Redo Replacement Picking (Irish Life Sciences 96 Well Plate 2200 \u00b5L)](./121d15-2-96-Irish-2200)\n
\n
\n* [Redo Replacement Picking (Greiner Masterblock 384 Well Plate 225 \u00b5L)](./121d15-2-384)\n
\n
\n* [Aliquoting - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-aliquot)\n
\n
\n* [Aliquoting - 15ml Tuberack to 2ml Tuberack](./121d15-7-2ml-15ml-aliquot)\n
\n
\n* [Pooling - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-pool)\n
\n
\n* [Pooling - 2ml Tuberack to 15ml Tuberack](./121d15-7-2ml-15ml-pool)\n
\n
\n\nThis protocol performs a custom pooling protocol from a worklist. The worklist should be specified as follows:\n\n```\nNumber\n8\npos TB_RCK_1,pos MTP_1,transfer_vol\nTUBE BAR 1,TUBE BAR 1\n1,1,200\n2,35,200\n3,2,150\n4,90,100\n5,84\n6,62\n7,42\n8,18\n...\n```\n\n---\n\n",
diff --git a/protoBuilds/121d15-repeat/README.json b/protoBuilds/121d15-repeat/README.json
index 1e07a5bba0..2390475713 100644
--- a/protoBuilds/121d15-repeat/README.json
+++ b/protoBuilds/121d15-repeat/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "121d15-repeat",
"labware": "\nCustom 96-tuberack\nNEST 1 Reservoir 195ml\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Manual Cleave](./121d15)\n
\n
\n* [Manual Cleave with Repeat for 2nd EDA](./121d15-repeat)\n
\n
\n* [Manual Cleave Elution (Off-deck Vacuum)](./121d15-4)\n
\n
\n* [Manual Cleave, ACN + Elution (Off-deck Vacuum)](./121d15-5)\n
\n
\n* [Manual Cleave, ACN + Elution (On-deck Vacuum)](./121d15-6)\n
\n
\n* [HPLC Picking](./121d15-3)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 500 \u00b5L)](./121d15-2-96-Greiner-500)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 1000 \u00b5L)](./121d15-2-96-Greiner-1000)\n
\n
\n* [Redo Replacement Picking (Irish Life Sciences 96 Well Plate 2200 \u00b5L)](./121d15-2-96-Irish-2200)\n
\n
\n* [Redo Replacement Picking (Greiner Masterblock 384 Well Plate 225 \u00b5L)](./121d15-2-384)\n
\n
\n* [Aliquoting - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-aliquot)\n
\n
\n* [Aliquoting - 15ml Tuberack to 2ml Tuberack](./121d15-7-2ml-15ml-aliquot)\n
\n
\n* [Pooling - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-pool)\n
\n
\n* [Pooling - 2ml Tuberack to 15ml Tuberack](./121d15-7-2ml-15ml-pool)\n
\n
\n\nThis protocol performs a custom plate filling for up to 4x 96-well plates from a single reagent reservoir. The user can input the number of rows and columns to be filled for each plate. Liquid handling parameters are automatically determined from the user's selection of which reagent will be added during the run.\n\n---\n\n",
diff --git a/protoBuilds/121d15/README.json b/protoBuilds/121d15/README.json
index 884039d91d..4489caf8dc 100644
--- a/protoBuilds/121d15/README.json
+++ b/protoBuilds/121d15/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "121d15",
"labware": "\nCustom 96-tuberack\nNEST 1 Reservoir 195ml\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Manual Cleave](./121d15)\n
\n
\n* [Manual Cleave with Repeat for 2nd EDA](./121d15-repeat)\n
\n
\n* [Manual Cleave Elution (Off-deck Vacuum)](./121d15-4)\n
\n
\n* [Manual Cleave, ACN + Elution (Off-deck Vacuum)](./121d15-5)\n
\n
\n* [Manual Cleave, ACN + Elution (On-deck Vacuum)](./121d15-6)\n
\n
\n* [HPLC Picking](./121d15-3)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 500 \u00b5L)](./121d15-2-96-Greiner-500)\n
\n
\n* [Redo Replacement Picking (Greiner MASTERBLOCK 96 Well Plate 1000 \u00b5L)](./121d15-2-96-Greiner-1000)\n
\n
\n* [Redo Replacement Picking (Irish Life Sciences 96 Well Plate 2200 \u00b5L)](./121d15-2-96-Irish-2200)\n
\n
\n* [Redo Replacement Picking (Greiner Masterblock 384 Well Plate 225 \u00b5L)](./121d15-2-384)\n
\n
\n* [Aliquoting - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-aliquot)\n
\n
\n* [Aliquoting - 15ml Tuberack to 2ml Tuberack](./121d15-7-2ml-15ml-aliquot)\n
\n
\n* [Pooling - 2ml Tuberack to 2ml Tuberack](./121d15-7-2ml-2ml-pool)\n
\n
\n* [Pooling - 2ml Tuberack to 15ml Tuberack](./121d15-7-2ml-15ml-pool)\n
\n
\n\nThis protocol performs a custom plate filling for up to 4x 96-well plates from a single reagent reservoir. The user can input the number of rows and columns to be filled for each plate. Liquid handling parameters are automatically determined from the user's selection of which reagent will be added during the run.\n\n---\n\n",
diff --git a/protoBuilds/12213d/README.json b/protoBuilds/12213d/README.json
index b2bf669103..dd8aa8022d 100644
--- a/protoBuilds/12213d/README.json
+++ b/protoBuilds/12213d/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol removes ethanol from a plate on the MagDeck and requires manual intervention by the user.\n\nAfter incubating on the MagDeck for 5 minutes, 150\u00b5L will be removed from each well. The user will then be prompted to remove the plate and centrifuge. After centrifuging, the user will replace the plate and a second incubation and liquid transfer will occur.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons Magnetic Module\nOpentrons P300 Multi-Channel Pipette\nOpentrons 300\u00b5L Tips\nOpentrons P20 Multi-Channel Pipette\nOpentrons 20\u00b5L Tips\nNEST 96-Well Plates\nUSA Scientific 12-Well Reservoir\n\n\n\nSlot 1: USA Scientific 12-Well Reservoir with ethanol in slots 1-4\nSlot 3: Opentrons 20\u00b5L Tips\nSlot 4: Opentrons Magnetic Module with NEST 96-Well Plates\nSlot 6: Opentrons 300\u00b5L Tips\n\n\nUsing the customizations field (below), set up your protocol.\n P300-Multi Pipette Mount: Select which mount (left or right) the P300-Multi is attached to.\n P20-Multi Pipette Mount: Select which mount (left or right) the P20-Multi is attached to.",
"internal": "12213d",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol removes ethanol from a plate on the MagDeck and requires manual intervention by the user.\n\nAfter incubating on the MagDeck for 5 minutes, 150\u00b5L will be removed from each well. The user will then be prompted to remove the plate and centrifuge. After centrifuging, the user will replace the plate and a second incubation and liquid transfer will occur.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons P20 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [USA Scientific 12-Well Reservoir](https://labware.opentrons.com/usascientific_12_reservoir_22ml?category=reservoir)\n\n\n---\n\n\nSlot 1: [USA Scientific 12-Well Reservoir](https://labware.opentrons.com/usascientific_12_reservoir_22ml?category=reservoir) with ethanol in slots 1-4\n\nSlot 3: [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\nSlot 4: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n\nSlot 6: [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n**Using the customizations field (below), set up your protocol.**\n* **P300-Multi Pipette Mount**: Select which mount (left or right) the P300-Multi is attached to.\n* **P20-Multi Pipette Mount**: Select which mount (left or right) the P20-Multi is attached to.\n\n\n\n",
"internal": "12213d\n",
diff --git a/protoBuilds/131de0/README.json b/protoBuilds/131de0/README.json
index b770a937b9..322903a109 100644
--- a/protoBuilds/131de0/README.json
+++ b/protoBuilds/131de0/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs mastermix and DNA sample transfer to a custom 384-well plate. All DNA samples are transferred in quadruplicate in adjacent wells forming 2x2 well squares on the 384-well plate (12x8 squares mapping to the 12x8 input from the 96-well DNA plate), as in the following image: \n\n\n\n\nMicroAmp Optical 384-well plate\nUSA Scientific 96 Well Plate 100 \u00b5L\nCustom single-channel reservoir 25ml (will span slots 4 and 1)\nOpentrons P10 multi-channel GEN1 pipette\nOpentrons 10\u00b5l tiprack\n\n\n",
"internal": "131de0",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol performs mastermix and DNA sample transfer to a custom 384-well plate. All DNA samples are transferred in quadruplicate in adjacent wells forming 2x2 well squares on the 384-well plate (12x8 squares mapping to the 12x8 input from the 96-well DNA plate), as in the following image: \n\n\n\n---\n\n\n* MicroAmp Optical 384-well plate\n* USA Scientific 96 Well Plate 100 \u00b5L\n* Custom single-channel reservoir 25ml (will span slots 4 and 1)\n* [Opentrons P10 multi-channel GEN1 pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 10\u00b5l tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n---\n\n\n",
"internal": "131de0\n",
diff --git a/protoBuilds/139815/README.json b/protoBuilds/139815/README.json
index 4c1cb4fc37..73a78bd12b 100644
--- a/protoBuilds/139815/README.json
+++ b/protoBuilds/139815/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Consolidation"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom pooling and consolidation protocol from a 96-well PCR plate to an 8-tube strip, and then from the strip to a microcentrifuge tube.\n\n\n\nEppendorf twin.tec 96-well PCR plate 200ul seated in aluminum block\nUSA Scientific 12x8-tube strips 300ul seated in aluminum block\n1.5ml Eppendorf safelock snapcap microcentrifuge tube seated in Opentrons 4-in-1 tuberack with 4x6 microcentrifuge tube insert\nOpentrons GEN1 P10-single channel electronic pipette\nOpentrons GEN1 P300-single channel electronic pipette\nOpentrons 10ul filter tip rack\nOpentrons 200ul filter tip rack\n\n\n\nseated 8-tube strips (slot 2)\n* strip 1: 8-tube strip for initial transfers from PCR plate\n4x6 insert on Opentrons tuberack (slot 3)\n* tube A1: tube for pooling",
"internal": "139815",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Consolidation\n\n",
"description": "This protocol performs a custom pooling and consolidation protocol from a 96-well PCR plate to an 8-tube strip, and then from the strip to a microcentrifuge tube.\n\n---\n\n\n* [Eppendorf twin.tec 96-well PCR plate 200ul](https://online-shop.eppendorf.us/US-en/Laboratory-Consumables-44512/Plates-44516/Eppendorf-twin.tec-PCR-Plates-PF-8180.html) seated in aluminum block\n* USA Scientific 12x8-tube strips 300ul seated in aluminum block\n* [1.5ml Eppendorf safelock snapcap microcentrifuge tube](https://online-shop.eppendorf.us/US-en/Laboratory-Consumables-44512/Tubes-44515/Eppendorf-Safe-Lock-Tubes-PF-8863.html) seated in [Opentrons 4-in-1 tuberack with 4x6 microcentrifuge tube insert](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [Opentrons GEN1 P10-single channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons GEN1 P300-single channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 10ul filter tip rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-filter-tip)\n* [Opentrons 200ul filter tip rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n\n---\n\n\nseated 8-tube strips (slot 2)\n* strip 1: 8-tube strip for initial transfers from PCR plate\n\n4x6 insert on Opentrons tuberack (slot 3)\n* tube A1: tube for pooling\n\n",
"internal": "139815\n",
diff --git a/protoBuilds/13ac78/README.json b/protoBuilds/13ac78/README.json
index a021506d81..50c7e57924 100644
--- a/protoBuilds/13ac78/README.json
+++ b/protoBuilds/13ac78/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "13ac78",
"labware": "\nNEST PCR 8-Strip Tubes 0.2ml #403022 seated in SSbio IsoFreeze PCR Rack #5650-T4\nThermoFisher MicroAmp\u2122 Optical 384-Well Reaction Plate #4343370\nOpentrons 20uL Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * PCR Prep\n\n",
"deck-setup": "\n\n\nReagents:\n* green: mastermix\n* blue: samples\n* pink: positive control\n* purple: molecular grade water\n\n---\n\n",
"description": "\nThis protocol performs a custom PCR Prep from strip tubes seated in a custom cooling rack into a 384-well plate for up to 16 samples. The transfer scheme is follows:\n\n\n\n---\n\n",
diff --git a/protoBuilds/13c0b8/README.json b/protoBuilds/13c0b8/README.json
index 5d0b0e3457..72c44779af 100644
--- a/protoBuilds/13c0b8/README.json
+++ b/protoBuilds/13c0b8/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Staining Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs the cell staining protocol as outlined in the 'Cell staining automation' document. This protocol uses a P20 Single (attached to the left mount) and a P300 (attached to the right mount).\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Temperature Module\nOpentrons P20 Single-Channel Pipette\nOpentrons P300 Multi-Channel Pipette\nOpentrons 20\u00b5L Tips\nOpentrons 300\u00b5L Tips\n96-Well BRANDplates with V-Bottom\nNEST 12-Well Reservoir\nUSA Scientific 12-Well Reservoir, 22mL\nReagents\nSamples\n\n\n\nSlot 1: Opentrons Temperature Module with 96-Well Plate\nSlot 2: 96-Well Plate with Reagents (in lieu of Tube Rack)\nThese should be loaded in the same well number as tube rack\n A1\n A2\n A3\n A4\n A5\n A6\n B1\n B2\n B3\n B4\n C1\n C2\n C3\n C4\n C5\n C6\n D1\n D2\n D3\n D4\nSlot 3: NEST 12-Well Reservoir\n A1: FACS Buffer\n A3: 'B6' Reagent (for step 5)\n* A12: Empty (for liquid waste)\nSlot 4: Opentrons 20\u00b5L Tips\nSlot 5: Opentrons 20\u00b5L Tips\nSlot 6: Opentrons 300\u00b5L Tips\nSlot 7: Opentrons 20\u00b5L Tips",
"internal": "13c0b8",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Staining Prep\n\n\n",
"description": "This protocol performs the cell staining protocol as outlined in the 'Cell staining automation' document. This protocol uses a P20 Single (attached to the left mount) and a P300 (attached to the right mount).\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* 96-Well BRANDplates with V-Bottom\n* [NEST 12-Well Reservoir](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* [USA Scientific 12-Well Reservoir, 22mL](https://labware.opentrons.com/usascientific_12_reservoir_22ml/)\n* Reagents\n* Samples\n\n\n---\n\n\nSlot 1: [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with 96-Well Plate\n\nSlot 2: 96-Well Plate with Reagents (in lieu of Tube Rack)\n*These should be loaded in the same well number as tube rack*\n* A1\n* A2\n* A3\n* A4\n* A5\n* A6\n* B1\n* B2\n* B3\n* B4\n* C1\n* C2\n* C3\n* C4\n* C5\n* C6\n* D1\n* D2\n* D3\n* D4\n\n\nSlot 3: [NEST 12-Well Reservoir](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* A1: FACS Buffer\n* A3: 'B6' Reagent (for step 5)\n* A12: Empty (for liquid waste)\n\nSlot 4: [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\nSlot 5: [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\nSlot 6: [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 7: [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n\n\n",
"internal": "13c0b8\n",
diff --git a/protoBuilds/13ea1e-part2/README.json b/protoBuilds/13ea1e-part2/README.json
index e1f4d63421..22f2682b82 100644
--- a/protoBuilds/13ea1e-part2/README.json
+++ b/protoBuilds/13ea1e-part2/README.json
@@ -17,7 +17,7 @@
"internal": "13ea1e-part2\n",
"labware": "* Kingfisher 96 Well Plate\n* 384 Well Plate\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 4-in-1 tube rack with 1.5mL tubes](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Partner Name](partner website link)\n\n",
+ "partner": "[Partner Name](partner website link)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P20 GEN2 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P300 GEN2 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n\n\n---\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "\n1. Mastermix is distributed to the 384 well plate depending on which wells will eventually have sample.\n2. Sample is added to these wells and mixed.\n3. Positive control is added to the last well in the plate.\n\n",
@@ -25,7 +25,7 @@
"title": "PCR Plate Prep with 384 Well Plate"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Partner Name",
+ "partner": "Partner Name\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP20 GEN2 Single Channel Pipette\nP300 GEN2 Multi-Channel Pipette\n\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nMastermix is distributed to the 384 well plate depending on which wells will eventually have sample.\nSample is added to these wells and mixed.\nPositive control is added to the last well in the plate.\n",
diff --git a/protoBuilds/13ea1e/README.json b/protoBuilds/13ea1e/README.json
index 022def7780..8f2f777bff 100644
--- a/protoBuilds/13ea1e/README.json
+++ b/protoBuilds/13ea1e/README.json
@@ -17,7 +17,7 @@
"internal": "13ea1e\n",
"labware": "* Kingfisher 96 Well Plate\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 4-in-1 tube rack with 1.5mL tubes](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Partner Name](partner website link)\n\n",
+ "partner": "[Partner Name](partner website link)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P1000 GEN2 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P300 GEN2 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n\n\n---\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Controls are added to sample plate\n2. Proteinase K is added to samples\n3. Magnetic beads are added to samples\n4. Incubate 15 minutes\n5. Ethanol block is prepped.\n6. NPW3 block is prepped.\n7. NPW4 block is prepped.\n10. Elution buffer block is prepped.\n\n",
@@ -25,7 +25,7 @@
"title": "Extraction Prep with Kingfisher Flex Extractor"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Partner Name",
+ "partner": "Partner Name\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP1000 GEN2 Single Channel Pipette\nP300 GEN2 Multi-Channel Pipette\n\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nControls are added to sample plate\nProteinase K is added to samples\nMagnetic beads are added to samples\nIncubate 15 minutes\nEthanol block is prepped.\nNPW3 block is prepped.\nNPW4 block is prepped.\nElution buffer block is prepped.\n",
diff --git a/protoBuilds/13fd88/README.json b/protoBuilds/13fd88/README.json
index 4b24819a47..53413afe3d 100644
--- a/protoBuilds/13fd88/README.json
+++ b/protoBuilds/13fd88/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Purification"
@@ -8,7 +8,7 @@
"description": "This protocol automates the purification of samples, using magnetic NI resin, ThermoFisher B-PER reagent, and homemade buffers.\n\nThis protocol requires the user to perform some off-deck steps (centrifuge) and replace tips midway. Both actions will be prompted by flashing lights a message in the Opentrons app.\n\nThis protocol uses custom labware definitions for the NEST deep well plate and the Chrom Tech 96-well filter plate. When downloading the protocol, the labware definitions (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: Using labware in your protocols\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P300 Multi-Channel Pipette\n(6) Opentrons 300\u00b5L Tips\nOpentrons Magnetic Module\n(4) NEST 96-Well Plates\nNEST 12-Well Reservoir\nNEST Deep Well Plate\nChrom Tech 96-Well Filter Plate\nSamples\nReagents\n\n\n\nPart 1: B-PER Addition, Incubation, and Transfer\nSlot 10: Opentrons 300\u00b5L Tips\nSlot 11: Opentrons 300\u00b5L Tips\nSlot 8: Opentrons 300\u00b5L Tips\nSlot 9: Opentrons 300\u00b5L Tips\nSlot 7: Opentrons Magnetic Module with NEST 96-Well Plates\nSlot 4: NEST 12-Well Reservoir\n A1: B-PER Reagent\n A3: Magnetic NI Resin\n A5: Wash Buffer (first wash)\n A7: Wash Buffer (second wash)\n* A9: Elution Buffer\n\nSlot 5: NEST 96-Well Plates\nSlot 1: NEST Deep Well Plate with Samples\n\nThis setup will be sufficient for the protocol up through the addition of the second wash buffer. After the second addition of the wash buffer, the OT-2 will pause and prompt the user to replace the tips in slots 8 and 9, and ensure that the remaining plates are in place (they can be loaded earlier if wanted)\n\nPart 2: 2nd Wash and Elution (in addition to previous labware...)\nSlot 6: NEST 96-Well Plates\nSlot 2: NEST 96-Well Plates\nSlot 3: Chrom Tech 96-Well Filter Plate\n\n",
"internal": "13fd88",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Purification\n\n\n",
"description": "This protocol automates the purification of samples, using magnetic NI resin, ThermoFisher B-PER reagent, and homemade buffers.\n\nThis protocol requires the user to perform some off-deck steps (centrifuge) and replace tips midway. Both actions will be prompted by flashing lights a message in the Opentrons app.\n\nThis protocol uses custom labware definitions for the NEST deep well plate and the Chrom Tech 96-well filter plate. When downloading the protocol, the labware definitions (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: [Using labware in your protocols](https://support.opentrons.com/en/articles/3136506-using-labware-in-your-protocols)\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* (6) [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* (4) [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* NEST Deep Well Plate\n* Chrom Tech 96-Well Filter Plate\n* Samples\n* Reagents\n\n\n\n---\n\n\n**Part 1: B-PER Addition, Incubation, and Transfer**\nSlot 10: [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 11: [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 8: [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 9: [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 7: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n\nSlot 4: [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* A1: B-PER Reagent\n* A3: Magnetic NI Resin\n* A5: Wash Buffer (first wash)\n* A7: Wash Buffer (second wash)\n* A9: Elution Buffer\n\n\nSlot 5: [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n\nSlot 1: NEST Deep Well Plate with Samples\n\n*This setup will be sufficient for the protocol up through the addition of the second wash buffer. After the second addition of the wash buffer, the OT-2 will pause and prompt the user to replace the tips in slots 8 and 9, and ensure that the remaining plates are in place (they can be loaded earlier if wanted)*\n\n**Part 2: 2nd Wash and Elution (in addition to previous labware...)**\nSlot 6: [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n\nSlot 2: [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n\nSlot 3: Chrom Tech 96-Well Filter Plate\n\n\n\n\n",
"internal": "13fd88\n",
diff --git a/protoBuilds/141a1a/README.json b/protoBuilds/141a1a/README.json
index 0197695b12..2aedaa1f87 100644
--- a/protoBuilds/141a1a/README.json
+++ b/protoBuilds/141a1a/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cell-Free Gene Expression"
@@ -8,7 +8,7 @@
"description": "This protocol automates the cell-free gene expression test as outlined by users at TU Munich. See below:\n\nThis protocol sets up a cell-free gene expression experiment (TXTL test) in a 384-well plate. A TXTL sample is composed of cell extract (E), buffer (B), and a plasmid coding for the desired genes. The cell extract and buffer volume in each sample is fixed, the volume of plasmid in each sample depends on the plasmid stock concentration and the desired sample concentration of the plasmid. The final volume of 15\u00b5l is reached by adding nuclease-free water to the sample.\n\nWith the parameters, the user is able to test separate samples for different plasmids coding for the fluorescent proteins (ex. RFP, YFP, GFP and CFP) and measure up to 3 replicates for each of them. In addition, up to two blank samples containing E+B and water are made. First, mastermixes of plasmid and water are made, then distributed to the corresponding wells in the 384-well plate. Then E+B mixes are prepared and also distributed to the corresponding wells.\n\nThis protocol uses a custom labware definition for the Brand 384-well plate. When downloading the protocol, the labware definition (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: Using labware in your protocols\n\nIn the case that the protocol needs more tips than can be accommodated, the robot will pause, flash the rail lights, and ask the user to replace the tiprack.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons Thermocycler Module\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P300 (GEN2) Single-Channel Pipette\nOpentrons P20 (GEN2) Single-Channel Pipette\nOpentrons 300\u00b5L Tips\nOpentrons 20\u00b5L Tips\n(2) Opentrons 4-in-1 Tube Rack Set, with 24-well top\n1.5mL Centrifuge Tubes\n2mL Centrifuge Tubes\n96-Well Aluminum Block\nPCR Strips\nBrand 384-Well Plate\nReagents\n\n\n\nSlot 1: Brand 384-Well Plate\nSlot 2: Opentrons 4-in-1 Tube Rack Set, with 24-well top with 1.5mL Centrifuge Tubes (for Cell Extracts + Buffer)\nSlot 3: 96-Well Aluminum Block with PCR Strips (containing Buffer aliquots)\nSlot 5: Opentrons 4-in-1 Tube Rack Set, with 24-well top with 2mL Centrifuge Tubes (for Plasmid stock and Nuclease-Free Water)\n A1: Plasmid 1 Stock (RFP)\n B1: Plasmid 2 Stock (YFP)\n C1: Plasmid 3 Stock (GFP)\n D1: Plasmid 4 Stock (CFP)\n A3: (Empty) Plasmid 1 Master Mix\n B3: (Empty) Plasmid 2 Master Mix\n C3: (Empty) Plasmid 3 Master Mix\n D3: (Empty) Plasmid 4 Master Mix\n A5: Nuclease-Free Water\n D5: (Empty) Pooled Buffer\nSlot 6: Opentrons 20\u00b5L Tips\nSlot 9: Opentrons 300\u00b5L Tips\nSlot 7/8/10/11: Opentrons Thermocycler Module\n\n\nUsing the customizations field (below), set up your protocol.\n Plasmid Mix CSV: Upload a CSV containing headers (plasmid, molecular weight, concentration, and sample concentration).\n Number of Replicates (1-3): The number of replicates per plasmid.\n Number of Cell Extracts (1-24): The number of cell extracts (each extract will be dispensed in a column (1-24)).\n Number of Blanks per Cell Extract (1, 2): The number of blanks (no plasmid) per cell extract (either 1 or 2).\n P20 Mount: Select which mount (left or right) the P20 Single is attached to.\n P300 Mount: Select which mount (left or right) the P300 Single is attached to.",
"internal": "141a1a",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cell-Free Gene Expression\n\n\n",
"description": "This protocol automates the cell-free gene expression test as outlined by users at TU Munich. See below:\n\n*This protocol sets up a cell-free gene expression experiment (TXTL test) in a 384-well plate. A TXTL sample is composed of cell extract (E), buffer (B), and a plasmid coding for the desired genes. The cell extract and buffer volume in each sample is fixed, the volume of plasmid in each sample depends on the plasmid stock concentration and the desired sample concentration of the plasmid. The final volume of 15\u00b5l is reached by adding nuclease-free water to the sample.*\n\nWith the parameters, the user is able to test separate samples for different plasmids coding for the fluorescent proteins (ex. RFP, YFP, GFP and CFP) and measure up to 3 replicates for each of them. In addition, up to two blank samples containing E+B and water are made. First, mastermixes of plasmid and water are made, then distributed to the corresponding wells in the 384-well plate. Then E+B mixes are prepared and also distributed to the corresponding wells.\n\nThis protocol uses a custom labware definition for the Brand 384-well plate. When downloading the protocol, the labware definition (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: [Using labware in your protocols](https://support.opentrons.com/en/articles/3136506-using-labware-in-your-protocols)\n\nIn the case that the protocol needs more tips than can be accommodated, the robot will pause, flash the rail lights, and ask the user to replace the tiprack.\n\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 (GEN2) Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons P20 (GEN2) Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* (2) [Opentrons 4-in-1 Tube Rack Set, with 24-well top](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [1.5mL Centrifuge Tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes)\n* [2mL Centrifuge Tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-2-0-ml-microcentrifuge-tubes)\n* [96-Well Aluminum Block](https://shop.opentrons.com/collections/verified-labware/products/aluminum-block-set)\n* PCR Strips\n* Brand 384-Well Plate\n* Reagents\n\n\n\n---\n\n\nSlot 1: Brand 384-Well Plate\n\nSlot 2: [Opentrons 4-in-1 Tube Rack Set, with 24-well top](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with [1.5mL Centrifuge Tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes) (for Cell Extracts + Buffer)\n\nSlot 3: [96-Well Aluminum Block](https://shop.opentrons.com/collections/verified-labware/products/aluminum-block-set) with PCR Strips (containing Buffer aliquots)\n\nSlot 5: [Opentrons 4-in-1 Tube Rack Set, with 24-well top](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with [2mL Centrifuge Tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-2-0-ml-microcentrifuge-tubes) (for Plasmid stock and Nuclease-Free Water)\n* A1: Plasmid 1 Stock (RFP)\n* B1: Plasmid 2 Stock (YFP)\n* C1: Plasmid 3 Stock (GFP)\n* D1: Plasmid 4 Stock (CFP)\n* A3: (Empty) Plasmid 1 Master Mix\n* B3: (Empty) Plasmid 2 Master Mix\n* C3: (Empty) Plasmid 3 Master Mix\n* D3: (Empty) Plasmid 4 Master Mix\n* A5: Nuclease-Free Water\n* D5: (Empty) Pooled Buffer\n\nSlot 6: [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\nSlot 9: [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 7/8/10/11: [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module)\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Plasmid Mix CSV**: Upload a CSV containing headers (plasmid, molecular weight, concentration, and sample concentration).\n* **Number of Replicates (1-3)**: The number of replicates per plasmid.\n* **Number of Cell Extracts (1-24)**: The number of cell extracts (each extract will be dispensed in a column (1-24)).\n* **Number of Blanks per Cell Extract (1, 2)**: The number of blanks (no plasmid) per cell extract (either 1 or 2).\n* **P20 Mount**: Select which mount (left or right) the P20 Single is attached to.\n* **P300 Mount**: Select which mount (left or right) the P300 Single is attached to.\n\n\n\n",
"internal": "141a1a\n",
diff --git a/protoBuilds/1440ad/README.json b/protoBuilds/1440ad/README.json
index 34dff1a606..e42dea6d07 100644
--- a/protoBuilds/1440ad/README.json
+++ b/protoBuilds/1440ad/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"RNA Extraction"
@@ -10,7 +10,7 @@
"internal": "1440ad",
"labware": "\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\n4-in-1 Tube Rack Set\nOpentrons 300uL Tips\nOpentrons 20\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* RNA Extraction\n\n",
"deck-setup": "\n\n",
"description": "This protocol extracts RNA using two temperature modules and a magnetic module. The protocol can be considered in 5 main parts:\n\n* Reagent added to sample\n* Sample moved to magnetic plate\n* Mag Beads added to sample\n* Two ethanol washes with magnet engagement\n* Water added to samples\n* Samples added to final tube rack\n\n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify the number of samples for this run.\n* `Number of Mag Bead Tubes`: Specify the number of tubes to store the magnetic beads in column 6 of the room temperature reagent rack. Note: mag bead volume should be split equally between the number of tubes specified. \n* `Mag Bead Resuspend Volume`: Specify the volume to mix the beads for resuspension before they are added to a sample.\n* `Mag Bead Resuspend Mix Repetions`: Specify the number of repetitions to mix `Mag Bead Resuspend Volume` before the mag beads are added to the sample.\n* `RR1-TR10 Volume`: Specify the volume of each respective reagent to put into each sample.\n* `P20 Single Channel Mount`: Specify which mount (left or right) to place the P20 single channel pipette.\n* `P300 Single Channel Mount`: Specify which mount (left or right) to place the P20 single channel pipette.\n\n\n---\n\n",
diff --git a/protoBuilds/1464-2/README.json b/protoBuilds/1464-2/README.json
index 40dcc6f20c..50ae77f45c 100644
--- a/protoBuilds/1464-2/README.json
+++ b/protoBuilds/1464-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"ELISA"
@@ -8,7 +8,7 @@
"description": "Links:\n Part 1: Dilution\n Part 2-1: Antibody Addition\n* Part 2-2: Substrate and Stop Solution Addition\nThis is the second part of a ELISA protocol: Antibody Addition. The robot will first add Enzyme Conjugate Reagent to all of the columns required in the output plate in slot 9. Water, diluent, and HCP standards, and diluted samples will be transferred to the output plate in duplicate. User will need to upload the same CSV file used in part 1 of the protocol. See Additional Notes in Part 1 for more details on the CSV requirements.\nDownload full protocol details here.\n\nYou will need:\n P300 Multi-channel Pipette\n P300 Single-channel Pipette\n Opentrons Tube Racks (2 mL Eppendorf)\n Opentrons Tube Rack (15 + 50 mL)\n 2 mL 96-deep well Plate\n 96-well Plate\n 12-row Trough\n 300 uL Tip Racks",
"internal": "AwOUI09G\n1464",
"markdown": {
- "author": "[Opentrons](http://www.opentrons.com/)\n\n",
+ "author": "[Opentrons](http://www.opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n * ELISA\n\n",
"description": "Links:\n* [Part 1: Dilution](./1464-natrix-separations-inc-milliporesigma-part1)\n* [Part 2-1: Antibody Addition](./1464-natrix-separations-inc-milliporesigma-part2)\n* [Part 2-2: Substrate and Stop Solution Addition](1464-natrix-separations-inc-milliporesigma-part3)\n\nThis is the second part of a ELISA protocol: Antibody Addition. The robot will first add Enzyme Conjugate Reagent to all of the columns required in the output plate in slot 9. Water, diluent, and HCP standards, and diluted samples will be transferred to the output plate in duplicate. User will need to upload the same CSV file used in part 1 of the protocol. See Additional Notes in Part 1 for more details on the CSV requirements.\n\nDownload full protocol details [here](https://s3.amazonaws.com/opentrons-protocol-library-website/custom-README-images/1464-natrix-separations-inc-milliporesigma/NEW_ELISA_protocol.xlsx).\n\n---\n\nYou will need:\n* P300 Multi-channel Pipette\n* P300 Single-channel Pipette\n* [Opentrons Tube Racks (2 mL Eppendorf)](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* [Opentrons Tube Rack (15 + 50 mL)](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* [2 mL 96-deep well Plate](https://www.usascientific.com/2ml-deep96-well-plateone-bulk.aspx)\n* 96-well Plate\n* 12-row Trough\n* 300 uL Tip Racks\n\n",
"internal": "AwOUI09G\n1464\n",
diff --git a/protoBuilds/1464-3/README.json b/protoBuilds/1464-3/README.json
index 771a321a06..17f4ea92de 100644
--- a/protoBuilds/1464-3/README.json
+++ b/protoBuilds/1464-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"ELISA"
@@ -8,7 +8,7 @@
"description": "Links:\n Part 1: Dilution\n Part 2-1: Antibody Addition\n* Part 2-2: Substrate and Stop Solution Addtion\nThis is the third part of a ELISA protocol: Substrate and Stop Solution Addition. This should be run immediately after the incubation and wash steps.\nDownload full protocol details here.\n\nYou will need:\n P300 Multi-channel Pipette\n P300 Single-channel Pipette\n Opentrons Tube Racks (2 mL Eppendorf)\n Opentrons Tube Rack (15 + 50 mL)\n 96-well Plate\n 12-row Trough\n* 300 uL Tip Racks",
"internal": "AwOUI09G\n1464",
"markdown": {
- "author": "[Opentrons](http://www.opentrons.com/)\n\n",
+ "author": "[Opentrons](http://www.opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n * ELISA\n\n",
"description": "Links:\n* [Part 1: Dilution](./1464-natrix-separations-inc-milliporesigma-part1)\n* [Part 2-1: Antibody Addition](./1464-natrix-separations-inc-milliporesigma-part2)\n* [Part 2-2: Substrate and Stop Solution Addtion](1464-natrix-separations-inc-milliporesigma-part3)\n\nThis is the third part of a ELISA protocol: Substrate and Stop Solution Addition. This should be run immediately after the incubation and wash steps.\n\nDownload full protocol details [here](https://s3.amazonaws.com/opentrons-protocol-library-website/custom-README-images/1464-natrix-separations-inc-milliporesigma/NEW_ELISA_protocol.xlsx).\n\n---\n\nYou will need:\n* P300 Multi-channel Pipette\n* P300 Single-channel Pipette\n* [Opentrons Tube Racks (2 mL Eppendorf)](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* [Opentrons Tube Rack (15 + 50 mL)](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* 96-well Plate\n* 12-row Trough\n* 300 uL Tip Racks\n\n",
"internal": "AwOUI09G\n1464\n",
diff --git a/protoBuilds/1464/README.json b/protoBuilds/1464/README.json
index c08e9ee17f..0562026ba4 100644
--- a/protoBuilds/1464/README.json
+++ b/protoBuilds/1464/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"ELISA"
@@ -8,7 +8,7 @@
"description": "Links:\n Part 1: Dilution\n Part 2-1: Antibody Addition\n* Part 2-2: Substrate and Stop Solution Addition\nThis is the first part of a ELISA protocol: Dilution Procedure. The robot will perform serial dilutions samples located in the Opentrons 2 mL Eppendorf tube rack in the 1.2 mL tube plates. User will need to upload a CSV specifying the desired concentrations of each sample to be transferred to a separate 96-well plate in part 2 of the protocol. See more information on the CSV requirement in Additional Notes below.\nDownload full protocol details here.\n\nYou will need:\n P1000 Single-channel Pipette\n P300 Single-channel Pipette\n Opentrons Tube Racks (2 mL Eppendorf)\n Opentrons Tube Rack (15 + 50 mL)\n NEST 2 mL 96-deep well Plate\n 96-well Plate\n 12-row Trough\n 1000 uL Tip Rack\n* 300 uL Tip Rack",
"internal": "1464",
"markdown": {
- "author": "[Opentrons](http://www.opentrons.com/)\n\n",
+ "author": "[Opentrons](http://www.opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n * ELISA\n\n",
"description": "Links:\n* [Part 1: Dilution](./1464)\n* [Part 2-1: Antibody Addition](./1464-2)\n* [Part 2-2: Substrate and Stop Solution Addition](1464-3)\n\nThis is the first part of a ELISA protocol: Dilution Procedure. The robot will perform serial dilutions samples located in the Opentrons 2 mL Eppendorf tube rack in the 1.2 mL tube plates. User will need to upload a CSV specifying the desired concentrations of each sample to be transferred to a separate 96-well plate in part 2 of the protocol. See more information on the CSV requirement in Additional Notes below.\n\n\nDownload full protocol details [here](https://s3.amazonaws.com/opentrons-protocol-library-website/custom-README-images/1464-natrix-separations-inc-milliporesigma/NEW_ELISA_protocol.xlsx).\n\n---\n\nYou will need:\n* P1000 Single-channel Pipette\n* P300 Single-channel Pipette\n* [Opentrons Tube Racks (2 mL Eppendorf)](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* [Opentrons Tube Rack (15 + 50 mL)](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* NEST 2 mL 96-deep well Plate\n* 96-well Plate\n* 12-row Trough\n* 1000 uL Tip Rack\n* 300 uL Tip Rack\n\n",
"internal": "1464\n",
diff --git a/protoBuilds/1477ea/README.json b/protoBuilds/1477ea/README.json
index a0c0e2d692..17e490ca6d 100644
--- a/protoBuilds/1477ea/README.json
+++ b/protoBuilds/1477ea/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -9,7 +9,7 @@
"description": "This protocol cherrypicks from source plate wells specified in the first column of an input CSV file and transfers 2 ul of the well's contents to destination plate wells specified in the second column of the input CSV file.\nLinks:\n Custom Cherrypicking json Protocol\n Opentrons Protocol Designer No-Code Tool\n* Sample CSV File\nThis protocol is a python translation of the attached custom cherrypicking json protocol that can be downloaded from the link above (and then uploaded to the online Opentrons Protocol Designer tool). This python translation has an added feature: upload of an input CSV file to specify source and destination wells.",
"internal": "1477ea",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Cherrypicking\n\n",
"deck-setup": "\n\n* Opentrons p300 tips (Deck Slots 10, 11)\n* Opentrons p20 tips (Deck Slot 7)\n* 195 mL NEST 1 Well Reservoir (Deck Slot 4)\n* Source Plate (TPP 96 well tissue culture plate Deck Slot 8)\n* Destination Plate (NUNC WHITE 96F 236105 96 well plate Deck Slot 6)\n\n",
"description": "\nThis protocol cherrypicks from source plate wells specified in the first column of an input CSV file and transfers 2 ul of the well's contents to destination plate wells specified in the second column of the input CSV file.\n\nLinks:\n* [Custom Cherrypicking json Protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-06-09/cn23rrl/custom%20program.zip)\n* [Opentrons Protocol Designer No-Code Tool](https://designer.opentrons.com/)\n* [Sample CSV File](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-06-09/0y13r26/custom%20transfer.csv)\n\nThis protocol is a python translation of the attached custom cherrypicking json protocol that can be downloaded from the link above (and then uploaded to the online Opentrons Protocol Designer tool). This python translation has an added feature: upload of an input CSV file to specify source and destination wells.\n\n",
diff --git a/protoBuilds/1479/README.json b/protoBuilds/1479/README.json
index d6686b4036..5bd67e4d1d 100644
--- a/protoBuilds/1479/README.json
+++ b/protoBuilds/1479/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -8,7 +8,7 @@
"description": "With this protocol, your robot can normalize concentration of DNA samples from 2 mL Eppendorf tubes in PCR strips. Volumes of DNA and buffer for each normalization will be provided by the user as a CSV file. See Additional Notes for more details.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.20.0 or later)\nOpentrons P10 and P50 Single-Channel Pipette (GEN1) and corresponding Tips\n4-in-1 Tube Rack Set with 4x6 insert for 2ml Eppendorf tubes\nstandard PCR strips\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nCSV layout (keep the headers)\nSource tube rack,Source tube well,Destination well,Vol. of DNA (?gL),Vol. of diluent (?gL)\nSlot 3,A1,A1,7,8\nSlot 3,B1,B1,6.4,8.6\nSlot 3,C1,C1,9.2,5.8\nSlot 3,D1,D1,6,9",
"internal": "1479",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"description": "With this protocol, your robot can normalize concentration of DNA samples from 2 mL Eppendorf tubes in PCR strips. Volumes of DNA and buffer for each normalization will be provided by the user as a CSV file. See Additional Notes for more details.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.20.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P10 and P50 Single-Channel Pipette (GEN1)](https://shop.opentrons.com/collections/ot-2-pipettes) and corresponding [Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [4-in-1 Tube Rack Set with 4x6 insert for 2ml Eppendorf tubes](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* standard PCR strips\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n---\n\n\nCSV layout (keep the headers)\n\n```\nSource tube rack,Source tube well,Destination well,Vol. of DNA (?gL),Vol. of diluent (?gL)\nSlot 3,A1,A1,7,8\nSlot 3,B1,B1,6.4,8.6\nSlot 3,C1,C1,9.2,5.8\nSlot 3,D1,D1,6,9\n```\n\n",
"internal": "1479\n",
diff --git a/protoBuilds/14b685/README.json b/protoBuilds/14b685/README.json
index cab7fd1d83..6307764b68 100644
--- a/protoBuilds/14b685/README.json
+++ b/protoBuilds/14b685/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina 16S"
@@ -8,7 +8,7 @@
"description": "This protocol automates the PCR Clean-Up step of the Illumnia 16S kit with a slight modification to volumes.\n\nBeginning with a starting volume of sample (input by user), 20\u00b5L of AMPure XP beads is added to each well. After incubation on the Magnetic Module, the supernatant is removed and two ethanol wash steps occur (with 195\u00b5L added to each well). Finally, 30\u00b5L of 10 mM Tris pH 8.5 or molecular grade water is added to each well and after incubation, 25\u00b5L of supernatant is transferred to a clean PCR plate.\n\nThis protocol requires an Opentrons Magnetic Module and the GEN2 P300 and P20 8-Channel Pipettes, in addition to the Illumina 16S kit and labware (more information below).\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.21.0 or later)\nOpentrons Magnetic Module\nOpentrons P300 Multi-Channel Pipette (GEN2)\nOpentrons P20 Multi-Channel Pipette (GEN2)\nOpentrons 200\u00b5L Filter Tip Rack(s)\nOpentrons 20\u00b5L Filter Tip Rack\nNEST 1-Well Reservoir, 195mL\nNEST 12-Well Reservoir, 15mL\nNEST 96-Well PCR Plate\nSamples\nReagents (AMPure XP beads, 80% Ethanol, and 10 mM Tris pH 8.5 or molecular grade water)\n\n\n\nThis protocol utilizes a variable number of 200\u00b5L filter tips, depending on the number of samples input (below). Each column of samples requires six (6) columns of tips. The protocol will use 200\u00b5L filter tipracks from slots in the following order: 8, 9, 5, 6, 2, and 3. Given this, if running 8 samples, only the first 6 columns of tips in slot 8 would be used. Additionally, tips used for adding ethanol are used again to remove supernatant. Finally, tips used after the addition of beads and removal of first supernatant, are replaced in empty slots/tipracks for easier disposal and to prevent waste bin from getting too full.\n\nDeck Layout\n\nSlot 1: NEST 96-Well PCR Plate (clean and empty for elutes)\n\nSlot 2: Opentrons 200\u00b5L Filter Tip Rack (Rack 5)\n\nSlot 3: Opentrons 200\u00b5L Filter Tip Rack (Rack 6)\n\nSlot 4: Opentrons 20\u00b5L Filter Tip Rack\n\nSlot 5: Opentrons 200\u00b5L Filter Tip Rack (Rack 3)\n\nSlot 6: Opentrons 200\u00b5L Filter Tip Rack (Rack 4)\n\nSlot 7: NEST 12-Well Reservoir, 15mL (for reagents)\nA1: AMPure XP beads (20\u00b5L per sample)\nA2: 80% Ethanol, wash 1, samples 1-48 (195\u00b5L per sample)\nA3: 80% Ethanol, wash 1, samples 49-96 (195\u00b5L per sample)\nA4: 80% Ethanol, wash 2, samples 1-48 (195\u00b5L per sample)\nA5: 80% Ethanol, wash 2, samples 49-96 (195\u00b5L per sample)\nA6: 10 mM Tris pH 8.5 or molecular grade water (30\u00b5L per sample)\n\nSlot 8: Opentrons 200\u00b5L Filter Tip Rack (Rack 1)\n\nSlot 9: Opentrons 200\u00b5L Filter Tip Rack (Rack 2)\n\nSlot 10: Opentrons Magnetic Module with NEST 96-Well PCR Plate containing samples\n\nSlot 11: NEST 1-Well Reservoir, 195mL (for liquid waste)\n",
"internal": "14b685",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina 16S\n\n\n",
"description": "This protocol automates the PCR Clean-Up step of the [Illumnia 16S kit](https://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html) with a slight modification to volumes.\n\nBeginning with a starting volume of sample (input by user), 20\u00b5L of AMPure XP beads is added to each well. After incubation on the Magnetic Module, the supernatant is removed and two ethanol wash steps occur (with 195\u00b5L added to each well). Finally, 30\u00b5L of 10 mM Tris pH 8.5 or molecular grade water is added to each well and after incubation, 25\u00b5L of supernatant is transferred to a clean PCR plate.\n\nThis protocol requires an Opentrons [Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) and the [GEN2 P300 and P20 8-Channel Pipettes](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette), in addition to the Illumina 16S kit and labware (more information below).\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.21.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P300 Multi-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons P20 Multi-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 200\u00b5L Filter Tip Rack(s)](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons 20\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* Samples\n* Reagents (AMPure XP beads, 80% Ethanol, and 10 mM Tris pH 8.5 or molecular grade water)\n\n---\n\n\nThis protocol utilizes a variable number of 200\u00b5L filter tips, depending on the number of samples input (below). Each column of samples requires six (6) columns of tips. The protocol will use 200\u00b5L filter tipracks from slots in the following order: 8, 9, 5, 6, 2, and 3. Given this, if running 8 samples, only the first 6 columns of tips in slot 8 would be used. Additionally, tips used for adding ethanol are used again to remove supernatant. Finally, tips used after the addition of beads and removal of first supernatant, are replaced in empty slots/tipracks for easier disposal and to prevent waste bin from getting too full.\n\n**Deck Layout**\n\nSlot 1: [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) (clean and empty for elutes)\n\nSlot 2: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips) (Rack 5)\n\nSlot 3: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips) (Rack 6)\n\nSlot 4: [Opentrons 20\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 5: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips) (Rack 3)\n\nSlot 6: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips) (Rack 4)\n\nSlot 7: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) (for reagents)\nA1: AMPure XP beads (20\u00b5L per sample)\nA2: 80% Ethanol, wash 1, samples 1-48 (195\u00b5L per sample)\nA3: 80% Ethanol, wash 1, samples 49-96 (195\u00b5L per sample)\nA4: 80% Ethanol, wash 2, samples 1-48 (195\u00b5L per sample)\nA5: 80% Ethanol, wash 2, samples 49-96 (195\u00b5L per sample)\nA6: 10 mM Tris pH 8.5 or molecular grade water (30\u00b5L per sample)\n\nSlot 8: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips) (Rack 1)\n\nSlot 9: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips) (Rack 2)\n\nSlot 10: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) containing samples\n\nSlot 11: [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml) (for liquid waste)\n\n\n\n",
"internal": "14b685\n",
diff --git a/protoBuilds/154ec5/README.json b/protoBuilds/154ec5/README.json
index 88ee47cbc7..55c64b1e16 100644
--- a/protoBuilds/154ec5/README.json
+++ b/protoBuilds/154ec5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -9,7 +9,7 @@
"description": "This protocol uses a p20 single channel pipette to transfer custom volumes (2-10 uL) from source wells located in one of up to 10 pieces of 24-well, 96-well or 384-well labware on the OT-2 deck to a destination well in one of the ten pieces of loaded labware. The labware, deck slot locations, source plate, source well, destination plate, destination well and transfer volume are supplied by the user in a csv file uploaded at the time of protocol download (see example for file and data format).\nLinks:\n* example input csv\nThis protocol was developed to transfer custom volumes (2-10 uL) from source wells to destination wells as described above.",
"internal": "154ec5",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Cherrypicking\n\n",
"deck-setup": "\n\n* Opentrons p20 tips (Deck Slot 11)\n* 24, 96 and/or 384-Well Labware As Specified With Pulldown Lists and in Your Uploaded CSV File - see example for file and data format (Deck Slots 1-10)\n\n\n\n",
"description": "\nThis protocol uses a p20 single channel pipette to transfer custom volumes (2-10 uL) from source wells located in one of up to 10 pieces of 24-well, 96-well or 384-well labware on the OT-2 deck to a destination well in one of the ten pieces of loaded labware. The labware, deck slot locations, source plate, source well, destination plate, destination well and transfer volume are supplied by the user in a csv file uploaded at the time of protocol download (see example for file and data format).\n\nLinks:\n* [example input csv](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/154ec5/example%2BCSV.csv)\n\nThis protocol was developed to transfer custom volumes (2-10 uL) from source wells to destination wells as described above.\n\n",
diff --git a/protoBuilds/1577-v2/README.json b/protoBuilds/1577-v2/README.json
index 4337a3e19d..3eecb010db 100644
--- a/protoBuilds/1577-v2/README.json
+++ b/protoBuilds/1577-v2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs PCR preparation on a custom 3x8 membrane chip on 6 PCR strips. Ensure that the chip is seated so that the wells of the second (middle) row are slightly offset to the top of the first and third rows as shown in 'Additional Notes' below. Please calibrate the custom chip so that the pipette tip lands lands just barely above the membrane-- this will ensure accurate liquid transfer without disturbing the eggs or crashing into the chip. For reagent setup, see 'Additional Notes' below.\n\n\n\nP10 Multi-channel electronic pipette\n10\u00b5l Pipette tips\nP50 Single-channel electronic pipette\n300\u00b5l Pipette tips\nOpentrons 4-in-1 tube rack set\nTempassure 200ml PCR strips (seated)\n1.7ml Eppendorf tubes\nCustom 3x8 offset membrane chip on shaker (mounted on OT-2 deck)\n\n\n\n4x6 tuberack for 2ml Eppendorf tubes:\n tube A1: nanopure H2O\n tube B1: reaction buffer\n tube C1: primer\n tube D1: DNA polymerase\n* tube A2: mastermix tube (loaded empty)\nSeated PCR strips:\n* strip column 12: nanopure H2O (for additions with P10-multi channel pipette)",
"internal": "yYlOorFh\n1577",
"markdown": {
- "author": "[Opentrons](http://www.opentrons.com/)\n\n",
+ "author": "[Opentrons](http://www.opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol performs PCR preparation on a custom 3x8 membrane chip on 6 PCR strips. Ensure that the chip is seated so that the wells of the second (middle) row are slightly offset to the **top** of the first and third rows as shown in 'Additional Notes' below. **Please calibrate the custom chip so that the pipette tip lands lands just barely above the membrane**-- this will ensure accurate liquid transfer without disturbing the eggs or crashing into the chip. For reagent setup, see 'Additional Notes' below.\n\n---\n\n\n\n* [P10 Multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette?variant=5978967113757)\n* [10\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [P50 Single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette?variant=5984549077021)\n* [300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 4-in-1 tube rack set](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [Tempassure 200ml PCR strips (seated)](https://www.usascientific.com/0.2ml-flex-free-pcr-8-tube-attached-clear-flat-caps.aspx)\n* 1.7ml Eppendorf tubes\n* Custom 3x8 offset membrane chip on shaker (mounted on OT-2 deck)\n\n---\n\n\n4x6 tuberack for 2ml Eppendorf tubes:\n* tube A1: nanopure H2O\n* tube B1: reaction buffer\n* tube C1: primer\n* tube D1: DNA polymerase\n* tube A2: mastermix tube (loaded empty)\n\nSeated PCR strips:\n* strip column 12: nanopure H2O (for additions with P10-multi channel pipette)\n\n",
"internal": "yYlOorFh\n1577\n",
diff --git a/protoBuilds/158d42/README.json b/protoBuilds/158d42/README.json
index fb9e997d69..d2bf3f7209 100644
--- a/protoBuilds/158d42/README.json
+++ b/protoBuilds/158d42/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "1584d2",
"labware": "\nOpentrons 4-in-1 24 tube tube rack with 1.5mL tubes\nNEST 100ul PCR Plate, Full Skirt\nOpentrons 200ul Filter tips\nCustom 32 tube tube racklink to labware on shop.opentrons.com when applicable\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "* 24 tube racks should be placed in order of slots 9, 6. Tubes by row.\n* 32 tube racks should be placed in order of slots 7 and 4. Tubes by row.\n\n\n---\n\n",
"description": "\nThis protocol preps a 96 well PCR plate with up to 46 samples. The order of operations is: transfer negative control to A1 of the well plate, transfer samples down plate columns starting from C1, transfer positive control to A12 of well plate, then B1. Resuspension steps are included after most transfers. For wells with powdered reagent, roughly half of the necessary volume is dispensed into the well from half of the well depth, then the pipette tip proceeds to travel to the bottom of the well to dispense the rest of the required volume and mix. Sample in buffer is transferred from the 32-tube rack to the 24 tube rack which contains sterile normalization buffer, and the resulting mixture is then added to the plate.\n\nExplanation of complex parameters below:\n* `Number of tubes (1-46)`: Specify the number of tubes placed in the custom 32-tube tube racks. Tubes should be placed by row, and up down (from slot 7 to slot 4).\n* `Use Temperature Module`: Specify whether or not to use the temperature module at 12C this run. If not, you can place the plate directly on slot 1.\n* `P300 Mount`: Specify which mount (left or right) to host the P300 single-channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/15dc4c/README.json b/protoBuilds/15dc4c/README.json
index cfa6b3a7dd..5067622949 100644
--- a/protoBuilds/15dc4c/README.json
+++ b/protoBuilds/15dc4c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"ELISA"
@@ -10,7 +10,7 @@
"internal": "15dc4c",
"labware": "\nNEST 96 Deepwell Plate 2mL #503001\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* ELISA\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs the [SMC IL-17A High Sensitivity Immunoassay Kit](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/15dc4c/SMC+IL-17A+Protocol+kit+03-0159-00.pdf) protocol. The user is prompted to pause for washes and incubations on other devices.\n\n\n",
diff --git a/protoBuilds/1601a9/README.json b/protoBuilds/1601a9/README.json
index d7f5077989..67526884d2 100644
--- a/protoBuilds/1601a9/README.json
+++ b/protoBuilds/1601a9/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Illumina Beachip Amplification"
@@ -8,7 +8,7 @@
"description": "This protocol automates sample distribution and the addition of MA1 and 0.1 N NaOH.\nUsing the customizations fields, below set up your protocol.\n* P10 Multi Mount: Select which mount (left or right) the P10 Multi is attached to.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons P10 Multi-Channel Pipette\nOpentrons 10\u00b5L Tips\nNEST 12-Well Reservoir, 15mL\nAxygen 96-Well Plate (PCR-96-FS-C)\nAbgene 96-Deep Well Plate (AB0765)\nReagents\nSamples\n\n\n\nSlot 1: NEST 12-Well Reservoir\n A5: MA1\n A9: 0.1 N NaOH\nSlot 2: Abgene 96-Deep Well Plate (AB0765)\nSlot 3: Axygen 96-Well Plate (PCR-96-FS-C)\nSlot 4: Opentrons 10\u00b5L Tips\nSlot 5: Opentrons 10\u00b5L Tips",
"internal": "1601a9",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Illumina Beachip Amplification\n\n\n",
"description": "This protocol automates sample distribution and the addition of [MA1](https://www.illumina.com/products/by-type/microarray-kits/infinium-global-screening.html) and [0.1 N NaOH](https://www.sigmaaldrich.com/catalog/product/sigma/s2770?lang=en\u00aeion=US).\n\n\n**Using the customizations fields, below set up your protocol.**\n* **P10 Multi Mount**: Select which mount (left or right) the P10 Multi is attached to.\n\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P10 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [NEST 12-Well Reservoir, 15mL](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* Axygen 96-Well Plate (PCR-96-FS-C)\n* Abgene 96-Deep Well Plate (AB0765)\n* Reagents\n* Samples\n\n\n---\n\n\nSlot 1: [NEST 12-Well Reservoir](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* A5: [MA1](https://www.illumina.com/products/by-type/microarray-kits/infinium-global-screening.html)\n* A9: [0.1 N NaOH](https://www.sigmaaldrich.com/catalog/product/sigma/s2770?lang=en\u00aeion=US)\n\nSlot 2: Abgene 96-Deep Well Plate (AB0765)\n\nSlot 3: Axygen 96-Well Plate (PCR-96-FS-C)\n\nSlot 4: [Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\nSlot 5: [Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n\n\n",
"internal": "1601a9\n",
diff --git a/protoBuilds/1649b1/README.json b/protoBuilds/1649b1/README.json
index db9c81cf4b..b36331acc9 100644
--- a/protoBuilds/1649b1/README.json
+++ b/protoBuilds/1649b1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Extraction"
@@ -8,7 +8,7 @@
"description": "This protocol automates a nucleic acid purification for the LGC Sbeadex Plant Maxi Kit using the Opentrons Magnetic Module and Opentrons Temperature Module\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons Magnetic Module\nOpentrons P300 Multi-Channel Pipette, GEN2\nOpentrons Tips\nNEST 1-Well Reservoir, 195mL\nReagents (magnetic beads, ethanol, elution buffer)\n\n\n\n\nSlot 1: Magnetic Module + NEST 96 Deep Well Plate\nSlot 2: SPEX 96 Well Plate\nSlot 3: Temperature Module + Elution Plate\nSlot 4: Lysis Buffer\nSlot 5: Binding Buffer\nSlot 6: Elution Buffer\nSlot 7: Wash 1 Buffer\nSlot 8: Wash 2 Buffer\nSlots 9-11: Opentrons 300 uL Tips\n",
"internal": "1649b1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Nucleic Acid Extraction\n\n\n",
"description": "This protocol automates a nucleic acid purification for the LGC Sbeadex Plant Maxi Kit using the [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) and [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P300 Multi-Channel Pipette, GEN2](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* Reagents (magnetic beads, ethanol, elution buffer)\n\n---\n\n\n* Slot 1: Magnetic Module + NEST 96 Deep Well Plate\n* Slot 2: SPEX 96 Well Plate\n* Slot 3: Temperature Module + Elution Plate\n* Slot 4: Lysis Buffer\n* Slot 5: Binding Buffer\n* Slot 6: Elution Buffer\n* Slot 7: Wash 1 Buffer\n* Slot 8: Wash 2 Buffer\n* Slots 9-11: Opentrons 300 uL Tips\n\n\n",
"internal": "1649b1",
diff --git a/protoBuilds/165a77/README.json b/protoBuilds/165a77/README.json
index 6776d38bc4..059c89f73b 100644
--- a/protoBuilds/165a77/README.json
+++ b/protoBuilds/165a77/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"qPCR setup"
@@ -8,7 +8,7 @@
"description": "This protocol automates the distribution of patient samples to the wells of up to three 384-well plates in a specific arrangement shown in the attached plate map. The number of columns filled with patient sample for each plate can be specified (as a single comma-separated string of values between 1 and 12 like \"8,8,12\") in the parameters below.\nLinks:\n* PCR/qPCR prep: distribution of primers to 384-well plates\n\nplate map\n\n\n\n\nOpentrons P20-multi channel electronic pipettes (GEN2)\n\n\nOpentrons 20ul Filter Tips\n\n",
"internal": "165a77",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * qPCR setup\n\n",
"description": "This protocol automates the distribution of patient samples to the wells of up to three 384-well plates in a specific arrangement shown in the attached plate map. The number of columns filled with patient sample for each plate can be specified (as a single comma-separated string of values between 1 and 12 like \"8,8,12\") in the parameters below.\n\nLinks:\n* [PCR/qPCR prep: distribution of primers to 384-well plates](http://protocols.opentrons.com/protocol/559aa0)\n\n* [plate map](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-03-08/e373l2s/384%20plate%20map.png)\n\n\n\n* [Opentrons P20-multi channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n\n* [Opentrons 20ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n\n\n",
"internal": "165a77\n",
diff --git a/protoBuilds/16b3fb/README.json b/protoBuilds/16b3fb/README.json
index 518fd226d7..d4796f225e 100644
--- a/protoBuilds/16b3fb/README.json
+++ b/protoBuilds/16b3fb/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "16b3fb",
"labware": "\nAG Filtration 48 Tube Rack with Custom Bulb 0.15 mL\nNEST 1 Well Reservoir 195 mL #360103\nOpentrons 96 Tip Rack 1000 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a custom tube filling protoocl for up to 432 custom tubes per run.\n\n\n",
diff --git a/protoBuilds/17cb2d/README.json b/protoBuilds/17cb2d/README.json
index b4a6cc7538..89b14a198f 100644
--- a/protoBuilds/17cb2d/README.json
+++ b/protoBuilds/17cb2d/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol fills Applied Biosystems MicroAmp 384-well plates with mastermix. Update: this protocol can now accomodate the SSI 384-well plate as well.\n\nThe protocol can fill up to 9 plates and will dispense 7.5\u00b5L of mastermix 1 into all the odd rows (A/C/E...) and 7.5\u00b5L of mastermix 2 into all the even rows (B/D/F...).\n\nThis protocol has been updated to use the GEN2 Multi-Channel Pipettes - please use corresponding tips.\n\nUpdate 09-21-20: The protocol will now do a blow out in the well and aspirate 2\u00b5L of air at the top of the well after each dispense step to ensure all liquid has been expelled from pipette and minimize contamination risk due to droplet hang.\n\nUpdate 09-25-20: The protocol has been updated in two ways. First, the mastermix should be split between two wells (A1/A2 and A11/A12). Second, after aspirating the mastermix, the pipette will move to the sides of the well to remove liquid attached to the tips.\n\nUpdate 10-29-20: The protocol will now transfer 10\u00b5L of mastermix.\n\nUpdate 10-30-20: The protocol will now multi-dispense mastermix instead of 1-to-1 transfer. \n\nUpdate 11-12-20: The protocol now has a new parameter for master mix volume (default is 7.5\u00b5L)\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons Multi-Channel Pipette\nOpentrons Tips\nNEST 12-Well Reservoir\nApplied Biosystems MicroAmp 384-Well Plate or SSI 384-Well Plate\nReagents (mastermix 1, mastermix 2)\n\n\n\nSlots 1 - 9: Applied Biosystems MicroAmp 384-Well Plate or SSI 384-Well Plate\nSlot 10: Opentrons Tips\nSlot 11: NEST 12-Well Reservoir\nA1: Mastermix 1 (for plates 1-5)\nA2: Mastermix 1 (for plates 6-9)\nA11: Mastermix 2 (for plates 1-5)\nA12: Mastermix 2 (for plates 6-9)\n\n\nUsing the customizations field (below), set up your protocol.\n Pipette Type: Select which GEN2 Multi-Channel Pipette (p300 or p20) will be used\n Pipette Mount: Select which mount (left or right) the Multi-Channel Pipette is attached to\n Number of Plates (1-9): Specify the number of plates to fill\n Plate (384-Well) Type: Select which plate (Applied Biosystems or SSI) to use\n* Master Mix Volume (uL): Input the volume of master mix to be transferred to each well",
"internal": "17cb2d",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol fills [Applied Biosystems MicroAmp 384-well plates]([https://www.thermofisher.com/order/catalog/product/4343370#/4343370) with mastermix. **Update**: this protocol can now accomodate the SSI 384-well plate as well.\n\nThe protocol can fill up to 9 plates and will dispense 7.5\u00b5L of mastermix 1 into all the odd rows (A/C/E...) and 7.5\u00b5L of mastermix 2 into all the even rows (B/D/F...).\n\nThis protocol has been updated to use the GEN2 Multi-Channel Pipettes - please use corresponding tips.\n\n**Update 09-21-20:** The protocol will now do a blow out in the well and aspirate 2\u00b5L of air at the top of the well after each dispense step to ensure all liquid has been expelled from pipette and minimize contamination risk due to droplet hang.\n\n**Update 09-25-20:** The protocol has been updated in two ways. First, the mastermix should be split between two wells (A1/A2 and A11/A12). Second, after aspirating the mastermix, the pipette will move to the sides of the well to remove liquid attached to the tips.\n\n**Update 10-29-20:** The protocol will now transfer 10\u00b5L of mastermix.\n\n**Update 10-30-20:** The protocol will now multi-dispense mastermix instead of 1-to-1 transfer. \n\n**Update 11-12-20:** The protocol now has a new parameter for master mix volume (default is 7.5\u00b5L)\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [Applied Biosystems MicroAmp 384-Well Plate](https://www.thermofisher.com/order/catalog/product/4343370#/4343370) or SSI 384-Well Plate\n* Reagents (mastermix 1, mastermix 2)\n\n\n---\n\n\nSlots 1 - 9: [Applied Biosystems MicroAmp 384-Well Plate](https://www.thermofisher.com/order/catalog/product/4343370#/4343370) or SSI 384-Well Plate\n\nSlot 10: [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\nSlot 11: [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\nA1: Mastermix 1 (for plates 1-5)\nA2: Mastermix 1 (for plates 6-9)\nA11: Mastermix 2 (for plates 1-5)\nA12: Mastermix 2 (for plates 6-9)\n\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Pipette Type**: Select which GEN2 Multi-Channel Pipette (p300 or p20) will be used\n* **Pipette Mount**: Select which mount (left or right) the Multi-Channel Pipette is attached to\n* **Number of Plates (1-9)**: Specify the number of plates to fill\n* **Plate (384-Well) Type**: Select which plate (Applied Biosystems or SSI) to use\n* **Master Mix Volume (uL)**: Input the volume of master mix to be transferred to each well\n\n\n\n",
"internal": "17cb2d\n",
diff --git a/protoBuilds/17d210-part-2/README.json b/protoBuilds/17d210-part-2/README.json
index 762eb9df5c..99f2f8a06f 100644
--- a/protoBuilds/17d210-part-2/README.json
+++ b/protoBuilds/17d210-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Verogen ForenSeq DNA Signature Prep Kit"
@@ -10,7 +10,7 @@
"internal": "17d210",
"labware": "\nAbgene Midi 96 Well Plate 800 \u00b5L\nAmplifyt 96 Well Plate 200 \u00b5L\nOpentrons 20\u00b5l and 300\u00b5l Tipracks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Verogen ForenSeq DNA Signature Prep Kit\n\n",
"deck-setup": "* blue: PCR2 buffer \n* pink: samples \n* green: indexes \n\n\n---\n\n",
"description": "\nLinks: \n* [Part 1](./17d210)\n
\n* [Part 2](./17d210-part-2)\n
\n* [Part 3](./17d210-part-3)\n
\n* [Part 4](./17d210-part-4)\n
\n* [Part 5](./17d210-part-5)\n\nThis custom indexing protocol is part 2/5 of the [Verogen ForenSeq DNA Signature Prep kit](https://verogen.com/products/forenseq-dna-signature-prep-kit/?utm_term=&utm_campaign=Product+Campaigns&utm_source=adwords&utm_medium=ppc&hsa_acc=2964416997&hsa_cam=12070402317&hsa_grp=115534580817&hsa_ad=544522374879&hsa_src=g&hsa_tgt=dsa-19959388920&hsa_kw=&hsa_mt=b&hsa_net=adwords&hsa_ver=3&gclid=CjwKCAjw4qCKBhAVEiwAkTYsPP4JakJA06WcfvubM80x5gzv7kIFucad6jw9WrACitcG6qERBSAU1xoCaOEQAvD_BwE). In this protocol, indexes are transferred to their corresponding wells in a PCR plate containing samples. Then, PCR2 buffer is added to each sample and mixed using fresh tips for each transfer.\n\n---\n\n",
diff --git a/protoBuilds/17d210-part-3/README.json b/protoBuilds/17d210-part-3/README.json
index 52f30c0a7e..2ce3bb389b 100644
--- a/protoBuilds/17d210-part-3/README.json
+++ b/protoBuilds/17d210-part-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Verogen ForenSeq DNA Signature Prep Kit"
@@ -10,7 +10,7 @@
"internal": "17d210",
"labware": "\nAbgene Midi 96 Well Plate 800 \u00b5L\nAmplifyt 96 Well Plate 200 \u00b5L\nNEST 12 Well Reservoir 15 mL\nOpentrons 20\u00b5l and 300\u00b5l Tipracks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Verogen ForenSeq DNA Signature Prep Kit\n\n",
"deck-setup": "* green: samples\n* blue: binding bead buffer\n* pink: 80% EtOH \n* purple: elution buffer\n\n\n---\n\n",
"description": "\nLinks: \n* [Part 1](./17d210)\n
\n* [Part 2](./17d210-part-2)\n
\n* [Part 3](./17d210-part-3)\n
\n* [Part 4](./17d210-part-4)\n
\n* [Part 5](./17d210-part-5)\n\nThis custom Library Purification protocol is part 3/5 of the [Verogen ForenSeq DNA Signature Prep kit](https://verogen.com/products/forenseq-dna-signature-prep-kit/?utm_term=&utm_campaign=Product+Campaigns&utm_source=adwords&utm_medium=ppc&hsa_acc=2964416997&hsa_cam=12070402317&hsa_grp=115534580817&hsa_ad=544522374879&hsa_src=g&hsa_tgt=dsa-19959388920&hsa_kw=&hsa_mt=b&hsa_net=adwords&hsa_ver=3&gclid=CjwKCAjw4qCKBhAVEiwAkTYsPP4JakJA06WcfvubM80x5gzv7kIFucad6jw9WrACitcG6qERBSAU1xoCaOEQAvD_BwE). The protocol is broken down into 4 main parts:\n* binding buffer addition to samples\n* bead wash 2x using magnetic module\n* final elution to chilled PCR plate\n\nSamples should be loaded on the magnetic module in an Abgene Midi plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 11) to the elution PCR plate (temperature module, slot 3). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\nExplanation of complex parameters below:\n* `park tips`: If set to `yes` (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to `no`, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n\n---\n\n",
diff --git a/protoBuilds/17d210-part-4/README.json b/protoBuilds/17d210-part-4/README.json
index 26b3a1026d..f67496bb0a 100644
--- a/protoBuilds/17d210-part-4/README.json
+++ b/protoBuilds/17d210-part-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Verogen ForenSeq DNA Signature Prep Kit"
@@ -10,7 +10,7 @@
"internal": "17d210",
"labware": "\nAbgene Midi 96 Well Plate 800 \u00b5L\nAmplifyt 96 Well Plate 200 \u00b5L\nNEST 12 Well Reservoir 15 mL\nOpentrons 20\u00b5l and 300\u00b5l Tipracks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Verogen ForenSeq DNA Signature Prep Kit\n\n",
"deck-setup": "* green: samples\n* blue: binding bead buffer\n* pink: wash solution \n* purple: HP3 dilution\n* orange: LNS2\n\n\n---\n\n",
"description": "\nLinks: \n* [Part 1](./17d210)\n
\n* [Part 2](./17d210-part-2)\n
\n* [Part 3](./17d210-part-3)\n
\n* [Part 4](./17d210-part-4)\n
\n* [Part 5](./17d210-part-5)\n\nThis custom extraction protocol is part 4/5 of the [Verogen ForenSeq DNA Signature Prep kit](https://verogen.com/products/forenseq-dna-signature-prep-kit/?utm_term=&utm_campaign=Product+Campaigns&utm_source=adwords&utm_medium=ppc&hsa_acc=2964416997&hsa_cam=12070402317&hsa_grp=115534580817&hsa_ad=544522374879&hsa_src=g&hsa_tgt=dsa-19959388920&hsa_kw=&hsa_mt=b&hsa_net=adwords&hsa_ver=3&gclid=CjwKCAjw4qCKBhAVEiwAkTYsPP4JakJA06WcfvubM80x5gzv7kIFucad6jw9WrACitcG6qERBSAU1xoCaOEQAvD_BwE). The protocol is broken down into 4 main parts:\n* binding buffer addition to samples\n* bead wash 2x using magnetic module\n* final elution to chilled PCR plate\n\nSamples should be loaded on the magnetic module in an Abgene Midi plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 11) to the elution PCR plate (temperature module, slot 3). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\nExplanation of complex parameters below:\n* `park tips`: If set to `yes` (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to `no`, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n\n---\n\n",
diff --git a/protoBuilds/17d210-part-5/README.json b/protoBuilds/17d210-part-5/README.json
index a9aec1cdac..ce2038c86a 100644
--- a/protoBuilds/17d210-part-5/README.json
+++ b/protoBuilds/17d210-part-5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Verogen ForenSeq DNA Signature Prep Kit"
@@ -10,7 +10,7 @@
"internal": "17d210",
"labware": "\nAmplifyt 96 Well Plate 200 \u00b5L\nOpentrons 20\u00b5l Tiprack\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Verogen ForenSeq DNA Signature Prep Kit\n\n",
"deck-setup": "* blue: pooling strip \n* green: samples \n\n\n---\n\n",
"description": "\nLinks: \n* [Part 1](./17d210)\n
\n* [Part 2](./17d210-part-2)\n
\n* [Part 3](./17d210-part-3)\n
\n* [Part 4](./17d210-part-4)\n
\n* [Part 5](./17d210-part-5)\n\nThis custom pooling protocol is part 5/5 of the [Verogen ForenSeq DNA Signature Prep kit](https://verogen.com/products/forenseq-dna-signature-prep-kit/?utm_term=&utm_campaign=Product+Campaigns&utm_source=adwords&utm_medium=ppc&hsa_acc=2964416997&hsa_cam=12070402317&hsa_grp=115534580817&hsa_ad=544522374879&hsa_src=g&hsa_tgt=dsa-19959388920&hsa_kw=&hsa_mt=b&hsa_net=adwords&hsa_ver=3&gclid=CjwKCAjw4qCKBhAVEiwAkTYsPP4JakJA06WcfvubM80x5gzv7kIFucad6jw9WrACitcG6qERBSAU1xoCaOEQAvD_BwE). In this protocol, samples are pooled from a PCR plate to a final pooling strip.\n\n---\n\n",
diff --git a/protoBuilds/17d210/README.json b/protoBuilds/17d210/README.json
index 38e0a5e968..912704e0d3 100644
--- a/protoBuilds/17d210/README.json
+++ b/protoBuilds/17d210/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Verogen ForenSeq DNA Signature Prep Kit"
@@ -10,7 +10,7 @@
"internal": "17d210",
"labware": "\nAmplifyt 96 Well Plate 200 \u00b5L\nOpentrons 20\u00b5l Tipracks\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Verogen ForenSeq DNA Signature Prep Kit\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks: \n* [Part 1](./17d210)\n
\n* [Part 2](./17d210-part-2)\n
\n* [Part 3](./17d210-part-3)\n
\n* [Part 4](./17d210-part-4)\n
\n* [Part 5](./17d210-part-5)\n\nThis custom PCR1 Setup protocol is part 1/5 of the [Verogen ForenSeq DNA Signature Prep kit](https://verogen.com/products/forenseq-dna-signature-prep-kit/?utm_term=&utm_campaign=Product+Campaigns&utm_source=adwords&utm_medium=ppc&hsa_acc=2964416997&hsa_cam=12070402317&hsa_grp=115534580817&hsa_ad=544522374879&hsa_src=g&hsa_tgt=dsa-19959388920&hsa_kw=&hsa_mt=b&hsa_net=adwords&hsa_ver=3&gclid=CjwKCAjw4qCKBhAVEiwAkTYsPP4JakJA06WcfvubM80x5gzv7kIFucad6jw9WrACitcG6qERBSAU1xoCaOEQAvD_BwE). In this protocol, mastermix is pre-added to a clean PCR plate. Then, samples from up to 4 source plates specified in the `.csv` file are transferred to the plate containing mastermix and mixed (optionally).\n\nThe .csv file should be formatted as shown below (**including header line**):\n```\nvolume,source plate # (1-4),source column (1-12),destination column(1-12)\n8,1,2,1\n8,2,3,2\n8,2,12,3\n```\n\n---\n\n",
diff --git a/protoBuilds/18050b/README.json b/protoBuilds/18050b/README.json
index 022bc7cd24..fe58242065 100644
--- a/protoBuilds/18050b/README.json
+++ b/protoBuilds/18050b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom drug screening assay through plate mapping from one mother plate to up to 6 daughter plates using a P10-multi channel pipette. All plates used are Corning 384-flatwell plates.\n\n\n\nCorning 384-flatwell plates 112 \u00b5L\nOpentrons P10 multi-channel electronic pipette\nOpentrons 10\u00b5l tipracks\n",
"internal": "18050b",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "This protocol performs a custom drug screening assay through plate mapping from one mother plate to up to 6 daughter plates using a P10-multi channel pipette. All plates used are Corning 384-flatwell plates.\n\n---\n\n\n* [Corning 384-flatwell plates 112 \u00b5L](https://labware.opentrons.com/corning_384_wellplate_112ul_flat?category=wellPlate)\n* [Opentrons P10 multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons 10\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n",
"internal": "18050b\n",
diff --git a/protoBuilds/18d6a1/README.json b/protoBuilds/18d6a1/README.json
index d8e35f0116..ec3a33432c 100644
--- a/protoBuilds/18d6a1/README.json
+++ b/protoBuilds/18d6a1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Extraction"
@@ -8,7 +8,7 @@
"description": "This protocol automates the MGI Easy Nucleic Acid Extraction Kit and offers the user the ability to modify parameters such as the number of samples and the types of labware used.\n\nThis protocol has been optimized for use on the OT-2 and begins with an empty deepwell plate on the Magnetic Module. Lysis mix is first added to the plate, then samples are transferred to the lysis mix and mixed. Three wash steps occur after this and the protocol concludes with an elution step that results in 75\u00b5L of elutes transferred to a clean plate.\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons Magnetic Module, GEN2\nOpentrons OT-2 Run App (Version 3.21.0 or later)\nOpentrons P300 Multi-Channel Pipette\nOpentrons Tip Racks for P300\nNEST 1-Well Reservoir, 195mL\nNEST 12-Well Reservoir, 15mL\n96-Well Plate Containing Samples (such as the NEST 96-Well Deep Well Plate)\n96-Deepwell Plate for Extraction on the Magnetic Module (such as the NEST 96-Well Deep Well Plate)\n96-Well Plate for Elutes (such as the NEST 96-Well PCR Plate)\nMGI Easy Nucleic Acid Extraction Kit\n\n\n\nDeck Layout\n\nSlot 1: 96-Well Plate for Elutes (such as the NEST 96-Well PCR Plate)\n\n\nSlot 2: Opentrons Tip Racks for P300 - Tip rack 5\n\n\nSlot 3: NEST 12-Well Reservoir, 15mL\nA1: Lysis Mix (Samples 1-32)\n\nA2: Lysis Mix (Samples 33-64)\n\nA3: Lysis Mix (Samples 65-96)\n\nA4: Wash Buffer 1 (Samples 1-48)\n\nA5: Wash Buffer 1 (Samples 49-96)\n\nA6: Wash Buffer 2 (Samples 1-48)\n\nA7: Wash Buffer 2 (Samples 49-96)\n\nA8: Wash Buffer 3 (Samples 1-48)\n\nA9: Wash Buffer 3 (Samples 49-96)\n\nA10: Empty\n\nA11: Empty\n\nA12: Nuclease-Free Water (Samples 1-96)\n\n\nSlot 4: 96-Well Plate Containing Samples (such as the NEST 96-Well Deep Well Plate)\n\n\nSlot 5: Opentrons Tip Racks for P300 - Tip rack 3\n\n\nSlot 6: Opentrons Tip Racks for P300 - Tip rack 4\n\n\nSlot 7: Opentrons Tip Racks for P300 - Tip rack for Samples\n\n\nSlot 8: Opentrons Tip Racks for P300 - Tip rack 1\n\n\nSlot 9: Opentrons Tip Racks for P300 - Tip rack 2\n\n\nSlot 10: Opentrons Magnetic Module, GEN2 with 96-Deepwell Plate for Extraction (such as the NEST 96-Well Deep Well Plate)\n\n\nSlot 11: NEST 1-Well Reservoir, 195mL (liquid waste)\n\n\nNote about Tip Racks: Each column of samples (8) will require five columns of tips + the tips needed for the initial sample transfer. \n\n\nUsing the customizations field (below), set up your protocol.\n Number of Samples: Specify the number of samples to run.\n Sample Plate (input) Type: Select the type of plate used.\n Extraction Plate (MagPlate) Type: Select the type of plate used.\n Elution Plate (output) Type: Select the type of plate used.\n P300-Multi Mount: Select which mount (left, right) the P300-Multi is attached to.\n Tip Type: Specify the type of tips (filtered, non-filtered) used.\n* Used Tips Destination: Specify the destination of used tips. \"Empty Tipracks\" is best suited for runs of more than 24 samples.",
"internal": "18d6a1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Nucleic Acid Extraction\n\n\n",
"description": "This protocol automates the [MGI Easy Nucleic Acid Extraction Kit](https://en.mgitech.cn/Uploads/Temp/file/20200416/5e97edc3ef00b.pdf) and offers the user the ability to modify parameters such as the number of samples and the types of labware used.\n\nThis protocol has been optimized for use on the OT-2 and begins with an empty deepwell plate on the Magnetic Module. Lysis mix is first added to the plate, then samples are transferred to the lysis mix and mixed. Three wash steps occur after this and the protocol concludes with an elution step that results in 75\u00b5L of elutes transferred to a clean plate.\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons Magnetic Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons OT-2 Run App (Version 3.21.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons Tip Racks for P300](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* 96-Well Plate Containing Samples (such as the [NEST 96-Well Deep Well Plate](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate))\n* 96-Deepwell Plate for Extraction on the Magnetic Module (such as the [NEST 96-Well Deep Well Plate](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate))\n* 96-Well Plate for Elutes (such as the [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt))\n* MGI Easy Nucleic Acid Extraction Kit\n\n\n\n---\n\n\n**Deck Layout**\n\n**Slot 1**: 96-Well Plate for Elutes (such as the [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt))\n\n\n**Slot 2**: [Opentrons Tip Racks for P300](https://shop.opentrons.com/collections/opentrons-tips) - Tip rack 5\n\n\n**Slot 3**: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\nA1: Lysis Mix (Samples 1-32)\n\nA2: Lysis Mix (Samples 33-64)\n\nA3: Lysis Mix (Samples 65-96)\n\nA4: Wash Buffer 1 (Samples 1-48)\n\nA5: Wash Buffer 1 (Samples 49-96)\n\nA6: Wash Buffer 2 (Samples 1-48)\n\nA7: Wash Buffer 2 (Samples 49-96)\n\nA8: Wash Buffer 3 (Samples 1-48)\n\nA9: Wash Buffer 3 (Samples 49-96)\n\nA10: *Empty*\n\nA11: *Empty*\n\nA12: Nuclease-Free Water (Samples 1-96)\n\n\n**Slot 4**: 96-Well Plate Containing Samples (such as the [NEST 96-Well Deep Well Plate](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate))\n\n\n**Slot 5**: [Opentrons Tip Racks for P300](https://shop.opentrons.com/collections/opentrons-tips) - Tip rack 3\n\n\n**Slot 6**: [Opentrons Tip Racks for P300](https://shop.opentrons.com/collections/opentrons-tips) - Tip rack 4\n\n\n**Slot 7**: [Opentrons Tip Racks for P300](https://shop.opentrons.com/collections/opentrons-tips) - Tip rack for Samples\n\n\n**Slot 8**: [Opentrons Tip Racks for P300](https://shop.opentrons.com/collections/opentrons-tips) - Tip rack 1\n\n\n**Slot 9**: [Opentrons Tip Racks for P300](https://shop.opentrons.com/collections/opentrons-tips) - Tip rack 2\n\n\n**Slot 10**: [Opentrons Magnetic Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with 96-Deepwell Plate for Extraction (such as the [NEST 96-Well Deep Well Plate](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate))\n\n\n**Slot 11**: [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml) (liquid waste)\n\n\n*Note about Tip Racks*: Each column of samples (8) will require five columns of tips + the tips needed for the initial sample transfer. \n\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Samples**: Specify the number of samples to run.\n* **Sample Plate (input) Type**: Select the type of plate used.\n* **Extraction Plate (MagPlate) Type**: Select the type of plate used.\n* **Elution Plate (output) Type**: Select the type of plate used.\n* **P300-Multi Mount**: Select which mount (left, right) the P300-Multi is attached to.\n* **Tip Type**: Specify the type of tips (filtered, non-filtered) used.\n* **Used Tips Destination**: Specify the destination of used tips. \"Empty Tipracks\" is best suited for runs of more than 24 samples.\n\n\n\n\n",
"internal": "18d6a1\n",
diff --git a/protoBuilds/18e62e/README.json b/protoBuilds/18e62e/README.json
index 06c56bb7d8..6da2b5e877 100644
--- a/protoBuilds/18e62e/README.json
+++ b/protoBuilds/18e62e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "18e62e",
"labware": "\nOpentrons 20\u00b5L Filter Tips\nKingFisher 96-Deepwell Plate\nMicroAmp\u2122 EnduraPlate\u2122 Optical 384-Well Plate\nOpentrons 96-Well Aluminum Block + PCR Strips (200\u00b5L)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n",
"description": "This protocol automates a Multiplex LDT 384-Well Plate PCR prep. Using up to four KingFisher 96-Well Plates as inputs and an [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) to keep reagents cool, this protocol transfers water, primer/probes, and master mix to necessary wells in 384-Well Plate before transferring samples from the KingFisher plates.\n\nThis protocol is still a work in progress and will be updated.\n\n**Update 2021-10-25:** A new parameter was added (*Only Transfer Samples*) that will skip the addition of reaction mix components and use of the temperature module; instead, simply transferring samples from 96-well plates to 384-well plate.\n\n\nExplanation of complex parameters below:\n* **Plate 1 Number of Samples**: Specify the number of samples in Plate 1 (up to 96). The plate can be skipped by setting this value to 0 (zero).\n* **Plate 2 Number of Samples**: Specify the number of samples in Plate 2 (up to 96). The plate can be skipped by setting this value to 0 (zero).\n* **Plate 3 Number of Samples**: Specify the number of samples in Plate 3 (up to 96). The plate can be skipped by setting this value to 0 (zero).\n* **Plate 4 Number of Samples**: Specify the number of samples in Plate 4 (up to 96). The plate can be skipped by setting this value to 0 (zero).\n* **Pipette Mount**: Select which mount the [P20 Multi-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette) is attached to.\n* **Used Tip Location**: Select where used tips should be dropped after transferring samples. *Empty Tip Rack* is recommended due to space constraints in the waste bin.\n\n\n---\n\n",
diff --git a/protoBuilds/19313a/README.json b/protoBuilds/19313a/README.json
index 28b8bba654..68e37d95e3 100644
--- a/protoBuilds/19313a/README.json
+++ b/protoBuilds/19313a/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Purification"
@@ -10,7 +10,7 @@
"internal": "19313a",
"labware": "\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nOpentrons 4-in-1 Tube Rack Set\nOpentrons 200uL Filter Tips\nOpentrons 300uL Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Nucleic Acid Purification\n\n",
"deck-setup": "\n\n",
"description": "This protocol extracts RNA with a temperature module and magnetic module. User has the choice to include a \"Waste water mode\" which consists of adding and mixing binding buffer on magnetically engaged beads. Tips are tracked between protocols with the protocol pausing and prompting the user to replace tip racks when a tip rack is depleted.\n\nExplanation of complex parameters below:\n* Most customizable variables below are in reference to the original protocol request coded by the user. All additional parameters are explained:\n* `Reset Tipracks`: Select `yes` to pick up the tip from A1 on both the 200ul and 300ul tip racks. Select `no` to pick up the tips for both tip racks from where the previous run left off.\n* `Remove supernatant aspiration height (mm)`: Select aspiration height from the bottom of the well to remove supernatant.\n* `Length from side of the well opposite magnetically engaged beads (mm)`: Specify the distance to aspirate from the side of the well opposite the magnetically engaged beads. A values of 1 is the side of the well, a values of 4.1 is the middle of the well.\n* `P300 Single GEN2 Mount`: Select which side (left or right) to mount P300 single channel pipette.\n\n\n---\n\n",
diff --git a/protoBuilds/1980cd/README.json b/protoBuilds/1980cd/README.json
index 5b71fbf783..c9f4d52468 100644
--- a/protoBuilds/1980cd/README.json
+++ b/protoBuilds/1980cd/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Plant DNA"
@@ -8,7 +8,7 @@
"description": "This protocol automates {mesoscale kit} in 4 steps over 2 days. Those steps are:\n\n(Day 1) Load blocking solution into test plates\n(Day 2) Load standards and up to 10 sample columns from sample plates into 2 test plates\n(Day 2) Load detection antibody into test plates\n(Day 2) Load read buffer into test plates\n\nThe initial Day 1 step can be done with up to 9 test plates. The subsequent steps are done with 1 sample plate and then 2 test plates. To increase throughput, run the step again with a separate plate. \nNote all volumes requested in process are the amount required + 10%. You may need more or less given your particular robot and calibrations.\n\n\n\nOpentrons P300-multi channel electronic pipettes (GEN2)\nOpentrons 200ul Filter Tips\nNEST 1 Well Reservoir 195 mL\nNEST 12-Well Reservoirs, 15 mL\n{mesoscale kit{\n",
"internal": "1980cd",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Plant DNA\n\n",
"description": "This protocol automates {mesoscale kit} in 4 steps over 2 days. Those steps are:\n\n1. (Day 1) Load blocking solution into test plates\n2. (Day 2) Load standards and up to 10 sample columns from sample plates into 2 test plates\n3. (Day 2) Load detection antibody into test plates\n4. (Day 2) Load read buffer into test plates\n\nThe initial Day 1 step can be done with up to 9 test plates. The subsequent steps are done with 1 sample plate and then 2 test plates. To increase throughput, run the step again with a separate plate. \n\nNote all volumes requested in process are the amount required + 10%. You may need more or less given your particular robot and calibrations.\n\n---\n\n\n* [Opentrons P300-multi channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [NEST 1 Well Reservoir 195 mL](http://www.cell-nest.com/page94?_l=en&product_id=102)\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [{mesoscale kit{](https://example.com)\n\n",
"internal": "1980cd\n",
diff --git a/protoBuilds/199721/README.json b/protoBuilds/199721/README.json
index 9082c4beb8..a56396d1a5 100644
--- a/protoBuilds/199721/README.json
+++ b/protoBuilds/199721/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Pooling"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom sample preparation through pooling up to 3 samples in primary racks into secondary racks.\n\n\n\nOpentrons P1000 GEN2 single-channel pipette\nOpentrons 1000ul tipracks\nMercedes Scientific Tubes Flat Base 7ml #60550050\nMercedes Scientific Tubes 5ml round base #GLO110470\nCustom Alphaqua racks\n\n\n\nTransfer Scheme\n\nTransfer Order\n",
"internal": "199721",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Pooling\n\n",
"description": "This protocol performs a custom sample preparation through pooling up to 3 samples in primary racks into secondary racks.\n\n---\n\n\n* [Opentrons P1000 GEN2 single-channel pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette?variant=5984549109789)\n* [Opentrons 1000ul tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* [Mercedes Scientific Tubes Flat Base 7ml #60550050](https://www.mercedesscientific.com/buy/product/screw-cap-tube/CLENC2fcbbe9ab2407f2b7db3c5ca4455f575?text=SRS+60550050&lsi=true)\n* [Mercedes Scientific Tubes 5ml round base #GLO110470](https://www.mercedesscientific.com/buy/product/test-tube/CLENC9a8cb62386ae3ba2bc4ba50fb708dc8b?text=GLO+110470&lsi=true)\n* [Custom Alphaqua racks](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2020-08-19/wg23biy/Alpaqua%20Custom%20Rack%20for%20CorePlus.pdf)\n\n---\n\n\n\nTransfer Scheme \n\n\nTransfer Order \n\n\n",
"internal": "199721\n",
diff --git a/protoBuilds/19fc32/README.json b/protoBuilds/19fc32/README.json
index 2c2dadb031..934c656411 100644
--- a/protoBuilds/19fc32/README.json
+++ b/protoBuilds/19fc32/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Clean Up"
@@ -8,7 +8,7 @@
"description": "This protocol is a slightly modified version of a clean up protocol. With this protocol, the user selects which mount their P50-Multi is attached to and the protocol adds the necessary reagents and (dis)engages the magnetic module to accomplish the clean up. To conclude this protocol, 4\u00b5L of supernatant is added to a 384-well plate and 20\u00b5L of supernatant is added to a 96-well plate.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Magnetic Module\nOpentrons P50 Multi-Channel Pipette\nOpentrons 50/300\u00b5L Tips\nBio-Rad 96-Well Plate, 200\u00b5L\nBio-Rad 384-Well Plate\nNEST 12-Well Reservoir\nReagents\nSamples\n\n\n\nSlot 1: NEST 12-Well Reservoir\n A1: Magnetic Beads\n A2: 80% Fresh Ethanol (~10mL)\n A3: 80% Fresh Ethanol (~10mL)\n A4: 80% Fresh Ethanol (~10mL)\n A5: 80% Fresh Ethanol (~10mL)\n A6: RSB Buffer\n A7: Empty\n A8: Empty\n A9: Empty, for liquid waste\n A10: Empty, for liquid waste\n A11: Empty, for liquid waste\n A12: Empty, for liquid waste\nSlot 2: Bio-Rad 96-Well Plate, clean and empty\nSlot 3: Bio-Rad 384-Well Plate\nSlot 4: Opentrons Magnetic Module with a Bio-Rad 96-Well Plate, filled with 25\u00b5L of sample\nSlot 5: Opentrons 50/300\u00b5L Tips\nSlot 6: Opentrons 50/300\u00b5L Tips\nSlot 7: Opentrons 50/300\u00b5L Tips\nSlot 8: Opentrons 50/300\u00b5L Tips\nSlot 9: Opentrons 50/300\u00b5L Tips\nSlot 10: Opentrons 50/300\u00b5L Tips\nUsing the customizations fields, below set up your protocol.\n* P50 Multi Mount: Select which mount (left or right) the P50 Multi is attached to.",
"internal": "19fc32",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Clean Up\n\n\n",
"description": "This protocol is a slightly modified version of a clean up protocol. With this protocol, the user selects which mount their P50-Multi is attached to and the protocol adds the necessary reagents and (dis)engages the magnetic module to accomplish the clean up. To conclude this protocol, 4\u00b5L of supernatant is added to a 384-well plate and 20\u00b5L of supernatant is added to a 96-well plate.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P50 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Bio-Rad 96-Well Plate, 200\u00b5L](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr)\n* [Bio-Rad 384-Well Plate](https://www.bio-rad.com/en-au/product/384-well-pcr-plates?ID=OCH4UM15)\n* [NEST 12-Well Reservoir](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* Reagents\n* Samples\n\n\n---\n\n\nSlot 1: [NEST 12-Well Reservoir](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* A1: Magnetic Beads\n* A2: 80% Fresh Ethanol (~10mL)\n* A3: 80% Fresh Ethanol (~10mL)\n* A4: 80% Fresh Ethanol (~10mL)\n* A5: 80% Fresh Ethanol (~10mL)\n* A6: RSB Buffer\n* A7: Empty\n* A8: Empty\n* A9: Empty, for liquid waste\n* A10: Empty, for liquid waste\n* A11: Empty, for liquid waste\n* A12: Empty, for liquid waste\n\nSlot 2: [Bio-Rad 96-Well Plate](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr), clean and empty\n\nSlot 3: [Bio-Rad 384-Well Plate](https://www.bio-rad.com/en-au/product/384-well-pcr-plates?ID=OCH4UM15)\n\nSlot 4: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with a [Bio-Rad 96-Well Plate](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr), filled with 25\u00b5L of sample\n\nSlot 5: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 6: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 7: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 8: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 9: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 10: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n\n\n**Using the customizations fields, below set up your protocol.**\n* **P50 Multi Mount**: Select which mount (left or right) the P50 Multi is attached to.\n\n\n\n",
"internal": "19fc32\n",
diff --git a/protoBuilds/1a2343/README.json b/protoBuilds/1a2343/README.json
index 32329750f2..134f8081c0 100644
--- a/protoBuilds/1a2343/README.json
+++ b/protoBuilds/1a2343/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom PCR preparation for gel samples using an on-deck Opentrons Thermocycler module.\n\n\n\nOpentrons Thermocycler (occupying slots 7, 8, 10, 11) with NEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOpentrons Temperature Module with USA Scientific 96 Deep Well Plate 2.4 mL\nCorning 96 Well Plate 360 \u00b5L Flat\nOpentrons 20\u00b5l and 50/300\u00b5l tipracks\n\n\n\n96-deepwell reagent block on temperature module (slot 4)\n well A1: DNTPs\n well B1: polymerase\n column 2: PCR buffer\n wells A3-H3: templates 1-8\n* column 4: water",
"internal": "1a2343",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol performs a custom PCR preparation for gel samples using an on-deck Opentrons Thermocycler module.\n\n---\n\n\n* [Opentrons Thermocycler (occupying slots 7, 8, 10, 11)](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module) with [NEST 0.1 mL 96-Well PCR Plate, Full Skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [USA Scientific 96 Deep Well Plate 2.4 mL](https://www.usascientific.com/plateone-96-deep-well-2ml/p/PlateOne-96-Deep-Well-2mL)\n* [Corning 96 Well Plate 360 \u00b5L Flat](https://ecatalog.corning.com/life-sciences/b2c/US/en/Microplates/Assay-Microplates/96-Well-Microplates/Corning%C2%AE-96-well-Solid-Black-and-White-Polystyrene-Microplates/p/corning96WellSolidBlackAndWhitePolystyreneMicroplates)\n* [Opentrons 20\u00b5l and 50/300\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n\n---\n\n\n96-deepwell reagent block on temperature module (slot 4)\n* well A1: DNTPs\n* well B1: polymerase\n* column 2: PCR buffer\n* wells A3-H3: templates 1-8\n* column 4: water\n\n",
"internal": "1a2343\n",
diff --git a/protoBuilds/1adec6-2/README.json b/protoBuilds/1adec6-2/README.json
index f2e11fe9d5..a03c5e1fa5 100644
--- a/protoBuilds/1adec6-2/README.json
+++ b/protoBuilds/1adec6-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Assay"
@@ -10,7 +10,7 @@
"internal": "1adec6-2",
"labware": "\nOpentrons 300\u00b5L Tipracks\nNEST 12-Well Reservoir, 15mL\nSPL 96-Well Cell Culture Plates (Round Bottom)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Assay\n\n",
"deck-setup": "**Slot 1**: Destination Plate 1 (SPL 96-Well Cell Culture Plate)\n\n**Slot 2**: Destination Plate 2 (SPL 96-Well Cell Culture Plate)\n\n**Slot 3**: Destination Plate 3 (SPL 96-Well Cell Culture Plate)\n\n**Slot 4**: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n\n**Slot 7**: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n\n\n",
"description": "**Updated**\nThis protocol has been updated based on feedback from the user.\n\nThis protocol is part two of a larger workflow. The entire workflow can be found below\n\nPart 1: [Small Molecule Library Prep](./1adec6)\nPart 2: [Seed Cells](./1adec6-2)\nPart 3: [Transfer Small Molecules](./1adec6-3)\nPart 4: [Transfer Small Molecules - CSV Input](./1adec6-4)\nPart 5: [Custom Supernatant Removal](./1adec6-5)\nPart 6: [ProcartaPlex Protocol-1](./1adec6-6)\nPart 7: [ProcartaPlex Protocol-2](./1adec6-7)\n\nIn this protocol, 250\u00b5L of cells are transferred from the [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) to the SPL 96-Well Cell Culture Plates.\n\nExplanation of complex parameters below:\n* **P300-Multi Mount**: Select which mount the P300-Multi Pipette is attached to.\n* **Number of Destination Plates**: Select the number of destination plates.\n\n---\n\n",
diff --git a/protoBuilds/1adec6-3/README.json b/protoBuilds/1adec6-3/README.json
index 9b14c8aed9..5466f81f8f 100644
--- a/protoBuilds/1adec6-3/README.json
+++ b/protoBuilds/1adec6-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Assay"
@@ -10,7 +10,7 @@
"internal": "1adec6-3",
"labware": "\nOpentrons 20\u00b5L Tipracks\n\n\nOpentrons 300\u00b5L Tipracks (optional)\n\n\nThermo-Fast 96-Well, Fully Skirted Plate\nSPL 96-Well Cell Culture Plates\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Assay\n\n",
"deck-setup": "**Slot 1**: Destination Plate 1 (SPL 96-Well Cell Culture Plat)\n\n**Slot 2**: Destination Plate 2 (SPL 96-Well Cell Culture Plate)\n\n**Slot 3**: Destination Plate 3 (SPL 96-Well Cell Culture Plate)\n\n**Slot 4**: [Opentrons 20\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-tips)\n\n**Slot 5**: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips) (optional, for mixing)\n\n**Slot 6**: Small Molecule Library Plate (Thermo-Fast 96-Well, Fully Skirted Plate)\n\n**Slot 7**: [Opentrons 20\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-tips)\n\n**Slot 8**: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips) (optional, for mixing)\n\n**Slot 10**: [Opentrons 20\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-tips)\n\n**Slot 11**: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips) (optional, for mixing)\n\n\n\n---\n\n",
"description": "**Updated**\nThis protocol has been updated based on feedback from the user.\n\nThis protocol is part three of a larger workflow. The entire workflow can be found below\n\nPart 1: [Small Molecule Library Prep](./1adec6)\nPart 2: [Seed Cells](./1adec6-2)\nPart 3: [Transfer Small Molecules](./1adec6-3)\nPart 4: [Transfer Small Molecules - CSV Input](./1adec6-4)\nPart 5: [Custom Supernatant Removal](./1adec6-5)\nPart 6: [ProcartaPlex Protocol-1](./1adec6-6)\nPart 7: [ProcartaPlex Protocol-2](./1adec6-7)\n\nIn this protocol, 1\u00b5L is transferred from the small molecule library plate to the experiment plate seeded with cells.\n\nExplanation of complex parameters below:\n* **P20-Multi Mount**: Select which mount the P20-Multi Pipette is attached to.\n* **Number of Destination Plates**: Select the number of destination plates.\n* **Transfer volume (in \u00b5L)**: Specify the volume to be transferred.\n* **Mix with P300-Multi**: Select whether or not to add an optional mix step with the P300-Multi Pipette (200\u00b5L, 3 times) after the transfer occurs\n\n---\n\n",
diff --git a/protoBuilds/1adec6-4/README.json b/protoBuilds/1adec6-4/README.json
index 08e05cfc46..f5414a67ca 100644
--- a/protoBuilds/1adec6-4/README.json
+++ b/protoBuilds/1adec6-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Assay"
@@ -10,7 +10,7 @@
"internal": "1adec6-4",
"labware": "\nOpentrons 20\u00b5L Tipracks\nOpentrons 300\u00b5L Tipracks\nThermo-Fast 96-Well, Fully Skirted Plate\nSPL 96-Well Cell Culture Plates\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Assay\n\n",
"deck-setup": "**Slot 1**: Small Molecule Library Plate (Thermo-Fast 96-Well, Fully Skirted Plate)\n\n**Slot 2**: Destination Plate (SPL 96-Well Cell Culture Plate)\n\n**Slot 3**: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-tips)\n\n**Slot 4**: [Opentrons 20\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-tips)\n\n**Slot 5**: [Opentrons 20\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-tips)\n\n**Slot 7**: [Opentrons 20\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-tips)\n\n**Slot 8**: [Opentrons 20\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-tips)\n\n**Slot 10**: [Opentrons 20\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-tips)\n\n**Slot 11**: [Opentrons 20\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-tips)\n\n\n\n---\n\n",
"description": "**Updated**\nThis protocol has been updated based on feedback from the user.\n\nThis protocol is part four of a larger workflow. The entire workflow can be found below\n\nPart 1: [Small Molecule Library Prep](./1adec6)\nPart 2: [Seed Cells](./1adec6-2)\nPart 3: [Transfer Small Molecules](./1adec6-3)\nPart 4: [Transfer Small Molecules - CSV Input](./1adec6-4)\nPart 5: [Custom Supernatant Removal](./1adec6-5)\nPart 6: [ProcartaPlex Protocol-1](./1adec6-6)\nPart 7: [ProcartaPlex Protocol-2](./1adec6-7)\n\nIn this protocol, a CSV is used to dictate the volume and destination of liquid transfers using the [P20 Single-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/Single-Channel-electronic-pipette).\n\nExplanation of complex parameters below:\n* **P20 Mount**: Select which mount the P20 Pipette is attached to.\n* **P20 Selection (Single or Multi)**: Select which pipette is used (please see note below)\n* **Transfer CSV**: Upload the CSV containing the liquid transfers. The CSV should be formatted as follows:\n\n\n| Source Well | Volume | Destination Well |\n| ----------- | ------ | ---------------- |\n| A1 | 3 | C2 |\n| A1 | 2 | B4 |\n| A2 | 5 | D7 |\n\n**Note**: If using the [P20 Multi-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-Channel-electronic-pipette), you should only use Row A when selecting a *destination well* in the CSV.\n\n---\n\n",
diff --git a/protoBuilds/1adec6-5/README.json b/protoBuilds/1adec6-5/README.json
index f316469bc5..3fbd15200c 100644
--- a/protoBuilds/1adec6-5/README.json
+++ b/protoBuilds/1adec6-5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Assay"
@@ -10,7 +10,7 @@
"internal": "1adec6-5",
"labware": "\nOpentrons 300\u00b5L Tipracks\nSPL 96-Well Cell Culture Plates\nNEST 12-Well Reservoir, 15mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Assay\n\n",
"deck-setup": "**Slot 1**: Source Plate, containing Cells (SPL 96-Well Cell Culture Plate)\n\n**Slot 4**: Destination Plate (SPL 96-Well Cell Culture Plate)\n\n**Slot 5**: **Optional**, Second Destination Plate (SPL 96-Well Cell Culture Plate)\n\n**Slot 6**: [[NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n\n**Slot 7**: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n\n",
"description": "**Updated**\nThis protocol has been updated based on feedback from the user.\n\nThis protocol is part five of a larger workflow. The entire workflow can be found below\n\nPart 1: [Small Molecule Library Prep](./1adec6)\nPart 2: [Seed Cells](./1adec6-2)\nPart 3: [Transfer Small Molecules](./1adec6-3)\nPart 4: [Transfer Small Molecules - CSV Input](./1adec6-4)\nPart 5: [Custom Supernatant Removal](./1adec6-5)\nPart 6: [ProcartaPlex Protocol-1](./1adec6-6)\nPart 7: [ProcartaPlex Protocol-2](./1adec6-7)\n\nIn this protocol, the [P300 8-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-Channel-electronic-pipette) transfers 220\u00b5L of supernatant from source plate containing cells to a destination plate. Once complete, the [P300 8-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-Channel-electronic-pipette) will transfer 100\u00b5L of Cell medium+10% PrestoBlue to the original source plate.\n\nExplanation of complex parameters below:\n* **P300-Multi Mount**: Select which mount the P300-Multi Pipette is attached to.\n* **Aspirate from Well Bottom Height**: Specify the height from the bottom of the well (in mm) that the pipette will aspirate supernatant (note: default height is 1mm from bottom).\n* **Perform 2nd aliquot**: Specify whether or not to perform a second aliquot (110\u00b5L transferred to plate in slot 5)\n\n\n---\n\n",
diff --git a/protoBuilds/1adec6-6/README.json b/protoBuilds/1adec6-6/README.json
index ed74beb96d..e4f564c8c3 100644
--- a/protoBuilds/1adec6-6/README.json
+++ b/protoBuilds/1adec6-6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Assay"
@@ -10,7 +10,7 @@
"internal": "1adec6-6",
"labware": "\nOpentrons 300\u00b5L Tipracks\nProcartaPlex 96-Well Cell Culture Plates\nNEST 12-Well Reservoir, 15mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Assay\n\n",
"deck-setup": "**Slot 1**: **Optional**, Destination Plate 2 (ProcartaPlex 96-Well Cell Culture Plate)\n\n**Slot 2**: **Optional**, Sample Plate 2 (ProcartaPlex 96-Well Cell Culture Plate)\n\n**Slot 3**: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n\n**Slot 4**: Destination Plate 1 (ProcartaPlex 96-Well Cell Culture Plate)\n\n**Slot 5**: Sample Plate 1 (ProcartaPlex 96-Well Cell Culture Plate)\n\n**Slot 6**: Standards Plate (ProcartaPlex 96-Well Cell Culture Plate)\n\n**Slot 7**: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n**Slot 8**: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n**Slot 10**: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n",
"description": "**Updated**\nThis protocol has been updated based on feedback from the user.\n\nThis protocol is part six of a larger workflow. The entire workflow can be found below\n\nPart 1: [Small Molecule Library Prep](./1adec6)\nPart 2: [Seed Cells](./1adec6-2)\nPart 3: [Transfer Small Molecules](./1adec6-3)\nPart 4: [Transfer Small Molecules - CSV Input](./1adec6-4)\nPart 5: [Custom Supernatant Removal](./1adec6-5)\nPart 6: [ProcartaPlex Protocol-1](./1adec6-6)\nPart 7: [ProcartaPlex Protocol-2](./1adec6-7)\n\nThis protocol is the first half of a custom, ProcartaPlex protocol. In this protocol, 50\u00b5L of Magnetic Beads are added to all the wells of the Destination Plate. The user is then prompted to remove the plate and incubate off the deck before returning to the robot. Once the Destination Plate is replaced, Wash Buffer is added and then 50\u00b5L of samples (columns 1-10 of Sample Plate) and standards (columns 1 and 2 of Standards Plate) are added to the Destination Plate. The user is then prompted to perform some off-deck steps before incubating overnight. Once incubation is complete, users can move on to [ProcartaPlex Protocol-2](./1adec6-7).\n\nExplanation of complex parameters below:\n* **P300-Multi Mount**: Select which mount the P300-Multi Pipette is attached to.\n* **Number of destination plates**: Select how many destination plates will be used in the protocol (1 or 2).\n\n\n---\n\n",
diff --git a/protoBuilds/1adec6-7/README.json b/protoBuilds/1adec6-7/README.json
index 48c5c4683d..3df4d353e4 100644
--- a/protoBuilds/1adec6-7/README.json
+++ b/protoBuilds/1adec6-7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Assay"
@@ -10,7 +10,7 @@
"internal": "1adec6-7",
"labware": "\nOpentrons 300\u00b5L Tipracks\nProcartaPlex 96-Well Cell Culture Plates\nNEST 12-Well Reservoir, 15mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Assay\n\n",
"deck-setup": "**Slot 1**: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n\n**Slot 2**: **Optional**, [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) (2)\n\n**Slot 3**: **Optional**, [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) (3)\n\n**Slot 4**: Destination Plate (ProcartaPlex 96-Well Cell Culture Plate)\n\n**Slot 5**: **Optional**, Destination Plate (2) (ProcartaPlex 96-Well Cell Culture Plate)\n\n**Slot 6**: **Optional**, Destination Plate (3) (ProcartaPlex 96-Well Cell Culture Plate)\n\n**Slot 7**: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n\n",
"description": "**Updated**\nThis protocol has been updated based on feedback from the user.\n\nThis protocol is part seven of a larger workflow. The entire workflow can be found below\n\nPart 1: [Small Molecule Library Prep](./1adec6)\nPart 2: [Seed Cells](./1adec6-2)\nPart 3: [Transfer Small Molecules](./1adec6-3)\nPart 4: [Transfer Small Molecules - CSV Input](./1adec6-4)\nPart 5: [Custom Supernatant Removal](./1adec6-5)\nPart 6: [ProcartaPlex Protocol-1](./1adec6-6)\nPart 7: [ProcartaPlex Protocol-2](./1adec6-7)\n\nThis protocol is the second half of a custom, ProcartaPlex protocol - the first half can be found [here](./1adec6-6). This protocol begins after the overnight incubation from the first half with two wash steps. After the washes, 25\u00b5L of Detection Antibodies are added to the wells and the user is prompted to remove the plate for off-deck processing. Once the plate is returned, two more wash steps occur. Once the washes are complete, 50\u00b5L of SAP is added to the wells and the user is again prompted to remove the deck for off-deck processing. Once the plate is returned, the final two wash steps occur, before 120\u00b5L of Reading Buffer is added to the wells - completing the automated portion of this workflow.\n\nExplanation of complex parameters below:\n* **P300-Multi Mount**: Select which mount the P300-Multi Pipette is attached to.\n* **Number of destination plates**: Select how many destination plates will be used in the protocol (1, 2, or 3).\n\n\n---\n\n",
diff --git a/protoBuilds/1adec6/README.json b/protoBuilds/1adec6/README.json
index 1b389fab9a..7a0c377e2a 100644
--- a/protoBuilds/1adec6/README.json
+++ b/protoBuilds/1adec6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Assay"
@@ -10,7 +10,7 @@
"internal": "1adec6",
"labware": "\nOpentrons 20\u00b5L Tipracks\nNEST 12-Well Reservoir, 15mL\nThermo-Fast 96-Well, Fully Skirted Plates\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Assay\n\n",
"deck-setup": "**Slot 1**: Aliquot Plate 1 (Thermo-Fast 96-Well, Fully Skirted Plates)\n\n**Slot 2**: Aliquot Plate 2 (Thermo-Fast 96-Well, Fully Skirted Plates)\n\n**Slot 3**: Aliquot Plate 3 (Thermo-Fast 96-Well, Fully Skirted Plates)\n\n**Slot 4**: Sample Plate (Thermo-Fast 96-Well, Fully Skirted Plate)\n\n**Slot 5**: Destination Plate (Thermo-Fast 96-Well, Fully Skirted Plate)\n\n**Slot 6**: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n\n**Slot 7**: [Opentrons 20\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-tips)\n\n**Slot 8**: [Opentrons 20\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-tips)\n\n\n\n",
"description": "This protocol now utilizes the [P20 8-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette) (instead of the P300), uses additional plates, and has modified liquid handling. The steps of this updated version can be found below.\n\n**Updated Sept 29, 2021**\nThis protocol has been updated based on feedback from the user. User can now select **None** when selecting a \"**Number of Destination Plates**\" (skipping the aliquoting step). Additionally, this update fixes an issue with the way tips are handled when using less than 8 tips with the multi-channel pipette.\n\nPart 1: [Small Molecule Library Prep](./1adec6)\nPart 2: [Seed Cells](./1adec6-2)\nPart 3: [Transfer Small Molecules](./1adec6-3)\nPart 4: [Transfer Small Molecules - CSV Input](./1adec6-4)\nPart 5: [Custom Supernatant Removal](./1adec6-5)\nPart 6: [ProcartaPlex Protocol-1](./1adec6-6)\nPart 7: [ProcartaPlex Protocol-2](./1adec6-7)\n\nExplanation of complex parameters below:\n* **P20-Multi Mount**: Select which mount the P20-Multi Pipette is attached to.\n* **Number of Destination Plates**: Select the number of destination plates.\n* **Destination plate PBS volume (\u00b5l)**: Specify the volume of PBS to transfer to Destination plate (step 1).\n* **Destination plate 66% DMSO volume (\u00b5l)**: Specify the volume of 66% DMSO to transfer to Destination plate (step 2).\n* **Source plate volume for dilution (\u00b5l)**: Specify the volume for first dilution transfer (step 3).\n* **Destination plate volume for dilution (\u00b5l)**: Specify the volume for second dilution transfer (step 5).\n* **Library aliquot volume (\u00b5l)**: Specify the volume for the library aliquot (step 7).\n\n---\n\n",
diff --git a/protoBuilds/1af856/README.json b/protoBuilds/1af856/README.json
index b46c59c812..0a0aea4270 100644
--- a/protoBuilds/1af856/README.json
+++ b/protoBuilds/1af856/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Lexogen"
@@ -10,7 +10,7 @@
"internal": "1af856",
"labware": "\n[Opentrons Filter Tips for the P20 and P300] (https://shop.opentrons.com)\n[Opentrons Temperature Module] (https://shop.opentrons.com/modules/)\n[Opentrons Magnetic Module] (https://shop.opentrons.com/modules/)\n[Opentrons 4-in-1 Tuberack, Nest Well Plates, Nest Reservoir] (https://shop.opentrons.com)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Lexogen\n\n",
"deck-setup": "\n\n\n**Slot 1**: Final Output Plate (nest_96_wellplate_100ul_pcr_full_skirt) \n**Slot 2**: Tube Rack for Reagents (opentrons_24_tuberack_nest_1.5ml_snapcap) \n\n\n\n**Slot 3**: 80 Percent Ethanol Reservoir (nest_12_reservoir_15ml) \n**Slot 4**: Opentrons Magnetic Module with 96-Deep Well Plate (nest_96_wellplate_2ml_deep) \n**Slot 5**: PCR Plate (nest_96_wellplate_100ul_pcr_full_skirt) \n**Slot 7**: Opentrons Temperature Module with 24-Well Aluminum Block holding 1.5 mL Nest SnapCap Tubes \n\n\n\n**Slot 8**: Index Plate \n**Slots 10,11**: Opentrons 20 uL filter tips \n**Slots 6,9**: Opentrons 200 uL filter tips \n\n\n---\n\n",
"description": "This protocol uses single-channel P20 and P300 pipettes to perform library prep steps on 8-96 input samples according to the attached experimental protocol and Lexogen user guide. User-determined parameters are available to specify the number of samples, the bead drying time and the magnet engage time.\n\n\n---\n\n\n",
diff --git a/protoBuilds/1b2788/README.json b/protoBuilds/1b2788/README.json
index 4b4bc3d7ca..733c87ae85 100644
--- a/protoBuilds/1b2788/README.json
+++ b/protoBuilds/1b2788/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"DNA Extraction"
@@ -10,7 +10,7 @@
"internal": "1b2788",
"labware": "\nOpentrons 200ul Filter tips\nNEST 12-Well Reservoirs, 15 mL\nNEST 1-Well Reservoirs, 195 mL\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nZymo 96 well plate 1.2mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* DNA Extraction\n\n",
"deck-setup": "\n\n",
"description": "This protocol performs the Zymo Quick-DNA Fecal/Soil Microbe 96 Magbead Kit. Please find a description of the kit below from [Zymo's website](https://www.zymoresearch.com/collections/quick-dna-fecal-soil-microbe-kits/products/quick-dna-fecal-soil-microbe-96-magbead-kit):\n\nThe Quick-DNA Fecal/Soil Microbe 96 Magbead Kits are designed for the simple and rapid isolation of inhibitor-free, PCR-quality host cell and microbial DNA from a variety of sample sources, including humans, birds, rats, mice, cattle, etc. The procedure is easy and can be completed in as little as 90 minutes: fecal samples are rapidly and efficiently lysed by bead beating with our state of the art, ultra-high density BashingBeads. Zymo MagBinding Bead technology, which features Zymo Research's Inhibitor Removal technology, is then used to isolate the DNA. Eluted DNA is ideal for downstream molecular-based applications including PCR, arrays, genotyping, methylation detection, etc.\n\nExplanation of complex parameters below:\n* `Number of Columns`: Specify the number of sample columns in this run (1-12).\n* `Pre-Wash buffer volume (ul)`: Specify wash volumes for both the pre-wash buffer, and gDNA wash buffer in microliters. Volumes can be greater than 200ul, and will be split if need be over multiple transfers.\n* `Magnetic Module Engage Height`: Specify the magnetic module engage height in mm.\n* `Elute Buffer Volume`: Specify the elute buffer volume in microliters.\n* `P300 Multi-Channel Pipette Mount`: Specify which mount (left or right) to host the P300 multi-channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/1bcd67/README.json b/protoBuilds/1bcd67/README.json
index b718f6ddcf..3f005c3936 100644
--- a/protoBuilds/1bcd67/README.json
+++ b/protoBuilds/1bcd67/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "1bcd67",
"labware": "\nThermoFisher Abgene\u2122 96 Well 2.2mL Polypropylene Deepwell Storage Plate\nThermoFisher Nalgene\u2122 Disposable Polypropylene Robotic Reservoirs 300ml\nNunc\u2122 96-Well Polypropylene Storage Microplates\nOpentrons 300\u00b5l and 1000\u00b5l pipette tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"deck-setup": "\n\n---\n\n",
"description": "With this flexible protocol, you can normalize the concentrations of up to 32 samples. Just upload your properly formatted .json files (keep scrolling for an example), customize your parameters, and download your ready-to-run protocol.\n\n---\n\n",
diff --git a/protoBuilds/1c086c/README.json b/protoBuilds/1c086c/README.json
index a2167b8c2e..c0da15f179 100644
--- a/protoBuilds/1c086c/README.json
+++ b/protoBuilds/1c086c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Distribution"
@@ -8,7 +8,7 @@
"description": "The distribute function in our Python API allows volumes from the same source well to be combined within the same tip and allow for multiple dispenses to multiple locations. However, sometimes additional parameters may be necessary such as overage volume, custom blowout locations, and air gaps. This protocol provides a custom distribute function that can be used to implement additional parameters and custom functionalities that fall outside of the default Python API distribute function.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Single-Channel Pipette and corresponding Tips\nCorning 24 Well Plate 3.4 mL Flat\nCorning 6 Well Plate 16.8 mL Flat\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nUsage: distribute_custom(pipette, vol, source, dest, overage, blowout, air_gap)\nInput:\ndistribute_custom(p300, 50, plate1['A1'], plate2.rows_by_name()['A'], 30, trash_plate['A1'], 10)\nOutput:\nPicking up tip from A1 of Opentrons 96 Tip Rack 300 \u00b5L on 4\nAspirating 230.0 uL from A1 of Plate 1 on 1 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A1 of Plate 1 on 1 at 92.86 uL/sec\nDispensing 60.0 uL into A1 of Plate 2 on 2 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A1 of Plate 2 on 2 at 92.86 uL/sec\nDispensing 60.0 uL into A2 of Plate 2 on 2 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A2 of Plate 2 on 2 at 92.86 uL/sec\nDispensing 60.0 uL into A3 of Plate 2 on 2 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A3 of Plate 2 on 2 at 92.86 uL/sec\nDispensing 60.0 uL into A4 of Plate 2 on 2 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A4 of Plate 2 on 2 at 92.86 uL/sec\nBlowout at A1 of Trash Plate on 3\nDispensing 300.0 uL into A1 of Trash Plate on 3 at 92.86 uL/sec\nAspirating 130.0 uL from A1 of Plate 1 on 1 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A1 of Plate 1 on 1 at 92.86 uL/sec\nDispensing 60.0 uL into A5 of Plate 2 on 2 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A5 of Plate 2 on 2 at 92.86 uL/sec\nDispensing 60.0 uL into A6 of Plate 2 on 2 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A6 of Plate 2 on 2 at 92.86 uL/sec\nBlowout at A1 of Trash Plate on 3\nDispensing 300.0 uL into A1 of Trash Plate on 3 at 92.86 uL/sec\nDropping tip into A1 of Opentrons Fixed Trash on 12\nIn this example 50 uL are distributed to wells across row A on plate2 from well A1 in plate1. The function will account for the 30 uL overage volume as well as the 10 uL air gaps after each dispense step. It will take those parameters into consideration and determine the maximum volume to aspirate before performing multiple dispenses. Once it reaches the maximum dispenses, it will blowout into the specified blowout location: trash_plate['A1'] (A1 of the Trash Plate)",
"internal": "1c086c",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Distribution\n\n",
"description": "The distribute function in our Python API allows volumes from the same source well to be combined within the same tip and allow for multiple dispenses to multiple locations. However, sometimes additional parameters may be necessary such as overage volume, custom blowout locations, and air gaps. This protocol provides a custom distribute function that can be used to implement additional parameters and custom functionalities that fall outside of the default Python API distribute function.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes) and corresponding [Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Corning 24 Well Plate 3.4 mL Flat](https://ecatalog.corning.com/life-sciences/b2c/US/en/Microplates/Assay-Microplates/96-Well-Microplates/Costar%C2%AE-Multiple-Well-Cell-Culture-Plates/p/3738)\n* [Corning 6 Well Plate 16.8 mL Flat](https://ecatalog.corning.com/life-sciences/b2c/US/en/Microplates/Assay-Microplates/96-Well-Microplates/Costar%C2%AE-Multiple-Well-Cell-Culture-Plates/p/3335)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n---\n\n\n**Usage:** `distribute_custom(pipette, vol, source, dest, overage, blowout, air_gap)`\n\nInput:\n```\ndistribute_custom(p300, 50, plate1['A1'], plate2.rows_by_name()['A'], 30, trash_plate['A1'], 10)\n```\n\nOutput:\n```\nPicking up tip from A1 of Opentrons 96 Tip Rack 300 \u00b5L on 4\nAspirating 230.0 uL from A1 of Plate 1 on 1 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A1 of Plate 1 on 1 at 92.86 uL/sec\nDispensing 60.0 uL into A1 of Plate 2 on 2 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A1 of Plate 2 on 2 at 92.86 uL/sec\nDispensing 60.0 uL into A2 of Plate 2 on 2 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A2 of Plate 2 on 2 at 92.86 uL/sec\nDispensing 60.0 uL into A3 of Plate 2 on 2 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A3 of Plate 2 on 2 at 92.86 uL/sec\nDispensing 60.0 uL into A4 of Plate 2 on 2 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A4 of Plate 2 on 2 at 92.86 uL/sec\nBlowout at A1 of Trash Plate on 3\nDispensing 300.0 uL into A1 of Trash Plate on 3 at 92.86 uL/sec\nAspirating 130.0 uL from A1 of Plate 1 on 1 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A1 of Plate 1 on 1 at 92.86 uL/sec\nDispensing 60.0 uL into A5 of Plate 2 on 2 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A5 of Plate 2 on 2 at 92.86 uL/sec\nDispensing 60.0 uL into A6 of Plate 2 on 2 at 92.86 uL/sec\nAir Gap\nAspirating 10.0 uL from A6 of Plate 2 on 2 at 92.86 uL/sec\nBlowout at A1 of Trash Plate on 3\nDispensing 300.0 uL into A1 of Trash Plate on 3 at 92.86 uL/sec\nDropping tip into A1 of Opentrons Fixed Trash on 12\n```\n\nIn this example 50 uL are distributed to wells across row A on `plate2` from well A1 in `plate1`. The function will account for the 30 uL overage volume as well as the 10 uL air gaps after each dispense step. It will take those parameters into consideration and determine the maximum volume to aspirate before performing multiple dispenses. Once it reaches the maximum dispenses, it will blowout into the specified blowout location: `trash_plate['A1']` (A1 of the Trash Plate)\n\n\n",
"internal": "1c086c",
diff --git a/protoBuilds/1c3611/README.json b/protoBuilds/1c3611/README.json
index ba6a1b8f86..d13895c870 100644
--- a/protoBuilds/1c3611/README.json
+++ b/protoBuilds/1c3611/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "1c3611",
"labware": "\nOpentrons 24 Tube Rack with Eppendorf 2 mL Safe-Lock Snapcap\nCorning 384 Well Plate 112 \u00b5L Flat #3640\nBio-Rad 96 Well Plate 200 \u00b5L PCR #hsp9601\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a custom 384-well PCR preparation. The user has the option to enter the following parameters:\n* number of samples\n* volume of sample\n* number of mastermixes\n* volume of mastermix\n* number of replicates per sample + mastermix combination\n\nMastermixes should be pre-mixed and loaded into individual tubes as shown below. For efficiency, the mastermixes will first be plated from their source tubes into distribution columns using a single channel pipette, before being loaded into the 384-well plate with an 8-channel pipette.\n\nFor sample traceability, an output .csv file will be written to the robot's Jupyter notebook server for each protocol run. The user has the option to input a `run ID` to distinguish each run. To find these output files, please refer to [this support article](https://support.opentrons.com/s/article/Uploading-files-through-Jupyter-Notebook).\n\n\n",
diff --git a/protoBuilds/1c8468/README.json b/protoBuilds/1c8468/README.json
index c07f998efc..cd28191602 100644
--- a/protoBuilds/1c8468/README.json
+++ b/protoBuilds/1c8468/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol transfers samples from a source plate to one of two target plates (to later undergo PCR) as specified in the uploaded CSV file. Samples from each well in the source plate are distributed to its corresponding target plate 6 columns apart. In the case that one of the target plates is filled, the user is prompted to replace it and resume the protocol. In the case that the source plate is depleted, the user is prompted to replace the source plate and upload its respective CSV file to this webpage, download, and run again. In the latter case, the OT-2 \"remembers\" what wells on the target plate are already filled when a new source plate is introduced (unless user resets the counter - see below). User will also be prompted to replace tip racks during a run if applicable.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP20 Single Pipette\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nThermoScientific 96-well Plate 200\u00b5l\n\n\n\nFor this protocol, be sure that the pipettes (P20 and P300) are attached.\nUsing the customization fields below, set up your protocol.\n Source Plate CSV: Upload a CSV corresponding with the source plate loaded onto the deck.\n Transfer Volume: Specify the volume (\u00b5L) desired for each well in target plates\n P20 Single Mount: Specify which mount the P10 is on (left or right).\n Reset Counter: Reset the counter if running with empty target plates. Do not reset the counter if wells in target plate from last run are to be tracked.\nNote about CSV\nThe CSV should be formatted like so:\nSamples ID | Sample Type | ct_value | Plate Name (ex. A:1) | Plate Position | Target Plate (25 or 30) | Transfer (\"done\" or leave blank)\nThe first row should contain headers (like above). All of the following rows should just have the necessary information.\nLabware Setup\nSlots 1: Custom Endura Source Plate on Opentrons 96 well Aluminum Block\nSlot 2: ThermoScientific 96-well 25-Cycle Target Plate\nSlot 4: ThermoScientific 96-well 30-Cycle Target Plate\nSlot 5: Opentrons 96 Filter Tip Rack 20 \u00b5L",
"internal": "1c8468",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol transfers samples from a source plate to one of two target plates (to later undergo PCR) as specified in the uploaded CSV file. Samples from each well in the source plate are distributed to its corresponding target plate 6 columns apart. In the case that one of the target plates is filled, the user is prompted to replace it and resume the protocol. In the case that the source plate is depleted, the user is prompted to replace the source plate and upload its respective CSV file to this webpage, download, and run again. In the latter case, the OT-2 \"remembers\" what wells on the target plate are already filled when a new source plate is introduced (unless user resets the counter - see below). User will also be prompted to replace tip racks during a run if applicable.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P20 Single Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [Opentrons 96 Filter Tip Rack 20 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_20ul?category=tipRack)\n* ThermoScientific 96-well Plate 200\u00b5l\n\n\n\n\n\n\n---\n\n\nFor this protocol, be sure that the pipettes (P20 and P300) are attached.\n\nUsing the customization fields below, set up your protocol.\n* Source Plate CSV: Upload a CSV corresponding with the source plate loaded onto the deck.\n* Transfer Volume: Specify the volume (\u00b5L) desired for each well in target plates\n* P20 Single Mount: Specify which mount the P10 is on (left or right).\n* Reset Counter: Reset the counter if running with empty target plates. Do not reset the counter if wells in target plate from last run are to be tracked.\n\n**Note about CSV**\n\nThe CSV should be formatted like so:\n\n`Samples ID` | `Sample Type` | `ct_value` | `Plate Name (ex. A:1)` | `Plate Position` | `Target Plate (25 or 30)` | `Transfer (\"done\" or leave blank)`\n\nThe first row should contain headers (like above). All of the following rows should just have the necessary information.\n\n**Labware Setup**\n\nSlots 1: Custom Endura Source Plate on Opentrons 96 well Aluminum Block\n\nSlot 2: ThermoScientific 96-well 25-Cycle Target Plate\n\nSlot 4: ThermoScientific 96-well 30-Cycle Target Plate\n\nSlot 5: Opentrons 96 Filter Tip Rack 20 \u00b5L\n\n\n",
"internal": "1c8468\n",
diff --git a/protoBuilds/1ccd23-station-A/README.json b/protoBuilds/1ccd23-station-A/README.json
index 64cbb406aa..6ebdd6aeee 100644
--- a/protoBuilds/1ccd23-station-A/README.json
+++ b/protoBuilds/1ccd23-station-A/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Covid Workstation": [
"Sample Plating"
@@ -8,7 +8,7 @@
"description": "This protocol plates samples from custom or standard tuberacks to a NEST 96-deepwell plate. 10\u00b5l proteinase K and 10\u00b5l internal control are then added to each sample. Samples are transferred to the plate first down each column, then across each row (A1, B1, C1, ...H1, A2, B2, C2, etc.)\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons Single-Channel Pipette\nOpentrons Filter Tips\nOpentrons 4-in-1 Tube Rack Set or custom tuberack\nNEST 96 Deepwell Plate 2mL\nSample Tubes\n\n\n\n2ml tubeblock on temperature module (slot 1):\n proteinase K: A1\n internal control: B1",
"internal": "generic_station_A",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Covid Workstation\n\t* Sample Plating\n\n\n",
"description": "This protocol plates samples from custom or standard tuberacks to a NEST 96-deepwell plate. 10\u00b5l proteinase K and 10\u00b5l internal control are then added to each sample. Samples are transferred to the plate first down each column, then across each row (A1, B1, C1, ...H1, A2, B2, C2, etc.)\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons Filter Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) or custom tuberack\n* [NEST 96 Deepwell Plate 2mL](http://www.cell-nest.com/page94?product_id=101&_l=en)\n* Sample Tubes\n\n\n---\n\n\n2ml tubeblock on temperature module (slot 1): \n* proteinase K: A1\n* internal control: B1\n\n",
"internal": "generic_station_A\n",
diff --git a/protoBuilds/1ccd23-station-B/README.json b/protoBuilds/1ccd23-station-B/README.json
index ede79fcffd..2ea3fdbf13 100644
--- a/protoBuilds/1ccd23-station-B/README.json
+++ b/protoBuilds/1ccd23-station-B/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons (verified)",
+ "author": "Opentrons (verified)\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Covid Workstation": [
"RNA Extraction"
@@ -8,7 +8,7 @@
"description": "This is a flexible protocol accommodating a wide range of commercial RNA extraction workflows for COVID-19 sample processing. The protocol is broken down into 5 main parts:\n binding buffer addition to samples\n bead wash 3x using magnetic module\n* final elution to chilled PCR plate\nLysed samples should be loaded on the magnetic module in a NEST 96-deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 4) to the elution PCR plate (temperature module, slot 1). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\nExplanation of complex parameters below:\n park tips: If set to yes (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to no, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n track tips across protocol runs: If set to yes, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to no, tips will be picked up from column 1 of the 1st tiprack.\n* flash: If set to yes, the robot rail lights will flash during any automatic pauses in the protocol. If set to no, the lights will not flash.\n\n \nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons Magnetic Module GEN2\nOpentrons Temperature Module GEN2\nNEST 12 Well Reservoir 15 mL\nNEST 1 Well Reservoir 195 mL\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt\nNEST 96 Deepwell Plate 2mL\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n\n\n\n\nReservoir 1: slot 5\n\nReservoir 2: slot 2\n\n",
"internal": "covid-19-rna-extraction",
"markdown": {
- "author": "[Opentrons (verified)](https://opentrons.com/)\n\n",
+ "author": "[Opentrons (verified)](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Covid Workstation\n * RNA Extraction\n\n",
"description": "This is a flexible protocol accommodating a wide range of commercial RNA extraction workflows for COVID-19 sample processing. The protocol is broken down into 5 main parts:\n* binding buffer addition to samples\n* bead wash 3x using magnetic module\n* final elution to chilled PCR plate\n\nLysed samples should be loaded on the magnetic module in a NEST 96-deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 4) to the elution PCR plate (temperature module, slot 1). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\nExplanation of complex parameters below:\n* `park tips`: If set to `yes` (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to `no`, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n* `flash`: If set to `yes`, the robot rail lights will flash during any automatic pauses in the protocol. If set to `no`, the lights will not flash.\n\n---\n\n \n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons Magnetic Module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons Temperature Module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [NEST 12 Well Reservoir 15 mL](https://labware.opentrons.com/nest_12_reservoir_15ml)\n* [NEST 1 Well Reservoir 195 mL](https://labware.opentrons.com/nest_1_reservoir_195ml)\n* [NEST 96 Well Plate 100 \u00b5L PCR Full Skirt](https://labware.opentrons.com/nest_96_wellplate_100ul_pcr_full_skirt)\n* [NEST 96 Deepwell Plate 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep)\n* [Opentrons 96 Filter Tip Rack 200 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n\n---\n\n\n* Reservoir 1: slot 5 \n\n* Reservoir 2: slot 2 \n\n\n",
"internal": "covid-19-rna-extraction\n",
diff --git a/protoBuilds/1ccd23-station-C/README.json b/protoBuilds/1ccd23-station-C/README.json
index 6ecdc9bec4..26bb592a4e 100644
--- a/protoBuilds/1ccd23-station-C/README.json
+++ b/protoBuilds/1ccd23-station-C/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Covid Workstation": [
"qPCR Setup"
@@ -8,7 +8,7 @@
"description": "This protocol creates a custom qPCR prep protocol for Covid-19 diagnostics. The input into this protocol is an elution plate of purified RNA, and the output is a PCR plate containing the samples mixed with mastermix.\nUsing a Single-Channel Pipette, this protocol will begin by creating reaction mix in a 2ml tube. The mix is then distributed to a PCR strip. Then, using the Multi-Channel Pipette, samples will be transferred from their plate to the qPCR plate and mixed with the reaction mix.\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.21.0 or later)\nOpentrons Temperature Module with Aluminum Block Set)\nOpentrons P300 Single-Channel GEN2 Pipette\nOpentrons P20 Multi-Channel GEN2 Pipette\nOpentrons Filter Tips\nNEST PCR Plate containing purified nucleic acid samples\n2ml NEST screwcap tubes or equivalent containing reaction mix(es)\n\n\n\nchilled 24-well aluminum block for 2ml tubes (slot 5):\n\nAllplex 2019-nCoV Assay (17\u00b5l/sample total):\n reagent 1: 2019-nCov MOM (5\u00b5l/sample)\n reagent 2: RNase-free water (5\u00b5l/sample)\n reagent 3: 5X Real-time One-step Buffer (5\u00b5l/sample)\n reagent 4: Real-time One-step Enzyme (2\u00b5l/sample)\nAllplex SARS-CoV-2 Assay (15\u00b5l/sample total):\n reagent 1: SARS2 MOM (5\u00b5l/sample)\n reagent 2: EM8 (5\u00b5l/sample)\n* reagent 3: RNase-free water (5\u00b5l/sample)\nSeegene Real-time One-step RT-PCR:\n reagent 1: SC2FabR MOM (5\u00b5l/sample)\n reagent 2: EM8 (5\u00b5l/sample)",
"internal": "1ccd23",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Covid Workstation\n\t* qPCR Setup\n\n\n",
"description": "This protocol creates a custom qPCR prep protocol for Covid-19 diagnostics. The input into this protocol is an elution plate of purified RNA, and the output is a PCR plate containing the samples mixed with mastermix.\nUsing a [Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette), this protocol will begin by creating reaction mix in a 2ml tube. The mix is then distributed to a PCR strip. Then, using the [Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette), samples will be transferred from their plate to the qPCR plate and mixed with the reaction mix.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.21.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [Aluminum Block Set](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set))\n* [Opentrons P300 Single-Channel GEN2 Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons P20 Multi-Channel GEN2 Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons Filter Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) containing purified nucleic acid samples\n* [2ml NEST screwcap tubes or equivalent](https://shop.opentrons.com/collections/tubes/products/nest-microcentrifuge-tubes) containing reaction mix(es)\n\n\n---\n\n\nchilled 24-well aluminum block for 2ml tubes (slot 5): \n\n\nAllplex 2019-nCoV Assay (17\u00b5l/sample total): \n* reagent 1: 2019-nCov MOM (5\u00b5l/sample)\n* reagent 2: RNase-free water (5\u00b5l/sample)\n* reagent 3: 5X Real-time One-step Buffer (5\u00b5l/sample)\n* reagent 4: Real-time One-step Enzyme (2\u00b5l/sample)\n\nAllplex SARS-CoV-2 Assay (15\u00b5l/sample total): \n* reagent 1: SARS2 MOM (5\u00b5l/sample)\n* reagent 2: EM8 (5\u00b5l/sample)\n* reagent 3: RNase-free water (5\u00b5l/sample)\n\nSeegene Real-time One-step RT-PCR: \n* reagent 1: SC2FabR MOM (5\u00b5l/sample)\n* reagent 2: EM8 (5\u00b5l/sample)\n\n",
"internal": "1ccd23\n",
diff --git a/protoBuilds/1ce6aa/README.json b/protoBuilds/1ce6aa/README.json
index 05101fc6da..46861c2e2b 100644
--- a/protoBuilds/1ce6aa/README.json
+++ b/protoBuilds/1ce6aa/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol is designed for transferring buffered peptone water to BHI with P300 Multi-Channel Pipette. The user has the ability to select the number of samples to run.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP300 Multi Channel Pipette\nFisherbrand 2-20\u00b5L Tips\nSimport Scientific 500\u00b5L Tubes in Custom Holder\nReagents\n\n\n\nFor this protocol, be sure that the correct pipette (P300-Multi) is attached for the intended test case.\nLabware Setup\nSlot 1: Fisherbrand 2-20\u00b5L Tips\nSlot 2: Simport Scientific 500\u00b5L Tubes in Custom Holder (Source)\nSlot 3: Simport Scientific 500\u00b5L Tubes in Custom Holder (Destination)\nUsing the customization fields below, set up your protocol.\n Pipette Mount: Specify which mount the P300-Multi is on (left or right).\n Number of Samples: Specify how many samples should be transferred (1-96).\n",
"internal": "1ce6aa",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol is designed for transferring buffered peptone water to BHI with P300 Multi-Channel Pipette. The user has the ability to select the number of samples to run.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P300 Multi Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Fisherbrand 2-20\u00b5L Tips](https://www.fishersci.com/shop/products/fisherbrand-sureone-aerosol-barrier-pipette-tips-14/02707432)\n* [Simport Scientific 500\u00b5L Tubes in Custom Holder](http://www.simport.com/en/products/173-t100-1.html)\n* Reagents\n\n\n\n---\n\n\n\nFor this protocol, be sure that the correct pipette (P300-Multi) is attached for the intended test case.\n\n**Labware Setup**\n\nSlot 1: [Fisherbrand 2-20\u00b5L Tips](https://www.fishersci.com/shop/products/fisherbrand-sureone-aerosol-barrier-pipette-tips-14/02707432)\n\nSlot 2: [Simport Scientific 500\u00b5L Tubes in Custom Holder (Source)](http://www.simport.com/en/products/173-t100-1.html)\n\nSlot 3: [Simport Scientific 500\u00b5L Tubes in Custom Holder (Destination)](http://www.simport.com/en/products/173-t100-1.html)\n\n**Using the customization fields below, set up your protocol.**\n* Pipette Mount: Specify which mount the P300-Multi is on (left or right).\n* Number of Samples: Specify how many samples should be transferred (1-96).\n\n---\n",
"internal": "1ce6aa\n",
diff --git a/protoBuilds/1d37e5/README.json b/protoBuilds/1d37e5/README.json
index cd5dd92efa..1a96621836 100644
--- a/protoBuilds/1d37e5/README.json
+++ b/protoBuilds/1d37e5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol automates the addition of three different reagents to 96-well plate(s) with the p20 8-channel pipette.\n\nFirst, 10\u00b5L of Reagent 1 (Polymixin B 256) will be added to columns 1-6 of each plate. Next, 10\u00b5L of Reagent 2 (Polymixin B 128) will be added to columns 6-12 of each plate. Finally, 10\u00b5L of Reagent 3 (Growth Media Gamma) will be added to all of the wells.\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.21.0 or later)\nOpentrons p20 8-Channel Pipette (on the right mount)\nOpentrons 20\u00b5L Filter Tips\nNEST 96-Well Deep Well Plate\nHiMic 96-Well Plates, 400\u00b5L\nReagents\n\n\n\nDeck Layout\n\nSlot 11: NEST 96-Well Deep Well Plate\nA1: Reagent 1 (Polymixin B 256)\nA2: Reagent 2 (Polymixin B 128)\nA3: Reagent 3 (Growth Media Gamma)\n\nSlot 10: Opentrons 20\u00b5L Filter Tips\n\nSlots 1-9: HiMic 96-Well Plates, 400\u00b5L\n\nUsing the customizations field (below), set up your protocol.\n* Number of Plates: Specify the number of plates to fill during the protocol.",
"internal": "1d37e5",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol automates the addition of three different reagents to 96-well plate(s) with the p20 8-channel pipette.\n\nFirst, 10\u00b5L of Reagent 1 (Polymixin B 256) will be added to columns 1-6 of each plate. Next, 10\u00b5L of Reagent 2 (Polymixin B 128) will be added to columns 6-12 of each plate. Finally, 10\u00b5L of Reagent 3 (Growth Media Gamma) will be added to all of the wells.\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.21.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons p20 8-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette) (on the right mount)\n* [Opentrons 20\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 96-Well Deep Well Plate](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate)\n* HiMic 96-Well Plates, 400\u00b5L\n* Reagents\n\n\n\n---\n\n\n**Deck Layout**\n\nSlot 11: [NEST 96-Well Deep Well Plate](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate)\nA1: Reagent 1 (Polymixin B 256)\nA2: Reagent 2 (Polymixin B 128)\nA3: Reagent 3 (Growth Media Gamma)\n\nSlot 10: [Opentrons 20\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlots 1-9: HiMic 96-Well Plates, 400\u00b5L\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Plates**: Specify the number of plates to fill during the protocol.\n\n\n\n\n",
"internal": "1d37e5\n",
diff --git a/protoBuilds/1d6d1b/README.json b/protoBuilds/1d6d1b/README.json
index aa5f3793ea..173deb330b 100644
--- a/protoBuilds/1d6d1b/README.json
+++ b/protoBuilds/1d6d1b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Slide Spotting"
@@ -8,7 +8,7 @@
"description": "This is a protocol developed for automating a custom slide-spotting workflow.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP300 Multi-Channel Pipette (GEN2)\nAxygen 4-channel reservoir 73mL\nAbgene 96-wellplate 200\u00b5L\n\n",
"internal": "1d6d1b",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Slide Spotting\n\n\n",
"description": "This is a protocol developed for automating a custom slide-spotting workflow.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P300 Multi-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* Axygen 4-channel reservoir 73mL\n* Abgene 96-wellplate 200\u00b5L\n\n---\n",
"internal": "1d6d1b\n",
diff --git a/protoBuilds/1dbc24/README.json b/protoBuilds/1dbc24/README.json
index fcb764ba4e..56e17a7c9a 100644
--- a/protoBuilds/1dbc24/README.json
+++ b/protoBuilds/1dbc24/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "1dbc24",
"labware": "\nElectron Microscopy Sciences 1-Well Reservoir 130mL, #70314-01\nCorning 96 Well Plate 360 \u00b5L Flat #3650\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "\nThis protocol performs a simple reagent addition for WISTD and water to up to 96 samples in a 96-flat well plate.\n\n---\n\n",
diff --git a/protoBuilds/1dec68/README.json b/protoBuilds/1dec68/README.json
index a6278c2f4b..ffa10d0ad2 100644
--- a/protoBuilds/1dec68/README.json
+++ b/protoBuilds/1dec68/README.json
@@ -17,14 +17,14 @@
"internal": "1dec68\n",
"labware": "* Bio-Rad 50ul 96 Well Plate\n* (Opentrons 300uL tips)[https://shop.opentrons.com/universal-filter-tips/]\n* Custom 96 tube rack\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Partner Name](partner website link)\n\n",
+ "partner": "[Partner Name](partner website link)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P300 Single-Channel Pipette](https://shop.opentrons.com/pipettes/)\n\n---\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Mount the needle on position 10.\n2. Extract 50\u00b5l of sample (contain 5ml/sample).\n3. Pipette 50\u00b5l to standard BIORAD 96 plate (with 50\u00b5l).\n4. Discard the needle on trash.\n\n",
"title": "Covid Sample Prep with Custom 96 Tube Rack"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Partner Name",
+ "partner": "Partner Name\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP300 Single-Channel Pipette\n\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nMount the needle on position 10.\nExtract 50\u00b5l of sample (contain 5ml/sample).\nPipette 50\u00b5l to standard BIORAD 96 plate (with 50\u00b5l).\nDiscard the needle on trash.\n",
diff --git a/protoBuilds/1dfebe/README.json b/protoBuilds/1dfebe/README.json
index d42177ef56..9ec27f159d 100644
--- a/protoBuilds/1dfebe/README.json
+++ b/protoBuilds/1dfebe/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "1dfebe",
"labware": "\nOpentrons 4-in-1 Tube Rack\nOpentrons 300uL Tips\nNEST 100ul 96 well plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "This protocol preps a 96 well plate with urine and enzyme hydrolysis, as well as trichloroacetic acid and dilution buffer. The urine samples in the 4 tube racks exactly mimic the 96 well plate with 15 controls already put in by column. The sample pick-off begins from D2 in the tuberack on slot one, corresponding to H2 of the well plate in slot 3 for dispensing. Delays and pauses are included in the protocol to allow for incubation periods and mix steps.\n\nExplanation of complex parameters below:\n* `Use csv for this run?`: Specify whether a csv file will be uploaded for this run. If not, nothing in `.CSV File` below will be referenced.\n* `.CSV File`: Upload the csv file for this run. The csv file will map which samples the pipette will visit the bottom of the tube. Wells marked with `X` will be aspirated from the `Sample Aspiration Height` parameter (see below). Otherwise, wells marked with `O` (capital letter O, not zero) will be aspirated from the bottom of the tube. Refer to the csv sample below.\n\n* `Number of Samples`: Specify the number of samples (1-81) for this run.\n* `Tip Withdrawal Speed`: Specify the tip withdrawal speed for the tip to leave the urine samples.\n* `Sample Aspiration Height (mm)`: Specify the sample aspiration height (in mm) from the bottom of the urine samples to aspirate from. Any well marked `X` in the csv, if uploaded will use this height. If the csv is not uploaded, all urine samples will use this height.\n* `Sample Aspiration Height (mm)`: Specify the dispense height from the bottom of the 96 well plate to dispense sample. \n* `P300 Multi-Channel Mount`: Specify which mount (left or right) to host the P300 single channel pipette.\n\n\n---\n\n",
diff --git a/protoBuilds/1e744e/README.json b/protoBuilds/1e744e/README.json
index c3dfbb472c..8591d68ec5 100644
--- a/protoBuilds/1e744e/README.json
+++ b/protoBuilds/1e744e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -9,7 +9,7 @@
"description": "This protocol uses a p20 multi-channel pipette to distribute (15 uL by default, or optionally 10 uL) BioGX XFree mastermix BioGX XFree Mastermix from columns of a 96-well plate held at 4 degrees on an Opentrons Temperature Module to columns of a reaction plate (either 96 or 384-well plate) on a second Opentrons Temperature Module also at 4 degrees. The p20 multi is then used to transfer 5 uL viral RNA input sample (sample count can be anywhere between 1 and 384 samples) from up to four 96-well sample plates to the reaction plate wells containing mastermix.",
"internal": "1e744e",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * PCR Prep\n\n",
"deck-setup": "\n\n* Opentrons 20ul filter tips (deck slots 7-11)\n* Reaction Plate 96-well PCR plate (on Temperature Module deck slot 1)\n* Mastermix Plate 96-well PCR Plate (on Temperature Module deck slot 3)\n* Sample Plates 96-well PCR Plates (deck slots 2, 4, 5, 6)\n\n",
"description": "\nThis protocol uses a p20 multi-channel pipette to distribute (15 uL by default, or optionally 10 uL) BioGX XFree mastermix [BioGX XFree Mastermix](https://www.biogx.com/xfree/) from columns of a 96-well plate held at 4 degrees on an Opentrons Temperature Module to columns of a reaction plate (either 96 or 384-well plate) on a second Opentrons Temperature Module also at 4 degrees. The p20 multi is then used to transfer 5 uL viral RNA input sample (sample count can be anywhere between 1 and 384 samples) from up to four 96-well sample plates to the reaction plate wells containing mastermix.\n\n",
diff --git a/protoBuilds/1eeb01/README.json b/protoBuilds/1eeb01/README.json
index eae152738f..56b3952fc9 100644
--- a/protoBuilds/1eeb01/README.json
+++ b/protoBuilds/1eeb01/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Extraction"
@@ -8,7 +8,7 @@
"description": "This protocol is a custom nucleic acid extraction that begins with the samples in 1.5mL tubes. This protocol utilizes the Opentrons Magnetic Module and the Opentrons Temperature Module for this extraction.\nThe protocol begins by adding 500\u00b5L of Buffer 1 to all tubes that will contain samples on the temperature module. Once complete, 500\u00b5L of sample is transferred from 1.5mL tubes in an Opentrons 4-in-1 Tube Rack to the tubes containing Buffer 1.\nAfter a heating step, DNA and Magnetic Beads are added to the sample tubes on the temperature module and the samples are transferred to a deepwell plate on the magnetic module.\nOnce on the magnetic module, the magnet is engaged and after the beads settle, the supernatant is removed. Using Buffer 2, three consecutive wash steps occur with the samples on the magnetic module.\nAt this point, the user will be prompted to replace the tube(s) in the Tube Rack with Buffer 3 and the Reaction Mix (more info on this below). Buffer 3 will then be added to the beads, mixed with the beads, then transferred back to the temperature module for a 5 minute incubation at 65C.\nPost-incubation, the samples will be moved back to the deepwell plate on the magnetic module, but in a new, clean well. The magnet will engage after all of the samples have been transferred. Following this, the reaction mix will be distributed to all of destinations in PCR strips. The elution will then be transferred to a clean PCR strip, before 5\u00b5L gets transferred to two tubes containing the reaction mix.\nThis concludes the OT-2 portion of the protocol and the user will be prompted to move the samples to 65c for a 20 minute incubation to complete the protocol. \nAdjust the parameters and download the protocol below. \nNote: This protocol is still a work in progress and will likely need further optimizations; please keep this in mind when using this protocol. \n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nP20 Single-Channel Pipette (left mount)\nP300 or P1000 Single-Channel Pipette (right mount)\nOpentrons 20\u00b5L Filter Tips\nOpentrons Filters Tips for P300 or P1000\nOpentrons Magnetic Module, GEN2\nOpentrons Temperature Module, GEN2\nOpentrons 4-in-1 Tube Rack\nNEST 12-Well Reservoir, 15mL\nNEST 1-Well Reservoir, 195mL\nNEST 96-Deepwell Plate, 2mL\nNEST 1.5mL Centrifuge Tubes\nPCR Strips\nReagents\nSamples\n\n\n\nDeck Layout \nSlot 1: Opentrons 96-Well Aluminum Block with PCR Strips\nA1: Magnetic Beads\nB1: DNA\nColumns 2-4: Elution Destination (empty to begin)\nColumns 7-12: Reaction Mix + Elution Destination (empty to begin) \nSlot 2: Opentrons 20\u00b5L Filter Tips \nSlot 3: Opentrons Filters Tips for P300 or P1000 (Tiprack 3) \nSlot 4: Opentrons 4-in-1 Tube Rack with NEST 1.5mL Centrifuge Tubes\nNote: At the beginning of the protocol, samples should be loaded in the tube rack. Once transferred:\nA1: Buffer 3\nA4: Reaction Mix \nSlot 5: Opentrons 20\u00b5L Filter Tips \nSlot 6: Opentrons Filters Tips for P300 or P1000 (Tiprack 2) \nSlot 7: Opentrons Temperature Module, GEN2 with [Opentrons 24-Well Aluminum Block]((https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) containing NEST 1.5mL Centrifuge Tubes \nSlot 8: NEST 12-Well Reservoir, 15mL\nA1: Buffer 1\nA2: Buffer 2 (Wash 1, Samples 1-12)\nA3: Buffer 2 (Wash 1, Samples 13-24)\nA4: Buffer 2 (Wash 2, Samples 1-12)\nA5: Buffer 2 (Wash 2, Samples 13-24)\nA6: Buffer 2 (Wash 3, Samples 1-12)\nA7: Buffer 2 (Wash 3, Samples 13-24) \nSlot 9: Opentrons Filters Tips for P300 or P1000 (Tiprack 1) \nSlot 10: Opentrons Magnetic Module, GEN2 with NEST 96-Deepwell Plate, 2mL \nSlot 11: NEST 1-Well Reservoir, 195mL (for liquid waste) \nUsing the customizations field (below), set up your protocol.\n Pipette & Tip Combo: Select which pipette (right mount) and corresponding tips are being used in this protocol.\n Number of Samples (1-24): Specify the number of samples to run (1-24).",
"internal": "1eeb01",
"markdown": {
- "author": "[Opentrons](http://www.opentrons.com/)\n\n",
+ "author": "[Opentrons](http://www.opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Nucleic Acid Extraction\n\n",
"description": "This protocol is a custom nucleic acid extraction that begins with the samples in 1.5mL tubes. This protocol utilizes the [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) and the [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) for this extraction. \nThe protocol begins by adding 500\u00b5L of Buffer 1 to all tubes that will contain samples on the temperature module. Once complete, 500\u00b5L of sample is transferred from 1.5mL tubes in an [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) to the tubes containing Buffer 1.\nAfter a heating step, DNA and Magnetic Beads are added to the sample tubes on the temperature module and the samples are transferred to a deepwell plate on the magnetic module.\nOnce on the magnetic module, the magnet is engaged and after the beads settle, the supernatant is removed. Using Buffer 2, three consecutive wash steps occur with the samples on the magnetic module. \nAt this point, the user will be prompted to replace the tube(s) in the Tube Rack with Buffer 3 and the Reaction Mix (more info on this below). Buffer 3 will then be added to the beads, mixed with the beads, then transferred back to the temperature module for a 5 minute incubation at 65C. \nPost-incubation, the samples will be moved back to the deepwell plate on the magnetic module, but in a new, clean well. The magnet will engage after all of the samples have been transferred. Following this, the reaction mix will be distributed to all of destinations in PCR strips. The elution will then be transferred to a clean PCR strip, before 5\u00b5L gets transferred to two tubes containing the reaction mix. \nThis concludes the OT-2 portion of the protocol and the user will be prompted to move the samples to 65c for a 20 minute incubation to complete the protocol. \n\nAdjust the parameters and download the protocol below. \n\n*Note*: This protocol is still a work in progress and will likely need further optimizations; please keep this in mind when using this protocol. \n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) (left mount)\n* [P300 or P1000 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) (right mount)\n* [Opentrons 20\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* Opentrons Filters Tips for [P300](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips) or [P1000](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips)\n* [Opentrons Magnetic Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons Temperature Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* [NEST 96-Deepwell Plate, 2mL](https://shop.opentrons.com/collections/verified-labware/products/nest-0-2-ml-96-well-deep-well-plate-v-bottom)\n* [NEST 1.5mL Centrifuge Tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes)\n* PCR Strips\n* Reagents\n* Samples\n\n\n---\n\n\n**Deck Layout** \n\n**Slot 1**: [Opentrons 96-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) with PCR Strips \nA1: Magnetic Beads \nB1: DNA \nColumns 2-4: Elution Destination (empty to begin) \nColumns 7-12: Reaction Mix + Elution Destination (empty to begin) \n\n**Slot 2**: [Opentrons 20\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips) \n\n**Slot 3**: Opentrons Filters Tips for [P300](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips) or [P1000](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips) (Tiprack 3) \n\n**Slot 4**: [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with [NEST 1.5mL Centrifuge Tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes) \n*Note*: At the beginning of the protocol, samples should be loaded in the tube rack. Once transferred: \nA1: Buffer 3 \nA4: Reaction Mix \n\n**Slot 5**: [Opentrons 20\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips) \n\n**Slot 6**: Opentrons Filters Tips for [P300](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips) or [P1000](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips) (Tiprack 2) \n\n**Slot 7**: [Opentrons Temperature Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [Opentrons 24-Well Aluminum Block]((https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) containing [NEST 1.5mL Centrifuge Tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes) \n\n**Slot 8**: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) \nA1: Buffer 1 \nA2: Buffer 2 (Wash 1, Samples 1-12) \nA3: Buffer 2 (Wash 1, Samples 13-24) \nA4: Buffer 2 (Wash 2, Samples 1-12) \nA5: Buffer 2 (Wash 2, Samples 13-24) \nA6: Buffer 2 (Wash 3, Samples 1-12) \nA7: Buffer 2 (Wash 3, Samples 13-24) \n\n**Slot 9**: Opentrons Filters Tips for [P300](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips) or [P1000](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips) (Tiprack 1) \n\n**Slot 10**: [Opentrons Magnetic Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [NEST 96-Deepwell Plate, 2mL](https://shop.opentrons.com/collections/verified-labware/products/nest-0-2-ml-96-well-deep-well-plate-v-bottom) \n\n**Slot 11**: [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml) (for liquid waste) \n\n**Using the customizations field (below), set up your protocol.**\n* **Pipette & Tip Combo**: Select which pipette (right mount) and corresponding tips are being used in this protocol.\n* **Number of Samples (1-24)**: Specify the number of samples to run (1-24).\n\n\n",
"internal": "1eeb01\n",
diff --git a/protoBuilds/1ef67e/README.json b/protoBuilds/1ef67e/README.json
index 66e5111159..744b27bea8 100644
--- a/protoBuilds/1ef67e/README.json
+++ b/protoBuilds/1ef67e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Assay"
@@ -8,7 +8,7 @@
"description": "This protocol represents one of several protocols that prepares a panel of set dilutions of primary antibodies and fluorophores. This protocol creates Panel 1. The following antibodies and fluorophores dilutions are prepared in this panel:\nAntibodies\n CD4\n CD8\n FOXP3\n PD-L1 EL13N\n PAN CK\n KI-67\nFluorophores\n 520\n 570\n 540\n 620\n 650\n 690\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons P10 Single-Channel Pipette\nOpentrons P1000 Single-Channel Pipette\nOpentrons 10\u00b5L Tips\nOpentrons 1000\u00b5L Tips\nCustom Tube Holder for 6mL Tubes and Tubes\nBio-Rad 96 Well Plate\nOpentrons 4-in-1 Tube Rack\nOpentrons 4-in-1 Tube Rack\n50mL Tubes\n2mL Tubes\n15mL Tubes\nAntibodies\nFluorophores\n\n\n\nSlot 1: Opentrons 1000\u00b5L Tips\nSlot 2: Custom 6mL Tube Rack Holder and Tubes\nSlot 3: Bio-Rad 96 Well Plate, for sample aliquots\nSlot 4: Opentrons 10\u00b5L Tips\nSlot 5: Opentrons 4-in-1 Tube Rack; 6x15mL, 4x50mL\n A3: Antibody Dilutent (initial volume=50mL)\n A4: Fluorophore Dilutent (initial volume=50mL)\nSlot 6: Opentrons 4-in-1 Tube Rack; 24x2mL\nSetup with initial volumes\n A1: 520, 75\u00b5L\n B1: 540, 75\u00b5L\n C1: 570, 75\u00b5L\n D1: 620, 75\u00b5L\n A2: 650, 75\u00b5L\n B2: 690, 75\u00b5L\n A3: CD3, 1000\u00b5L\n B3: CD4, 100\u00b5L\n C3: CD8, 100\u00b5L\n D3: CD20, 250\u00b5L\n A4: CD56, 1000\u00b5L\n B4: CD68, 100\u00b5L\n C4: CD163, 100\u00b5L\n D4: PD1, 100\u00b5L\n A5: PD-L1 EL13N, 100\u00b5L\n B5: FOXP3, 500\u00b5L\n C5: PAN CK, 1000\u00b5L\n D5: KI-67, 2000\u00b5L (Source 1)\n A6: KI-67, 2000\u00b5L (Source 2)\n B6: KI-67, 2000\u00b5L (Source 3)\n C6: KI-67, 2000\u00b5L (Source 4)\n D6: KI-67, 2000\u00b5L (Source 5)",
"internal": "1ef67e",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Assay\n\n\n",
"description": "This protocol represents one of several protocols that prepares a panel of set dilutions of primary antibodies and fluorophores. This protocol creates Panel 1. The following antibodies and fluorophores dilutions are prepared in this panel:\n\n**Antibodies**\n* CD4\n* CD8\n* FOXP3\n* PD-L1 EL13N\n* PAN CK\n* KI-67\n\n**Fluorophores**\n* 520\n* 570\n* 540\n* 620\n* 650\n* 690\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P10 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons P1000 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 1000\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* Custom Tube Holder for 6mL Tubes and Tubes\n* [Bio-Rad 96 Well Plate](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate)\n* [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [50mL Tubes](https://shop.opentrons.com/collections/tubes/products/nest-50-ml-centrifuge-tube)\n* [2mL Tubes](https://shop.opentrons.com/collections/tubes/products/nest-2-0-ml-microcentrifuge-tubes)\n* [15mL Tubes](https://shop.opentrons.com/collections/tubes/products/nest-15-ml-centrifuge-tube)\n* Antibodies\n* Fluorophores\n\n\n\n---\n\n\nSlot 1: [Opentrons 1000\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n\nSlot 2: Custom 6mL Tube Rack Holder and Tubes\n\nSlot 3: [Bio-Rad 96 Well Plate](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate), for sample aliquots\n\nSlot 4: [Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\nSlot 5: [Opentrons 4-in-1 Tube Rack; 6x15mL, 4x50mL](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* A3: Antibody Dilutent (initial volume=50mL)\n* A4: Fluorophore Dilutent (initial volume=50mL)\n\nSlot 6: [Opentrons 4-in-1 Tube Rack; 24x2mL](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n*Setup with initial volumes*\n* A1: 520, 75\u00b5L\n* B1: 540, 75\u00b5L\n* C1: 570, 75\u00b5L\n* D1: 620, 75\u00b5L\n* A2: 650, 75\u00b5L\n* B2: 690, 75\u00b5L\n* A3: CD3, 1000\u00b5L\n* B3: CD4, 100\u00b5L\n* C3: CD8, 100\u00b5L\n* D3: CD20, 250\u00b5L\n* A4: CD56, 1000\u00b5L\n* B4: CD68, 100\u00b5L\n* C4: CD163, 100\u00b5L\n* D4: PD1, 100\u00b5L\n* A5: PD-L1 EL13N, 100\u00b5L\n* B5: FOXP3, 500\u00b5L\n* C5: PAN CK, 1000\u00b5L\n* D5: KI-67, 2000\u00b5L (Source 1)\n* A6: KI-67, 2000\u00b5L (Source 2)\n* B6: KI-67, 2000\u00b5L (Source 3)\n* C6: KI-67, 2000\u00b5L (Source 4)\n* D6: KI-67, 2000\u00b5L (Source 5)\n\n\n\n\n\n",
"internal": "1ef67e\n",
diff --git a/protoBuilds/1f62ba/README.json b/protoBuilds/1f62ba/README.json
index 8f26f32b43..96eceaf067 100644
--- a/protoBuilds/1f62ba/README.json
+++ b/protoBuilds/1f62ba/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "1f62ba",
"labware": "\nNest 12-well reservoir, 15mL\n20ul Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "![deck layout]()\n\n\n---\n\n",
"description": "This protocol preps one PCR plate with DNA, water, primer letter, primer number, and Kappa enzyme. New tips are granted for DNA and primers, whereas just one column of tips is used for the Kappa enzyme and water, respectively. The water is added via multi-dispensing.\n\n\nExplanation of complex parameters below:\n* `Number of Columns`: Specify the number of DNA columns for this protocol.\n* `P20 Multi-Channel Mount`: Specify which mount (left or right) to host the P20 multi-channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/1f8b55/README.json b/protoBuilds/1f8b55/README.json
index ebf14ebe64..16a2dd8288 100644
--- a/protoBuilds/1f8b55/README.json
+++ b/protoBuilds/1f8b55/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol automates spotting of slides, using a custom labware piece that holds 4 slides, each containing 21 spots.\n\nThe protocol begins by adding 1\u00b5L to each spot of the four slides, using solutions in A2, A3, A4, and A5 of a 1.5mL eppendorf tube in an Opentrons Tube Rack in slot 2 (changes tip between each solution).\n\nThe protocol concludes by adding 1\u00b5L of solution to each spot of the slides in triplicate. Cycling through tubes A6 of the tuberack in slot 2 and tubes in A1-A6 in slot 3, the p20 will make each transfer and discard tip after each transfer. This behavior continues with rows B, C, and D for slides 2, 3, and 4, respectively.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P20 Single-Channel Pipette\nOpentrons 10/20\u00b5L Tips\nOpentrons 4-in-1 Tube Rack Set\n1.5mL Microcentrifuge Tube\nCustom Adaptor that contains 4 PTFE Printed Slides\n\n\n\nDeck Setup\n\nSlot 1: Opentrons 20\u00b5L Tips\n\nSlot 2: Opentrons Tube Rack with 1.5mL eppendorf tubes\nA2: Solution for all spots of slide 1\nA3: Solution for all spots of slide 2\nA4: Solution for all spots of slide 3\nA5: Solution for all spots of slide 4\nA6: Solution for first 3 spots of slide 1\nB6: Solution for first 3 spots of slide 2\nC6: Solution for first 3 spots of slide 3\nD6: Solution for first 3 spots of slide 4\n\nSlot 3: Opentrons Tube Rack with 1.5mL eppendorf tubes\nA1-A6: Solution for slide 1\nB1-B6: Solution for slide 2\nC1-C6: Solution for slide 3\nD1-D6: Solution for slide 4\n\nSlot 5: Custom Adaptor that contains 4 PTFE Printed Slides",
"internal": "1f8b55",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol automates spotting of slides, using a custom labware piece that holds 4 slides, each containing 21 spots.\n\nThe protocol begins by adding 1\u00b5L to each spot of the four slides, using solutions in A2, A3, A4, and A5 of a 1.5mL eppendorf tube in an Opentrons Tube Rack in slot 2 (changes tip between each solution).\n\nThe protocol concludes by adding 1\u00b5L of solution to each spot of the slides in triplicate. Cycling through tubes A6 of the tuberack in slot 2 and tubes in A1-A6 in slot 3, the p20 will make each transfer and discard tip after each transfer. This behavior continues with rows B, C, and D for slides 2, 3, and 4, respectively.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 10/20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [1.5mL Microcentrifuge Tube](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes)\n* Custom Adaptor that contains 4 [PTFE Printed Slides](https://www.2spi.com/item/02289-ab/)\n\n\n\n---\n\n\n\n**Deck Setup**\n\nSlot 1: [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 2: [Opentrons Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 1.5mL eppendorf tubes\nA2: Solution for all spots of slide 1\nA3: Solution for all spots of slide 2\nA4: Solution for all spots of slide 3\nA5: Solution for all spots of slide 4\nA6: Solution for first 3 spots of slide 1\nB6: Solution for first 3 spots of slide 2\nC6: Solution for first 3 spots of slide 3\nD6: Solution for first 3 spots of slide 4\n\nSlot 3: [Opentrons Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 1.5mL eppendorf tubes\nA1-A6: Solution for slide 1\nB1-B6: Solution for slide 2\nC1-C6: Solution for slide 3\nD1-D6: Solution for slide 4\n\nSlot 5: Custom Adaptor that contains 4 [PTFE Printed Slides](https://www.2spi.com/item/02289-ab/)\n\n\n",
"internal": "1f8b55\n",
diff --git a/protoBuilds/1f96ca-part-2/README.json b/protoBuilds/1f96ca-part-2/README.json
index 77356257d5..8afb1a6ce0 100644
--- a/protoBuilds/1f96ca-part-2/README.json
+++ b/protoBuilds/1f96ca-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"KAPA Hyper Plus 96rx, cat 07962428001 ROCHE"
@@ -9,7 +9,7 @@
"description": "Part 2 of 2: Library Clean Up and Pooling\nLinks:\n Part 1: Fragmentation, End Repair, Adapter Ligation, Library Amplification\n Part 2: Clean Up Libraries\nWith this protocol, your robot can perform the ROCHE KAPA HyperPlus NGS library prep protocol described by the KAPA HyperPlus Kit.\nThis is part 2 of the protocol, which includes the steps (1) bead clean up (2) pooling.",
"internal": "1f96ca",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * KAPA Hyper Plus 96rx, cat 07962428001 ROCHE\n\n",
"deck-setup": "* Opentrons Magnetic Module (Deck Slot 9)\n* Opentrons p300 tips (Deck Slots 10, 11)\n* Opentrons p20 tips (Deck Slots 3, 4, 7, 8)\n* opentrons_96_aluminumblock_generic_pcr_strip_200ul (Deck Slot 5) with\nbeads, water, intermediate_pools in columns 1-3\n* post-PCR plate (Deck Slot 1)\n* nest_12_reservoir_15ml (Deck Slot 2)\n* pool tube and tube rack (Deck Slot 6) with pool tube in A1\n\n",
"description": "Part 2 of 2: Library Clean Up and Pooling\n\nLinks:\n* [Part 1: Fragmentation, End Repair, Adapter Ligation, Library Amplification](http://protocols.opentrons.com/protocol/1f96ca)\n* [Part 2: Clean Up Libraries](http://protocols.opentrons.com/protocol/1f96ca-part-2)\n\nWith this protocol, your robot can perform the ROCHE KAPA HyperPlus NGS library prep protocol described by the [KAPA HyperPlus Kit](https://www.n-genetics.com/products/1104/1023/17277.pdf).\n\nThis is part 2 of the protocol, which includes the steps (1) bead clean up (2) pooling.\n\n",
diff --git a/protoBuilds/1f96ca/README.json b/protoBuilds/1f96ca/README.json
index b8d6bfe496..8700b4ab44 100644
--- a/protoBuilds/1f96ca/README.json
+++ b/protoBuilds/1f96ca/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"KAPA Hyper Plus 96rx, cat 07962428001 ROCHE"
@@ -9,7 +9,7 @@
"description": "Part 1 of 2: Fragmentation, End Repair, Adapter Ligation, Library Amplification\nLinks:\n Part 1: Fragmentation, End Repair, Adapter Ligation, Library Amplification\n Part 2: Clean Up Libraries\nWith this protocol, your robot can perform the ROCHE KAPA HyperPlus NGS library prep protocol described by the KAPA HyperPlus Kit.\nThis is part 1 of the protocol, which includes the steps (1) enzymatic DNA fragmentation (2) end repair and A-tailing (3) adapter ligation and (4) library amplification.\nThe double stranded DNA fragments are end-repaired to generate 5'-phosphorylated, 3'-dA-tailed dsDNA. dsDNA adapters with 3'-dTMP overhangs are ligated to the 3'-dA-tailed DNA. These steps are followed by PCR amplification of the libraries.",
"internal": "1f96ca",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * KAPA Hyper Plus 96rx, cat 07962428001 ROCHE\n\n",
"deck-setup": "* Opentrons Thermocycler Module (Deck Slots 7, 8, 10, 11)\n* Opentrons Magnetic Module (Deck Slot 9)\n* Opentrons p300 tips (Deck Slot 4)\n* Opentrons p20 tips (Deck Slots 3, 6)\n* opentrons_96_aluminumblock_generic_pcr_strip_200ul (Deck Slot 5) with\nfrag_mm, erat_mm, liga_mm, beads, water, kapa mix in columns 1-6\n* DNA sample plate (Deck Slot 1)\n* nest_12_reservoir_15ml (Deck Slot 2)\n\n",
"description": "Part 1 of 2: Fragmentation, End Repair, Adapter Ligation, Library Amplification\n\nLinks:\n* [Part 1: Fragmentation, End Repair, Adapter Ligation, Library Amplification](http://protocols.opentrons.com/protocol/1f96ca)\n* [Part 2: Clean Up Libraries](http://protocols.opentrons.com/protocol/1f96ca-part-2)\n\nWith this protocol, your robot can perform the ROCHE KAPA HyperPlus NGS library prep protocol described by the [KAPA HyperPlus Kit](https://www.n-genetics.com/products/1104/1023/17277.pdf).\n\nThis is part 1 of the protocol, which includes the steps (1) enzymatic DNA fragmentation (2) end repair and A-tailing (3) adapter ligation and (4) library amplification.\n\nThe double stranded DNA fragments are end-repaired to generate 5'-phosphorylated, 3'-dA-tailed dsDNA. dsDNA adapters with 3'-dTMP overhangs are ligated to the 3'-dA-tailed DNA. These steps are followed by PCR amplification of the libraries.\n\n",
diff --git a/protoBuilds/1fcf02/README.json b/protoBuilds/1fcf02/README.json
index 41df82e5a7..0fb4ce9af5 100644
--- a/protoBuilds/1fcf02/README.json
+++ b/protoBuilds/1fcf02/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "1fcf02",
"labware": "\nOpentrons 4-in-1 Tube Rack\nGreiner 384 Well Plate\nOpentrons 20ul Tips\nOpentrons 300ul Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "This protocol preps (3) 384 plates with urea, buffer and sample. One tip is used for each the urea and buffer reagents, with changing tips for the sample. Pipettes are selected dependent on the volume passed to the csv. If the protocol runs out of tips, it will pause and prompt the user to refill tip racks and resume. After sample is added, the well is mixed at 20ul for 3 repetitions.\n\nExplanation of complex parameters below:\n* `Number of 384 plates`: Specify how many 384 well plates there are for this run. Note that for plates less than 3, plates should be placed in order of slot number (1, 2, then 3), and csvs should also be uploaded in that order (from the top down below).\n* `.CSV`: Find below the format for the urea/buffer, and sample csvs. NOTE: Always put an \"x\" in the top left well of the csv, and include the header line. All csvs should be seperate. For wells which receive no volume, input an \"x\" instead. Please see the csvs below for reference.\n\n* Urea/buffer map:\n\n* Sample map:\n\n\n* `Urea/Buffer Initial Volume (mL)`: Specify the initial volume in mL in each the urea and buffer tubes.\n* `P20/P300 Mount`: Specify which mount (left or right) to host the single-channel pipettes.\n\n\n---\n\n",
diff --git a/protoBuilds/203136/README.json b/protoBuilds/203136/README.json
index 9e3e849403..feec020d4a 100644
--- a/protoBuilds/203136/README.json
+++ b/protoBuilds/203136/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Invitrogen ChargeSwitch"
@@ -8,7 +8,7 @@
"description": "This protocol performs the Invitrogen ChargeSwitch gDNA Kit, using the P50 Single-Channel Pipette and the P300 Multi-Channel Pipette.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Magnetic Module\nOpentrons P50 Single-Channel Pipette\nOpentrons P300 Multi-Channel Pipette\nOpentrons 50/300\u00b5L Tips\nBio-Rad 96-Well Plate, 200\u00b5L\nUSA Scientific 96-Deep Well Plate, 2.4mL\nUSA Scientific 12-Well Reservoir, 22mL\nReagents\nSamples\n\n\n\nSlot 1: Bio-Rad 96-Well Plate, clean and empty (for final elution)\nSlot 2: Opentrons 50/300\u00b5L Tips\nSlot 3: Opentrons 50/300\u00b5L Tips\nSlot 4: Opentrons Magnetic Module with a 96-Deep Well Plate, filled with 200\u00b5L of sample\nSlot 5: Opentrons 50/300\u00b5L Tips\nSlot 6: Opentrons 50/300\u00b5L Tips\nSlot 7: USA Scientific 12-Well Reservoir\n A1: Lysis Buffer\n A2: Proteinase K\n A3: Purification Buffer\n A4: Mag Beads\n A5: Wash Buffer (Wash 1)\n A6: Wash Buffer (Wash 2)\n A7: Elution Buffer\n A8: Empty, for liquid waste\n A9: Empty, for liquid waste\n A10: Empty, for liquid waste\n A11: Empty, for liquid waste\n A12: Empty, for liquid waste\nSlot 8: Opentrons 50/300\u00b5L Tips\nSlot 9: Opentrons 50/300\u00b5L Tips\nSlot 10: Opentrons 50/300\u00b5L Tips\nSlot 11: Opentrons 50/300\u00b5L Tips\nUsing the customizations fields, below set up your protocol.\n P300 Multi Mount: Select which mount (left or right) the P300 Multi is attached to.\n P50 Single Mount: Select which mount (left or right) the P50 Single is attached to.\n* Number of Samples: Specify the number of samples you'd like to run.\nNote: The number of tips needed, will change according to the number of samples you select. For example, if running 96 samples, you'd need 8 tip racks, but if running 8 samples, only 1 tip rack is required. When you upload the protocol, you should see an updated deck layout with the necessary tip racks shown.",
"internal": "203136",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Invitrogen ChargeSwitch\n\n\n",
"description": "This protocol performs the Invitrogen ChargeSwitch gDNA Kit, using the P50 Single-Channel Pipette and the P300 Multi-Channel Pipette.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P50 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Bio-Rad 96-Well Plate, 200\u00b5L](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr)\n* [USA Scientific 96-Deep Well Plate, 2.4mL](https://labware.opentrons.com/usascientific_96_wellplate_2.4ml_deep?category=wellPlate)\n* [USA Scientific 12-Well Reservoir, 22mL](https://labware.opentrons.com/usascientific_12_reservoir_22ml/)\n* Reagents\n* Samples\n\n\n---\n\n\nSlot 1: [Bio-Rad 96-Well Plate](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr), clean and empty (for final elution)\n\nSlot 2: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 3: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 4: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with a [96-Deep Well Plate](https://labware.opentrons.com/usascientific_96_wellplate_2.4ml_deep?category=wellPlate), filled with 200\u00b5L of sample\n\nSlot 5: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 6: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 7: [USA Scientific 12-Well Reservoir](https://labware.opentrons.com/usascientific_12_reservoir_22ml/)\n* A1: Lysis Buffer\n* A2: Proteinase K\n* A3: Purification Buffer\n* A4: Mag Beads\n* A5: Wash Buffer (Wash 1)\n* A6: Wash Buffer (Wash 2)\n* A7: Elution Buffer\n* A8: Empty, for liquid waste\n* A9: Empty, for liquid waste\n* A10: Empty, for liquid waste\n* A11: Empty, for liquid waste\n* A12: Empty, for liquid waste\n\nSlot 8: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 9: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 10: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 11: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n**Using the customizations fields, below set up your protocol.**\n* **P300 Multi Mount**: Select which mount (left or right) the P300 Multi is attached to.\n* **P50 Single Mount**: Select which mount (left or right) the P50 Single is attached to.\n* **Number of Samples**: Specify the number of samples you'd like to run.\n\n\n\n**Note**: The number of tips needed, will change according to the number of samples you select. For example, if running 96 samples, you'd need 8 tip racks, but if running 8 samples, only 1 tip rack is required. When you upload the protocol, you should see an updated deck layout with the necessary tip racks shown.\n\n",
"internal": "203136\n",
diff --git a/protoBuilds/2082dd/README.json b/protoBuilds/2082dd/README.json
index a38132b14b..4d78ac91e3 100644
--- a/protoBuilds/2082dd/README.json
+++ b/protoBuilds/2082dd/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -8,7 +8,7 @@
"description": "This protocol performs a NGS library prep through 2 fold dilutions and final pooling. All volumes are specified in a .csv file.\n\n\n\nBio-Rad Hardshell 96-well PCR plate 200ul #HSP9601\nOpentrons 4-in-1 tuberack with 4x6 insert\nOpentrons 4-in-1 tuberack with 3x5 insert holding 15ml NEST centrifuge tubes or equivalent\nOpentrons P20 and P300 GEN2 single-channel electronic pipettes\nOpentrons 20ul and 300ul tipracks\n\n\n\n3x5 tuberack (slot 4)\n* tube A1: TE buffer (filled to at least ~3cm below tube opening)\n4x6 tuberack (slot 5)\n tubes A1: 1.5ml snapcap tube for pooling\n tube A2: 1.5ml screwcap tube for final pooled libraries",
"internal": "2082dd",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"description": "This protocol performs a NGS library prep through 2 fold dilutions and final pooling. All volumes are specified in a `.csv` file.\n\n---\n\n\n* [Bio-Rad Hardshell 96-well PCR plate 200ul #HSP9601](https://www.bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [Opentrons 4-in-1 tuberack with 4x6 insert](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [Opentrons 4-in-1 tuberack with 3x5 insert](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) holding [15ml NEST centrifuge tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-15-ml-centrifuge-tube) or equivalent\n* [Opentrons P20 and P300 GEN2 single-channel electronic pipettes](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 20ul and 300ul tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n\n---\n\n\n3x5 tuberack (slot 4)\n* tube A1: TE buffer (filled to at least ~3cm below tube opening)\n\n4x6 tuberack (slot 5)\n* tubes A1: 1.5ml snapcap tube for pooling\n* tube A2: 1.5ml screwcap tube for final pooled libraries\n\n",
"internal": "2082dd\n",
diff --git a/protoBuilds/211a24/README.json b/protoBuilds/211a24/README.json
index 97bd4e7a8b..18bbf93266 100644
--- a/protoBuilds/211a24/README.json
+++ b/protoBuilds/211a24/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Serial Dilution"
@@ -9,7 +9,7 @@
"internal": "211a24",
"labware": "\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt\nNEST 1 Well Reservoir 195 mL\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Serial Dilution\n\n",
"description": "This protocol allows for a customizable serial dilution workflow based on 4 text files containing worklist information. This protocol uses a P300 GEN2 single-channel pipette. The 4 steps, each corresponding to its own input text file, are as follows:\n* diluent addition to serial dilution plate\n* sample addition to serial dilution plate\n* serial dilution across plate\n* cherrypicking to load final plate\n\n",
"internal": "211a24\n",
diff --git a/protoBuilds/211fe1/README.json b/protoBuilds/211fe1/README.json
index e70a0907f5..02847b2091 100644
--- a/protoBuilds/211fe1/README.json
+++ b/protoBuilds/211fe1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Mass Spec"
@@ -8,7 +8,7 @@
"description": "This protocol performs mass spec sample prep on up to 96 samples using an Opentrons thermocycler for incubation steps. Samples should be specified as comma-separated values on the first line of a .csv file, as in the following example:\nA1,B1,C1,D1,E1,G1,B2\nThe samples are filled according to the order of the .csv, and invalid wells are ignored (valid wells must be A1-H12)\n\n\n\nNEST 96-well PCR plate 100\u00b5L #402501 mounted in Opentrons Thermocycler\nNEST 1.5ml snapcap microcentrifuge tubes (or equivalent) mounted in Opentrons 4-in-1 tuberack with 4x6 insert\nOpentrons P20 GEN2 electronic pipette\nOpentrons 10/20ul tiprack\n\n\n\nOpentrons 4-in-1 tuberack with 4x6 1.5ml tube insert\n tube A1: enzyme\n tube B1: reagent 1\n* tube C1: reagent 2",
"internal": "211fe1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Mass Spec\n\n",
"description": "This protocol performs mass spec sample prep on up to 96 samples using an Opentrons thermocycler for incubation steps. Samples should be specified as comma-separated values on the first line of a `.csv` file, as in the following example:\n```\nA1,B1,C1,D1,E1,G1,B2\n```\n\nThe samples are filled according to the order of the `.csv`, and invalid wells are ignored (valid wells must be `A1`-`H12`)\n\n---\n\n\n* [NEST 96-well PCR plate 100\u00b5L #402501](http://www.cell-nest.com/page94?_l=en&product_id=97&product_category=96) mounted in [Opentrons Thermocycler](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module)\n* [NEST 1.5ml snapcap microcentrifuge tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes) (or equivalent) mounted in [Opentrons 4-in-1 tuberack with 4x6 insert](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [Opentrons P20 GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 10/20ul tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n\n---\n\n\nOpentrons 4-in-1 tuberack with 4x6 1.5ml tube insert\n* tube A1: enzyme\n* tube B1: reagent 1\n* tube C1: reagent 2\n\n",
"internal": "211fe1\n",
diff --git a/protoBuilds/21e4d8-pt1/README.json b/protoBuilds/21e4d8-pt1/README.json
index a8373fe180..50be016f63 100644
--- a/protoBuilds/21e4d8-pt1/README.json
+++ b/protoBuilds/21e4d8-pt1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Twist Library Prep"
@@ -8,7 +8,7 @@
"description": "This protocol is part one of a three-part series to automate the Twist Library Prep protocol. See below for all three parts:\n\nPart 1: Fragmentation & Repair\nPart 2: Ligate Adapters\nPart 3: PCR Amplification\n\nThis protocol begins with samples diluted to 5ng/\u00b5L (if you still need to normalize your sample concentration, try our Normalization protocol) in either PCR tubes or a 96-well plate. Enzymatic Fragmentation Mastermix (40\u00b5L) is added to empty PCR tubes/96-well plate first, before 10\u00b5L of normalized sample is added. The PCR tubes/96-well plate containing sample and mastermix should then be moved off deck to thermocycler and run on the program listed in the Twist Library Prep protocol. After the thermocycler, the samples can be returned to the OT-2 for part two of this protocol.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons p20 (Single- or Multi-Channel) Pipette\nOpentrons p300 (Single- or Multi-Channel) Pipette\nOpentrons Tips\noptional: Opentrons Thermocycler or Opentrons Temperature Module, for active cooling during protocol\nNEST 96-Well PCR Plate\nOpentrons Tube Rack or Aluminum Block Set\nMicrocentrifuge Tube, 1.5mL or PCR strips\nSamples\nTwist Enzymatic Fragmentation and Twist Universal Adapter System\n\n\n\nDeck Layout\n\nSlot 1: Normalized samples in NEST 96-Well PCR Plate or PCR tubes on 96-Well Aluminum Block\n\nSlot 2 (if using P300 Single-Channel): Enyzmatic Fragmentation Master Mix in Microcentrifuge Tube, 1.5mL in Opentrons Tube Rack or 24-Well Aluminum Block\nTubes should be loaded in A1, A2, and A3; each tube can accommodate up to 32 samples and will be used sequentially\nSlot 2 (if using P300 Multi-Channel): Enyzmatic Fragmentation Master Mix in NEST 96-Well PCR Plate or PCR tubes on 96-Well Aluminum Block\nColumns 1, 2, and 3 should be loaded; each column can accommodate up to 32 samples and will be used sequentially\n\nSlot 3: Opentrons Tips for P300\n\nSlot 6: Opentrons Tips for P20\n\nSlot 7: NEST 96-Well PCR Plate or PCR tubes on 96-Well Aluminum Block with optional Opentrons Thermocycler or Opentrons Temperature Module, for active cooling during protocol\n\nUsing the customizations field (below), set up your protocol.\n P20 Type: Select the type (Single- or Multi-Channel) to use; P20 should should be mounted on the left mount\n P300 Type: Select the type (Single- or Multi-Channel) to use; P300 should should be mounted on the right mount\n Number of Samples: Specify the number of samples to run (1-96).\n Module (for cooling): Select whether or not an Opentrons module will be used for active cooling during protocol\n Destination Plate: Select the labware that will be used to combine the samples and the master mix\n Sample Plate: Select the labware that will contain the gDNA samples\n* Master Mix Labware: Select the labware that will contain the enzymatic fragmentation master mix\n\n",
"internal": "21e4d8-pt1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Twist Library Prep\n\n\n",
"description": "This protocol is [part one](https://develop.protocols.opentrons.com/protocol/21e4d8-pt1) of a three-part series to automate the [Twist Library Prep protocol](https://www.twistbioscience.com/sites/default/files/resources/2019-09/Protocol_NGS_EnzymaticFragUniversalAdapterSystem_11Sep19_Rev1.pdf). See below for all three parts:\n\nPart 1: [Fragmentation & Repair](https://develop.protocols.opentrons.com/protocol/21e4d8-pt1)\nPart 2: [Ligate Adapters](https://develop.protocols.opentrons.com/protocol/21e4d8-pt2)\nPart 3: [PCR Amplification](https://develop.protocols.opentrons.com/protocol/21e4d8-pt3)\n\nThis protocol begins with samples diluted to 5ng/\u00b5L (if you still need to normalize your sample concentration, try our [Normalization protocol](https://protocols.opentrons.com/protocol/normalization)) in either PCR tubes or a 96-well plate. Enzymatic Fragmentation Mastermix (40\u00b5L) is added to empty PCR tubes/96-well plate first, before 10\u00b5L of normalized sample is added. The PCR tubes/96-well plate containing sample and mastermix should then be moved off deck to thermocycler and run on the program listed in the [Twist Library Prep protocol](https://www.twistbioscience.com/sites/default/files/resources/2019-09/Protocol_NGS_EnzymaticFragUniversalAdapterSystem_11Sep19_Rev1.pdf). After the thermocycler, the samples can be returned to the OT-2 for [part two](https://develop.protocols.opentrons.com/protocol/21e4d8-pt2) of this protocol.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons p20 (Single- or Multi-Channel) Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [Opentrons p300 (Single- or Multi-Channel) Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* *optional*: [Opentrons Thermocycler](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module) or [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck), for active cooling during protocol\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) or [Aluminum Block Set](https://shop.opentrons.com/collections/verified-labware/products/aluminum-block-set)\n* [Microcentrifuge Tube, 1.5mL](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes) or PCR strips\n* Samples\n* [Twist Enzymatic Fragmentation and Twist Universal Adapter System](https://www.twistbioscience.com/resources/protocol/enzymatic-fragmentation-and-twist-universal-adapter-system-use-twist-ngs)\n\n\n\n---\n\n\n**Deck Layout**\n\nSlot 1: Normalized samples in [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) or PCR tubes on [96-Well Aluminum Block](https://shop.opentrons.com/collections/verified-labware/products/aluminum-block-set)\n\nSlot 2 (if using P300 Single-Channel): Enyzmatic Fragmentation Master Mix in [Microcentrifuge Tube, 1.5mL](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes) in [Opentrons Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) or [24-Well Aluminum Block](https://shop.opentrons.com/collections/verified-labware/products/aluminum-block-set)\n*Tubes should be loaded in A1, A2, and A3; each tube can accommodate up to 32 samples and will be used sequentially*\nSlot 2 (if using P300 Multi-Channel): Enyzmatic Fragmentation Master Mix in [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) or PCR tubes on [96-Well Aluminum Block](https://shop.opentrons.com/collections/verified-labware/products/aluminum-block-set)\n*Columns 1, 2, and 3 should be loaded; each column can accommodate up to 32 samples and will be used sequentially*\n\nSlot 3: [Opentrons Tips for P300](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 6: [Opentrons Tips for P20](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 7: [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) or PCR tubes on [96-Well Aluminum Block](https://shop.opentrons.com/collections/verified-labware/products/aluminum-block-set) with optional [Opentrons Thermocycler](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module) or [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck), for active cooling during protocol\n\n\n**Using the customizations field (below), set up your protocol.**\n* **P20 Type**: Select the type (Single- or Multi-Channel) to use; P20 should should be mounted on the left mount\n* **P300 Type**: Select the type (Single- or Multi-Channel) to use; P300 should should be mounted on the right mount\n* **Number of Samples**: Specify the number of samples to run (1-96).\n* **Module (for cooling)**: Select whether or not an Opentrons module will be used for active cooling during protocol\n* **Destination Plate**: Select the labware that will be used to combine the samples and the master mix\n* **Sample Plate**: Select the labware that will contain the gDNA samples\n* **Master Mix Labware**: Select the labware that will contain the enzymatic fragmentation master mix\n\n\n\n",
"internal": "21e4d8-pt1\n",
diff --git a/protoBuilds/21e4d8-pt2/README.json b/protoBuilds/21e4d8-pt2/README.json
index cf3c1902d6..ec8ff00a5a 100644
--- a/protoBuilds/21e4d8-pt2/README.json
+++ b/protoBuilds/21e4d8-pt2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Twist Library Prep"
@@ -8,7 +8,7 @@
"description": "This protocol is part two of a three-part series to automate the Twist Library Prep protocol. See below for all three parts:\n\nPart 1: Fragmentation & Repair\nPart 2: Ligate Adapters\nPart 3: PCR Amplification\n\nThe final plate from part one should be run through the thermocycler program outlined in the Twist Library Prep protocol before being moved back to the OT-2 for this protocol.\n\nThis protocol begins by adding 5\u00b5L of the Twist Universal Adapters to each well containing samples from part one, followed by the addition of 45\u00b5L of Ligation Master Mix. The user is then prompted to remove plate and incubate the samples on a thermocycler. Following incubation, the user will return the plate to the OT-2 for a fully-automated magbead-based wash process (2 times) that will result in the 15\u00b5L of supernatant transferred to labware selected by the user. The user should then take the labware containing the supernatant and run on the thermocycler before moving to part three.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons p20 (Single- or Multi-Channel) Pipette\nOpentrons p300 (Single- or Multi-Channel) Pipette\nOpentrons Tips\nOpentrons Magnetic Module\nOptional, Opentrons Temperature Module, for active cooling during protocol\nMulti-Well Reservoir, such as the NEST 12-Well Reservoir or USA Scientific 12-Well Reservoir\nNEST 96-Well PCR Plate\nOpentrons Tube Rack or Aluminum Block Set\nMicrocentrifuge Tube, 1.5mL or PCR strips\nSamples\nTwist Enzymatic Fragmentation and Twist Universal Adapter System\nOptional, Single-Well Reservoir, such as the NEST 1-Well Reservoir, 195mL or Agilent 1-Well Reservoir, 290mL, for liquid waste\n\n\n\nDeck Layout\n\nSlot 1: Empty Destination PCR Labware (such as NEST 96-Well PCR Plate or PCR Strips in 96-Well Aluminum Block)\n\nSlot 2: Plate containing Twist Universal Adapters\n\nSlot 3: Opentrons Tips for P300\n\nSlot 4: Multi-Well Reservoir\nLayout for 12-well reservoir:\nA1: DNA Purification Beads (80\u00b5L per sample)\nA3: Freshly prepared 80% ethanol for wash 1, samples 1-48 (200\u00b5L per sample)\nA4: Freshly prepared 80% ethanol for wash 1, samples 48-96 (200\u00b5L per sample)\nA5: Freshly prepared 80% ethanol for wash 2, samples 1-48 (200\u00b5L per sample)\nA6: Freshly prepared 80% ethanol for wash 2, samples 48-96 (200\u00b5L per sample)\nA12: Elution solution (water, 10mM Tris-HCl pH, or Buffer EB) (17\u00b5L per sample)\nLayout for 4-well reservoir:\nA1: DNA Purification Beads (80\u00b5L per sample)\nA2: Freshly prepared 80% ethanol for wash 1, samples 1-96 (200\u00b5L per sample)\nA3: Freshly prepared 80% ethanol for wash 2, samples 1-96 (200\u00b5L per sample)\nA4: Elution solution (water, 10mM Tris-HCl pH, or Buffer EB) (17\u00b5L per sample)\n\nSlot 5: Opentrons Tips for P20\n\nSlot 6: Opentrons Tips for P300\n\nSlot 7: Labware containing ligation master mix (on Opentrons Temperature Module, if active cooling during protocol)\n\nSlot 10: Opentrons Magnetic Module with plate from part one\n\nOptional, Slot 11: Single-Well Reservoir for liquid waste\n\nUsing the customizations field (below), set up your protocol.\n P20 Type: Select the type (Single- or Multi-Channel) to use; P20 should should be mounted on the left mount\n P300 Type: Select the type (Single- or Multi-Channel) to use; P300 should should be mounted on the right mount\n Number of Samples: Specify the number of samples to run (1-96)\n Type of MagDeck: Select which generation (1, 2) magnetic module to use\n Plate on MagDeck: Select which labware type contains samples and is placed on magnetic module\n Module (for cooling): Select whether or not an Opentrons module will be used for active cooling during protocol\n Destination Plate: Select the labware that will be used for the elutions\n Reservor for Reagents: Select which reservoir will be used for reagents\n Sample Plate: Select the labware that will contain the gDNA samples\n Master Mix Labware: Select the labware that will contain the ligation master mix\n* Liquid Waste Destination: Select where liquid waste should be disposed of\n\n",
"internal": "21e4d8-pt2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Twist Library Prep\n\n\n",
"description": "This protocol is [part two](https://develop.protocols.opentrons.com/protocol/21e4d8-pt2) of a three-part series to automate the [Twist Library Prep protocol](https://www.twistbioscience.com/sites/default/files/resources/2019-09/Protocol_NGS_EnzymaticFragUniversalAdapterSystem_11Sep19_Rev1.pdf). See below for all three parts:\n\nPart 1: [Fragmentation & Repair](https://develop.protocols.opentrons.com/protocol/21e4d8-pt1)\nPart 2: [Ligate Adapters](https://develop.protocols.opentrons.com/protocol/21e4d8-pt2)\nPart 3: [PCR Amplification](https://develop.protocols.opentrons.com/protocol/21e4d8-pt3)\n\nThe final plate from [part one](https://develop.protocols.opentrons.com/protocol/21e4d8-pt1) should be run through the thermocycler program outlined in the [Twist Library Prep protocol](https://www.twistbioscience.com/sites/default/files/resources/2019-09/Protocol_NGS_EnzymaticFragUniversalAdapterSystem_11Sep19_Rev1.pdf) before being moved back to the OT-2 for this protocol.\n\nThis protocol begins by adding 5\u00b5L of the Twist Universal Adapters to each well containing samples from [part one](https://develop.protocols.opentrons.com/protocol/21e4d8-pt1), followed by the addition of 45\u00b5L of Ligation Master Mix. The user is then prompted to remove plate and incubate the samples on a thermocycler. Following incubation, the user will return the plate to the OT-2 for a fully-automated magbead-based wash process (2 times) that will result in the 15\u00b5L of supernatant transferred to labware selected by the user. The user should then take the labware containing the supernatant and run on the thermocycler before moving to [part three](https://develop.protocols.opentrons.com/protocol/21e4d8-pt3).\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons p20 (Single- or Multi-Channel) Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [Opentrons p300 (Single- or Multi-Channel) Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* *Optional*, [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck), for active cooling during protocol\n* Multi-Well Reservoir, such as the [NEST 12-Well Reservoir](https://labware.opentrons.com/nest_12_reservoir_15ml/) or [USA Scientific 12-Well Reservoir](https://labware.opentrons.com/usascientific_12_reservoir_22ml/)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) or [Aluminum Block Set](https://shop.opentrons.com/collections/verified-labware/products/aluminum-block-set)\n* [Microcentrifuge Tube, 1.5mL](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes) or PCR strips\n* Samples\n* [Twist Enzymatic Fragmentation and Twist Universal Adapter System](https://www.twistbioscience.com/resources/protocol/enzymatic-fragmentation-and-twist-universal-adapter-system-use-twist-ngs)\n* *Optional*, Single-Well Reservoir, such as the [NEST 1-Well Reservoir, 195mL](https://labware.opentrons.com/nest_1_reservoir_195ml?category=reservoir) or [Agilent 1-Well Reservoir, 290mL](https://labware.opentrons.com/agilent_1_reservoir_290ml?category=reservoir), for liquid waste\n\n\n\n---\n\n\n**Deck Layout**\n\nSlot 1: Empty Destination PCR Labware (such as [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) or PCR Strips in [96-Well Aluminum Block](https://shop.opentrons.com/collections/verified-labware/products/aluminum-block-set))\n\nSlot 2: Plate containing Twist Universal Adapters\n\nSlot 3: [Opentrons Tips for P300](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 4: Multi-Well Reservoir\n*Layout for 12-well reservoir:*\nA1: DNA Purification Beads (80\u00b5L per sample)\nA3: Freshly prepared 80% ethanol for wash 1, samples 1-48 (200\u00b5L per sample)\nA4: Freshly prepared 80% ethanol for wash 1, samples 48-96 (200\u00b5L per sample)\nA5: Freshly prepared 80% ethanol for wash 2, samples 1-48 (200\u00b5L per sample)\nA6: Freshly prepared 80% ethanol for wash 2, samples 48-96 (200\u00b5L per sample)\nA12: Elution solution (water, 10mM Tris-HCl pH, or Buffer EB) (17\u00b5L per sample)\n*Layout for 4-well reservoir:*\nA1: DNA Purification Beads (80\u00b5L per sample)\nA2: Freshly prepared 80% ethanol for wash 1, samples 1-96 (200\u00b5L per sample)\nA3: Freshly prepared 80% ethanol for wash 2, samples 1-96 (200\u00b5L per sample)\nA4: Elution solution (water, 10mM Tris-HCl pH, or Buffer EB) (17\u00b5L per sample)\n\nSlot 5: [Opentrons Tips for P20](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 6: [Opentrons Tips for P300](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 7: Labware containing ligation master mix (on [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck), if active cooling during protocol)\n\nSlot 10: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with plate from part one\n\n*Optional*, Slot 11: Single-Well Reservoir for liquid waste\n\n**Using the customizations field (below), set up your protocol.**\n* **P20 Type**: Select the type (Single- or Multi-Channel) to use; P20 should should be mounted on the left mount\n* **P300 Type**: Select the type (Single- or Multi-Channel) to use; P300 should should be mounted on the right mount\n* **Number of Samples**: Specify the number of samples to run (1-96)\n* **Type of MagDeck**: Select which generation (1, 2) magnetic module to use\n* **Plate on MagDeck**: Select which labware type contains samples and is placed on magnetic module\n* **Module (for cooling)**: Select whether or not an Opentrons module will be used for active cooling during protocol\n* **Destination Plate**: Select the labware that will be used for the elutions\n* **Reservor for Reagents**: Select which reservoir will be used for reagents\n* **Sample Plate**: Select the labware that will contain the gDNA samples\n* **Master Mix Labware**: Select the labware that will contain the ligation master mix\n* **Liquid Waste Destination**: Select where liquid waste should be disposed of\n\n\n\n",
"internal": "21e4d8-pt2\n",
diff --git a/protoBuilds/21e4d8-pt3/README.json b/protoBuilds/21e4d8-pt3/README.json
index 424d97e28f..57516eff09 100644
--- a/protoBuilds/21e4d8-pt3/README.json
+++ b/protoBuilds/21e4d8-pt3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Twist Library Prep"
@@ -8,7 +8,7 @@
"description": "This protocol is part three of a three-part series to automate the Twist Library Prep protocol. See below for all three parts:\n\nPart 1: Fragmentation & Repair\nPart 2: Ligate Adapters\nPart 3: PCR Amplification\n\nThe final plate from part two should be moved back to the OT-2 for this protocol.\n\nThis protocol begins by adding 10\u00b5L of the Twist UDI Primers to each well containing samples from part two, followed by the addition of 25\u00b5L of KAPA HiFi HotStart ReadyMix. The user is then prompted to remove plate and incubate the samples on a thermocycler. Following incubation, the user will return the plate to the OT-2 for a fully-automated magbead-based wash process (2 times) that will result in the 20\u00b5L of supernatant transferred to labware selected by the user. This completes the three-part Twist NGS Library Prep. The user can then perform QC or downstream applications on the elutes.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons p20 (Single- or Multi-Channel) Pipette\nOpentrons p300 (Single- or Multi-Channel) Pipette\nOpentrons Tips\nOpentrons Magnetic Module\nOptional, Opentrons Temperature Module, for active cooling during protocol\nMulti-Well Reservoir, such as the NEST 12-Well Reservoir or USA Scientific 12-Well Reservoir\nNEST 96-Well PCR Plate\nOpentrons Tube Rack or Aluminum Block Set\nMicrocentrifuge Tube, 1.5mL or PCR strips\nSamples\nTwist Enzymatic Fragmentation and Twist Universal Adapter System\nOptional, Single-Well Reservoir, such as the NEST 1-Well Reservoir, 195mL or Agilent 1-Well Reservoir, 290mL, for liquid waste\n\n\n\nDeck Layout\n\nSlot 1: Empty Destination PCR Labware (such as NEST 96-Well PCR Plate or PCR Strips in 96-Well Aluminum Block)\n\nSlot 2: Opentrons Tips for P20\n\nSlot 3: Opentrons Tips for P300\n\nSlot 4: Plate containing Twist UDI Prmer (on Opentrons Temperature Module, if active cooling during protocol)\nSlot 5: Labware containing KAPA HiFi HotStart ReadyMix\n\nSlot 6: Opentrons Tips for P300\n\nSlot 8: Multi-Well Reservoir\nLayout for 12-well reservoir:\nA1: DNA Purification Beads (80\u00b5L per sample)\nA3: Freshly prepared 80% ethanol for wash 1, samples 1-48 (200\u00b5L per sample)\nA4: Freshly prepared 80% ethanol for wash 1, samples 48-96 (200\u00b5L per sample)\nA5: Freshly prepared 80% ethanol for wash 2, samples 1-48 (200\u00b5L per sample)\nA6: Freshly prepared 80% ethanol for wash 2, samples 48-96 (200\u00b5L per sample)\nA12: Elution solution (water, 10mM Tris-HCl pH, or Buffer EB) (17\u00b5L per sample)\nLayout for 4-well reservoir:\nA1: DNA Purification Beads (80\u00b5L per sample)\nA2: Freshly prepared 80% ethanol for wash 1, samples 1-96 (200\u00b5L per sample)\nA3: Freshly prepared 80% ethanol for wash 2, samples 1-96 (200\u00b5L per sample)\nA4: Elution solution (water, 10mM Tris-HCl pH, or Buffer EB) (17\u00b5L per sample)\n\nSlot 9: Opentrons Tips for P300\n\nSlot 10: Opentrons Magnetic Module with plate from part two\n\nOptional, Slot 11: Single-Well Reservoir for liquid waste\n\nUsing the customizations field (below), set up your protocol.\n P20 Type: Select the type (Single- or Multi-Channel) to use; P20 should should be mounted on the left mount\n P300 Type: Select the type (Single- or Multi-Channel) to use; P300 should should be mounted on the right mount\n Number of Samples: Specify the number of samples to run (1-96)\n Type of MagDeck: Select which generation (1, 2) magnetic module to use\n Plate on MagDeck: Select which labware type contains samples and is placed on magnetic module\n Module (for cooling): Select whether or not an Opentrons module will be used for active cooling during protocol\n Destination Plate: Select the labware that will be used for the elutions\n Reservor for Reagents: Select which reservoir will be used for reagents\n Kapa Mix Labware: Select the labware that will contain the ligation master mix\n Liquid Waste Destination: Select where liquid waste should be disposed of\n\n",
"internal": "21e4d8-pt3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Twist Library Prep\n\n\n",
"description": "This protocol is [part three](https://develop.protocols.opentrons.com/protocol/21e4d8-pt3) of a three-part series to automate the [Twist Library Prep protocol](https://www.twistbioscience.com/sites/default/files/resources/2019-09/Protocol_NGS_EnzymaticFragUniversalAdapterSystem_11Sep19_Rev1.pdf). See below for all three parts:\n\nPart 1: [Fragmentation & Repair](https://develop.protocols.opentrons.com/protocol/21e4d8-pt1)\nPart 2: [Ligate Adapters](https://develop.protocols.opentrons.com/protocol/21e4d8-pt2)\nPart 3: [PCR Amplification](https://develop.protocols.opentrons.com/protocol/21e4d8-pt3)\n\nThe final plate from [part two](https://develop.protocols.opentrons.com/protocol/21e4d8-pt2) should be moved back to the OT-2 for this protocol.\n\nThis protocol begins by adding 10\u00b5L of the Twist UDI Primers to each well containing samples from [part two](https://develop.protocols.opentrons.com/protocol/21e4d8-pt2), followed by the addition of 25\u00b5L of KAPA HiFi HotStart ReadyMix. The user is then prompted to remove plate and incubate the samples on a thermocycler. Following incubation, the user will return the plate to the OT-2 for a fully-automated magbead-based wash process (2 times) that will result in the 20\u00b5L of supernatant transferred to labware selected by the user. This completes the three-part Twist NGS Library Prep. The user can then perform QC or downstream applications on the elutes.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons p20 (Single- or Multi-Channel) Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [Opentrons p300 (Single- or Multi-Channel) Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* *Optional*, [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck), for active cooling during protocol\n* Multi-Well Reservoir, such as the [NEST 12-Well Reservoir](https://labware.opentrons.com/nest_12_reservoir_15ml/) or [USA Scientific 12-Well Reservoir](https://labware.opentrons.com/usascientific_12_reservoir_22ml/)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) or [Aluminum Block Set](https://shop.opentrons.com/collections/verified-labware/products/aluminum-block-set)\n* [Microcentrifuge Tube, 1.5mL](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes) or PCR strips\n* Samples\n* [Twist Enzymatic Fragmentation and Twist Universal Adapter System](https://www.twistbioscience.com/resources/protocol/enzymatic-fragmentation-and-twist-universal-adapter-system-use-twist-ngs)\n* *Optional*, Single-Well Reservoir, such as the [NEST 1-Well Reservoir, 195mL](https://labware.opentrons.com/nest_1_reservoir_195ml?category=reservoir) or [Agilent 1-Well Reservoir, 290mL](https://labware.opentrons.com/agilent_1_reservoir_290ml?category=reservoir), for liquid waste\n\n\n\n---\n\n\n**Deck Layout**\n\nSlot 1: Empty Destination PCR Labware (such as [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) or PCR Strips in [96-Well Aluminum Block](https://shop.opentrons.com/collections/verified-labware/products/aluminum-block-set))\n\nSlot 2: [Opentrons Tips for P20](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 3: [Opentrons Tips for P300](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 4: Plate containing Twist UDI Prmer (on [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck), if active cooling during protocol)\nSlot 5: Labware containing KAPA HiFi HotStart ReadyMix\n\nSlot 6: [Opentrons Tips for P300](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 8: Multi-Well Reservoir\n*Layout for 12-well reservoir:*\nA1: DNA Purification Beads (80\u00b5L per sample)\nA3: Freshly prepared 80% ethanol for wash 1, samples 1-48 (200\u00b5L per sample)\nA4: Freshly prepared 80% ethanol for wash 1, samples 48-96 (200\u00b5L per sample)\nA5: Freshly prepared 80% ethanol for wash 2, samples 1-48 (200\u00b5L per sample)\nA6: Freshly prepared 80% ethanol for wash 2, samples 48-96 (200\u00b5L per sample)\nA12: Elution solution (water, 10mM Tris-HCl pH, or Buffer EB) (17\u00b5L per sample)\n*Layout for 4-well reservoir:*\nA1: DNA Purification Beads (80\u00b5L per sample)\nA2: Freshly prepared 80% ethanol for wash 1, samples 1-96 (200\u00b5L per sample)\nA3: Freshly prepared 80% ethanol for wash 2, samples 1-96 (200\u00b5L per sample)\nA4: Elution solution (water, 10mM Tris-HCl pH, or Buffer EB) (17\u00b5L per sample)\n\nSlot 9: [Opentrons Tips for P300](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 10: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with plate from part two\n\n*Optional*, Slot 11: Single-Well Reservoir for liquid waste\n\n**Using the customizations field (below), set up your protocol.**\n* **P20 Type**: Select the type (Single- or Multi-Channel) to use; P20 should should be mounted on the left mount\n* **P300 Type**: Select the type (Single- or Multi-Channel) to use; P300 should should be mounted on the right mount\n* **Number of Samples**: Specify the number of samples to run (1-96)\n* **Type of MagDeck**: Select which generation (1, 2) magnetic module to use\n* **Plate on MagDeck**: Select which labware type contains samples and is placed on magnetic module\n* **Module (for cooling)**: Select whether or not an Opentrons module will be used for active cooling during protocol\n* **Destination Plate**: Select the labware that will be used for the elutions\n* **Reservor for Reagents**: Select which reservoir will be used for reagents\n* **Kapa Mix Labware**: Select the labware that will contain the ligation master mix\n* **Liquid Waste Destination**: Select where liquid waste should be disposed of\n\n\n\n",
"internal": "21e4d8-pt3\n",
diff --git a/protoBuilds/21u968/README.json b/protoBuilds/21u968/README.json
index 482126edb9..70759424fe 100644
--- a/protoBuilds/21u968/README.json
+++ b/protoBuilds/21u968/README.json
@@ -1,5 +1,5 @@
{
- "author": "Iva Pitelkova",
+ "author": "Iva Pitelkova\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"DNA/RNA": [
"DNA/RNA"
@@ -10,7 +10,7 @@
"internal": "21u968",
"labware": "\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 6 Tube Rack with Falcon 50 mL Conical\nBio-Rad 96 Well Plate 200 \u00b5L PCR #hsp9601\n",
"markdown": {
- "author": "Iva Pitelkova\n\n\n",
+ "author": "Iva Pitelkova\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* DNA/RNA\n\t* DNA/RNA\n\n\n",
"deck-setup": "[deck](https://drive.google.com/open?id=1rM_4kiPlj5B-P6rYrz1pR8ZCOglsa5IK)\n\n\n",
"description": "This is a basic protocol that can be used for distributing master mix for PCR.\n\n\n",
diff --git a/protoBuilds/226d24/README.json b/protoBuilds/226d24/README.json
index 69ef1b1703..fd83ed34b6 100644
--- a/protoBuilds/226d24/README.json
+++ b/protoBuilds/226d24/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol automates a PCR prep involving uniquely barcoded forward primers and can accommodate 1-96 samples.\n\nThe protocol begins with the creation of a reaction mix (water, mastermix, and reverse primers) and distribution of that reaction mix. Following this, uniquely barcoded forward primers are transferred to corresponding wells of the PCR plate containing the reaction mix. Finally, samples are transferred in a similar manner from a deep well plate to the PCR plate.\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.21.0 or later)\nOpentrons P300 Multi-Channel Pipette (GEN2)\nOpentrons P20 Multi-Channel Pipette (GEN2)\nOpentrons 200\u00b5L Filter Tip Rack(s)\nOpentrons 20\u00b5L Filter Tip Rack\nNEST 12-Well Reservoir, 15mL\nNEST 96-Well PCR Plate\nKingFisher Deep Well Plate, 2mL\nPlateOne Deep Well Plate, 1mL\nSamples\nReagents (Water, Mastermix, Primers)\n\n\n\nDeck Layout\n\nSlot 1: KingFisher Deep Well Plate containing Samples\n\nSlot 2: Opentrons 200\u00b5L Filter Tip Rack\n\nSlot 3: Opentrons 20\u00b5L Filter Tip Rack\n\nSlot 4: NEST 96-Well PCR Plate (clean and empty)\n\nSlot 5: NEST 12-Well Reservoir, 15mL (for reagents)\nA1: Water (26\u00b5L per sample, plus overage)\nA2: Mastermix (20\u00b5L per sample, plus overage)\nA3: Reverse Primers (1\u00b5L per sample, plus overage)\nA4: Empty; will be used for reaction mix\n\nSlot 6: Opentrons 20\u00b5L Filter Tip Rack\n\nSlot 7: PlateOne Deep Well Plate containing Forward Primers\n",
"internal": "226d24",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"description": "This protocol automates a PCR prep involving uniquely barcoded forward primers and can accommodate 1-96 samples.\n\nThe protocol begins with the creation of a reaction mix (water, mastermix, and reverse primers) and distribution of that reaction mix. Following this, uniquely barcoded forward primers are transferred to corresponding wells of the PCR plate containing the reaction mix. Finally, samples are transferred in a similar manner from a deep well plate to the PCR plate.\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.21.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 Multi-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons P20 Multi-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 200\u00b5L Filter Tip Rack(s)](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons 20\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* KingFisher Deep Well Plate, 2mL\n* PlateOne Deep Well Plate, 1mL\n* Samples\n* Reagents (Water, Mastermix, Primers)\n\n---\n\n\n\n**Deck Layout**\n\nSlot 1: KingFisher Deep Well Plate containing Samples\n\nSlot 2: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 3: [Opentrons 20\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 4: [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) (clean and empty)\n\nSlot 5: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) (for reagents)\nA1: Water (26\u00b5L per sample, plus overage)\nA2: Mastermix (20\u00b5L per sample, plus overage)\nA3: Reverse Primers (1\u00b5L per sample, plus overage)\nA4: Empty; will be used for reaction mix\n\nSlot 6: [Opentrons 20\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 7: PlateOne Deep Well Plate containing Forward Primers\n\n\n\n\n",
"internal": "226d24\n",
diff --git a/protoBuilds/234495/README.json b/protoBuilds/234495/README.json
index 41a9d65be3..e1119a47ea 100644
--- a/protoBuilds/234495/README.json
+++ b/protoBuilds/234495/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Mass Spec"
@@ -9,7 +9,7 @@
"description": "Prepare Standard Samples for Calibration Curves. Prepare Pooled and or Patient Samples.\nWith this protocol, your robot can perform sample preparation for LC-MS as described experimental protocol\nThis step prepares standards for calibration curves followed by patient samples.",
"internal": "234495",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Mass Spec\n\n",
"deck-setup": "* Opentrons p300 tips (Deck Slots 6, 9)\n* Opentrons p20 tips (Deck Slots 2, 3)\n* nest_12_reservoir_15ml (Deck Slot 5)\n* opentrons_24_tuberack_nest_1.5ml_snapcap (Deck Slots 1 and 4)\n\n",
"description": "Prepare Standard Samples for Calibration Curves. Prepare Pooled and or Patient Samples.\n\nWith this protocol, your robot can perform sample preparation for LC-MS as described [experimental protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-05-12/eq03qff/Protocol-90%20ul-tube.pdf)\n\nThis step prepares standards for calibration curves followed by patient samples.\n\n",
diff --git a/protoBuilds/243973/README.json b/protoBuilds/243973/README.json
index fade1cf51d..d025acc63e 100644
--- a/protoBuilds/243973/README.json
+++ b/protoBuilds/243973/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "243973",
"labware": "\nOpentrons 20\u00b5L Tips\nOpentrons 300\u00b5L Tips\nNEST 12-Well Reservoir, 15mL\nGreiner Bio-One 96-Well Plate, #650161\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"deck-setup": "The deck should be setup as follows:\n**[Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-tips)**: Slot 11\n**[Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)**: Slot 10\n**[NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)**: Slot 9\n**Protein Plate (Source)**: Slot 8, containing proteins\n**Destination Plate(s)**: Slots 1-7; plate numbers match slot numbers (Plate 1 in Slot 1, etc.)\n\n",
"description": "This protocol automates protein normalization with a CSV input. Additionally, it gives the user the ability to send and receive email notifications at points requiring user intervention.\n\nUsing the [P20 8-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette) and the [P300 8-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette), diluents are added (based on volume) on up to seven plates with a single set of tips. After the diluents are added, the proteins are transferred with a new tip each time and again based on the volume outlined in the CSV. \n\nBefore the OT-2 transfers the proteins, the robot will pause and prompt the user to add the plate containing proteins to the deck. If selected, the user will also receive an email when this step occurs. The same process will occur at the end of the protocol, letting the user know that they can remove the samples for downstream applications.\n\nExplanation of complex parameters below:\n* **P300-Multi Mount**: Select which mount the [P300 8-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette) is attached to.\n* **P20-Multi Mount**: Select which mount the [P20 8-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette) is attached to.\n* **Number of Destination Plates**: Specify the number of destination plates that are used in the protocol. Up to 7 plates can be used.\n* **Transfer CSV**: Upload the CSV containing the information for normalization. The CSV should have a header row and the following columns (in this order): `Source Well`, `Destination Plate`, `Destination Well`, `Sample Volume`, `Diluent Volume`.\n* **Email (if notifying)**: If receiving notifications via email, which email would you like to notify?\n* **Email Password**: This is the password for the notification email (not your e-mail). The applications engineer that developed the protocol can supply this.\n\n---\n\n",
diff --git a/protoBuilds/245eb2/README.json b/protoBuilds/245eb2/README.json
index 2ca6217a73..78dcd32fcf 100644
--- a/protoBuilds/245eb2/README.json
+++ b/protoBuilds/245eb2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Extraction"
@@ -8,7 +8,7 @@
"description": "This protocol automates a nucleic acid extraction using the Opentrons Magnetic Module and incorporates the Axygen 300\u00b5L plate (holding samples and elutes).\n\nThis protocol begins with 20\u00b5L of sample in the Axygen plate and 20\u00b5L of magnetic beads are added and mixed. After removing the supernatant, there are two subsequent washes with ethanol (150\u00b5L per sample). Finally, 50\u00b5L of elution buffer is added to the sample wells and after incubation, the elution is transferred to a second Axygen plate.\n\nBetween 1 and 96 samples can be processed with this protocol using the GEN2 p300 Multi-Channel Pipette. Because the 8-channel pipette is used, the protocol will behave as if a multiple of eight is used (ex. if you select 10 samples, the protocol will behave in the same way as if you selected 16 samples) - please plan reagent consumption accordingly.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons Magnetic Module\nOpentrons p300 Multi-Channel Pipette, GEN2\nOpentrons Tips\nNEST 12-Well Reservoir, 15mL\nNEST 1-Well Reservoir, 195mL\nAxygen 300\u00b5L Plate\nReagents (magnetic beads, ethanol, elution buffer)\nSamples\n\n\n\nThe Opentrons Magnetic Module should be placed in slot 6 with an Axygen plate containing samples on top.\n\nThe NEST 12-Well Reservoir should be placed in slot 5 with the following reagnets:\n Magnetic Beads: Slot 8\n Ethanol (for wash 1): slot 6 (for samples 1-48), slot 5 (for 48+ samples)\n Ethanol (for wash 2): slot 4 (for samples 1-48), slot 3 (for 48+ samples)\n Elution Buffer: slot 1\n\n\nThe NEST 1-Well Reservoir should be placed in slot 9 for liquid waste.\n\nAn empty Axygen plate should be placed in slot 3, for elutes.\n\nThe Opentrons Tiprack(s) will be accessed in the following order: slot 1, slot 2, slot 4, slot 7, slot 10, slot 11. Each tiprack contains 12 columns of tips and this protocol will use 6 columns of tips per column of sample.\n\n\nUsing the customizations field (below), set up your protocol.\n p300 Multi Mount: Select which mount (left, right) the p300 Multi is attached to.\n Number of Samples: Specify the number of samples to run (1-96).\n\n",
"internal": "245eb2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Nucleic Acid Extraction\n\n\n",
"description": "This protocol automates a nucleic acid extraction using the [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) and incorporates the Axygen 300\u00b5L plate (holding samples and elutes).\n\nThis protocol begins with 20\u00b5L of sample in the Axygen plate and 20\u00b5L of magnetic beads are added and mixed. After removing the supernatant, there are two subsequent washes with ethanol (150\u00b5L per sample). Finally, 50\u00b5L of elution buffer is added to the sample wells and after incubation, the elution is transferred to a second Axygen plate.\n\nBetween 1 and 96 samples can be processed with this protocol using the [GEN2 p300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette). Because the 8-channel pipette is used, the protocol will behave as if a multiple of eight is used (ex. if you select 10 samples, the protocol will behave in the same way as if you selected 16 samples) - please plan reagent consumption accordingly.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons p300 Multi-Channel Pipette, GEN2](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* Axygen 300\u00b5L Plate\n* Reagents (magnetic beads, ethanol, elution buffer)\n* Samples\n\n\n\n---\n\n\nThe [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) should be placed in **slot 6** with an Axygen plate containing samples on top.\n\nThe [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) should be placed in **slot 5** with the following reagnets:\n* Magnetic Beads: **Slot 8**\n* Ethanol (for wash 1): **slot 6** (for samples 1-48), **slot 5** (for 48+ samples)\n* Ethanol (for wash 2): **slot 4** (for samples 1-48), **slot 3** (for 48+ samples)\n* Elution Buffer: **slot 1**\n\n\n\nThe [NEST 1-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml) should be placed in **slot 9** for liquid waste.\n\nAn empty Axygen plate should be placed **in slot 3**, for elutes.\n\nThe [Opentrons Tiprack(s)](https://shop.opentrons.com/collections/opentrons-tips) will be accessed in the following order: **slot 1**, **slot 2**, **slot 4**, **slot 7**, **slot 10**, **slot 11**. Each tiprack contains 12 columns of tips and this protocol will use 6 columns of tips per column of sample.\n\n\n**Using the customizations field (below), set up your protocol.**\n* **p300 Multi Mount**: Select which mount (left, right) the p300 Multi is attached to.\n* **Number of Samples**: Specify the number of samples to run (1-96).\n\n\n\n",
"internal": "245eb2\n",
diff --git a/protoBuilds/25a03d/README.json b/protoBuilds/25a03d/README.json
index bfa6905358..1743a1d326 100644
--- a/protoBuilds/25a03d/README.json
+++ b/protoBuilds/25a03d/README.json
@@ -17,14 +17,14 @@
"internal": "25a03d\n",
"labware": "* [Opentrons 96 well aluminum block with tube strips](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n* [Opentrons 24 well aluminum block](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n* Custom tip racks\n* Custom 96 well plate\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Partner Name](partner website link)\n\n",
+ "partner": "[Partner Name](partner website link)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [Opentrons P20 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [P300 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n\n\n---\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Use p300 single channel pipette to transfer 48ul of MasterMix (MM) from 2.0 ml tube (slot 1) to columns 1, 4, 7, and 10 of the plate/strip tubes in slot 4 (final qPCR plate). Want to be able to use either strip tubes or a plate for the final qPCR reactions in slot 4. Solution will be viscous so want to make sure all is expelled from tips.\n2. Use p20 multi-channel pipette to transfer 12ul of DNA from strip tubes in slot 7 to the final qPCR plate in slot 4. Transfer column 1 of DNA samples to column 1 of final plate; column 2 of DNA samples to column 4 of final plate; and column 3 of DNA samples to column 7 of final plate.\n3. Use p20 multi-channel pipette to transfer 12ul of DNA from strip tubes in slot 9 to the final qPCR plate in slot 4. Transfer column 12 of strip tubes to column 10 of plate. (Could possibly put this strip tube with the DNA in slot 7 but would want it to remain in column 12 so it is not near the other DNA samples. This is the control DNA and we do not want to contaminate the test samples).\n4. Use p20 multi-channel pipette to mix column 1 of final qPCR plate (slot 4) 5 times (set a 20ul). Then transfer 20ul of column 1 to column 2 and column 3 of the final qPCR plate. Repeat with MM/DNA sample in column 4 and transfer 20ul to columns 5 and 6. Repeat with MM/DNA sample in column 7 and transfer 20ul to columns 8 and 9. Repeat with MM/DNA sample in column 10 and transfer 20ul to columns 11 and 12. Solution will be viscous so want to make sure all is expelled from tips.\n\n\n\n",
"title": "qPCR Prep in Triplicates"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Partner Name",
+ "partner": "Partner Name\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nOpentrons P20 Multi-Channel Pipette\nP300 Single-Channel Pipette\n\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nUse p300 single channel pipette to transfer 48ul of MasterMix (MM) from 2.0 ml tube (slot 1) to columns 1, 4, 7, and 10 of the plate/strip tubes in slot 4 (final qPCR plate). Want to be able to use either strip tubes or a plate for the final qPCR reactions in slot 4. Solution will be viscous so want to make sure all is expelled from tips.\nUse p20 multi-channel pipette to transfer 12ul of DNA from strip tubes in slot 7 to the final qPCR plate in slot 4. Transfer column 1 of DNA samples to column 1 of final plate; column 2 of DNA samples to column 4 of final plate; and column 3 of DNA samples to column 7 of final plate.\nUse p20 multi-channel pipette to transfer 12ul of DNA from strip tubes in slot 9 to the final qPCR plate in slot 4. Transfer column 12 of strip tubes to column 10 of plate. (Could possibly put this strip tube with the DNA in slot 7 but would want it to remain in column 12 so it is not near the other DNA samples. This is the control DNA and we do not want to contaminate the test samples).\nUse p20 multi-channel pipette to mix column 1 of final qPCR plate (slot 4) 5 times (set a 20ul). Then transfer 20ul of column 1 to column 2 and column 3 of the final qPCR plate. Repeat with MM/DNA sample in column 4 and transfer 20ul to columns 5 and 6. Repeat with MM/DNA sample in column 7 and transfer 20ul to columns 8 and 9. Repeat with MM/DNA sample in column 10 and transfer 20ul to columns 11 and 12. Solution will be viscous so want to make sure all is expelled from tips.\n",
diff --git a/protoBuilds/269b21/README.json b/protoBuilds/269b21/README.json
index 3454bb7c47..8321fee628 100644
--- a/protoBuilds/269b21/README.json
+++ b/protoBuilds/269b21/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom PCR preparation by transferring mastermix and sample from 1.5ml snapcap tubes and 8-strip PCR tubes, respectively. Mastermix tubes are actively cooled on a temperature module, and samples are passively cooled on a pre-frozen 96-well aluminum block.\nMastermix tubes should be aligned down the aluminum block on the temperature module before moving over a column (i.e. A1, B1, C1, D1, A2, etc.)\n\n\n\nOpentrons temperature module with 4x6 aluminum block for VWR 1.5ml snapcap microtubes #20170-038\nOpentrons 96-well aluminum block with VWR 8-strip microtubes #490004-440 (pre-frozen)\nBio-Rad Hardshell 384-well PCR plate 50\u00b5l #HSP3805\nOpentrons P20 GEN2 single-channel electronic pipette\nOpentrons 20\u00b5l tiprack\n\n\n\n4x6 aluminum block on temperature module\n* tubes A1-D1: buffer",
"internal": "269b21",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol performs a custom PCR preparation by transferring mastermix and sample from 1.5ml snapcap tubes and 8-strip PCR tubes, respectively. Mastermix tubes are actively cooled on a temperature module, and samples are passively cooled on a pre-frozen 96-well aluminum block.\n\nMastermix tubes should be aligned *down* the aluminum block on the temperature module before moving *over* a column (i.e. A1, B1, C1, D1, A2, etc.)\n\n---\n\n\n* [Opentrons temperature module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [4x6 aluminum block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) for [VWR 1.5ml snapcap microtubes #20170-038](https://us.vwr.com/store/product?keyword=20170-038)\n* [Opentrons 96-well aluminum block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) with [VWR 8-strip microtubes #490004-440](https://us.vwr.com/store/product/16208471/genemate-combination-packs-8-strip-standard-tubes-with-8-strip-optically-clear-flat-caps) (pre-frozen)\n* [Bio-Rad Hardshell 384-well PCR plate 50\u00b5l #HSP3805](https://www.bio-rad.com/en-us/sku/hsp3805-hard-shell-384-well-pcr-plates-thin-wall-skirted-clear-white?ID=HSP3805)\n* [Opentrons P20 GEN2 single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 20\u00b5l tiprack](opentrons_96_tiprack_20ul)\n\n---\n\n\n4x6 aluminum block on temperature module\n* tubes A1-D1: buffer\n\n",
"internal": "269b21\n",
diff --git a/protoBuilds/26a414/README.json b/protoBuilds/26a414/README.json
index 21c095e519..74cc721847 100644
--- a/protoBuilds/26a414/README.json
+++ b/protoBuilds/26a414/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol utilizes a CSV (.csv) file to dictate transfers of two reagents (Guanidine and PBS) to a 384-well plate. Simply upload the properly formatted CSV (examples below), set parameters, then download your protocol for use with the OT-2.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP10 Single Pipette\nP50 Single Pipette\nOpentrons 10\u00b5L Tiprack\nOpentrons 50/300\u00b5L Tiprack\nUSA Scientific 12-Well Reservoir, 22mL\nCorning 384-Well Plate\n\n\n\nFor this protocol, be sure that the pipettes (P10 and P50) are attached.\nUsing the customization fields below, set up your protocol.\n Transfer CSV: Upload your properly formatted (see below) CSV.\n P10 Single Mount: Specify which mount the P10 is on (left or right).\n* P50 Single Mount: Specify which mount the P50 is on (left or right).\nNote about CSV\nThe CSV should be formatted like so:\nWell | Guanidine Amount (\u00b5L) | PBS Amount (\u00b5L)\nThe first row (A1, B1, C1) can contain headers (like above) or simply have the desired information. All of the following rows should just have the necessary information.\nLabware Setup\nSlot 1: Opentrons 10\u00b5L Tiprack\nSlot 2: 12-Well Reservoir\n Column 1: Guanidine Solution\n Column 2: PBS Solution\nSlot 3: 384-Well Plate\nSlot 4: Opentrons 50/300\u00b5L Tiprack",
"internal": "26a414",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol utilizes a CSV (.csv) file to dictate transfers of two reagents (Guanidine and PBS) to a 384-well plate. Simply upload the properly formatted CSV (examples below), set parameters, then download your protocol for use with the OT-2.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P10 Single Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P50 Single Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [Opentrons 10\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 50/300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [USA Scientific 12-Well Reservoir, 22mL](https://labware.opentrons.com/usascientific_12_reservoir_22ml?category=reservoir)\n* [Corning 384-Well Plate](https://labware.opentrons.com/corning_384_wellplate_112ul_flat?category=wellPlate)\n\n\n\n---\n\n\nFor this protocol, be sure that the pipettes (P10 and P50) are attached.\n\nUsing the customization fields below, set up your protocol.\n* Transfer CSV: Upload your properly formatted (see below) CSV.\n* P10 Single Mount: Specify which mount the P10 is on (left or right).\n* P50 Single Mount: Specify which mount the P50 is on (left or right).\n\n**Note about CSV**\n\nThe CSV should be formatted like so:\n\n`Well` | `Guanidine Amount (\u00b5L)` | `PBS Amount (\u00b5L)`\n\nThe first row (A1, B1, C1) can contain headers (like above) or simply have the desired information. All of the following rows should just have the necessary information.\n\n**Labware Setup**\n\nSlot 1: [Opentrons 10\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\nSlot 2: [12-Well Reservoir](https://labware.opentrons.com/usascientific_12_reservoir_22ml?category=reservoir)\n* Column 1: Guanidine Solution\n* Column 2: PBS Solution\n\nSlot 3: [384-Well Plate](https://labware.opentrons.com/corning_384_wellplate_112ul_flat?category=wellPlate)\n\nSlot 4: [Opentrons 50/300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n",
"internal": "26a414\n",
diff --git a/protoBuilds/26c304/README.json b/protoBuilds/26c304/README.json
index ce1c7a9e49..a14d37e005 100644
--- a/protoBuilds/26c304/README.json
+++ b/protoBuilds/26c304/README.json
@@ -17,7 +17,7 @@
"internal": "26c304\n",
"labware": "* Greiner 384 well plate 200ul\n* Opentrons 4-in-1 tube rack - 2x3 grid with Appleton 50mL tubes\n* Opentrons 4-in-1 tube rack - 4x6 grid with Axygen 1.7mL tubes\n* Greiner 96 well plate 2000ul\n* [Opentrons 20ul tip rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 1000ul tip rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Partner Name](partner website link)\n\n",
+ "partner": "[Partner Name](partner website link)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P1000 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P20 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Transfer diluent to 300ug/mL tubes for CR3022, mAb45, mAb269 antibodies.\n2. Step down to 30ug/mL, 10ug/mL.\n3. Dispense diluent to A1, A2, A3, A4, A5, A6, B1, B2, B3, B4, B5, B6, C1, C2, C3, C4, C5, C6, D1, D2, D6, E1, G1, F1, H1, E2, E3, E4, E5, F2, F3, F4, F5, G2, G3, G4, G5, H2, H3, H4 and H5\n4. Dispense 30ug/mL and 10ug/mL CR3022, mAb45, and mAb269 into masterblock\n5. Add negative to masterblock\n6. Add TF to masterblock\n7. Mix and transfer solutions across masterblock (x4)\n8. Transfer 180ul A1, B1, C1, D1 masterblock to A1, C1, E1, G1 and I23, L23, M23 and O23 of control plates 1, 2 and 3\n9. Transfer 180ul A2, B2, C2, D2 masterblock to B1, D1, F1, H1 and J23, K23, N23 and P23 of control plates 1, 2 and 3\n10. Transfer 180ul A3 and B3 masterblock to A2, C2 and L24, N24 of control plates 1, 2 and 3\n11. Transfer 180ul A3 and B3 masterblock to A2, C2 and L24, N24 of control plates 1, 2 and 3\n\n",
@@ -26,7 +26,7 @@
"title": "Sample Prep for ELISA Test of Monoclonal Antibodies"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Partner Name",
+ "partner": "Partner Name\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP1000 Single Channel Pipette\nP20 Single Channel Pipette\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nTransfer diluent to 300ug/mL tubes for CR3022, mAb45, mAb269 antibodies.\nStep down to 30ug/mL, 10ug/mL.\nDispense diluent to A1, A2, A3, A4, A5, A6, B1, B2, B3, B4, B5, B6, C1, C2, C3, C4, C5, C6, D1, D2, D6, E1, G1, F1, H1, E2, E3, E4, E5, F2, F3, F4, F5, G2, G3, G4, G5, H2, H3, H4 and H5\nDispense 30ug/mL and 10ug/mL CR3022, mAb45, and mAb269 into masterblock\nAdd negative to masterblock\nAdd TF to masterblock\nMix and transfer solutions across masterblock (x4)\nTransfer 180ul A1, B1, C1, D1 masterblock to A1, C1, E1, G1 and I23, L23, M23 and O23 of control plates 1, 2 and 3\nTransfer 180ul A2, B2, C2, D2 masterblock to B1, D1, F1, H1 and J23, K23, N23 and P23 of control plates 1, 2 and 3\nTransfer 180ul A3 and B3 masterblock to A2, C2 and L24, N24 of control plates 1, 2 and 3\nTransfer 180ul A3 and B3 masterblock to A2, C2 and L24, N24 of control plates 1, 2 and 3\n",
diff --git a/protoBuilds/272c63/README.json b/protoBuilds/272c63/README.json
index 2b350e4dc3..a836509263 100644
--- a/protoBuilds/272c63/README.json
+++ b/protoBuilds/272c63/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Tube Filling"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom tube filling protocol through transferring sample from custom 100ml tubes to custom test tubes.\nDepth from the lip of the sample tube from which to aspirate, as well as aspiration and dispense flow rates, should be input as a .csv in the following format including the header line:\n```\ndistance down tube to aspirate (in mm),aspiration speed (in ul/s), dispense speed (in ul/s)\n20,100,100\n20,100,100\n```\nEmpty lines will be ignored. You can download a template for this .csv file here.\n\n\n\nOpentrons P1000 GEN2 single-channel pipette\nOpentrons 1000ul tipracks\nCustom tubes and tuberacks\n\n\n\nTransfer Scheme\n",
"internal": "272c63",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Tube Filling\n\n",
"description": "This protocol performs a custom tube filling protocol through transferring sample from custom 100ml tubes to custom test tubes.\n\nDepth from the lip of the sample tube from which to aspirate, as well as aspiration and dispense flow rates, should be input as a .csv in the following format **including the header line**:\n\n```\ndistance down tube to aspirate (in mm),aspiration speed (in ul/s), dispense speed (in ul/s)\n20,100,100\n20,100,100\n\n```\n\nEmpty lines will be ignored. You can download a template for this .csv file [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/272c63/template.csv).\n\n---\n\n\n* [Opentrons P1000 GEN2 single-channel pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette?variant=5984549109789)\n* [Opentrons 1000ul tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* Custom tubes and tuberacks\n\n---\n\n\n\nTransfer Scheme \n\n\n",
"internal": "272c63\n",
diff --git a/protoBuilds/274d2a/README.json b/protoBuilds/274d2a/README.json
index 450990cdcf..15d626fac0 100644
--- a/protoBuilds/274d2a/README.json
+++ b/protoBuilds/274d2a/README.json
@@ -8,7 +8,7 @@
"description": "\nCherrypicking, or hit-picking, is a key component of many workflows from high-throughput screening to microbial transfections. With this protocol, you can easily select specific wells in any labware without worrying about missing or selecting the wrong well. Just upload your properly formatted CSV file (keep scrolling for an example), customize your parameters, and download your ready-to-run protocol.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Single-Channel Pipette and corresponding Tips\nMicroplates (96-well or 384-well)\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nLabware will be loaded automatically by specifying the labware loadname and labware slot in the .csv file. All available empty slots will be filled with the necessary tipracks, and the user will be prompted to refill the tipracks if all are emptied in the middle of the protocol.\nCSV Format\nYour cherrypicking transfers must be saved as a comma separated value (.csv) file type. Your CSV must contain values corresponding to volumes in microliters (\u03bcL). Note that the header line (first row of the .csv file) should also be included!\nHere's an example of how a short cherrypicking protocol should be properly formatted:\nNew Tip,Source Labware,Source Slot,Source Well,Source Aspiration Height Above Bottom (in mm),Dest Labware,Dest Slot,Dest Well,Dest Dispense Height Above Bottom (in mm),Volume (in ul)\nyes,agilent_1_reservoir_290ml,1,A1,1,nest_96_wellplate_100ul_pcr_full_skirt,4,A11,2,1\n,nest_12_reservoir_15ml,2,A1,1,corning_384_wellplate_112ul_flat,5,B11,2,3\nyes,nest_1_reservoir_195ml,3,A1,1,corning_384_wellplate_112ul_flat,5,A12,2,7\nIn the above example, 1\u03bcL will be transferred from 1mm above the bottom of well A1 in an Agilent 1-well 290ml reservoir (slot 1) to well A11 in the destination NEST 96-well plate 100\u00b5l (slot 4) with a new tip. After this, 3\u03bcL will be transferred from 1mm above the bottom of well A1 in a NEST 12-well 15ml reservoir (slot 2) to well A5 in the destination NEST 96-well plate 100\u00b5l (slot 5) with the same tip. Last, 7\u03bcL will be transferred from 1mm above the bottom of well A1 in a NEST 1-well 195ml reservoir (slot 3) to well H12 in the destination NEST 96-well plate 100\u00b5l (slot 5) with a new tip.\nIf you\u2019d like to follow our template, you can make a copy of this spreadsheet, fill out your values, and export as CSV for use with this protocol.\nUsing the customizations fields, below set up your protocol.\n Transfer .csv File: Upload the .csv file containing your well locations, volumes, and source plate (optional).\n Pipette Model: Select which pipette you will use for this protocol.\n Pipette Mount: Specify which mount your single-channel pipette is on (left or right)\n Tip Type: Specify whether you want to use filter tips.\n* Tip Usage Strategy: Specify whether you'd like to use a new tip for each transfer, or keep the same tip throughout the protocol.",
"internal": "274d2a",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/274d2a). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/274d2a). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"description": "\n\nCherrypicking, or hit-picking, is a key component of many workflows from high-throughput screening to microbial transfections. With this protocol, you can easily select specific wells in any labware without worrying about missing or selecting the wrong well. Just upload your properly formatted CSV file (keep scrolling for an example), customize your parameters, and download your ready-to-run protocol.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes) and corresponding [Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Microplates (96-well or 384-well)](https://labware.opentrons.com/?category=wellPlate)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n\n---\n\n\nLabware will be loaded automatically by specifying the labware loadname and labware slot in the .csv file. All available empty slots will be filled with the necessary [tipracks](https://shop.opentrons.com/collections/opentrons-tips), and the user will be prompted to refill the tipracks if all are emptied in the middle of the protocol.\n\n**CSV Format**\n\nYour cherrypicking transfers must be saved as a comma separated value (.csv) file type. Your CSV must contain values corresponding to volumes in microliters (\u03bcL). Note that the header line (first row of the .csv file) should also be included!\n\nHere's an example of how a short cherrypicking protocol should be properly formatted:\n\n```\nNew Tip,Source Labware,Source Slot,Source Well,Source Aspiration Height Above Bottom (in mm),Dest Labware,Dest Slot,Dest Well,Dest Dispense Height Above Bottom (in mm),Volume (in ul)\nyes,agilent_1_reservoir_290ml,1,A1,1,nest_96_wellplate_100ul_pcr_full_skirt,4,A11,2,1\n,nest_12_reservoir_15ml,2,A1,1,corning_384_wellplate_112ul_flat,5,B11,2,3\nyes,nest_1_reservoir_195ml,3,A1,1,corning_384_wellplate_112ul_flat,5,A12,2,7\n```\n\nIn the above example, 1\u03bcL will be transferred from 1mm above the bottom of well A1 in an Agilent 1-well 290ml reservoir (slot 1) to well A11 in the destination NEST 96-well plate 100\u00b5l (slot 4) with a new tip. After this, 3\u03bcL will be transferred from 1mm above the bottom of well A1 in a NEST 12-well 15ml reservoir (slot 2) to well A5 in the destination NEST 96-well plate 100\u00b5l (slot 5) with the same tip. Last, 7\u03bcL will be transferred from 1mm above the bottom of well A1 in a NEST 1-well 195ml reservoir (slot 3) to well H12 in the destination NEST 96-well plate 100\u00b5l (slot 5) with a new tip.\n\nIf you\u2019d like to follow our template, you can make a copy of [this spreadsheet](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/274d2a/ex.csv), fill out your values, and export as CSV for use with this protocol.\n\nUsing the customizations fields, below set up your protocol.\n* Transfer .csv File: Upload the .csv file containing your well locations, volumes, and source plate (optional).\n* Pipette Model: Select which pipette you will use for this protocol.\n* Pipette Mount: Specify which mount your single-channel pipette is on (left or right)\n* Tip Type: Specify whether you want to use filter tips.\n* Tip Usage Strategy: Specify whether you'd like to use a new tip for each transfer, or keep the same tip throughout the protocol.\n\n\n",
"internal": "274d2a\n",
diff --git a/protoBuilds/277a3d/README.json b/protoBuilds/277a3d/README.json
index 0c57997da5..0252e64edd 100644
--- a/protoBuilds/277a3d/README.json
+++ b/protoBuilds/277a3d/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Oxford Nanopore 16S Barcoding Kit"
@@ -10,7 +10,7 @@
"internal": "277a3d",
"labware": "\nOpentrons 4-in-1 Tube Rack Set with 4x6 insert for 1.5ml Eppendorf tubes\nApplied Biosystems MicroAmp\u2122 Optical 96-Well Reaction Plate #N8010560 seated in MicroAmp\u2122 Splash-Free 96-Well Base #4312063\nOpentrons 20\u00b5L Filter Tips\nOpentrons 200\u00b5L Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Oxford Nanopore 16S Barcoding Kit\n\n",
"deck-setup": "\n \n* blue: water\n* pink: longamp taq\n* purple: barcodes\n* green: starting samples\n* orange: EtOH\n\n---\n\n",
"description": "\nThis protocol performs a semi-automated solution for the [Oxford Nanopore 16S Barcoding NGS Prep](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2022-02-22/yf03wql/16S%20Barcoding%20Kit%20SQK-RAB204-minion.pdf)\n\n---\n\n",
diff --git a/protoBuilds/280ea4/README.json b/protoBuilds/280ea4/README.json
index ab0c50453a..4b2753d3a7 100644
--- a/protoBuilds/280ea4/README.json
+++ b/protoBuilds/280ea4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol creates a custom PCR prep protocol. The input into this protocol is an elution plate of purified DNA, and the output is PCR strips containing the samples mixed with mastermix.\nUsing a Single-Channel Pipette, this protocol will begin by creating reaction mix in a 2ml tube. Samples will then be transferred from their plate to the qPCR plate and mixed with the reaction mix.\nYou can select any of the 5 mastermix compositions shown below:\n\n\n \nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nTemperature Module (GEN2)\nNEST PCR Plate containing purified nucleic acid samples\nOpentrons Tips\n2ml NEST screwcap tubes or equivalent in Opentrons aluminum block containing PCR reagents\nOpentrons P300 Single-Channel GEN2 Pipette\n\n\n\n\n",
"internal": "280ea4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol creates a custom PCR prep protocol. The input into this protocol is an elution plate of purified DNA, and the output is PCR strips containing the samples mixed with mastermix.\nUsing a [Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette), this protocol will begin by creating reaction mix in a 2ml tube. Samples will then be transferred from their plate to the qPCR plate and mixed with the reaction mix.\n\nYou can select any of the 5 mastermix compositions shown below: \n\n\n---\n\n \n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Temperature Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [NEST PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) containing purified nucleic acid samples\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [2ml NEST screwcap tubes or equivalent](https://shop.opentrons.com/collections/tubes/products/nest-microcentrifuge-tubes) in Opentrons aluminum block containing PCR reagents\n* [Opentrons P300 Single-Channel GEN2 Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n\n---\n\n\n\n\n\n\n\n",
"internal": "280ea4\n",
diff --git a/protoBuilds/2905a4/README.json b/protoBuilds/2905a4/README.json
index d86db32729..44fe707df7 100644
--- a/protoBuilds/2905a4/README.json
+++ b/protoBuilds/2905a4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"RNA Purification"
@@ -9,7 +9,7 @@
"description": "This protocol automates the purification of RNA using magnetic beads. It can support up to 94 samples and can have a custom starting well position. The protocol is also optimized for tip conservation and reuses tips for various steps.\nExplanation of parameters below:\n\nNumber of Samples: The total number of samples per run. The maximum samples supported is 94 samples. This value is also used to determine additional wells needed when setting a Starting Well below. For example, if there are 3 samples and the starting well is C7, then the sample wells for all plates will be C7, D7, E7.\nStarting Well Name: The well name samples should begin with for each 96 well plate in the protocol.\nP300 Single GEN2 Mount Position: Specify which mount (left or right) to load the P300 single channel pipette.\nThermocycler Hold Temperature (\u00b0C): The temperature the thermocycler will be set to and held for the entire protocol.\nMagnetic Module Engage Height (mm): The height the magnets should raise.\nSupernatant Removal Radius (X-dimension) (mm): The radius in the X-dimension of how far the pipette should move before aspirating supernatant. Note: Only enter positive values. The protocol will automatically determine the side (+/-) based on the well/column.\nSupernatant Removal Aspirate Flow Rate (uL/s): The aspiration flow rate at which the pipette will aspirate supernatant.\nSupernatant Removal Dispense Flow Rate (uL/s): The aspiration flow rate at which the pipette will dispense supernatant.\n\n",
"internal": "2905a4",
"markdown": {
- "author": "\n[Opentrons](https://opentrons.com/)\n\n",
+ "author": "\n[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "\n* Nucleic Acid Extraction & Purification\n * RNA Purification\n\n",
"deck-setup": "\n\n\n",
"description": "\nThis protocol automates the purification of RNA using magnetic beads. It can support up to 94 samples and can have a custom starting well position. The protocol is also optimized for tip conservation and reuses tips for various steps.\n\nExplanation of parameters below:\n\n- `Number of Samples`: The total number of samples per run. The maximum samples supported is 94 samples. This value is also used to determine additional wells needed when setting a `Starting Well` below. For example, if there are 3 samples and the starting well is C7, then the sample wells for all plates will be C7, D7, E7.\n- `Starting Well Name`: The well name samples should begin with for each 96 well plate in the protocol.\n- `P300 Single GEN2 Mount Position`: Specify which mount (left or right) to load the P300 single channel pipette.\n- `Thermocycler Hold Temperature (\u00b0C)`: The temperature the thermocycler will be set to and held for the entire protocol.\n- `Magnetic Module Engage Height (mm)`: The height the magnets should raise.\n- `Supernatant Removal Radius (X-dimension) (mm)`: The radius in the X-dimension of how far the pipette should move before aspirating supernatant. **Note: Only enter positive values. The protocol will automatically determine the side (+/-) based on the well/column.**\n- `Supernatant Removal Aspirate Flow Rate (uL/s)`: The aspiration flow rate at which the pipette will aspirate supernatant.\n- `Supernatant Removal Dispense Flow Rate (uL/s)`: The aspiration flow rate at which the pipette will dispense supernatant.\n\n---\n\n",
diff --git a/protoBuilds/29225e-part-2/README.json b/protoBuilds/29225e-part-2/README.json
index d3b566148a..9e7ed8de8e 100644
--- a/protoBuilds/29225e-part-2/README.json
+++ b/protoBuilds/29225e-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -9,7 +9,7 @@
"description": "This protocol uses p300 single and p20 (single or multi channel) pipettes to distribute custom volumes of water (0-2 uL from a single tube) and master mix (up to 19 uL from any single well or tube within up to 4 racks or plates) and then transfer normalized RNA samples (2-7 uL with either single or multi channel p20 from wells or columns of up to 4 racks or plates) to a single 96-well PCR plate held at 4 degrees C on the temperature module (water, master mix and normalized RNA volumes, locations etc. specified by user in two csv files uploaded at the time of protocol download). Valid source and destination well locations for the multi-channel pipette must be in row A of a 96-well formatted labware.\nLinks:\n* example master mix input csv\n\n\nexample transfers input csv for p20 single\n\n\nexample transfers input csv for p20 multi\n\n\nPart 1 - RNA Normalization\n\n\nThis protocol was developed to distribute custom volumes (0-2 uL) of water (to specific wells), (up to 19 uL) master mix (to specific wells), and then transfer (2-7 uL) normalized RNA (to specific wells or columns) to a single 96-well PCR plate held at 4 degrees on the temperature module.",
"internal": "29225e-part-2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * PCR Prep\n\n",
"deck-setup": "\n\n\n* Opentrons 200ul filter tips (Deck Slot 11)\n* Opentrons 20ul filter tips (Deck Slot 10)\n* Selected Labware for Normalized RNA Samples (Deck Slots 2, 3, 5, 6)\n* Selected Labware for Water and Master Mixes (Deck Slots 4, 7, 8, 9)\n* Opentrons Temperature Module with 96-well PCR Plate (Deck Slot 1)\n\n\n\n\n\n",
"description": "\nThis protocol uses p300 single and p20 (single or multi channel) pipettes to distribute custom volumes of water (0-2 uL from a single tube) and master mix (up to 19 uL from any single well or tube within up to 4 racks or plates) and then transfer normalized RNA samples (2-7 uL with either single or multi channel p20 from wells or columns of up to 4 racks or plates) to a single 96-well PCR plate held at 4 degrees C on the temperature module (water, master mix and normalized RNA volumes, locations etc. specified by user in two csv files uploaded at the time of protocol download). Valid source and destination well locations for the multi-channel pipette must be in row A of a 96-well formatted labware.\n\nLinks:\n* [example master mix input csv](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/29225e/mastermix.csv)\n\n* [example transfers input csv for p20 single](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/29225e/qRT-PCR_for_p20_single.csv)\n\n* [example transfers input csv for p20 multi](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/29225e/qRT-PCR_for_p20_multi.csv)\n\n* [Part 1 - RNA Normalization](https://protocols.opentrons.com/protocol/29225e)\n\nThis protocol was developed to distribute custom volumes (0-2 uL) of water (to specific wells), (up to 19 uL) master mix (to specific wells), and then transfer (2-7 uL) normalized RNA (to specific wells or columns) to a single 96-well PCR plate held at 4 degrees on the temperature module.\n\n",
diff --git a/protoBuilds/29225e/README.json b/protoBuilds/29225e/README.json
index 19e15ce74c..b6ad49e553 100644
--- a/protoBuilds/29225e/README.json
+++ b/protoBuilds/29225e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -9,7 +9,7 @@
"description": "This protocol uses p300 and p20 single channel pipettes to transfer custom volumes of water (1-50 uL from a single tube) and then RNA (1-50 uL) from a single 96-well source plate or from up to four 24-tube racks to a single 96-well destination plate (RNA volume and water volume, rack number (1-4) or plate number (1) for the input RNA source, tube (A1-D6) or well (A1-H12) location for the RNA source, and destination well (A1-H12) specified by user in a csv file uploaded at the time of protocol download).\nLinks:\n* example input csv\n\nPart 2 - qRT-PCR Setup\n\nThis protocol was developed to transfer custom volumes of up to 96 input RNA samples from one of up to four 24-tube racks (or alternatively from a single 96-well RNA-source plate) to a single 96-well destination plate according to volumes, rack number (1-4) or plate number (1), and tube locations (A1-D6) or well locations (A1-H12) in the uploaded csv file.",
"internal": "29225e",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Normalization\n\n",
"deck-setup": "\n\n* Opentrons 200ul filter tips (Deck Slot 11)\n* Opentrons 20ul filter tips (Deck Slot 10)\n* Selected Labware for Input RNA Samples (Deck Slots 2, 3, 5, 6)\n* Opentrons 24-tube rack with Water Tubes in A1, B1, C1, D1, A2 (Deck Slot 4)\n* Opentrons Temperature Module with 96-well Destination Plate (Deck Slot 1)\n\n\n\n",
"description": "\nThis protocol uses p300 and p20 single channel pipettes to transfer custom volumes of water (1-50 uL from a single tube) and then RNA (1-50 uL) from a single 96-well source plate or from up to four 24-tube racks to a single 96-well destination plate (RNA volume and water volume, rack number (1-4) or plate number (1) for the input RNA source, tube (A1-D6) or well (A1-H12) location for the RNA source, and destination well (A1-H12) specified by user in a csv file uploaded at the time of protocol download).\n\nLinks:\n* [example input csv](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/29225e/RNA+normalization+123456.csv)\n\n* [Part 2 - qRT-PCR Setup](https://protocols.opentrons.com/protocol/29225e-part-2)\n\nThis protocol was developed to transfer custom volumes of up to 96 input RNA samples from one of up to four 24-tube racks (or alternatively from a single 96-well RNA-source plate) to a single 96-well destination plate according to volumes, rack number (1-4) or plate number (1), and tube locations (A1-D6) or well locations (A1-H12) in the uploaded csv file.\n\n",
diff --git a/protoBuilds/29414b-ot/README.json b/protoBuilds/29414b-ot/README.json
index d447033747..23b90211a8 100644
--- a/protoBuilds/29414b-ot/README.json
+++ b/protoBuilds/29414b-ot/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Getting Started": [
"Calibration"
@@ -8,7 +8,7 @@
"description": "This protocol is designed for testing calibration of the OT-2 with a custom tube holder, holding 500\u00b5L tubes from Simport Scientific, with Opentrons Tips. This protocol can be used for testing the P300 Single Channel Pipette or the P300 Multi Channel Pipette.\nIf testing the P300 Single Channel Pipette, the robot will transfer 5\u00b5L from A1 to A2, A3, and A4 with a 200\u00b5L filter tip. The robot will then make transfers with 200\u00b5L filter tips, first 50\u00b5L from B1 to B2, B3, and B4; then 200\u00b5L from C1, to C2, C3, and C4.\nIf testing the P300 Multi Channel Pipette, the robot will transfer 5\u00b5L from column 1 to column 2, 3, and 4 with 200\u00b5L filter tips. The robot will then make transfers with 200\u00b5L filter tips, first 50\u00b5L from column 5 to column 6, 7, and 8; then 200\u00b5L from column 9 to column 10, 11, and 12.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP300 Single Channel Pipette\nP300 Multi Channel Pipette\nOpentrons 200\u00b5L Filter Tips\nSimport Scientific 500\u00b5L Tubes in Custom Holder\n\n\n\nFor this protocol, be sure that the correct pipette (P300 Single or Multi) is attached for the intended test case.\nLabware Setup\nSlot 1: Opentrons 200\u00b5L Filter Tips\nSlot 3: Simport Scientific 500\u00b5L Tubes in Custom Holder\nUsing the customization fields below, set up your protocol.\n Pipette Type: Select either Single Channel or Multi Channel. This will also modify the protocol behavior to the corresponding test protocol.\n Pipette Mount: Specify which mount the P300 is on (left or right).\n Touch Tip (After Aspiration): Select yes to perform a touch-tip after aspirating from source well.\n Touch Tip (After Dispense): Select yes to perform a touch-tip after dispensing into source well.\n",
"internal": "29414b-ot",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Getting Started\n\t* Calibration\n\n\n",
"description": "This protocol is designed for testing calibration of the OT-2 with a custom tube holder, holding 500\u00b5L tubes from Simport Scientific, with Opentrons Tips. This protocol can be used for testing the **P300 Single Channel Pipette** or the **P300 Multi Channel Pipette**.\n\nIf testing the P300 Single Channel Pipette, the robot will transfer 5\u00b5L from A1 to A2, A3, and A4 with a 200\u00b5L filter tip. The robot will then make transfers with 200\u00b5L filter tips, first 50\u00b5L from B1 to B2, B3, and B4; then 200\u00b5L from C1, to C2, C3, and C4.\n\nIf testing the P300 Multi Channel Pipette, the robot will transfer 5\u00b5L from column 1 to column 2, 3, and 4 with 200\u00b5L filter tips. The robot will then make transfers with 200\u00b5L filter tips, first 50\u00b5L from column 5 to column 6, 7, and 8; then 200\u00b5L from column 9 to column 10, 11, and 12.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P300 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P300 Multi Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Simport Scientific 500\u00b5L Tubes in Custom Holder](http://www.simport.com/en/products/173-t100-1.html)\n\n\n\n---\n\n\n\nFor this protocol, be sure that the correct pipette (P300 Single or Multi) is attached for the intended test case.\n\n**Labware Setup**\n\nSlot 1: [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n\nSlot 3: [Simport Scientific 500\u00b5L Tubes in Custom Holder](http://www.simport.com/en/products/173-t100-1.html)\n\n**Using the customization fields below, set up your protocol.**\n* Pipette Type: Select either `Single Channel` or `Multi Channel`. This will also modify the protocol behavior to the corresponding test protocol.\n* Pipette Mount: Specify which mount the P300 is on (left or right).\n* Touch Tip (After Aspiration): Select `yes` to perform a touch-tip after aspirating from source well.\n* Touch Tip (After Dispense): Select `yes` to perform a touch-tip after dispensing into source well.\n\n---\n",
"internal": "29414b-ot\n",
diff --git a/protoBuilds/29414b/README.json b/protoBuilds/29414b/README.json
index 2f273e62b4..87cfd1464b 100644
--- a/protoBuilds/29414b/README.json
+++ b/protoBuilds/29414b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Getting Started": [
"Calibration"
@@ -8,7 +8,7 @@
"description": "This protocol is designed for testing calibration of the OT-2 with a custom tube holder, holding 500\u00b5L tubes from Simport Scientific, and non-standard tips. This protocol can be used for testing the P300 Single Channel Pipette or the P300 Multi Channel Pipette.\nIf testing the P300 Single Channel Pipette, the robot will transfer 5\u00b5L from A1 to A2, A3, and A4 with a 2-20\u00b5L tip. The robot will then make transfers with 20-200\u00b5L tips, first 50\u00b5L from B1 to B2, B3, and B4; then 200\u00b5L from C1, to C2, C3, and C4.\nIf testing the P300 Multi Channel Pipette, the robot will transfer 5\u00b5L from column 1 to column 2, 3, and 4 with 2-20\u00b5L tips. The robot will then make transfers with 20-200\u00b5L tips, first 50\u00b5L from column 5 to column 6, 7, and 8; then 200\u00b5L from column 9 to column 10, 11, and 12.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP300 Single Channel Pipette\nP300 Multi Channel Pipette\nFisherbrand 2-20\u00b5L Tips\nFisherbrand 20-200\u00b5L Tips\nSimport Scientific 500\u00b5L Tubes in Custom Holder\n\n\n\nFor this protocol, be sure that the correct pipette (P300 Single or Multi) is attached for the intended test case.\nLabware Setup\nSlot 1: Fisherbrand 2-20\u00b5L Tips\nSlot 2: Fisherbrand 20-200\u00b5L Tips\nSlot 3: Simport Scientific 500\u00b5L Tubes in Custom Holder\nUsing the customization fields below, set up your protocol.\n Pipette Type: Select either Single Channel or Multi Channel. This will also modify the protocol behavior to the corresponding test protocol.\n Pipette Mount: Specify which mount the P300 is on (left or right).\n",
"internal": "29414b",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Getting Started\n\t* Calibration\n\n\n",
"description": "This protocol is designed for testing calibration of the OT-2 with a custom tube holder, holding 500\u00b5L tubes from Simport Scientific, and non-standard tips. This protocol can be used for testing the **P300 Single Channel Pipette** or the **P300 Multi Channel Pipette**.\n\nIf testing the P300 Single Channel Pipette, the robot will transfer 5\u00b5L from A1 to A2, A3, and A4 with a 2-20\u00b5L tip. The robot will then make transfers with 20-200\u00b5L tips, first 50\u00b5L from B1 to B2, B3, and B4; then 200\u00b5L from C1, to C2, C3, and C4.\n\nIf testing the P300 Multi Channel Pipette, the robot will transfer 5\u00b5L from column 1 to column 2, 3, and 4 with 2-20\u00b5L tips. The robot will then make transfers with 20-200\u00b5L tips, first 50\u00b5L from column 5 to column 6, 7, and 8; then 200\u00b5L from column 9 to column 10, 11, and 12.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P300 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P300 Multi Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Fisherbrand 2-20\u00b5L Tips](https://www.fishersci.com/shop/products/fisherbrand-sureone-aerosol-barrier-pipette-tips-14/02707432)\n* [Fisherbrand 20-200\u00b5L Tips](https://www.fishersci.com/shop/products/fisherbrand-sureone-aerosol-barrier-pipette-tips-20-200-l-beveled-tip-graduated-at-10-50-100-l/02707430)\n* [Simport Scientific 500\u00b5L Tubes in Custom Holder](http://www.simport.com/en/products/173-t100-1.html)\n\n\n\n---\n\n\n\nFor this protocol, be sure that the correct pipette (P300 Single or Multi) is attached for the intended test case.\n\n**Labware Setup**\n\nSlot 1: [Fisherbrand 2-20\u00b5L Tips](https://www.fishersci.com/shop/products/fisherbrand-sureone-aerosol-barrier-pipette-tips-14/02707432)\n\nSlot 2: [Fisherbrand 20-200\u00b5L Tips](https://www.fishersci.com/shop/products/fisherbrand-sureone-aerosol-barrier-pipette-tips-20-200-l-beveled-tip-graduated-at-10-50-100-l/02707430)\n\nSlot 3: [Simport Scientific 500\u00b5L Tubes in Custom Holder](http://www.simport.com/en/products/173-t100-1.html)\n\n**Using the customization fields below, set up your protocol.**\n* Pipette Type: Select either `Single Channel` or `Multi Channel`. This will also modify the protocol behavior to the corresponding test protocol.\n* Pipette Mount: Specify which mount the P300 is on (left or right).\n\n---\n",
"internal": "29414b\n",
diff --git a/protoBuilds/2942dd/README.json b/protoBuilds/2942dd/README.json
index e6f80e14b8..1410898594 100644
--- a/protoBuilds/2942dd/README.json
+++ b/protoBuilds/2942dd/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -9,7 +9,7 @@
"description": "This protocol uses a p300 single-channel pipette to transfer custom volumes of serum reagent from specified locations in four Opentrons 24-tube racks into specified tubes held in a 96-well rack. A p20 single-channel pipette is then used to tranfer a custom volume of RNase A and 20 percent Tween 20 to the same 96-tube rack locations. Finally, destination tube contents are mixed and the tubes are held at room temperature for 30 minutes. Volumes, rack locations and well locations are specified in a csv file uploaded at the time of protocol download.\nLinks:\n example input csv\n Fluidx rack and 1 mL tubes\n* MTC Bio MTC-3220-SG tubes",
"internal": "2942dd",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Plate Filling\n\n",
"deck-setup": "\n\n* Opentrons 200ul filter tips (Deck Slot 10)\n* Opentrons 20ul filter tips (Deck Slots 5,11)\n* 96-tube Fluidx rack with 1 mL Fluidx tubes (Deck Slot 9)\n* 24-tube Opentrons rack with MTC-3220-SG 2 mL tubes (Deck Slots 7,4,1,8)\n\n\n\n\n\n",
"description": "\nThis protocol uses a p300 single-channel pipette to transfer custom volumes of serum reagent from specified locations in four Opentrons 24-tube racks into specified tubes held in a 96-well rack. A p20 single-channel pipette is then used to tranfer a custom volume of RNase A and 20 percent Tween 20 to the same 96-tube rack locations. Finally, destination tube contents are mixed and the tubes are held at room temperature for 30 minutes. Volumes, rack locations and well locations are specified in a csv file uploaded at the time of protocol download.\n\nLinks:\n* [example input csv](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/2942dd/example.csv)\n* [Fluidx rack and 1 mL tubes](https://www.azenta.com/products/1.0ml-tri-coded-tube-96-format-external-thread#specifications)\n* [MTC Bio MTC-3220-SG tubes](http://mtcbiotech.com/product/clearseal-screw-cap-microcentrifuge-tubes/)\n\n",
diff --git a/protoBuilds/298069/README.json b/protoBuilds/298069/README.json
index e281c24ba6..2fc5beba9e 100644
--- a/protoBuilds/298069/README.json
+++ b/protoBuilds/298069/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Aliquoting"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom sample aliquoting workflow by transferring the contents of 2 15ml tubes to 12 2ml tubes each. 300\u00b5l is transferred to each tube, and height tracking is performed automatically so that the pipette does not submerge into the solution in the 15ml tubes.\n\n\n\nOpentrons 4-in-1 tuberack with 3x5 insert for 15ml tubes\nOpentrons 4-in-1 tuberack with 4x6 insert for 2ml tubes\nOpentrons P1000 GEN2 single-channel electronic pipette\nOpentrons 1000\u00b5l tiprack\n\n\n\n3x5 tuberack for 15ml tubes (slot 1):\n tube A1: solution 1 (filled to at least 10ml mark)\n tube B1: solution 2 (filled to at least 10ml mark)\n4x6 tuberack for 2ml tubes (slot 2):\n rows A and B: tubes to receive solution 1\n rows C and D: tubes to receive solution 2",
"internal": "298069",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Aliquoting\n\n",
"description": "This protocol performs a custom sample aliquoting workflow by transferring the contents of 2 15ml tubes to 12 2ml tubes each. 300\u00b5l is transferred to each tube, and height tracking is performed automatically so that the pipette does not submerge into the solution in the 15ml tubes.\n\n---\n\n\n* [Opentrons 4-in-1 tuberack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 3x5 insert for 15ml tubes\n* [Opentrons 4-in-1 tuberack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 4x6 insert for 2ml tubes\n* [Opentrons P1000 GEN2 single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette?variant=5984549142557)\n* [Opentrons 1000\u00b5l tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n\n---\n\n\n3x5 tuberack for 15ml tubes (slot 1):\n* tube A1: solution 1 (filled to at least 10ml mark)\n* tube B1: solution 2 (filled to at least 10ml mark)\n\n4x6 tuberack for 2ml tubes (slot 2):\n* rows A and B: tubes to receive solution 1\n* rows C and D: tubes to receive solution 2\n\n",
"internal": "298069\n",
diff --git a/protoBuilds/29effa-buffer/README.json b/protoBuilds/29effa-buffer/README.json
index bfad82e445..9751e29add 100644
--- a/protoBuilds/29effa-buffer/README.json
+++ b/protoBuilds/29effa-buffer/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol automates the distribution of process buffer from the Lyra Direct SARS-CoV-2 Assay into a 96-well format.\nUsing an Opentrons P300 8-Channel Pipette, this protocol will transfer 400\u00b5L of process buffer from a 12-well reservoir to the specified number of wells in a 96-well plate.\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.21.0 or later)\nOpentrons P300 8-Channel Pipette, GEN2\nOpentrons 300\u00b5L Tip Rack\nNEST 12-Well Reservoir\nEppendorf 96-Well Plate\nLyra Direct SARS-CoV-2 Assay Kit\n\n\n\nDeck Layout\n\nSlot 1: Opentrons 300\u00b5L Tip Rack\n\nSlot 4: Eppendorf 96-Well Plate\n\nSlot 7: NEST 12-Well Reservoir\nProcess Buffer Loading: Each column of the reservoir can contain enough buffer for up to 24 wells. Ex: if running 96 wells, columns 1-4 should be loaded with buffer\n",
"internal": "29effa-buffer",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol automates the distribution of process buffer from the [Lyra Direct SARS-CoV-2 Assay](https://www.quidel.com/molecular-diagnostics/lyra-direct-sars-cov-2-assay) into a 96-well format.\nUsing an [Opentrons P300 8-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette), this protocol will transfer 400\u00b5L of process buffer from a 12-well reservoir to the specified number of wells in a 96-well plate.\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.21.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 8-Channel Pipette, GEN2](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons 300\u00b5L Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* Eppendorf 96-Well Plate\n* [Lyra Direct SARS-CoV-2 Assay Kit](https://www.quidel.com/molecular-diagnostics/lyra-direct-sars-cov-2-assay)\n\n\n---\n\n\n**Deck Layout**\n\nSlot 1: [Opentrons 300\u00b5L Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 4: Eppendorf 96-Well Plate\n\nSlot 7: [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n**Process Buffer Loading:** Each column of the reservoir can contain enough buffer for up to 24 wells. Ex: if running 96 wells, columns 1-4 should be loaded with buffer\n\n\n\n",
"internal": "29effa-buffer\n",
diff --git a/protoBuilds/29effa-mastermix/README.json b/protoBuilds/29effa-mastermix/README.json
index 65d7e842c6..1208f12c5a 100644
--- a/protoBuilds/29effa-mastermix/README.json
+++ b/protoBuilds/29effa-mastermix/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol automates the mastermix distribution portion of the Lyra Direct SARS-CoV-2 Assay.\nUsing an Opentrons P20 Single-Channel Pipette, this protocol will transfer 15\u00b5L of mastermix from a 1.5mL eppendorf tube to the specified number of wells in a standard 96-well PCR plate.\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.21.0 or later)\nOpentrons Temperature Module, GEN2\n96-Well Aluminum Block for Temperature Module\nOpentrons P20 Single-Channel Pipette\nOpentrons 20\u00b5L Filter Tip Rack\nOpentrons 4-in-1 Tube Rack Set\nNEST 96-Well PCR Plate\nEppendorf 1.5mL Microcentrifuge Tube\nLyra Direct SARS-CoV-2 Assay Kit\n\n\n\nDeck Layout\n\nSlot 2: Opentrons 20\u00b5L Filter Tip Rack\n\nSlot 3: Opentrons Temperature Module with 96-Well Aluminum Block and NEST 96-Well PCR Plate\n\nSlot 6: Opentrons 4-in-1 Tube Rack Set with 24-Tube Topper, containing eppendorf 1.5mL microcentrifuge tube containing mastermix in position D1",
"internal": "29effa-mastermix",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol automates the mastermix distribution portion of the [Lyra Direct SARS-CoV-2 Assay](https://www.quidel.com/molecular-diagnostics/lyra-direct-sars-cov-2-assay).\nUsing an [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette), this protocol will transfer 15\u00b5L of mastermix from a 1.5mL eppendorf tube to the specified number of wells in a standard 96-well PCR plate.\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.21.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Temperature Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [96-Well Aluminum Block for Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set)\n* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 20\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* Eppendorf 1.5mL Microcentrifuge Tube\n* [Lyra Direct SARS-CoV-2 Assay Kit](https://www.quidel.com/molecular-diagnostics/lyra-direct-sars-cov-2-assay)\n\n\n---\n\n\n**Deck Layout**\n\nSlot 2: [Opentrons 20\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 3: [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [96-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) and [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n\nSlot 6: [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 24-Tube Topper, containing eppendorf 1.5mL microcentrifuge tube containing mastermix in position D1\n\n\n",
"internal": "29effa-mastermix\n",
diff --git a/protoBuilds/29f643/README.json b/protoBuilds/29f643/README.json
index 2f080ea8c0..cbe593ca2d 100644
--- a/protoBuilds/29f643/README.json
+++ b/protoBuilds/29f643/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"ELISA"
@@ -10,7 +10,7 @@
"internal": "29f643",
"labware": "\nNEST 12-Well Reservoir, 15 mL\nOpentrons Tip Rack, 300 \u00b5L\nOpentrons 24-Well Aluminum Block\nNunc MaxiSorp 96 Well Plate, 250\u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* ELISA\n\n\n",
"deck-setup": "\n\n\nStandards and Controls will be transferred to the first column of each 96-well plate and their duplicate in the second column. After the Standards and Controls have been added, each sample will be added in duplicate (to the well immediately to the right of the initial target destination). Please see below for an example of the resulting plate layout with one sample.\n\n\n",
"description": "This protocol automates the Alpco Insulin ELISA protocol. The protocol begins by transferring aliquots of samples, standards, and controls. The rest of the protocol can be automated or include manual interventions (for wash steps and off-deck incubations). Please note this protocol is currently being optimized.\n\n**Update:** This protocol was updated on September 8, 2022\n\n\nExplanation of complex parameters below:\n* **Number of Samples**: Specify the number of samples\n* **P300 Multi-Channel Mount**: Select which mount the P300 Multi-Channel Pipette is attached to (the Single-Channel Pipette should be attached to the opposite mount)\n\n\n",
diff --git a/protoBuilds/2a370c/README.json b/protoBuilds/2a370c/README.json
index 8ac8a3ed3a..6ef5577415 100644
--- a/protoBuilds/2a370c/README.json
+++ b/protoBuilds/2a370c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Omega Mag-Bind Bacterial DNA 96 Kit"
@@ -10,7 +10,7 @@
"internal": "2a370c",
"labware": "\nOpentrons Filter Tips\nPlateOne\u00ae Deep 96-Well 2 mL Polypropylene Plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Omega Mag-Bind Bacterial DNA 96 Kit\n\n",
"deck-setup": "\n\n",
"description": "This is a custom protocol that automates certain steps of the Omega Mag-Bind Bacterial DNA 96 Kit on the OT-2 robot. This kit allows rapid isolation of genomic DNA from bacterial samples.\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P300 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `P1000 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `Magnetic Module Engage Height`: The height the magnets will raise on the magnetic module.\n\n---\n\n",
diff --git a/protoBuilds/2aee74-48-2/README.json b/protoBuilds/2aee74-48-2/README.json
index adf6fd1fcf..733797539d 100644
--- a/protoBuilds/2aee74-48-2/README.json
+++ b/protoBuilds/2aee74-48-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Olink\u00ae Target 48"
@@ -10,7 +10,7 @@
"internal": "2aee74",
"labware": "\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nNEST 12-Well Reservoir, 15 mL\nOpentrons Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Olink\u00ae Target 48\n\n",
"deck-setup": "\nNote: all volumes for 48 samples (including controls)\n* green on reservoir (slot 5): 10562.0\u00b5l extension mix\n* blue on incubation plate (slot 2): samples\n\n---\n\n",
"description": "\nLinks:\n* [Part 1: Incubation](./2aee74-48)\n* [Part 2: Extension](./2aee74-48-2)\n* [Part 3: Detection](./2aee74-48-3)\n\nThis protocol accomplishes part 2/3: Extension for use with the [Olink\u00ae Target 48 protocol](https://www.olink.com/products-services/target/) for protein biomarker discovery.\n\nOlink\u00ae is a registered trademark of Olink Proteomics AB. Opentrons is not affiliated with or endorsed by Olink Proteomics AB.\n\n---\n\n",
diff --git a/protoBuilds/2aee74-48-3/README.json b/protoBuilds/2aee74-48-3/README.json
index 96724fb08a..3f43484a22 100644
--- a/protoBuilds/2aee74-48-3/README.json
+++ b/protoBuilds/2aee74-48-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Olink\u00ae Target 48"
@@ -10,7 +10,7 @@
"internal": "2aee74",
"labware": "\nFluidigm 96.96 Dynamic Array\u2122 IFC for Gene Expression\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOpentrons 4-in-1 Tube Rack Set with NEST 1.5 mL Screwcap Tubes or equivalent\nOpentrons Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Olink\u00ae Target 48\n\n",
"deck-setup": "\nNote: all volumes for 48 samples (including controls)\n* purple on primer plate (slot 1): primers\n* blue on sample plate (slot 5): samples from extension\n* green on tuberack (slot 9): 790.9\u00b5l detection mix\n\n---\n\n",
"description": "\nLinks:\n* [Part 1: Incubation](./2aee74-48)\n* [Part 2: Extension](./2aee74-48-2)\n* [Part 3: Detection](./2aee74-48-3)\n\nThis protocol accomplishes part 3/3: Detection for use with the [Olink\u00ae Target 48 protocol](https://www.olink.com/products-services/target/) for protein biomarker discovery. Primers are transferred to the left 48 wells of the Fluidigm detection plate, and samples are transferred to the right 48 wells. The transfer mapping for these plates is shown in the following image: \n\n\nOlink\u00ae is a registered trademark of Olink Proteomics AB. Opentrons is not affiliated with or endorsed by Olink Proteomics AB.\n\n---\n\n",
diff --git a/protoBuilds/2aee74-48/README.json b/protoBuilds/2aee74-48/README.json
index 293cf33c07..b54df09b92 100644
--- a/protoBuilds/2aee74-48/README.json
+++ b/protoBuilds/2aee74-48/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Olink\u00ae Target 48"
@@ -10,7 +10,7 @@
"internal": "2aee74",
"labware": "\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOpentrons 4-in-1 Tube Rack Set with NEST 1.5 mL Screwcap Tubes or equivalent\nOpentrons Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Olink\u00ae Target 48\n\n",
"deck-setup": "\nNote: all volumes for 48 samples (including controls)\n* blue on sample plate (slot 7): starting samples\n* green on tuberack (slot 8): 400.0\u00b5l incubation mix\n\n---\n\n",
"description": "\nLinks:\n* [Part 1: Incubation](./2aee74-48)\n* [Part 2: Extension](./2aee74-48-2)\n* [Part 3: Detection](./2aee74-48-3)\n\nThis protocol accomplishes part 1/3: Incubation for use with the [Olink\u00ae Target 48 protocol](https://www.olink.com/products-services/target/) for protein biomarker discovery.\n\nOlink\u00ae is a registered trademark of Olink Proteomics AB. Opentrons is not affiliated with or endorsed by Olink Proteomics AB.\n---\n\n",
diff --git a/protoBuilds/2aee74-96-2/README.json b/protoBuilds/2aee74-96-2/README.json
index e53094c797..6af243274a 100644
--- a/protoBuilds/2aee74-96-2/README.json
+++ b/protoBuilds/2aee74-96-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Olink\u00ae Target 96"
@@ -10,7 +10,7 @@
"internal": "2aee74",
"labware": "\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nNEST 12-Well Reservoir, 15 mL\nOpentrons Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Olink\u00ae Target 96\n\n",
"deck-setup": "\nNote: all volumes for 96 samples (including controls)\n* green on reservoir (slot 5): 10562.0\u00b5l extension mix\n* blue on incubation plate (slot 2): samples\n\n---\n\n",
"description": "\nLinks:\n* [Part 1: Incubation](./2aee74)\n* [Part 2: Extension](./2aee74-2)\n* [Part 3: Detection](./2aee74-3)\n\nThis protocol accomplishes part 2/3: Extension for use with the [Olink\u00ae Target 96 protocol](https://www.olink.com/products-services/target/) for protein biomarker discovery.\n\nOlink\u00ae is a registered trademark of Olink Proteomics AB. Opentrons is not affiliated with or endorsed by Olink Proteomics AB.\n\n---\n\n",
diff --git a/protoBuilds/2aee74-96-3/README.json b/protoBuilds/2aee74-96-3/README.json
index 121b954207..4f54271db1 100644
--- a/protoBuilds/2aee74-96-3/README.json
+++ b/protoBuilds/2aee74-96-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Olink\u00ae Target 96"
@@ -10,7 +10,7 @@
"internal": "2aee74",
"labware": "\nFluidigm 96.96 Dynamic Array\u2122 IFC for Gene Expression\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOpentrons 4-in-1 Tube Rack Set with NEST 1.5 mL Screwcap Tubes or equivalent\nOpentrons Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Olink\u00ae Target 96\n\n",
"deck-setup": "\nNote: all volumes for 96 samples (including controls)\n* purple on primer plate (slot 1): samples\n* blue on sample plate (slot 5): samples from extension\n* green on tuberack (slot 9): 790.9\u00b5l detection mix\n\n---\n\n",
"description": "\nLinks:\n* [Part 1: Incubation](./2aee74)\n* [Part 2: Extension](./2aee74-2)\n* [Part 3: Detection](./2aee74-3)\n\nThis protocol accomplishes part 3/3: Detection for use with the [Olink\u00ae Target 96 protocol](https://www.olink.com/products-services/target/) for protein biomarker discovery. Primers are transferred to the left 96 wells of the Fluidigm detection plate, and samples are transferred to the right 96 wells. The transfer mapping for these plates is shown in the following images: \n\n\n\nOlink\u00ae is a registered trademark of Olink Proteomics AB. Opentrons is not affiliated with or endorsed by Olink Proteomics AB.\n---\n\n",
diff --git a/protoBuilds/2aee74-96/README.json b/protoBuilds/2aee74-96/README.json
index 0df88e48fb..a73a1a77a2 100644
--- a/protoBuilds/2aee74-96/README.json
+++ b/protoBuilds/2aee74-96/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Olink\u00ae Target 96"
@@ -10,7 +10,7 @@
"internal": "2aee74",
"labware": "\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOpentrons 4-in-1 Tube Rack Set with NEST 1.5 mL Screwcap Tubes or equivalent\nOpentrons Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Olink\u00ae Target 96\n\n",
"deck-setup": "\nNote: all volumes for 96 samples (including controls)\n* blue on sample plate (slot 7): starting samples\n* green on tuberack (slot 8): 400.0\u00b5l incubation mix\n\n---\n\n",
"description": "\nLinks:\n* [Part 1: Incubation](./2aee74)\n* [Part 2: Extension](./2aee74-2)\n* [Part 3: Detection](./2aee74-3)\n\nThis protocol accomplishes part 1/3: Incubation for use with [Olink\u00ae Target 96 protocol](https://www.olink.com/products-services/target/) for protein biomarker discovery.\n\nOlink\u00ae is a registered trademark of Olink Proteomics AB. Opentrons is not affiliated with or endorsed by Olink Proteomics AB.\n\n---\n\n",
diff --git a/protoBuilds/2b3642/README.json b/protoBuilds/2b3642/README.json
index dbe8a95230..bcb05fd152 100644
--- a/protoBuilds/2b3642/README.json
+++ b/protoBuilds/2b3642/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II DNA Library Preparation Kit for Illumina E7645S"
@@ -10,7 +10,7 @@
"internal": "2b3642",
"labware": "\nOpentrons Aluminum Block Set\nNEST 12-well 15 mL Reservoir\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOpentrons 200uL Filter Tips\nOpentrons 20uL Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext Ultra II DNA Library Preparation Kit for Illumina E7645S\n\n",
"deck-setup": "\n**Note:** The deck layout shown in the image above will change throughout the protocol as labware is moved around. The image above represents the starting layout of the protocol. Please refer to the protocol steps below for more detailed instructions.\n\n",
"description": "This protocol automates various parts of the NEBNext Ultra II DNA Library Preparation Kit for Illumina E7645S using the OT-2 and additional hardware modules. The protocol is divided into five sections which includes the following: End Preparation, Adaptor Ligation, Cleanup of Adaptor-Ligated DNA, PCR Amplication and Cleanup of PCR Reaction.\n\nThe protocol will also pause at various stages prompting for user intervention to replace reagents, tip racks, and perform centrifugation of reaction mixtures.\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of samples or reactions being run. This kit supports a maximum of 24 samples. Please enter sample numbers in multiples of 8 (Ex: 8, 16, 24).\n* `P300 Multichannel GEN2 Mount Position`: The position your P300 Multichannel GEN2 pipette is mounted (Left or Right).\n* `P20 Single Channel GEN2 Mount Position`: The position your P300 Multichannel GEN2 pipette is mounted (Left or Right).\n* `Initial Denaturation Cycles`: Number of cycles at 98C for 30 seconds.\n* `Denaturation and Annealing/Extension Cycles`: Number of cycles at 98C for 10 seconds and 65C for 75 seconds.\n* `Final Extension Cycles`: Number of cycles at 65C for 5 minutes.\n\n---\n\n",
diff --git a/protoBuilds/2b9486/README.json b/protoBuilds/2b9486/README.json
index c77edaf285..6448b73aea 100644
--- a/protoBuilds/2b9486/README.json
+++ b/protoBuilds/2b9486/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom NGS library prep with the Opentrons Thermocycler module. The protocol requires 2 manual pauses for user intervention.\n\n\n\nOpentrons Thermocycler Module\nISOFREEZE cooling block\nThermo Scientific 96-well PCR plate 200\u00b5l\nNunc 96-deepwell plate 2ml\nUSA Scientific 2-200ul tips mounted on Opentrons tiprack base\n\n",
"internal": "2b9486",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Custom\n\n",
"description": "This protocol performs a custom NGS library prep with the Opentrons Thermocycler module. The protocol requires 2 manual pauses for user intervention.\n\n---\n\n\n* [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module)\n* [ISOFREEZE cooling block](https://www.fishersci.com/shop/products/isofreeze-pcr-sbs-rack-2pk/50999415)\n* [Thermo Scientific 96-well PCR plate 200\u00b5l](https://www.fishersci.com/shop/products/pcr-plate-96-well-low-profile-skirted-12/p-7184552#?keyword=ab+0800)\n* [Nunc 96-deepwell plate 2ml](https://www.thermofisher.com/order/catalog/product/260251#/260251)\n* USA Scientific 2-200ul tips mounted on Opentrons tiprack base\n\n---\n",
"internal": "2b9486\n",
diff --git a/protoBuilds/2bba96/README.json b/protoBuilds/2bba96/README.json
index 7c93a5de69..cbdca5aeb1 100644
--- a/protoBuilds/2bba96/README.json
+++ b/protoBuilds/2bba96/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -9,7 +9,7 @@
"description": "This protocol uses a p300 single-channel pipette to distribute custom volumes of buffer from a 12-well reservoir to specified wells of a 96-well child plate followed by transfer of well contents from up to five 96-well parent plates to specified wells in the child plate (volumes, plate locations and well locations are specified in a csv file uploaded at the time of protocol download).\nLinks:\n* example input csv\nThis protocol was developed to distribute custom volumes of buffer from a 12-well reservoir to specific wells of a child plate and then transfer contents from specific wells of parent plates to specified wells of the child plate.",
"internal": "2bba96",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Normalization\n\n",
"deck-setup": "\n\n* Opentrons 200ul filter tips (Deck Slot 6)\n* Opentrons 20ul filter tips (Deck Slots 1, 2, 3)\n* 96-well Child Plate (Deck Slot 5)\n* 96-well Parent Plates (Deck Slots 4, 7, 8, 10, 11)\n* Reservoir (Deck Slot 9)\n\n\n\n",
"description": "\nThis protocol uses a p300 single-channel pipette to distribute custom volumes of buffer from a 12-well reservoir to specified wells of a 96-well child plate followed by transfer of well contents from up to five 96-well parent plates to specified wells in the child plate (volumes, plate locations and well locations are specified in a csv file uploaded at the time of protocol download).\n\nLinks:\n* [example input csv](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/2bba96/ExampleCSV-cherrypicking+and+normalization+1.csv)\n\n\nThis protocol was developed to distribute custom volumes of buffer from a 12-well reservoir to specific wells of a child plate and then transfer contents from specific wells of parent plates to specified wells of the child plate.\n\n",
diff --git a/protoBuilds/2bdff3/README.json b/protoBuilds/2bdff3/README.json
index 1abea0ae33..a76cc017a8 100644
--- a/protoBuilds/2bdff3/README.json
+++ b/protoBuilds/2bdff3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Pooling"
@@ -8,7 +8,7 @@
"description": "This protocol performs the post RC-PCR pooling of the Nimagen SARS CoV-2 Library Prep. It has the option to pool samples from a variety of 384 well plates. The 384 well plate is divided into four quadrants and each quadrant can have up to 12 columns of samples, yielding a total of 48 columns worth of samples on a full plate.\n\nNote: The diagram above color coordinates the four quadrant positions. Each quadrant can take up to 12 columns worth of samples. The diagram above shows 1 column worth of samples.\n\n\n\nOpentrons 200uL Filter Tips\nOpentrons 20uL Filter Tips\n4-titude FrameStar 384 Well Plate\nBIOplastics 384 Well Plate\nEppendorf Twin.tec LoBind 384 Well\nEppendorf Twin.tec LoBind 96 Well\nP300 Single Channel GEN2\nP20 Multichannel GEN2\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nDeck Setup\n\nNote: Slots 9, 10, 11 contain empty tip racks for the Opentrons 20 uL tips.\nProtocol Steps\n\nMix column in the 384 well donor plate, then transfer sample to the first column of the 96 well recipient plate. This step should proceed for all available columns in all four quadrants.\nMix well A1 and then transfer the total volume of samples in A1 of the 96 well recipient plate to the 1.5 mL Eppendorf tube in position A1 of the tube rack. This step should proceed for all wells in column 1 (A1-H1).\n",
"internal": "2bdff3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Pooling\n\n",
"description": "This protocol performs the post RC-PCR pooling of the [Nimagen SARS CoV-2 Library Prep](https://www.nimagen.com/shop/products/rc-cov096/easyseq-sars-cov-2-novel-coronavirus-whole-genome-sequencing-kit). It has the option to pool samples from a variety of 384 well plates. The 384 well plate is divided into four quadrants and each quadrant can have up to 12 columns of samples, yielding a total of 48 columns worth of samples on a full plate.\n\n\n\n**Note:** The diagram above color coordinates the four quadrant positions. Each quadrant can take up to 12 columns worth of samples. The diagram above shows 1 column worth of samples.\n\n---\n\n\n* [Opentrons 200uL Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 20uL Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [4-titude FrameStar 384 Well Plate](https://www.thermofisher.com/order/catalog/product/95040450#/95040450)\n* [BIOplastics 384 Well Plate](https://www.bioplastics.com/productdetails.aspx?code=B70515)\n* [Eppendorf Twin.tec LoBind 384 Well](https://online-shop.eppendorf.co.uk/UK-en/Laboratory-Consumables-44512/Plates-44516/Eppendorf-twin.tec-PCR-Plates-LoBind-PF-58208.html#Accessory)\n* [Eppendorf Twin.tec LoBind 96 Well](https://online-shop.eppendorf.co.uk/UK-en/Laboratory-Consumables-44512/Plates-44516/Eppendorf-twin.tec-PCR-Plates-LoBind-PF-58208.html#Accessory)\n* [P300 Single Channel GEN2](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=5984549109789)\n* [P20 Multichannel GEN2](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n---\n\n\n**Deck Setup**\n\n\n\n**Note: Slots 9, 10, 11 contain empty tip racks for the Opentrons 20 uL tips.**\n\n**Protocol Steps**\n\n1. Mix column in the 384 well donor plate, then transfer sample to the first column of the 96 well recipient plate. This step should proceed for all available columns in all four quadrants.\n2. Mix well A1 and then transfer the total volume of samples in A1 of the 96 well recipient plate to the 1.5 mL Eppendorf tube in position A1 of the tube rack. This step should proceed for all wells in column 1 (A1-H1).\n\n",
"internal": "2bdff3",
diff --git a/protoBuilds/2c19aa/README.json b/protoBuilds/2c19aa/README.json
index 574b0cd05e..fae68af276 100644
--- a/protoBuilds/2c19aa/README.json
+++ b/protoBuilds/2c19aa/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol creates a custom plate-filling protocol from 1 source plate to up to 9 destination plate using a P300-multi channel pipette. The transfer scheme is shown here.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP300 Multi Channel Pipette\nNEST 96 Deepwell Plate 2mL\nGreiner Microlon 96 Well Plate 340 \u00b5L\n",
"internal": "2c19aa",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "This protocol creates a custom plate-filling protocol from 1 source plate to up to 9 destination plate using a P300-multi channel pipette. The transfer scheme is shown [here](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2020-11-24/ek13tro/image.png).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P300 Multi Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [NEST 96 Deepwell Plate 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep)\n* Greiner Microlon 96 Well Plate 340 \u00b5L\n\n",
"internal": "2c19aa\n",
diff --git a/protoBuilds/2c62b7/README.json b/protoBuilds/2c62b7/README.json
index efda82bf61..ae632cbdd8 100644
--- a/protoBuilds/2c62b7/README.json
+++ b/protoBuilds/2c62b7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Omega Mag-Bind Environmental DNA 96 Kit"
@@ -8,7 +8,7 @@
"description": "This protocol automates the Soil Protocol from the Mag-Bind\u00ae Environmental DNA 96 Kit from Omega Bio-tek.\nThe protocol on the OT-2 begins at Step 14 with the addition of the XP1 Buffer and Mag-Bind\u00ae Particles RQ. These two reagents should be prepared as a master mix (referred to as Master Mix 2) in the OT-2 protocol. The first 13 steps of the protocol should be prepared and performed manually. Please refer to the protocol steps section below for corresponding OT-2 steps.\n\n\n\nTemperature Module (GEN2)\nMagnetic Module (GEN2)\nNEST 2 mL 96-Well Deep Well Plate\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt\nOpentrons 96-well Aluminum Block\nNEST 12-Well Reservoirs, 15 mL\nOpentrons 300uL Tips\nOpentrons 200uL Filter Tips\nP300 8-Channel Electronic Pipette (GEN2)\nMag-Bind\u00ae Environmental DNA 96 Kit\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nDeck Setup\n\nNote: The Tip Isolator is simply an empty Opentrons tip rack that is filled with water. The water should be filled up to the level that is enough for the tips to be slightly touching the surface of the water. The top of a standard Opentrons ti rack is detachable from the base (looks like a deep well plate). The tip isolator is used to conserve tips at specific steps.\nReagent Setup\n\nThis section can contain finer detail and images describing reagent volumes and positioning in their respective labware. Examples:\nReservoir 1: Slot 5\n\nReservoir 2: Slot 2\n\n\nReagent Steps\nNote: Steps 1-13 are performed manually as instructed in the kit manual. The protocol starts on the OT-2 from step 14 and has modified steps that are written below.\n\nAdd 1 volume XP1 Buffer and 20 \u00b5L Mag-Bind\u00ae Particles RQ (Prepared as Master Mix 2). Mixing is performed 10 times thoroughly, referred to as tip mixing (start mixing from the middle, then the bottom, then back to the top)\n\nNote: The master mix is thoroughly mixed and then transferred to 3 columns at a time. It is then mixed again to prevent the beads from settling in the reservoir (repeats every 3 columns of samples)\n\n\nIncubate at Room Temp for 10 minutes while mixing (increase yields). Each column is mixed 2 times via tip mixing. Park tips in the Tip Isolator.\n\n\nEngage Magnetic Module and Delay for 5 minutes (settling time parameter) to allow mag beads to settle on the walls. \n\n\nAspirate supernatant and discard without touching the magnetic beads. Reuse tips from step 15 and discard tips.\n\n\nDisengage Magnetic Module\n\n\nAdd max volume (300uL on P300 Pipette) of VHB Buffer to NEST 2 mL 96-Well Deep Well Plate, V Bottom (Dispensed at a fast rate to agitate the beads).\n\n\nTip Mixing is performed and tips are conserved in the tip isolator. (Vortexing steps are substituted with Tip Mixing)\n\n\nEngage Magnetic Module and Delay for 5 minutes.\n\n\nAspirate supernatant and discard without touching the magnetic beads.\n\n\nDisengage Magnetic Module\n\n\nAdd 500 uL of 70% Ethanol to NEST 2 mL 96-Well Deep Well Plate, V Bottom\n\n\nTip Mixing is performed (reuse tips from step 20).\n\n\nEngage Magnetic Module and Delay for 5 minutes.\n\n\nAspirate supernatant and discard without touching the magnetic beads. \n\n\nRepeat 23-27 for a second ethanol wash\n\n\nWhile the plate is on top of the Magnetic Module and is engaged, delay 1 minute and then remove any liquid.\n\n\nWhile Magnetic Module is engaged, delay for 10 minutes to allow mag beads to dry.\n\n\nTransfer Elution buffer from A12 of Reservoir 1 to plate on temperature module. Heat elution buffer to 70C on the Temperature Module, then transfer 100 uL of elution buffer to samples.\n\n\nTip Mixing is performed (Tips are discarded).\n\n\nEngage Magnetic Module and Delay for 2 minutes to allow mag beads to settle on the walls.\n\n\nTransfer 100 uL of clear supernatant containing purified DNA to NEST 0.1 mL 96 Well PCR Plate, Full Skirt.\n\n",
"internal": "2c62b7",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Omega Mag-Bind Environmental DNA 96 Kit\n\n",
"description": "This protocol automates the Soil Protocol from the [Mag-Bind\u00ae Environmental DNA 96 Kit](https://www.omegabiotek.com/product/mag-bind-environmental-dna-kit/?cn-reloaded=1) from Omega Bio-tek.\n\nThe protocol on the OT-2 begins at Step 14 with the addition of the XP1 Buffer and Mag-Bind\u00ae Particles RQ. These two reagents should be prepared as a master mix (referred to as Master Mix 2) in the OT-2 protocol. The first 13 steps of the protocol should be prepared and performed manually. Please refer to the protocol steps section below for corresponding OT-2 steps.\n\n---\n\n\n* [Temperature Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Magnetic Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [NEST 2 mL 96-Well Deep Well Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-2-ml-96-well-deep-well-plate-v-bottom)\n* [NEST 96 Well Plate 100 \u00b5L PCR Full Skirt](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons 96-well Aluminum Block](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [Opentrons 300uL Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 200uL Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [P300 8-Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Mag-Bind\u00ae Environmental DNA 96 Kit](https://www.omegabiotek.com/product/mag-bind-environmental-dna-kit/?cn-reloaded=1)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n---\n\n\n**Deck Setup**\n\n\n**Note**: The **Tip Isolator** is simply an empty Opentrons tip rack that is filled with water. The water should be filled up to the level that is enough for the tips to be slightly touching the surface of the water. The top of a standard Opentrons ti rack is detachable from the base (looks like a deep well plate). The tip isolator is used to conserve tips at specific steps.\n\n**Reagent Setup**\n\n* This section can contain finer detail and images describing reagent volumes and positioning in their respective labware. Examples:\n* Reservoir 1: Slot 5\n\n* Reservoir 2: Slot 2 \n\n\n**Reagent Steps**\n**Note**: Steps 1-13 are performed manually as instructed in the [kit manual](https://ensur.omegabio.com/ensur/contentAction.aspx?key=Production.4023.S2R4E1A3.20190920.67.4679917). The protocol starts on the OT-2 from step 14 and has modified steps that are written below.\n\n14. Add 1 volume XP1 Buffer and 20 \u00b5L Mag-Bind\u00ae Particles RQ (Prepared as Master Mix 2). Mixing is performed 10 times thoroughly, referred to as `tip mixing` (start mixing from the middle, then the bottom, then back to the top)\n\n**Note**: The master mix is thoroughly mixed and then transferred to 3 columns at a time. It is then mixed again to prevent the beads from settling in the reservoir (repeats every 3 columns of samples)\n\n15. Incubate at Room Temp for 10 minutes while mixing (increase yields). Each column is mixed 2 times via tip mixing. Park tips in the Tip Isolator.\n\n16. Engage Magnetic Module and Delay for 5 minutes (settling time parameter) to allow mag beads to settle on the walls. \n\n17. Aspirate supernatant and discard without touching the magnetic beads. Reuse tips from step 15 and discard tips.\n\n18. Disengage Magnetic Module\n\n19. Add max volume (300uL on P300 Pipette) of VHB Buffer to NEST 2 mL 96-Well Deep Well Plate, V Bottom (Dispensed at a fast rate to agitate the beads).\n\n20. Tip Mixing is performed and tips are conserved in the tip isolator. (Vortexing steps are substituted with Tip Mixing)\n\n21. Engage Magnetic Module and Delay for 5 minutes.\n\n22. Aspirate supernatant and discard without touching the magnetic beads.\n\n23. Disengage Magnetic Module\n\n24. Add 500 uL of 70% Ethanol to NEST 2 mL 96-Well Deep Well Plate, V Bottom\n\n25. Tip Mixing is performed (reuse tips from step 20).\n\n26. Engage Magnetic Module and Delay for 5 minutes.\n\n27. Aspirate supernatant and discard without touching the magnetic beads. \n\n28. Repeat 23-27 for a second ethanol wash\n\n29. While the plate is on top of the Magnetic Module and is engaged, delay 1 minute and then remove any liquid.\n\n30. While Magnetic Module is engaged, delay for 10 minutes to allow mag beads to dry.\n\n31. Transfer Elution buffer from A12 of Reservoir 1 to plate on temperature module. Heat elution buffer to 70C on the Temperature Module, then transfer 100 uL of elution buffer to samples.\n\n32. Tip Mixing is performed (Tips are discarded).\n\n33. Engage Magnetic Module and Delay for 2 minutes to allow mag beads to settle on the walls.\n\n34. Transfer 100 uL of clear supernatant containing purified DNA to NEST 0.1 mL 96 Well PCR Plate, Full Skirt.\n\n",
"internal": "2c62b7",
diff --git a/protoBuilds/2c81c0/README.json b/protoBuilds/2c81c0/README.json
index 895b3eefd2..95977071bf 100644
--- a/protoBuilds/2c81c0/README.json
+++ b/protoBuilds/2c81c0/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Featured": [
"Cherrypicking"
@@ -10,7 +10,7 @@
"internal": "2c81c0",
"labware": "\nBio-Rad Hard-Shell\u00ae 96-Well PCR Plates 200\u00b5l #HSP9601\nNEST 96 Deepwell Plate 2mL\nOpentrons P10 GEN1 single-channel pipette\nOpentrons P300 GEN1 single-channel pipette\nBiotix 96 Filter Tiprack 10\u00b5l\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Featured\n\t* Cherrypicking\n\n",
"deck-setup": "* Example deck layout. Biotix and Opentrons tips should always be in below slots. Source and Destination labware and slots can be specified in the CSV.\n\n---\n\n",
"description": "This protocol performs a custom cherrypicking workflow from a .csv file using custom Biotix filtertips. This protocol also performs a mix step after transferring liquid (3 repetitions) with the ability to specify the aspirate and dispense height from the top of the source and destination wells.\n\n\n\nExplanation of complex parameters below:\n\n* `input .csv file`: Here, you should upload a .csv file formatted in the [following way](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/2c81c0/csv_template.csv), being sure to include the header line. Note that `source labware` and `destination labware` must be either `plate`, `tuberack`, or `deepplate`, which will be considered as the Bio-Rad 200ul plate, Opentrons 24 Eppendorf 1.5mL safelock snapcap tube rack, and the Nest 96 Deepwell Plate 2mL respectively. A value of 0 for the `Height Offset` columns returns the top of the well.\n* `P10 GEN1 single-channel pipette mount`: Specify which mount (left or right) to load the P10 GEN1 Single Channel pipette.\n* `P300 GEN1 single-channel pipette mount`: Specify which mount (left or right) to load the P300 GEN1 Single Channel pipette.\n* `Change Tips`: Specify whether the pipette should have a new tip either once per well transfer or in between well transfers.\n\n---\n\n",
diff --git a/protoBuilds/2c83f7/README.json b/protoBuilds/2c83f7/README.json
index c0e1070e1f..85fa850d33 100644
--- a/protoBuilds/2c83f7/README.json
+++ b/protoBuilds/2c83f7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Zymo Kit"
@@ -8,7 +8,7 @@
"description": "This protocol performs capsule nucleic acid purification according to the Zymo Research Direct-zol-96 RNA MagPrep kit. This protocol has been updated from this protocol (written with APIv1) to APIv2.\n\n\n\nOpentrons magnetic module\nP300 Multi-channel electronic pipette\nOpentrons 300ul tiprack\nBio-Rad Hardshell 96-Well PCR Plate 200ul #HSP9601\nAxygen 96-Deepwell Plate 2.0ml #P-DW-20-C-S\nAgilent 1 Well Reservoir 290mL #201252-100\nUSA Scientific 12-channel Reservoir 22ml #1061-8150\n\n\n\n12-channel reservoir (deck slot 5)\n channel 1: magbeads vortexed and added to the reservoir immediately before beginning the protocol run\n channel 2: DNAse\n channels 3-5: RNA buffer\n channels 6-8: magbead wash 1\n channels 9-11: magbead wash 2\n channel 12: DNAse/RNAse-free water",
"internal": "2c83f7",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Zymo Kit\n\n\n",
"description": "This protocol performs capsule nucleic acid purification according to the Zymo Research Direct-zol-96 RNA MagPrep kit. This protocol has been updated from [this protocol](https://protocol-delivery.protocols.opentrons.com/protocol/716efb) (written with APIv1) to APIv2.\n\n---\n\n\n* [Opentrons magnetic module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [P300 Multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 300ul tiprack](https://shop.opentrons.com/collections/opentrons-tips)\n* [Bio-Rad Hardshell 96-Well PCR Plate 200ul #HSP9601](https://www.bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [Axygen 96-Deepwell Plate 2.0ml #P-DW-20-C-S](https://ecatalog.corning.com/life-sciences/b2c/US/en/Genomics-&-Molecular-Biology/Automation-Consumables/Deep-Well-Plate/Axygen%C2%AE-Deep-Well-and-Assay-Plates/p/P-DW-20-C-S)\n* [Agilent 1 Well Reservoir 290mL #201252-100](https://www.agilent.com/store/en_US/Prod-201252-100/201252-100)\n* [USA Scientific 12-channel Reservoir 22ml #1061-8150](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n\n---\n\n\n12-channel reservoir (deck slot 5)\n* channel 1: magbeads **vortexed and added to the reservoir immediately before beginning the protocol run**\n* channel 2: DNAse\n* channels 3-5: RNA buffer\n* channels 6-8: magbead wash 1\n* channels 9-11: magbead wash 2\n* channel 12: DNAse/RNAse-free water\n\n",
"internal": "2c83f7\n",
diff --git a/protoBuilds/2cc59d/README.json b/protoBuilds/2cc59d/README.json
index ae0134013c..3579a83941 100644
--- a/protoBuilds/2cc59d/README.json
+++ b/protoBuilds/2cc59d/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "2cc59d",
"labware": "\nNEST 12 Well Reservoir\nNEST 1 Well Reservoir\nOpentrons 300ul Tips\nOpentrons 20ul Filter Tips\nOpentrons 4-in-1 tube rack with 1.5mL safe-lock Eppendorf tubes\nGreiner 96 Plate\nCostar 96 Plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n",
"description": "This protocol performs an ELISA assay between two 96 well plates. For detailed instruction, please see protocol steps below. Stop solution and TMB should only be added when prompted by the protocol upon a pause step. The protocol will automatically pause if it runs out of tips, prompting the user to replace. Tip boxes should always be placed in order of slot 7, 8, and 9 on the deck, even if running low sample numbers. Tip box on slot 9 is always used for 4 wash steps.\n\nExplanation of complex parameters below:\n* `Number of samples (1 to 38)`: Specify the number of samples for this run. The protocol will run as many columns rounding up on the number of samples provided.\n* `P20/P300 Pipette Mounts`: Specify which mount (left or right) to host the P20 and P300 pipettes.\n\n---\n\n",
diff --git a/protoBuilds/2d7126/README.json b/protoBuilds/2d7126/README.json
index 21ac4cfdd7..6cc6a62f2e 100644
--- a/protoBuilds/2d7126/README.json
+++ b/protoBuilds/2d7126/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Slide Spotting"
@@ -8,7 +8,7 @@
"description": "This protocol uses a custom 8-pin replicator and adapter to transfer samples from one Corning 96-flat well microplate to a second, column by column. The same set of pins is used for the entire process, and pins are returned at the end of the protocol.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nP300 Multi-Channel Pipette (GEN2)\nCorning 96 Well Plate 360 \u00b5L Flat\n\n",
"internal": "2d7126",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Slide Spotting\n\n\n",
"description": "This protocol uses a custom 8-pin replicator and adapter to transfer samples from one Corning 96-flat well microplate to a second, column by column. The same set of pins is used for the entire process, and pins are returned at the end of the protocol.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [P300 Multi-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Corning 96 Well Plate 360 \u00b5L Flat](https://ecatalog.corning.com/life-sciences/b2c/US/en/Microplates/Assay-Microplates/96-Well-Microplates/Corning\u00ae-96-well-Solid-Black-and-White-Polystyrene-Microplates/p/corning96WellSolidBlackAndWhitePolystyreneMicroplates)\n\n---\n",
"internal": "2d7126\n",
diff --git a/protoBuilds/2d7d86/README.json b/protoBuilds/2d7d86/README.json
index 0c2ae5212d..d9ef8c04f2 100644
--- a/protoBuilds/2d7d86/README.json
+++ b/protoBuilds/2d7d86/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"qpcr setup"
@@ -10,7 +10,7 @@
"internal": "2d7d86",
"labware": "\nOpentrons 20ul Filter tips\nThermofisher 96 well plate\nThermofisher 384 well plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* qpcr setup\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This protocols preps a 384 plate for qPCR processing. 1-14 samples can be selected. If running less than 14 samples, the protocol will pick up the correct number of tips (less than 8) to dispense across all columns in the 384 well plate. Tips are exchanged per source column in the 96 well plate.\n\nExplanation of complex parameters below:\n* `Tip pickup starting column (1-12)`: Specify which column (1-12) of the tiprack to start picking up tips. If \"3\" is selected, then the tip pickup will start on column 3 of the tip rack. Please ensure the starting tip column and the following tip column are full (16 tips between the selected column and the column after).\n* `P20 Multi-Channel Pipette Mount`: Specify which mount (left or right) to host the P20 Multi-Channel pipette.\n\n\n---\n\n",
diff --git a/protoBuilds/2d9a0c/README.json b/protoBuilds/2d9a0c/README.json
index b36c5cd2de..b5c8e7b493 100644
--- a/protoBuilds/2d9a0c/README.json
+++ b/protoBuilds/2d9a0c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"Complete PCR Workflow"
@@ -8,7 +8,7 @@
"description": "This protocol transfers buffer and sample to a PCR plate on an Opentrons Thermocycler. Buffer and mastermix is added in between temperature profile steps. Block temperature of the thermocycler is set to the temperature of the last sub-step in each profile incubation step. Lid temperature stays constant throughout the protocol and is specified by the user.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Thermocycler\nP20 Single Pipette\nP300 Single Pipette\nOpentrons 96 Tip Rack 20 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\nOpentrons 24 Tube Rack with Eppendorf 2 mL Safe-Lock Snapcap\nThermoFisher PCR Plate 96 Well Non Skirted\n\n\n\nFor this protocol, be sure that the pipettes (P20 and P300) are attached.\nUsing the customization fields below, set up your protocol.\n Lid Temperature: Specify lid temperature to be set for the duration of the protocol (default 105C).\n Tube rack used: Specify whether you are using 1.5mL or 2mL Eppendorf tube rack.\n Number of Samples in Plate B: Specify the number of samples to be transferred and ultimately cycled on the 96 well plate on the thermocycler.\n Number of tip racks: Specify the number of 20\u00b5L tip racks you will have loaded onto the deck.\n P20 Single Mount: Specify which mount the P10 is on (left or right).\n P300 Single Mount: Specify which mount the P50 is on (left or right).\nNote about 20\u00b5L tip racks\nYou can upload up to 3 Opentrons 20\u00b5L tip racks for this protocol in slots 1, 2, and 3, respectively. If only loading one tip rack, load it onto Slot 1. If loading two tip racks, load to Slots 1 and 2 and so forth. The protocol will prompt you to replace tips when tips run out - you will need to reload all tip racks to the original configuration at the start of the protocol.\nLabware Setup\nSlots 1, 2, 3: Opentrons 20\u00b5L tip rack(s)\nSlot 4: Thermo Fisher 96 well-plate\nSlot 5: Opentrons 24 Tube Rack with Eppendorf 1.5 or 2mL Safe-Lock Snapcap\nSlot 6: Opentrons 300\u00b5L tip rack\nSlot 7, 8, 10, 11: Opentrons Thermocycler loaded with Thermo Fisher 96 well-plate loaded",
"internal": "2d9a0c",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* Complete PCR Workflow\n\n",
"description": "This protocol transfers buffer and sample to a PCR plate on an Opentrons Thermocycler. Buffer and mastermix is added in between temperature profile steps. Block temperature of the thermocycler is set to the temperature of the last sub-step in each profile incubation step. Lid temperature stays constant throughout the protocol and is specified by the user.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Thermocycler](https://opentrons.com/modules/#thermocycler_module)\n* [P20 Single Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P300 Single Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [Opentrons 96 Tip Rack 20 \u00b5L](https://labware.opentrons.com/opentrons_96_tiprack_20ul?category=tipRack)\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://labware.opentrons.com/opentrons_96_tiprack_300ul?category=tipRack)\n* [Opentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap](https://labware.opentrons.com/opentrons_24_tuberack_eppendorf_1.5ml_safelock_snapcap?category=tubeRack)\n* [Opentrons 24 Tube Rack with Eppendorf 2 mL Safe-Lock Snapcap](https://labware.opentrons.com/opentrons_24_tuberack_eppendorf_2ml_safelock_snapcap?category=tubeRack)\n* [ThermoFisher PCR Plate 96 Well Non Skirted](https://www.thermofisher.com/order/catalog/product/AB0600?us&en#/AB0600?us&en)\n\n\n\n\n\n\n---\n\n\nFor this protocol, be sure that the pipettes (P20 and P300) are attached.\n\nUsing the customization fields below, set up your protocol.\n* Lid Temperature: Specify lid temperature to be set for the duration of the protocol (default 105C).\n* Tube rack used: Specify whether you are using 1.5mL or 2mL Eppendorf tube rack.\n* Number of Samples in Plate B: Specify the number of samples to be transferred and ultimately cycled on the 96 well plate on the thermocycler.\n* Number of tip racks: Specify the number of 20\u00b5L tip racks you will have loaded onto the deck.\n* P20 Single Mount: Specify which mount the P10 is on (left or right).\n* P300 Single Mount: Specify which mount the P50 is on (left or right).\n\n**Note about 20\u00b5L tip racks**\n\nYou can upload up to 3 Opentrons 20\u00b5L tip racks for this protocol in slots 1, 2, and 3, respectively. If only loading one tip rack, load it onto Slot 1. If loading two tip racks, load to Slots 1 and 2 and so forth. The protocol will prompt you to replace tips when tips run out - you will need to reload all tip racks to the original configuration at the start of the protocol.\n\n**Labware Setup**\n\nSlots 1, 2, 3: Opentrons 20\u00b5L tip rack(s)\n\nSlot 4: Thermo Fisher 96 well-plate\n\nSlot 5: Opentrons 24 Tube Rack with Eppendorf 1.5 or 2mL Safe-Lock Snapcap\n\nSlot 6: Opentrons 300\u00b5L tip rack\n\nSlot 7, 8, 10, 11: Opentrons Thermocycler loaded with Thermo Fisher 96 well-plate loaded\n\n\n",
"internal": "2d9a0c\n",
diff --git a/protoBuilds/2df9f8-pt2/README.json b/protoBuilds/2df9f8-pt2/README.json
index de694815a8..088a170831 100644
--- a/protoBuilds/2df9f8-pt2/README.json
+++ b/protoBuilds/2df9f8-pt2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "2df9f8-pt2",
"labware": "\nOpentrons 20ul tips\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "* This is the deck setup for a run with 94 samples (bottom two rows of slot 11 are not populated).\n\n\n\n\n---\n\n",
"description": "This protocol preps a 96 well plate by transferring 6ul from a source plate deepwell plate to a destination deepwell plate.\n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify the number of samples (1-94) to run on this protocol. Note: samples will be picked by row in the tube racks, in order of slots 7, 8, 10, and 11.\n* `P20 Mount`: Specify which side to mount your P20 Multi Channel Pipette.\n\n---\n\n",
diff --git a/protoBuilds/2df9f8-pt3/README.json b/protoBuilds/2df9f8-pt3/README.json
index 4114d4fc85..098dc36f18 100644
--- a/protoBuilds/2df9f8-pt3/README.json
+++ b/protoBuilds/2df9f8-pt3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "2df9f8-pt3",
"labware": "\nOpentrons 300ul tips\nNEST 2mL 96 well deep well plate\nNEST 195mL Reservoir\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "* This is the deck setup for a run with 94 samples (bottom two rows of slot 11 are not populated).\n\n\n\n\n---\n\n",
"description": "This protocol preps multiple 96 deep well plates with 600ul of saline up to the specified number of plates. All plates are fully filled with 600ul of saline. The left and right multi-channel pipettes aspirate and dispense two columns at a time. \n\nExplanation of complex parameters below:\n* `Number of Plates`: Specify the number of plates to fill starting from slot 4.\n\n\n---\n\n",
diff --git a/protoBuilds/2df9f8-pt4/README.json b/protoBuilds/2df9f8-pt4/README.json
index 7c226cebd3..69b18620d7 100644
--- a/protoBuilds/2df9f8-pt4/README.json
+++ b/protoBuilds/2df9f8-pt4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "2df9f8-pt4",
"labware": "\nNEST 2mL 96 well deep well plate\nOpentrons 300uL Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This protocol pools up to 5 plates by column to a final destination plate. 100ul of sample is taken from each column of each specified plate and dispensed into the final deep well plate on slot 11.\n\nExplanation of complex parameters below:\n* `Number of plates`: Specify the number of agar plates (1-5) to fill for this run.\n* `Number of columns in each plate`: Specify the number of columns (1-12) in each agar plate to fill.\n* `P300 Multi-Channel Pipette Mount`: Specify which mount (left or right) to host the P300 multi-channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/2df9f8/README.json b/protoBuilds/2df9f8/README.json
index 0ff72293f3..75e9390765 100644
--- a/protoBuilds/2df9f8/README.json
+++ b/protoBuilds/2df9f8/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "2df9f8",
"labware": "\nOpentrons 200ul tips\nMicroamp 200ul well plate\nOpentrons 4-in-1 tuberack with 4x6 tube insert (1.5mL tubes)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "* This is the deck setup for a run with 94 samples (bottom two rows of slot 11 are not populated).\n\n\n\n---\n\n",
"description": "This protocol preps a 96 well plate with heat inactivated Covid samples and mastermix, for further processing on a Quant-5. A1 and A2 are left for positive and negative conrols. The protocol can be considered in 2 main sections:\n\n* Mastermix added to 96 well plate\n* Samples added to 96 well plate\n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify the number of samples (1-94) to run on this protocol. Note: samples will be picked by row in the tube racks, in order of slots 7, 8, 10, and 11.\n* `Plate`: Specify whether loading the microamp (I48HO) 96 well plate, or another 96 well plate.\n* `P20 Mount`: Specify which side to mount your P20 Multi Channel Pipette.\n\n---\n\n",
diff --git a/protoBuilds/2e84cc/README.json b/protoBuilds/2e84cc/README.json
index cd1cac6dd3..82964f1a01 100644
--- a/protoBuilds/2e84cc/README.json
+++ b/protoBuilds/2e84cc/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina Nextera XT"
@@ -8,7 +8,7 @@
"description": "This protocol automates the TG Nextera XT DNA Sample Preparation Kit from illumina on up to 96 samples. While this protocol functions perfectly fine on up to 96 samples, it will take a significant amount of time to run that many samples. For most preparations, there will need to be a technician available to replace tip boxes as they are used.\n\n\n\nOpentrons P300-multi channel electronic pipettes (GEN2)\nOpentrons 200ul Filter Tips\nOpentrons P20-single channel electronic pipettes (GEN2)\nOpentrons 20ul Filter Tips\nOpentrons Thermocycler Module\nOpentrons Temperature Module (GEN2)\nOpentrons Magnetic Module (GEN2)\nNEST 12-Well Reservoirs, 15 mL\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nNextera XT DNA Library Preparation Kit\nPhusion High-Fidelity PCR Master Mix with HF Buffer\nKAPA HiFi HotStart ReadyMix\nOmega BioTek Mag-Bind TotalPure NGS\n",
"internal": "2e84cc",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina Nextera XT\n\n",
"description": "This protocol automates the TG Nextera XT DNA Sample Preparation Kit from illumina on up to 96 samples. While this protocol functions perfectly fine on up to 96 samples, it will take a significant amount of time to run that many samples. For most preparations, there will need to be a technician available to replace tip boxes as they are used.\n\n---\n\n\n* [Opentrons P300-multi channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons P20-single channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=31059478970462)\n* [Opentrons 20ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [Opentrons Thermocycler Module](https://shop.opentrons.com/products/thermocycler-module)\n* [Opentrons Temperature Module (GEN2)](https://shop.opentrons.com/products/tempdeck)\n* [Opentrons Magnetic Module (GEN2)](https://shop.opentrons.com/products/magdeck)\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 0.1 mL 96-Well PCR Plate, Full Skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Nextera XT DNA Library Preparation Kit](https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/nextera-xt-dna.html)\n* [Phusion High-Fidelity PCR Master Mix with HF Buffer](https://www.thermofisher.com/order/catalog/product/F531S#/F531S)\n* [KAPA HiFi HotStart ReadyMix](https://rochesequencingstore.com/catalog/kapa-hifi-hotstart-readymix/)\n* [Omega BioTek Mag-Bind TotalPure NGS](https://shop.opentrons.com/collections/verified-reagents/products/mag-bind-total-pure-ngs)\n\n",
"internal": "2e84cc\n",
diff --git a/protoBuilds/2ed4de-2/README.json b/protoBuilds/2ed4de-2/README.json
index cb49a73d22..03327d8418 100644
--- a/protoBuilds/2ed4de-2/README.json
+++ b/protoBuilds/2ed4de-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Ribogreen Assay"
@@ -10,7 +10,7 @@
"internal": "2ed4de",
"labware": "\nCorning 96 Well Plate 360 \u00b5L Flat\nNEST 12 Well Reservoir 15 mL\nNEST 2 mL 96-Well Deep Well Plate\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Screwcap Tubes or equivalent\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 1000 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Ribogreen Assay\n\n",
"deck-setup": "This example starting deck state shows the layout for 24 samples: \n\n\n* green on sample tuberack: samples\n* blue on reagent reservoir: working standard\n* pink on reagent reservoir: assay buffer (TE or TR)\n\n---\n\n",
"description": "\nLinks:\n* [Part 1](./2ed4de)\n* [Part 2](./2ed4de-2)\n\nThis is a protocol for the Flu Ribogreen Assay protocol. Samples are aligned in the sample tuberack (slot 1) in the following order:\n* A1 -> sample 1\n* B1 -> sample 2\n* C1 -> sample 3\n* D1 -> sample 4\n* A2 -> sample 5\n* B2 -> sample 6\n* C2 -> sample 7\n* D2 -> sample 8\n* etc.\n\n---\n\n",
diff --git a/protoBuilds/2ed4de/README.json b/protoBuilds/2ed4de/README.json
index b9c9ad4101..410fcf47f2 100644
--- a/protoBuilds/2ed4de/README.json
+++ b/protoBuilds/2ed4de/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Ribogreen Assay"
@@ -10,7 +10,7 @@
"internal": "2ed4de",
"labware": "\nCorning 96 Well Plate 360 \u00b5L Flat\nNEST 12 Well Reservoir 15 mL\nNEST 2 mL 96-Well Deep Well Plate\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Screwcap Tubes or equivalent\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 1000 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Ribogreen Assay\n\n",
"deck-setup": "This example starting deck state shows the layout for 8 samples: \n\n\n* green on sample tuberack: samples\n* blue on reagent reservoir: working standard 1\n* pink on reagent reservoir: assay buffer 1 (TE)\n* purple on reagent reservoir: working standard 2\n* orange on reagent reservoir: assay buffer 2 (TR)\n\n---\n\n",
"description": "\nLinks:\n* [Part 1](./2ed4de)\n* [Part 2](./2ed4de-2)\n\nThis is a protocol for the Flu Ribogreen Assay protocol. Samples are aligned in the sample tuberack (slot 1) in the following order:\n* A1 -> sample 1\n* B1 -> sample 2\n* C1 -> sample 3\n* D1 -> sample 4\n* A2 -> sample 5\n* B2 -> sample 6\n* C2 -> sample 7\n* D2 -> sample 8\n\n---\n\n",
diff --git a/protoBuilds/2ee5be/README.json b/protoBuilds/2ee5be/README.json
index 621666030b..db5a963dbe 100644
--- a/protoBuilds/2ee5be/README.json
+++ b/protoBuilds/2ee5be/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs PCR prep by reformatting 4 different 96 Well King Fisher plates into a MicroAmp 384 Well PCR plate. It begins by creating an adequate amount of mastermix depending on the number of samples.\n96 Well Plate Format\n\n384 Well Plate Format\n\n\n\n\nP300 single-channel GEN2 electronic pipette\nP20 multi-channel GEN2 electronic pipette\nOpentrons 300ul tiprack\nOpentrons 20ul tiprack\nKingFisher\u2122 Plastics for 96 deep-well format\nMicroAmp\u2122 Optical 384-Well Reaction Plate with Barcode\n\n\n\nDeck Setup\n KingFisher Deep Well Plates (Slots 1-4)\n MicroAmp 384 Well PCR Plate (Slot 5)\n NEST 12 Reservoir 15mL (Slot 6)\n - Water (A1)\n - Multiplex Reagent (A2)\n - Dye (A3)\n Opentrons 300 uL Tip Rack (Slot 7)\n* Opentrons 200 uL Tip Rack (Slots 8-11)",
"internal": "2ee5be",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol performs PCR prep by reformatting 4 different 96 Well King Fisher plates into a MicroAmp 384 Well PCR plate. It begins by creating an adequate amount of mastermix depending on the number of samples.\n\n**96 Well Plate Format**\n\n\n\n**384 Well Plate Format**\n\n\n\n---\n\n\n* [P300 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=5984549109789)\n* [P20 multi-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5978988707869)\n* [Opentrons 300ul tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 20ul tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [KingFisher\u2122 Plastics for 96 deep-well format](https://www.thermofisher.com/order/catalog/product/95040450#/95040450)\n* [MicroAmp\u2122 Optical 384-Well Reaction Plate with Barcode](https://www.thermofisher.com/order/catalog/product/4309849?SID=srch-srp-4309849#/4309849?SID=srch-srp-4309849)\n\n\n---\n\n\nDeck Setup\n* KingFisher Deep Well Plates (Slots 1-4)\n* MicroAmp 384 Well PCR Plate (Slot 5)\n* NEST 12 Reservoir 15mL (Slot 6)\n\t- Water (A1)\n\t- Multiplex Reagent (A2)\n\t- Dye (A3)\n* Opentrons 300 uL Tip Rack (Slot 7)\n* Opentrons 200 uL Tip Rack (Slots 8-11)\n\n",
"internal": "2ee5be",
diff --git a/protoBuilds/300073-2/README.json b/protoBuilds/300073-2/README.json
index 0746ae26d0..d8f66b36a3 100644
--- a/protoBuilds/300073-2/README.json
+++ b/protoBuilds/300073-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"PCR prep"
@@ -10,7 +10,7 @@
"internal": "300073-2",
"labware": "\nStellar Scientific 96 well plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* PCR prep\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This protocol transfers 5 \u00b5L of saliva samples from a source to a target 96 well plate using a 20 \u00b5L multi-channel pipette.\nThe protocol lets the user control the height of aspiration/dispension from the bottom of the samples and target wells, as well as the flow rate of aspiration and dispension.\n\n\nExplanation of parameters below:\n* `Number of samples to transfer`: How many samples to transfer from the source to the target plate\n* `Aspiration flow rate (\u00b5L/s)`: rate of aspiration in microliters per second\n* `Dispension flow rate (\u00b5L/s)`: rate of dispension in microliters per second\n* `Aspiration height from the bottom of the tubes (mm)`: Height from the bottom of the tubes to aspirate from in mm\n* `Dispension height from the bottom of the tubes (mm)`: Height from the bottom of the plate wells to dispense from in mm\n* `(Optional) Temperature module with aluminum block`: Parameter to indicate whether you are using a temperature module with an aluminum block to put your source plate on\n* `Set temperature of the temperature module`: Temperature to set on the temperature module in degrees C\n* `Amount of time to keep the pipette in the tube after aspiration (s)`: How many seconds to wait before withdrawing the pipette from the tube after aspiration\n\n---\n\n",
diff --git a/protoBuilds/300073/README.json b/protoBuilds/300073/README.json
index 52eff89e79..d21d56fe35 100644
--- a/protoBuilds/300073/README.json
+++ b/protoBuilds/300073/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"PCR prep"
@@ -10,7 +10,7 @@
"internal": "300073",
"labware": "\n4-in-1 tube racks\nStellar Scientific 96 well plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* PCR prep\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This protocol transfers saliva samples from 25 mL tubes to a 96 well plate. The samples are located in 6 4-in-4 tube racks on the left side of the deck. Optionally a second set of tube racks can replace the first one if more than 45 samples need to be distributed. Optionally the target plate can be placed on a temperature module with an aluminum block.\n\nThe destination plate is divided into four quadrants and the samples are distributed in a zig zag pattern from top left - top right - bottom left - bottom right (see below). Samples are not transferred to the three first wells of the target plate because they contain control samples, symmetrically the first 3 tube slots in Sample set 1:Tuberack quadrant 1 contain no sample tubes.\n\nThe tubes in the tuberacks in slot 10, 11, 7 and 8 constitute the 1st tuberack quadrant, and should be regarded as a single unit (Tuberack quadrant 1) consisting of 4 rows of tubes with 12 columns. Samples are transferred in row order to quadrant 1 of the target plate for Sample set 1, and to quadrant 3 for Sample set 2.\n\nSimilarly the tuberacks in slot 4, 5, 1 and 2 constitute Tuberack quadrant 2. Samples from Sample set 1 in Tuberack quadrant 2 are transferred to Target quadrant 2, and samples from Sample set 2 are transferred to Target quadrant 4.\n\n**96 well-plate layout**\n```\nTarget quadrant 1 (top left)\nA1-D1 to A6-D6 (A1-A3 are unused)\n\nTarget quadrant 2 (top right)\nA7-D7 to A12-D12\n\nTarget quadrant 3 (bottom left)\nE1-H1 to E6-H6\n\nTarget quadrant 4 (bottom right)\nE7-H7 to E12-H12\n```\n\nExplanation of parameters below:\n* `Is there a first set of tubes?`: Is there a first set of tubes? (Sometimes the user may have already transferred the first set of tubes and wants the protocol to run from the second set of tuberack samples)\n* `Number of tubes/samples in the first set of tubes`: How many tubes there are in the first set of tube racks. These samples will be transferred to quadrant 1 (and 2) of the destination plate\n* `Is there a second set of tubes?`: Whether to pause the protocol so that a second set of tube racks can be loaded onto the deck\n* `Number of tubes/samples in the second set of tubes`: How many samples there are in the second set of tuberacks (Only needs to be set if a second set of tube racks is loaded)\n* `Sample volume (\u00b5L) to aspirate`: Volume of sample to transfer (5 or 50 \u00b5L). This parameter controls which pipette and tiprack is used: 5 uL means that the 20 uL pipette will be used in in the right mount, and that the pipette tips should be filtered 20 uL tips. 50 uL means that the 300 uL pipette will be used in the left mount along with 200 uL filter tips.\n* `Aspiration flow rate (\u00b5L/s)`: rate of aspiration in microliters per second\n* `Dispension flow rate (\u00b5L/s)`: rate of dispension in microliters per second\n* `Aspiration height from the bottom of the tubes (mm)`: Height from the bottom of the tubes to aspirate from in mm\n* `Dispension height from the bottom of the tubes (mm)`: Height from the bottom of the plate wells to dispense from in mm\n* `(Optional) Temperature module with aluminum block`: Parameter to indicate whether you are using a temperature module with an aluminum block to put your target plate on\n* `Set temperature of the temperature module`: Temperature to set on the temperature module in degrees C\n* `Amount of time to keep the pipette in the tube after aspiration (s)`: How many seconds to wait before withdrawing the pipette from the tube after aspiration\n\n---\n\n",
diff --git a/protoBuilds/30174a/README.json b/protoBuilds/30174a/README.json
index b8fcb5d08d..d8d7ca64f8 100644
--- a/protoBuilds/30174a/README.json
+++ b/protoBuilds/30174a/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol automates dilution of DNA in wells in a plate (Plate 1) with TE in a second plate (Plate2). The protocol does this using an input .csv file with 3 columns: plate1 (for the well location to transfer), DNA (for volume of DNA to transfer), and TE Buffer (for volume of TE to transfer). TE is loaded from a trough initially into Plate 2, and then the DNA is loaded into Plate 2 from Plate 1.\nThe protocol can transfer at 1-20ul of DNA and 20-200ul of TE into Plate 2. \n\n\n\nOpentrons P300-single channel electronic pipettes (GEN2)\nOpentrons P20-single channel electronic pipettes (GEN2)\nOpentrons 200ul Filter Tips\nOpentrons 20ul Filter Tips\nNEST 1 Well Reservoir 195 mL\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\n",
"internal": "30174a",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol automates dilution of DNA in wells in a plate (Plate 1) with TE in a second plate (Plate2). The protocol does this using an input .csv file with 3 columns: plate1 (for the well location to transfer), DNA (for volume of DNA to transfer), and TE Buffer (for volume of TE to transfer). TE is loaded from a trough initially into Plate 2, and then the DNA is loaded into Plate 2 from Plate 1.\n\nThe protocol can transfer at 1-20ul of DNA and 20-200ul of TE into Plate 2. \n\n---\n\n\n* [Opentrons P300-single channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=5984549109789)\n* [Opentrons P20-single channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=31059478970462)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 20ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [NEST 1 Well Reservoir 195 mL](http://www.cell-nest.com/page94?_l=en&product_id=102)\n* [NEST 0.1 mL 96-Well PCR Plate, Full Skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n\n\n",
"internal": "30174a\n",
diff --git a/protoBuilds/30450c-part-2/README.json b/protoBuilds/30450c-part-2/README.json
index b692c49ab6..d36ba5ee11 100644
--- a/protoBuilds/30450c-part-2/README.json
+++ b/protoBuilds/30450c-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "30450c-part-2",
"labware": "\nOpentrons Tips for the p300 and p20 (https://shop.opentrons.com)\nNest 195 mL Reservoir\nCorning 96 Well Plate 360 \u00b5L Flat\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\n**Slots 1-5**: ELISA plates \n**Slot 7**: Reagent Reservoir (195 mL Nest reservoir) \n\n\n\n**Slot 8**: Sample Plate (96-Well PCR plate) \n\n\n\n**Slot 9**: Empty Opentrons 300 uL tip box \n**Slot 10**: Opentrons 20 uL tips \n**Slot 11**: Opentrons 300 uL tips \n\n\n---\n\n",
"description": "This protocol uses 8-channel P300 and p20 pipettes to transfer samples and standard dilutions to a batch of 1-5 ELISA plates for the attached BioLegend Human IL-2 ELISA. User-determined parameters are available to specify the number of ELISA plates (1-5), to include or not include a tip touch following dispenses, the aspiration position in the large reservoir (in millimeters above the labware bottom), and to automate or not automate the placement and replenishment of tips in row H of the empty tip box in slot 9 (these tips will be used to transfer standard from row H of the sample plate to row H of the ELISA plate).\n\nLinks:\n* [BioLegend Human IL-2 ELISA MAX Deluxe Set - user manual](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/30450c/431804_R7_Human_IL-2_Deluxe+1.pdf)\n\n\n---\n\n\n\n",
diff --git a/protoBuilds/30450c/README.json b/protoBuilds/30450c/README.json
index 328059f716..18382f0be2 100644
--- a/protoBuilds/30450c/README.json
+++ b/protoBuilds/30450c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "30450c",
"labware": "\nOpentrons Tips for the p300 (https://shop.opentrons.com)\nNest 195 mL Reservoir\nCorning 96 Well Plate 360 \u00b5L Flat\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\n**Slots 1-5**: ELISA plates \n**Slot 10**: Reagent Reservoir (195 mL Nest reservoir) \n**Slot 11**: Opentrons 300 uL tips \n\n\n---\n\n",
"description": "\nThis protocol uses an 8-channel P300 pipette to add 100 or 200 uL of reagent to a batch of 1-5 ELISA plates for steps of the attached BioLegend Human IL-2 ELISA. User-determined parameters are available to specify which step of the ELISA process is being performed (coating, blocking, detection antibody, avidin HRP, TMB substrate or stop step), the number of ELISA plates (1-5), the aspiration position in the large reservoir (in millimeters above the labware bottom), to optionally pause the robot with the P300 above the reagent reservoir to wait for off-deck plate washing steps, and to include or not include a tip touch following reagent dispenses.\n\nLinks:\n* [BioLegend Human IL-2 ELISA MAX Deluxe Set - user manual](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/30450c/431804_R7_Human_IL-2_Deluxe+1.pdf)\n\n\n---\n\n\n\n",
diff --git a/protoBuilds/313086-logixsmart-station-B/README.json b/protoBuilds/313086-logixsmart-station-B/README.json
index 812063a1ad..0dc27c7c15 100644
--- a/protoBuilds/313086-logixsmart-station-B/README.json
+++ b/protoBuilds/313086-logixsmart-station-B/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Covid Workstation": [
"qPCR Setup"
@@ -10,7 +10,7 @@
"internal": "313086",
"labware": "\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nOpentrons 24 tuberack with NEST 1.5 mL Microcentrifuge Tubes\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOpentrons 10\u00b5l tipracks\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Covid Workstation\n\t* qPCR Setup\n\n",
"deck-setup": "* green: mastermix\n* pink: positive control\n* purple: negative control \n* blue: starting samples\n\n\n---\n\n",
"description": "Links: \n* [Logix Smart Nasopharyngeal Covid-19 Plating (Station A)](./313086)\n* [Logix Smart Nasopharyngeal/Saliva Covid-19 PCR Prep (Station B)](./313086-logixsmart-station-B)\n\nThis protocol performs PCR prep in a NEST 96-well PCR plate. Samples with buffer pre-added should be arranged in a NEST 96-deepwell, and mastermix and controls should be loaded in 1.5ml microcentrifuge tubes in the Opentrons 4x6 tuberack. The transfer order is as shown below: \n\n\n---\n\n",
diff --git a/protoBuilds/313086/README.json b/protoBuilds/313086/README.json
index a1a9a1bb1e..db8bc455c4 100644
--- a/protoBuilds/313086/README.json
+++ b/protoBuilds/313086/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Covid Workstation": [
"Sample Plating"
@@ -10,7 +10,7 @@
"internal": "313086",
"labware": "\nNEST 12-Well Reservoirs, 15 mL\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nOpentrons 24 tuberack with NEST 1.5 mL Microcentrifuge Tubes\nOpentrons 300\u00b5l tipracks\nOpentrons 1000\u00b5l tipracks\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Covid Workstation\n\t* Sample Plating\n\n\n",
"deck-setup": "* green: lysis/binding buffer (13ml per channel)\n* pink: elution buffer: (9ml per channel)\n* blue: starting samples \n\n\n---\n\n",
"description": "Links: \n* [Logix Smart Nasopharyngeal Covid-19 Plating (Station A)](./313086)\n* [Logix Smart Nasopharyngeal/Saliva Covid-19 PCR Prep (Station B)](./313086-logixsmart-station-B)\n\nThis protocol plates lysis/binding buffer, samples, and elution buffer in a NEST 96-deepwell plate. Samples should be arranged in up to 4x Opentrons tuberacks, and buffers should be loaded in a NEST 12-channel reservoir. The transfer order is as shown below: \n\n\nDeepwells A1 and B1 are left empty for controls to be added later on.\n\n---\n\n",
diff --git a/protoBuilds/315118/README.json b/protoBuilds/315118/README.json
index 5da372b56e..5723a15d74 100644
--- a/protoBuilds/315118/README.json
+++ b/protoBuilds/315118/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Pooling"
@@ -9,7 +9,7 @@
"description": "This protocol automates the pooling of 20 ul aliquots taken from designated wells of a 2 mL 96-well plate and dispensed into a single tube.\nOnly the rear-most channel of the p300 multichannel pipette is used.\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons Multi-Channel p300 pipette\nOpentrons Tips for the p300 pipette\n\n\n",
"internal": "315118",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Pooling\n\n",
"deck-setup": "* Opentrons p300 tips (Deck Slot 4)\n* 'nest_96_wellplate_2ml_deep' (Deck Slot 5)\n* 'lvltechnologies_48_wellplate_2000ul' (Deck Slot 6)\n\n",
"description": "This protocol automates the pooling of 20 ul aliquots taken from designated wells of a 2 mL 96-well plate and dispensed into a single tube.\nOnly the rear-most channel of the p300 multichannel pipette is used.\n\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Multi-Channel p300 pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons Tips for the p300 pipette](https://shop.opentrons.com/collections/opentrons-tips)\n\n---\n\n\n",
diff --git a/protoBuilds/32207e/README.json b/protoBuilds/32207e/README.json
index 1f748d1f96..93c4805408 100644
--- a/protoBuilds/32207e/README.json
+++ b/protoBuilds/32207e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "32207e",
"labware": "\nOpentrons 4-in-1 Tube Rack with 50mL Falcon Tubes\nOpentrons 300ul Tips\nSarstedt PCR plate full skirt, 96 well, transparent, Low Profile, 100 \u00b5l\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This protocol preps a 96 well plate with up to 96 samples of DNA, normalizing all DNA samples to the same concentration with TE. TE should not be filled more than 20mL in the 50mL falcon tube to prevent pipette plunging. One tip is used to load TE onto the plate, whereas one tip per sample is used for loading DNA samples to plate. After DNA is loaded, DNA and TE are mixed at 80% of the total well volume for 3 repetitions. If a volume greater than 180ul is passed in to the TE volume (greater than well volume), an error is thrown and the protocol will stop. \n\nExplanation of complex parameters below:\n* `.csv file`: Please upload your csv file in the following format, header included (no commas in header!). \"Name\" column blocked out for proprietary reasons.\n\n* `DNA Aspiration Rate`: Specify the rate at which to aspirate DNA. A value of 1 is default, 0.5 being 50% of the default value, 1.2 being 20% faster than the default value, etc.\n* `Track tips?`: Specify whether to start at A1 of both tip racks, or to start picking up from where the last protocol left off.\n* `P20/P300 Mount`: Specify which mount (left or right) to host the P20 and P300 single channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/3308b4/README.json b/protoBuilds/3308b4/README.json
index 156dce4a1f..23222bef43 100644
--- a/protoBuilds/3308b4/README.json
+++ b/protoBuilds/3308b4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "3308b4",
"labware": "\n3x NEST 1 Well Reservoir 195ml\n3x NEST 96 Deepwell Plate 2ml\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n",
"deck-setup": "\n\n",
"description": "This is a specific protocol for aliquoting liquid from three reagent plates to three 96 deep well plates using a multi-channel p300 pipette. Tips are changed between reagents. Either pipette mount can be specified in setup.\n* 350ul SPR Wash Buffer is added to a 96 deep well plate\n* 350ul VHB Buffer is added to a second 96 deep well plate\n* 50ul nuclease free water is added to a third 96 deep well plate\n\n* 'P300-Multi Mount Side': Left or right refers to the pipette mount side\n\n",
diff --git a/protoBuilds/3359a5/README.json b/protoBuilds/3359a5/README.json
index 29f6e3fd5e..eebe858773 100644
--- a/protoBuilds/3359a5/README.json
+++ b/protoBuilds/3359a5/README.json
@@ -10,7 +10,7 @@
"internal": "3359a5",
"labware": "\nAny verified labware found in our Labware Library\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/3359a5). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/3359a5). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Featured\n\t* Cherrypicking\n\n",
"deck-setup": "Example Deck Setup - this is variable depending on the .csv uploaded.\n\n\n\n---\n\n",
"description": "With this protocol, your robot can perform multiple well-to-well liquid transfers using a single-channel pipette by parsing through a user-defined .csv file. The protocol can use Opentrons GEN1 or GEN2 pipettes.\n\nThis particular cherrypicking protocol allows you to specify the source plate labware and slot number, as well as the aspiration height above the bottom of the well (in mm). Appropriate tip racks will be placed in every slot that isn't already populated with a source or destination labware.\n\n\nExplanation of complex parameters below:\n* `Pipette Type`: Specify which single channel pipette you will be using for this protocol.\n* `input .csv file`: Here, you should upload a .csv file formatted in the [following way](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/3359a5/template.csv) being sure to include the header line. Refer to our [Labware Library](https://labware.opentrons.com/?category=wellPlate) to copy API names for labware to include in the `source_labware` and `destination_labware` columns of the .csv.\n\n---\n\n",
diff --git a/protoBuilds/33900b/README.json b/protoBuilds/33900b/README.json
index bc78d577d3..158dddfae4 100644
--- a/protoBuilds/33900b/README.json
+++ b/protoBuilds/33900b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"MagneHis\u2122 Protein Purification System"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom protein purification on the OT-2 using the Promega MagneHis\u2122 Protein Purification System in conjunction with the Opentrons magnetic module. The user is prompted to replace tipracks once in the protocol if necessary.\n\n\n\nOpentrons magnetic module with Greiner Bio-One 96-deepwell block 2ml #780270\nGreiner Bio-One 96-well PCR plate 200\u00b5l #651161\nAgilent 12-channel reservoir 21ml #201256-100\nAgilent 3-channel reservoir 95ml #204249-100\nOpentrons P300 GEN1 multi-channel electronic pipette\nOpentrons 300ul tipracks\n\n\n\n12-channel reservoir (slot 2, volumes for 96 samples)\n channel 1: 17ml 10x FastBreak\u2122 Cell Lysis Reagent\n channel 2: 17ml 10x Benzonase/Lysozyme Mix\n channel 3: 17ml 5M NaCl\n channel 4: 10ml MagneHis particles\n channel 5: 17ml MagneHis Elution Buffer\n channels 7-10: liquid waste (loaded empty)\n3-channel reservoir (slot 3, volumes for 96 samples)\n channel 1: 80ml MagneHis Wash Buffer\n channels 2-3: liquid waste (loaded empty)",
"internal": "33900b",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* MagneHis\u2122 Protein Purification System\n\n",
"description": "This protocol performs a custom protein purification on the OT-2 using the [Promega MagneHis\u2122 Protein Purification System](https://www.promega.co.uk/products/protein-purification/protein-purification-kits/magnehis-protein-purification-system/?catNum=V8550) in conjunction with the [Opentrons magnetic module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck). The user is prompted to replace tipracks once in the protocol if necessary.\n\n---\n\n\n* [Opentrons magnetic module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [Greiner Bio-One 96-deepwell block 2ml #780270](https://shop.gbo.com/en/usa/products/bioscience/microplates/polypropylene-storage-plates/96-well-masterblock-2ml/780270.html)\n* [Greiner Bio-One 96-well PCR plate 200\u00b5l #651161](https://shop.gbo.com/en/usa/products/bioscience/microplates/96-well-microplates/96-well-microplates-clear/651161.html)\n* [Agilent 12-channel reservoir 21ml #201256-100](https://www.agilent.com/store/productDetail.jsp?catalogId=201256-100)\n* [Agilent 3-channel reservoir 95ml #204249-100](https://www.agilent.com/store/productDetail.jsp?catalogId=204249-100)\n* [Opentrons P300 GEN1 multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 300ul tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n\n---\n\n\n12-channel reservoir (slot 2, volumes for 96 samples)\n* channel 1: 17ml 10x FastBreak\u2122 Cell Lysis Reagent\n* channel 2: 17ml 10x Benzonase/Lysozyme Mix\n* channel 3: 17ml 5M NaCl\n* channel 4: 10ml MagneHis particles\n* channel 5: 17ml MagneHis Elution Buffer\n* channels 7-10: liquid waste (loaded empty)\n\n3-channel reservoir (slot 3, volumes for 96 samples)\n* channel 1: 80ml MagneHis Wash Buffer\n* channels 2-3: liquid waste (loaded empty)\n\n",
"internal": "33900b\n",
diff --git a/protoBuilds/33a449/README.json b/protoBuilds/33a449/README.json
index fdb7139e28..d098c71fc3 100644
--- a/protoBuilds/33a449/README.json
+++ b/protoBuilds/33a449/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"DNA Extraction"
@@ -10,7 +10,7 @@
"internal": "33a449",
"labware": "\nOpentrons 200ul or 300ul tips\nNEST 12 Well Reservoir\nAbgene 96 Well Plate 700ul\nMacherey Nagel 96 Squarewell Block\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* DNA Extraction\n\n",
"deck-setup": "* Note: reagents in reservoirs are in the order of the colors from left to right.\n\n\n\n\n",
"description": "This protocol extracts DNA using the [Nucleomag blood DNA kit](https://www.mn-net.com/us/nucleomag-blood-200-l-for-dna-purification-from-blood-744501.4). Pro-k is added to sample, as well as bead buffer. Supernatant is removed with magnets engaged before a dispense of 200ul of mbl3. After supernatant is again removed, ethanol is dispensed and removed in wash before mbl5 is added to sample. The final product is moved to the elute plate on slot 2.\n\nExplanation of complex parameters below:\n* `Number of Columns`: Specify the number of sample columns to run.\n* `Use filter tips`: Specify whether this run will use filter tips or regular tips.\n* `mbl5 Volume`: Specify the mbl5 volume in microliters.\n* `Magnetic engage height (mm)`: Specify the engage height for the magnets to raise under the base of the magnetic plate. The engage height should be empirically tested.\n* `P300 Multi-Channel Mount`: Specify which mount (left or right) to host the P300 multi-channel pipette.\n\n\n\n---\n\n",
diff --git a/protoBuilds/33b12a/README.json b/protoBuilds/33b12a/README.json
index 7aacf43069..89c07c5084 100644
--- a/protoBuilds/33b12a/README.json
+++ b/protoBuilds/33b12a/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol automates the transfer of rehydration buffer, samples, positive control, and negative control from the CerTest VIASURE SARS-CoV-2 Real Time PCR Detection kit into a PCR plate. It requires a 96 deep well plate of samples (1-94), the positive and negative control tubes in a tube rack, and a new PCR plate. This protocol uses exactly 2 tip racks and takes advantage of manually programmed multichannel transfers so that it only requires a p20 multichannel pipette. \n\n\n\nOpentrons P20-multi channel electronic pipette (GEN2)\nOpentrons 20ul Filter Tips\nNEST 1 Well Reservoir 195 mL\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nVIASURE SARS-CoV-2 Real Time PCR Detection Kit\n",
"internal": "33b12a",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol automates the transfer of rehydration buffer, samples, positive control, and negative control from the CerTest VIASURE SARS-CoV-2 Real Time PCR Detection kit into a PCR plate. It requires a 96 deep well plate of samples (1-94), the positive and negative control tubes in a tube rack, and a new PCR plate. This protocol uses exactly 2 tip racks and takes advantage of manually programmed multichannel transfers so that it only requires a p20 multichannel pipette. \n\n---\n\n\n* [Opentrons P20-multi channel electronic pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5978988707869)\n* [Opentrons 20ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [NEST 1 Well Reservoir 195 mL](http://www.cell-nest.com/page94?_l=en&product_id=102)\n* [NEST 0.1 mL 96-Well PCR Plate, Full Skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [VIASURE SARS-CoV-2 Real Time PCR Detection Kit](https://www.certest.es/viasure/)\n\n\n",
"internal": "33b12a\n",
diff --git a/protoBuilds/33y0f3/README.json b/protoBuilds/33y0f3/README.json
index b3459e103f..0bc3b0bc64 100644
--- a/protoBuilds/33y0f3/README.json
+++ b/protoBuilds/33y0f3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Lachlan Munro",
+ "author": "Lachlan Munro\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Microbiology": [
"Microbiology"
@@ -10,7 +10,7 @@
"internal": "33y0f3",
"labware": "\nBio-Rad 96 Well Plate 200 \u00b5L PCR #hsp9601\nCorning 96 Well Plate 360 \u00b5L Flat #3650\nOpentrons 96 Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "Lachlan Munro\n\n\n",
+ "author": "Lachlan Munro\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Microbiology\n\t* Microbiology\n\n\n",
"deck-setup": "[deck]()\n\n\n",
"description": "This is a protocol to dilute and spot bacteria for isolation and quantification. Bacterial samples are passed through 10, 10x serial dilutions with each dilution spotted onto agar. This can process up to 40 samples at a time (5 plates x 8 wells (1 column) per plate). \n\nPlates should be preloaded with the bacterial samples in column 1 and 90 \u00b5L of dilution buffer or media in columns 2-12. Agar plates should be prepared by filling Nunc Omnitrays with 30 mL of melted agar. These should be dried on as flat a surface as possible to ensure consistent heights across the plate.\n\nCalibration on the agar plate should be performed so that the tips are just barely touching the agar as the \"top of the well\". \n\n\n",
diff --git a/protoBuilds/343001/README.json b/protoBuilds/343001/README.json
index 1e8bb1a511..88964b3245 100644
--- a/protoBuilds/343001/README.json
+++ b/protoBuilds/343001/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Zymo Extraction"
@@ -8,7 +8,7 @@
"description": "This protocol performs a Zymo extraction. The protocol splits elutes (up to 48) six columns apart (for ex, wells A1 and A7).\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Magnetic Module\nOpentrons P50 Single-Channel Pipette\nOpentrons P1000 Single-Channel Pipette\nOpentrons 50/300\u00b5L Tips\nOpentrons 1000\u00b5L Tips\nOpentrons Tube Rack, 24-well\nOpentrons Tube Rack, 6-Well\nNEST 1-Channel Reservoir, 195mL\nZymo Elution Plate\nZymo Block Plate\nReagents\nSamples\n\n\n\nSlot 1: Zymo Elution Plate, clean and empty\nSlot 2: Opentrons Tube Rack, 24-well with 2mL Tubes\n A1: Proteinase K\n A2: Magbeads\n B1: DNase/RNase-Free Water\n B2: DNase/RNase-Free Water\nSlot 3: Opentrons Tube Rack, 6-well with 50mL Falcon Tubes\n A1: Pathogen DNA/RNA Buffer\n A2: Wash Buffer 1\n B2: Wash Buffer 2\n A3: Ethanol\n* B3: Ethanol\nSlot 4: Opentrons Magnetic Module with 200\u00b5L Sample in Zymo Block Plate\nSlot 5: Opentrons 1000\u00b5L Tips\nSlot 6: Opentrons 1000\u00b5L Tips\nSlot 7: Opentrons 1000\u00b5L Tips\nSlot 8: Opentrons 1000\u00b5L Tips\nSlot 9: Opentrons 50/300\u00b5L Tips\nSlot 10: Opentrons 50/300\u00b5L Tips\nSlot 11: NEST 1-Channel Reservoir, 195mL (for liquid waste)\nUsing the customizations fields, below set up your protocol.\n Number of Samples: Specify the number of samples (1-48) you'd like to run.\n P50 Single Mount: Select which mount (left or right) the P50 Single is attached to.\n P1000 Single Mount: Select which mount (left or right) the P1000 Single is attached to.\n MagDeck Incubation Time (mins): Specify how long (in minutes) the MagDeck should be engaged to create pellet, before removing supernatant.\n* Water Volume (for elution): Specify how much DNase/RNase-Free Water (in microliters) to add to each sample. This elution volume will then be split in two different wells in equal amounts.",
"internal": "343001",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Zymo Extraction\n\n\n",
"description": "This protocol performs a [Zymo](https://www.zymoresearch.com/) extraction. The protocol splits elutes (up to 48) six columns apart (for ex, wells `A1` and `A7`).\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P50 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons P1000 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 1000\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* [Opentrons Tube Rack, 24-well](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [Opentrons Tube Rack, 6-Well](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [NEST 1-Channel Reservoir, 195mL](https://labware.opentrons.com/nest_1_reservoir_195ml?category=reservoir)\n* Zymo Elution Plate\n* Zymo Block Plate\n* Reagents\n* Samples\n\n\n---\n\n\nSlot 1: Zymo Elution Plate, clean and empty\n\nSlot 2: [Opentrons Tube Rack, 24-well](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 2mL Tubes\n* A1: Proteinase K\n* A2: Magbeads\n* B1: DNase/RNase-Free Water\n* B2: DNase/RNase-Free Water\n\nSlot 3: [Opentrons Tube Rack, 6-well](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 50mL Falcon Tubes\n* A1: Pathogen DNA/RNA Buffer\n* A2: Wash Buffer 1\n* B2: Wash Buffer 2\n* A3: Ethanol\n* B3: Ethanol\n\nSlot 4: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with 200\u00b5L Sample in Zymo Block Plate\n\nSlot 5: [Opentrons 1000\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n\nSlot 6: [Opentrons 1000\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n\nSlot 7: [Opentrons 1000\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n\nSlot 8: [Opentrons 1000\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n\nSlot 9: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 10: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 11: [NEST 1-Channel Reservoir, 195mL](https://labware.opentrons.com/nest_1_reservoir_195ml?category=reservoir) (for liquid waste)\n\n\n\n\n**Using the customizations fields, below set up your protocol.**\n* **Number of Samples**: Specify the number of samples (1-48) you'd like to run.\n* **P50 Single Mount**: Select which mount (left or right) the P50 Single is attached to.\n* **P1000 Single Mount**: Select which mount (left or right) the P1000 Single is attached to.\n* **MagDeck Incubation Time (mins)**: Specify how long (in minutes) the MagDeck should be engaged to create pellet, before removing supernatant.\n* **Water Volume (for elution)**: Specify how much DNase/RNase-Free Water (in microliters) to add to each sample. This elution volume will then be split in two different wells in equal amounts.\n\n\n\n",
"internal": "343001\n",
diff --git a/protoBuilds/35269b/README.json b/protoBuilds/35269b/README.json
index 71757976a8..e35de19eb9 100644
--- a/protoBuilds/35269b/README.json
+++ b/protoBuilds/35269b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Viral RNA"
@@ -8,7 +8,7 @@
"description": "This protocol automates a multi-wash virus isolation using magnetic beads and a magnetic module GEN2. It contains custom shakes in between transfers to release any droplets on the end of the pipette tips.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons P300 GEN2 Multi-Channel Pipette\nOpentrons 200\u00b5L Filter Tips\nUSA Scientific 12-Well Reservoir, 22ml\nOpentrons Magnetic Module (GEN2)\nNEST 96 Well Plate 2000 \u00b5L\n\n",
"internal": "35269b",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Viral RNA\n\n",
"description": "This protocol automates a multi-wash virus isolation using magnetic beads and a magnetic module GEN2. It contains custom shakes in between transfers to release any droplets on the end of the pipette tips.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons P300 GEN2 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [USA Scientific 12-Well Reservoir, 22ml](https://www.usascientific.com/12-channel-automation-reservoir/p/1061-8150)\n* [Opentrons Magnetic Module (GEN2)](https://shop.opentrons.com/products/magdeck)\n* NEST 96 Well Plate 2000 \u00b5L\n\n---\n\n",
"internal": "35269b",
diff --git a/protoBuilds/355b30/README.json b/protoBuilds/355b30/README.json
index f76aefa33c..d9fe022a56 100644
--- a/protoBuilds/355b30/README.json
+++ b/protoBuilds/355b30/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Pooling"
@@ -10,7 +10,7 @@
"internal": "355b30",
"labware": "\nOpentrons Tips for the Selected Pipette (https://shop.opentrons.com)\nOpentrons Temperature Modules (https://shop.opentrons.com/modules/)\nSelected Source and Destination Labware ([see parameter dropdown lists below] https://labware.opentrons.com/)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Pooling\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Temperature Module with dropdown-selected labware \n**Slot 4**: 2nd Opentrons Temperature Module with dropdown-selected labware \n**Slots 2-6,11**: dropdown-selected labware \n**Slots 7-9**: Opentrons Tips \n**Slot 10**: dropdown-selected reservoir \n\n\n---\n\n",
"description": "This protocol uses single-channel pipettes to prepare a number of sample mixtures composed of a number of samples from a number of sample groups according to an input CSV file. User-determined parameters are available for selection of the left and right pipettes (none, p20 single, p300 single or p1000), pipetting mode (one pipette operating alone, two pipettes operating independently with all steps of one transfer being completed before any steps of the next transfer are started, or two pipettes multi-tasking with coordinated tip pickups, aspirations, dispenses, and tip drops), source and destination labware (see dropdown lists below), flow rates, and filtered or standard pipette tips. The pipette used for each transfer will be one with a volume range inclusive of the transfer volume (p20 for transfer volume 1-20 uL, p300 for 20-200 uL, p1000 for 100 - 1000 uL) or the next smaller pipette if the former is not available. If pipettes are selected that are not compatible with the volumes specified (10 uL transfer volume with p300 and p1000 pipettes for example), an error will be thrown to notify the user prior to the start of the OT-2 run. If the aspiration volume + air gap volume exceeds tip capacity the transfer will be divided into multiple transfers having equal volume.\n\nLinks:\n* [example input csv](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/355b30/csv_example.csv)\n\n\n\n---\n\n\n\n",
diff --git a/protoBuilds/357404/README.json b/protoBuilds/357404/README.json
index 34a1d9d735..1cbf719939 100644
--- a/protoBuilds/357404/README.json
+++ b/protoBuilds/357404/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Microscopy Slide Antibody Staining"
@@ -10,7 +10,7 @@
"internal": "357404",
"labware": "\nOpentrons tuberacks\nOpentrons tubes & vials\nAgilent 1-Well Reservoir 290 mL\n1000 uL tipracks\nOpentrons aluminum block set (optional)\nAgilent 1-Well Reservoir 290 mL\nCustom 3D printed block for holding Shandon coverplates + slides.\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Microscopy Slide Antibody Staining\n\n",
"deck-setup": "\n\n",
"description": "This protocol performs immunostaining of slides in a custom 3D printed slide block with Shandon coverplates.\nUp to 7 slide blocks, each with 8 wells can be placed on the deck resulting in the ability to immunostain up to 56 slides simultaneously.\n\nThe protocol can be stopped after the addition of the block reagent if the user wants to incubate the slides overnight, and can be restarted at the next step which is the addition of the 1st antibody.\n\nPipette racks and reagents may have to be replaced during the run depending on the number of samples. If that happens the lights on the OT-2 will flash to indicate that the protocol requires the users attention.\n\nExplanation of parameters below:\n* `Number of slide blocks`: How many slide holding blocks there are on the deck. The maximum number that will fit on the deck are 7.\n* `Number of samples in the last block`: All slide blocks except the last one are assumed to be full, however the last block may have a number of samples between 1 and 8\n* `Volume in reagents containers`: How much volume (\u00b5L) there is in each reagent tube (meaning block, antibody 1, antibody 2, and nuclear counterstain tubes). It is a good idea to have some amount of excess in each tube, about 50-100 uL to account for pipetting variance.\n* `Sweep dispense steps`: How many discrete step motions + dispenses to do when dispensing reagents and PBS, this is designed to replicate a sweeping motion with a manual pipette while dispensing. For example if the parameter is set to 5 the pipette will cover the length of the Shandon coverplate's mouth in 5 steps and dispense a 5th of the total dispensation volume each time.\n* `Reagent tuberack`: What type of tuberack you wish to use for the reagents (block, the antibodies and the nuclear counterstain). The maximal number of tubes for any reagents is 4 (a full column, so dimension the tubes accordingly). The maximal required volume for each reagent is 4.9 mL for a full deck (7 blocks*8 samples each). This volume would fit for example in 4 1.5 mL tubes.\n* `Pipette offset`: Pipetting offset in `millimeter` when dispensing, increasing this parameter will mean that the pipette will dispense at a higher height in the wells, while making it negative will lower the height of dispenses. **This parameter must be adjusted carefully so that there are no collisions between the pipette tip and the Shandon coverplates!**\n* `Start protocol after 1st incubation step`: Starts the protocol at the step after the samples have already been incubated with `block` overnight.\n* `Stop protocol after 1st incubation step`: The protocol stops after adding the block reagent and the user is asked to incubate the samples at 4 degrees C overnight.\n* `Do a dry run?`: Skip all incubation pauses and return tips to their racks after use.\n* `Time per block (s)`: The amount of time it takes to transfer reagent per block. The user must measure the time it takes for the reagent dispensation steps to finish for each block using a stopwatch, for example by putting the maximum of of seven blocks on the deck, running the protocol and then calculating the average block time by dividing by seven. This time is used to subtract from the total incubation time such that each sample will have an hour of incubation instead of 1 hour plus the time it takes to transfer reagent or PBS.\n* `Multi-dispense reagents?`: Whether to aspirate a 100 uL of reagent at a time and dispense it in the next well or aspirate 900 uL and go from well to well dispensing 100 uL each before picking up more reagent.\n* `Reuse reagent tips?`: Use the same tip for each individual reagent.\n* `Reuse PBS wash tips?`: Use one tip to do one wash with PBS\n* `Use temperature module?`: Whether to use a temperature module to chill the reagents in the tuberack, or put the tuberack directly on the slot.\n* `P1000 slot`: Which mount to use for the P1000 single GEN2 pipette\n* `Reagent tuberack`: Which reagent tuberack you want to use in the protocol\n* `Well edge offset (mm)`: The offset from the well's edge when performing the dispensing while moving action. The pipette will start dispensing on one side of the well minus the offset and will travel to the other side of the well minus the same offset.\n* `Use a custom slide block`: Setting this parameter to Yes means that the protocol will try to load an alternative definition for the slide block. See `Custom labware loadName` parameter below.\n* `Custom labware loadName`: Put the labware load name of the alternative slide block labware definition you wish to use. To find the loadName: Open your labware_definition.json file, look for the section \"parameters\"\ne.g.\n```\n\"parameters\": {\n\t\t\"format\": \"irregular\",\n\t\t\"quirks\": [],\n\t\t\"isTiprack\": false,\n\t\t\"isMagneticModuleCompatible\": false,\n\t\t\"loadName\": \"customslideblockv2_8_wellplate\"\n```\nand copy the value of loadName (without the quotation marks). paste that value into this parameter field, i.e. in this case that would be: `customslideblockv2_8_wellplate`\n\nRemember to load the labware into the Opentrons app before running your protocol\n\n---\n\n",
diff --git a/protoBuilds/35b1a9/README.json b/protoBuilds/35b1a9/README.json
index 18b708a01f..70d354e52f 100644
--- a/protoBuilds/35b1a9/README.json
+++ b/protoBuilds/35b1a9/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol is designed for transferring BHI to Lysis Buffer with P300 Multi-Channel Pipette. The user has the ability to select the number of samples to run.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP300 Multi Channel Pipette\nFisherbrand 2-20\u00b5L Tips\nSimport Scientific 500\u00b5L Tubes in Custom Holder\nReagents\n\n\n\nFor this protocol, be sure that the correct pipette (P300-Multi) is attached for the intended test case.\nLabware Setup\nSlot 1: Fisherbrand 2-20\u00b5L Tips\nSlot 2: Simport Scientific 500\u00b5L Tubes in Custom Holder (Source)\nSlot 3: Simport Scientific 500\u00b5L Tubes in Custom Holder (Destination)\nUsing the customization fields below, set up your protocol.\n Pipette Mount: Specify which mount the P300-Multi is on (left or right).\n Number of Samples: Specify how many samples should be transferred (1-96).\n",
"internal": "35b1a9",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol is designed for transferring BHI to Lysis Buffer with P300 Multi-Channel Pipette. The user has the ability to select the number of samples to run.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P300 Multi Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Fisherbrand 2-20\u00b5L Tips](https://www.fishersci.com/shop/products/fisherbrand-sureone-aerosol-barrier-pipette-tips-14/02707432)\n* [Simport Scientific 500\u00b5L Tubes in Custom Holder](http://www.simport.com/en/products/173-t100-1.html)\n* Reagents\n\n\n\n---\n\n\n\nFor this protocol, be sure that the correct pipette (P300-Multi) is attached for the intended test case.\n\n**Labware Setup**\n\nSlot 1: [Fisherbrand 2-20\u00b5L Tips](https://www.fishersci.com/shop/products/fisherbrand-sureone-aerosol-barrier-pipette-tips-14/02707432)\n\nSlot 2: [Simport Scientific 500\u00b5L Tubes in Custom Holder (Source)](http://www.simport.com/en/products/173-t100-1.html)\n\nSlot 3: [Simport Scientific 500\u00b5L Tubes in Custom Holder (Destination)](http://www.simport.com/en/products/173-t100-1.html)\n\n**Using the customization fields below, set up your protocol.**\n* Pipette Mount: Specify which mount the P300-Multi is on (left or right).\n* Number of Samples: Specify how many samples should be transferred (1-96).\n\n---\n",
"internal": "35b1a9\n",
diff --git a/protoBuilds/3607d5-2/README.json b/protoBuilds/3607d5-2/README.json
index e59aaf9436..dcfc7dbb53 100644
--- a/protoBuilds/3607d5-2/README.json
+++ b/protoBuilds/3607d5-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Extraction"
@@ -10,7 +10,7 @@
"internal": "3607d5",
"labware": "\nAbgene Midi 96 Well Plate 800 \u00b5L\nAmplifyt 96 Well Plate 200 \u00b5L\nNEST 12 Well Reservoir 15 mL\nOpentrons 20\u00b5l and 300\u00b5l Tipracks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Nucleic Acid Extraction\n\n",
"deck-setup": "* blue: samples \n* green: binding beads \n* pink: EtOH \n* purple: RSB\n\n\n---\n\n",
"description": "\nLinks: \n* [SPRI 1 & 2](./3607d5-2)\n
\n
\n* [PCR2 Setup](./3607d5)\n
\n
\n* [SPRI 3](./3607d5-3)\n
\n
\n* [Normalization and Pooling](./3607d5-4)\n
\n
\n* [Rerack](./3607d5-5)\n\nThe protocol is broken down into 4 main parts:\n* binding buffer addition to samples\n* bead wash 2x using magnetic module\n* final elution to chilled PCR plate\n\nThe entire sequence is repeated for a total of 2x.\n\nSamples should be loaded on the magnetic module in an Abgene Midi plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 11) to the elution PCR plate (temperature module, slot 3). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\n---\n\n",
diff --git a/protoBuilds/3607d5-3/README.json b/protoBuilds/3607d5-3/README.json
index c5b813439b..274b9810c6 100644
--- a/protoBuilds/3607d5-3/README.json
+++ b/protoBuilds/3607d5-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Extraction"
@@ -10,7 +10,7 @@
"internal": "3607d5",
"labware": "\nAbgene Midi 96 Well Plate 800 \u00b5L\nAmplifyt 96 Well Plate 200 \u00b5L\nNEST 12 Well Reservoir 15 mL\nOpentrons 20\u00b5l and 300\u00b5l Tipracks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Nucleic Acid Extraction\n\n",
"deck-setup": "* blue: samples \n* green: binding beads \n* pink: EtOH \n* purple: RSB\n\n\n---\n\n",
"description": "\nLinks: \n* [SPRI 1 & 2](./3607d5-2)\n
\n
\n* [PCR2 Setup](./3607d5)\n
\n
\n* [SPRI 3](./3607d5-3)\n
\n
\n* [Normalization and Pooling](./3607d5-4)\n
\n
\n* [Rerack](./3607d5-5)\n\nThe protocol is broken down into 4 main parts:\n* binding buffer addition to samples\n* bead wash 2x using magnetic module\n* final elution to chilled PCR plate\n\nSamples should be loaded on the magnetic module in an Abgene Midi plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 11) to the elution PCR plate (temperature module, slot 3). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\n---\n\n",
diff --git a/protoBuilds/3607d5-4/README.json b/protoBuilds/3607d5-4/README.json
index 1604e2ba69..34ee134396 100644
--- a/protoBuilds/3607d5-4/README.json
+++ b/protoBuilds/3607d5-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "3607d5",
"labware": "\nOpentrons 20\u00b5l Tiprack\nOpentrons 300\u00b5l Tiprack\nOpentrons 96 Well Aluminum Block\nOpentrons 4-in-1 Tuberack Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"deck-setup": "* green: RSB \n* blue: samples \n\n\n---\n\n",
"description": "\nLinks: \n* [SPRI 1 & 2](./3607d5-2)\n
\n
\n* [PCR2 Setup](./3607d5)\n
\n
\n* [SPRI 3](./3607d5-3)\n
\n
\n* [Normalization and Pooling](./3607d5-4)\n
\n
\n* [Rerack](./3607d5-5)\n\n\nThis is a custom normalization pooling protocol from up to 12 sources to a pooling tube. The input .csv for RSB volume specification should be specified as follows:\n\n```\nwell,volume rsb\nA1,200\nB1,15\n...\n```\n\n---\n\n",
diff --git a/protoBuilds/3607d5-5/README.json b/protoBuilds/3607d5-5/README.json
index b221c8ff84..f1cc31fc35 100644
--- a/protoBuilds/3607d5-5/README.json
+++ b/protoBuilds/3607d5-5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -9,7 +9,7 @@
"internal": "3607d5",
"labware": "\nOpentrons 200ul Filtertipracks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"description": "\nLinks: \n* [SPRI 1 & 2](./3607d5-2)\n
\n
\n* [PCR2 Setup](./3607d5)\n
\n
\n* [SPRI 3](./3607d5-3)\n
\n
\n* [Normalization and Pooling](./3607d5-4)\n
\n
\n* [Rerack](./3607d5-5)\n\n\nThis protocol performs a tip reracking consolidation from 4x 200ul filter tipracks with **only rows A and B occupied** to one of the racks. A P300 multi-channel pipette is used with offset tip pickups to perform the consolidation.\n\n---\n\n",
"internal": "3607d5\n",
diff --git a/protoBuilds/3607d5/README.json b/protoBuilds/3607d5/README.json
index 042429165f..a26316c969 100644
--- a/protoBuilds/3607d5/README.json
+++ b/protoBuilds/3607d5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "3607d5",
"labware": "\nAbgene Midi 96 Well Plate 800 \u00b5L\nAmplifyt 96 Well Plate 200 \u00b5L\nOpentrons 20\u00b5l and 300\u00b5l Tipracks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "* green: PCR2 buffer \n* blue: samples \n* pink: UDI\n\n\n---\n\n",
"description": "\nLinks: \n* [SPRI 1 & 2](./3607d5-2)\n
\n
\n* [PCR2 Setup](./3607d5)\n
\n
\n* [SPRI 3](./3607d5-3)\n
\n
\n* [Normalization and Pooling](./3607d5-4)\n
\n
\n* [Rerack](./3607d5-5)\n\nIn this protocol, indexes are transferred to their corresponding wells in a PCR plate containing samples. Then, PCR2 buffer is added to each sample and mixed using fresh tips for each transfer.\n\n---\n\n",
diff --git a/protoBuilds/3633ca/README.json b/protoBuilds/3633ca/README.json
index 3641c880e4..4431842219 100644
--- a/protoBuilds/3633ca/README.json
+++ b/protoBuilds/3633ca/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol adds and removes ethanol to a plate on the Magnetic Module.\n\nThe protocol adds 150\u00b5L of ethanol to each well, using the P300 Multi-Channel pipette, waits 30 seconds, then removes the ethanol and discards the tips. After doing this a second time, the P20 Multi-Channel pipette aspirates 15\u00b5L of liquid from the wells and disposes of it.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons Magnetic Module\nOpentrons P300 Multi-Channel Pipette\n(2) Opentrons 300\u00b5L Tips\nOpentrons P20 Multi-Channel Pipette\nOpentrons 20\u00b5L Tips\nNEST 96-Well Plates\nUSA Scientific 12-Well Reservoir\n\n\n\nSlot 1: USA Scientific 12-Well Reservoir with ethanol in slots 1-4\nSlot 3: Opentrons 20\u00b5L Tips\nSlot 4: Opentrons Magnetic Module with NEST 96-Well Plates\nSlot 7: Opentrons 300\u00b5L Tips\nSlot 10: Opentrons 300\u00b5L Tips\n\n\nUsing the customizations field (below), set up your protocol.\n P300-Multi Pipette Mount: Select which mount (left or right) the P300-Multi is attached to.\n P20-Multi Pipette Mount: Select which mount (left or right) the P20-Multi is attached to.",
"internal": "3633ca",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol adds and removes ethanol to a plate on the Magnetic Module.\n\nThe protocol adds 150\u00b5L of ethanol to each well, using the P300 Multi-Channel pipette, waits 30 seconds, then removes the ethanol and discards the tips. After doing this a second time, the P20 Multi-Channel pipette aspirates 15\u00b5L of liquid from the wells and disposes of it.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* (2) [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons P20 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [USA Scientific 12-Well Reservoir](https://labware.opentrons.com/usascientific_12_reservoir_22ml?category=reservoir)\n\n\n---\n\n\nSlot 1: [USA Scientific 12-Well Reservoir](https://labware.opentrons.com/usascientific_12_reservoir_22ml?category=reservoir) with ethanol in slots 1-4\n\nSlot 3: [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\nSlot 4: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n\nSlot 7: [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 10: [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n**Using the customizations field (below), set up your protocol.**\n* **P300-Multi Pipette Mount**: Select which mount (left or right) the P300-Multi is attached to.\n* **P20-Multi Pipette Mount**: Select which mount (left or right) the P20-Multi is attached to.\n\n\n\n",
"internal": "3633ca\n",
diff --git a/protoBuilds/3662e6/README.json b/protoBuilds/3662e6/README.json
index 9fe82bc5b9..a599521bff 100644
--- a/protoBuilds/3662e6/README.json
+++ b/protoBuilds/3662e6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Serial Dilution"
@@ -8,7 +8,7 @@
"description": "This protocol is an updated version of a drug dilution protocol designed with Protocol Designer.\n\nIn this protocol, the p300-multi channel pipette (GEN1) is used to transfer 100\u00b5L media to all of the columns of the three dilution plates (10uM, 1uM, and 0.1uM). When transferring the media, the same tips (column 1) will be used for all transfers, but after every four transfers, the pipette will return the tips to the tiprack and pick up them up again to ensure a good seal with the tips.\nAfter the media is added, the p10-multi channel pipette is used for the dilutions, transferring 10\u00b5L from a stock plate in slot 10, to the first dilution plate and so on... (using the same set of tip for each column of dilutions).\nFinally, the p300-multi will be used again, this time transferring 100\u00b5L from the reservoir in slot 11 to the three dilution plates. These transfers will follow the same pattern for tip usage as the earlier transfer with the p300-multi, but instead use the tips from column 2.\n\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nP300 Multi-Channel Pipette (GEN1)\nP10 Multi-Channel Pipette\nOpentrons 300\u00b5l Pipette Tips\nOpentrons 10\u00b5L Pipette Tips\nAxygen 1-Well Reservoir, 90mL\nCorning 96-Well Plate, 360\u00b5L Flat\n\n\n\nDeck Layout\n\nSlot 1: Corning 96-Well Plate (Drug Dilution 0.1\u00b5M)\n\nSlot 2: Opentrons 300\u00b5l Pipette tips\n\nSlot 4: Corning 96-Well Plate (Drug Dilution 1\u00b5M)\n\nSlot 5: Opentrons 10\u00b5L Pipette Tips\n\nSlot 7: Corning 96-Well Plate (Drug Dilution 10\u00b5M)\n\nSlot 8: Axygen 1-Well Reservoir (containing Media for 1st transfer)\n\nSlot 10: Corning 96-Well Plate (Drug Stock)\n\nSlot 11: Axygen 1-Well Reservoir (containing Cells for 2nd transfer)\n",
"internal": "3662e6",
"markdown": {
- "author": "[Opentrons](http://www.opentrons.com/)\n\n",
+ "author": "[Opentrons](http://www.opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Serial Dilution\n\n",
"description": "This protocol is an updated version of a drug dilution protocol designed with Protocol Designer.\n\nIn this protocol, the p300-multi channel pipette (GEN1) is used to transfer 100\u00b5L media to all of the columns of the three dilution plates (10uM, 1uM, and 0.1uM). When transferring the media, the same tips (column 1) will be used for all transfers, but after every four transfers, the pipette will return the tips to the tiprack and pick up them up again to ensure a good seal with the tips.\nAfter the media is added, the p10-multi channel pipette is used for the dilutions, transferring 10\u00b5L from a stock plate in slot 10, to the first dilution plate and so on... (using the same set of tip for each column of dilutions).\nFinally, the p300-multi will be used again, this time transferring 100\u00b5L from the reservoir in slot 11 to the three dilution plates. These transfers will follow the same pattern for tip usage as the earlier transfer with the p300-multi, but instead use the tips from column 2.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* P300 Multi-Channel Pipette (GEN1)\n* P10 Multi-Channel Pipette\n* [Opentrons 300\u00b5l Pipette Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 10\u00b5L Pipette Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [Axygen 1-Well Reservoir, 90mL](https://labware.opentrons.com/axygen_1_reservoir_90ml/)\n* [Corning 96-Well Plate, 360\u00b5L Flat](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate)\n\n\n---\n\n\n**Deck Layout**\n\n**Slot 1**: [Corning 96-Well Plate](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (Drug Dilution 0.1\u00b5M)\n\n**Slot 2**: [Opentrons 300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n**Slot 4**: [Corning 96-Well Plate](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (Drug Dilution 1\u00b5M)\n\n**Slot 5**: [Opentrons 10\u00b5L Pipette Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n\n**Slot 7**: [Corning 96-Well Plate](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (Drug Dilution 10\u00b5M)\n\n**Slot 8**: [Axygen 1-Well Reservoir](https://labware.opentrons.com/axygen_1_reservoir_90ml/) (containing Media for 1st transfer)\n\n**Slot 10**: [Corning 96-Well Plate](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (Drug Stock)\n\n**Slot 11**: [Axygen 1-Well Reservoir](https://labware.opentrons.com/axygen_1_reservoir_90ml/) (containing Cells for 2nd transfer)\n\n\n",
"internal": "3662e6\n",
diff --git a/protoBuilds/37190e/README.json b/protoBuilds/37190e/README.json
index bdd3dcba9a..5ce4e01d12 100644
--- a/protoBuilds/37190e/README.json
+++ b/protoBuilds/37190e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sterile Workflows": [
"Cell Culture"
@@ -10,7 +10,7 @@
"internal": "37190e",
"labware": "\nCorning 96 Well Plate 360 \u00b5L Flat, 3650\nOpentrons 10 Tube Rack with NEST 4x50 mL, 6x15 mL Conical\nOpentrons 200\u00b5L Filter Tips\nOpentrons 20\u00b5L Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sterile Workflows\n\t* Cell Culture\n\n",
"deck-setup": "\n \n\nTuberack on slot 9:\n* tube A1: drug A\n* tube A2: drug B\n* tube A3: media\n\n---\n\n",
"description": "\nThis protocol performs a custom cell culture assay on up to 8 flat well plates. Please see [these instructions](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/37190e/opentrons_media_table.xlsx) for the exact transfer scheme.\n\n---\n\n",
diff --git a/protoBuilds/37de37/README.json b/protoBuilds/37de37/README.json
index 09b627ce30..e8f1d0cd51 100644
--- a/protoBuilds/37de37/README.json
+++ b/protoBuilds/37de37/README.json
@@ -17,14 +17,14 @@
"internal": "37de37\n",
"labware": "* [Corning 384 Well Plate 360 \u00b5L](https://www.corning.com/catalog/cls/documents/drawings/DWG00834.PDF)\n* [Brooks Life Sciences 21mL 15-well reservoir](https://www.brookslifesciences.com/products/reservoir-plate)\n* [Opentrons 20uL Tips](https://shop.opentrons.com/collections/opentrons-tips)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Partner Name](partner website link)\n\n",
+ "partner": "[Partner Name](partner website link)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P20 Single-Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n\n---\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Transfer volume of diluent as per CSV (1 tip only needed) up to 3 source plates\n2. Mix DNA and transfer from initial to the final plate (as per CSV), then mix after dispensing.\n3. Pool 5ul of each sample into H12 of the final plate.\n\n",
"title": "Cherrypicking DNA and Pooling with CSV input"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Partner Name",
+ "partner": "Partner Name\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP20 Single-Channel Electronic Pipette (GEN2)\n\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nTransfer volume of diluent as per CSV (1 tip only needed) up to 3 source plates\nMix DNA and transfer from initial to the final plate (as per CSV), then mix after dispensing.\nPool 5ul of each sample into H12 of the final plate.\n",
diff --git a/protoBuilds/37ffa5/README.json b/protoBuilds/37ffa5/README.json
index 57d1a42b45..727994bb69 100644
--- a/protoBuilds/37ffa5/README.json
+++ b/protoBuilds/37ffa5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "37ffa5",
"labware": "\n[Opentrons Tips for the P300 and P1000] (https://shop.opentrons.com)\n[Opentrons 4-in-1 Tuberack, Nest 1.5 mL Snap Cap Tubes, Corning 360 uL Flat 96-well Plate] (https://shop.opentrons.com)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons 1000 uL Tips \n**Slot 2**: Opentrons 300 uL Tips \n**Slot 3**: Test Plate 1 (Corning 360 uL Flat 96-Well Plate) \n**Slot 4**: Sample Plate (Corning 360 uL Flat 96-Well Plate) \n**Slot 5**: Test Plate 4 (Corning 360 uL Flat 96-Well Plate) \n**Slot 6**: Test Plate 2 (Corning 360 uL Flat 96-Well Plate) \n**Slot 7**: 25 ug/mL Sample Stock (Opentrons 24-Tube Rack holding 1.5 mL Nest SnapCap Tubes) \n\n\n\n**Slot 8**: Buffer Reservoir (USA Scientific 22 mL 12 Well Reservoir) \n\n\n\n**Slot 9**: Test Plate 3 (Corning 360 uL Flat 96-Well Plate) \n\n---\n\n",
"description": "This protocol uses single-channel P300 and P1000 pipettes to perform plate filling steps with 1-5 input samples for each of up to 4 test plates to be filled using the deck layout and source and destination locations displayed in the attached .json file [deck map and source and destination wells](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/37ffa5/Based_on_zoom_conf.json) (note: the attached json file is for visualization of the deck layout and location of source and destination wells using [Opentrons Protocol Designer](https://designer.opentrons.com/), it is not a fully-developed .json protocol to be run on the OT-2, the pipetting technique and transfer order have been modified for run time reasons). All transfers required for set up of the first test plate are completed prior to the start of the next test plate to minimize the per-plate run time. The user can pause the robot in order to remove completed plates during the run. Multi pre-air gaps with multiple aspirations are performed to enable repeat dispenses at the top of the well without need for blowout, tip touch or tip drops. User-determined parameters are available to specify the number of samples in each row of the tube rack.\n\n\n---\n\n\n",
diff --git a/protoBuilds/384b23-plate-transfer/README.json b/protoBuilds/384b23-plate-transfer/README.json
index 54c1eaa1f1..dcbd1df8bf 100644
--- a/protoBuilds/384b23-plate-transfer/README.json
+++ b/protoBuilds/384b23-plate-transfer/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol transfers 200\u00b5L from vials in 24 tube tuberacks to a 96-well plate. New tips are granted between each transfer. Transfers are completed by column in each tube rack starting at the tube rack in slot 1.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P1000 Single-Channel Pipette\nOpentrons 1000\u00b5L Filter Tip Rack\nOpentrons 200\u00b5L Filter Tips\nCustom Opentrons 24-tube tube rack\n\n\n\nSlots 1, 4, 7, 10: Custom 24-tube tube rack\nSlot 2: 96-well plate\nSlot 3: Opentrons 200\u00b5L Filter Tip Rack\nSlot 5: Opentrons 1000\u00b5L Filter Tip Rack\n\n\nUsing the customizations field (below), set up your protocol.\n Number of Samples: Specify number of samples in the source plate to be transferred. Note, samples should be put in tube racks by column starting from the tube rack in Slot 1. The tube rack in Slot 1 will be transferred down by column before the tube rack in Slot 4 is transferred, and so on.\n Volume Dispensed: Specify the volume (in microliters) to transfer from the tube rack to the plate. A value greater than 100 will use the P1000 pipette. A value less than 100 will use the P300 pipette. \n Delay Time after Aspirating (in seconds): Since saliva is being transferred, often times a delay after aspiration allows the pipette to achieve the full volume. Specify the amount of time in seconds after each aspiration (a value of 0 can be inputted).\n Aspiration Height: Specify the height (in mm) from the bottom of the tubes the pipette will aspirate from.\n* P1000 Single GEN2 Mount: Specify the mount side for the P300 Single GEN2 pipette",
"internal": "384b23-96-plate-transfer",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol transfers 200\u00b5L from vials in 24 tube tuberacks to a 96-well plate. New tips are granted between each transfer. Transfers are completed by column in each tube rack starting at the tube rack in slot 1.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P1000 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 1000\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips)\n* [Opentrons 200\u00b5L Filter Tips](hhttps://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* Custom Opentrons 24-tube tube rack\n\n\n\n---\n\n\nSlots 1, 4, 7, 10: Custom 24-tube tube rack\n\nSlot 2: 96-well plate\n\nSlot 3: Opentrons 200\u00b5L Filter Tip Rack\n\nSlot 5: Opentrons 1000\u00b5L Filter Tip Rack\n\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Samples**: Specify number of samples in the source plate to be transferred. Note, samples should be put in tube racks by column starting from the tube rack in Slot 1. The tube rack in Slot 1 will be transferred down by column before the tube rack in Slot 4 is transferred, and so on.\n* **Volume Dispensed**: Specify the volume (in microliters) to transfer from the tube rack to the plate. A value greater than 100 will use the P1000 pipette. A value less than 100 will use the P300 pipette. \n* **Delay Time after Aspirating (in seconds)**: Since saliva is being transferred, often times a delay after aspiration allows the pipette to achieve the full volume. Specify the amount of time in seconds after each aspiration (a value of 0 can be inputted).\n* **Aspiration Height**: Specify the height (in mm) from the bottom of the tubes the pipette will aspirate from. \n* **P1000 Single GEN2 Mount**: Specify the mount side for the P300 Single GEN2 pipette\n\n",
"internal": "384b23-96-plate-transfer\n",
diff --git a/protoBuilds/384b23-pooling/README.json b/protoBuilds/384b23-pooling/README.json
index bfb9da32a4..1be6bab072 100644
--- a/protoBuilds/384b23-pooling/README.json
+++ b/protoBuilds/384b23-pooling/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Normalization": [
"Normalization and Pooling"
@@ -8,7 +8,7 @@
"description": "This protocol pools 100\u00b5L from wells across each row to the recipient tube in column 6 of the respective row (total of 500\u00b5L in each recipient tube).\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P300 Single-Channel Pipette\nOpentrons 200\u00b5L Filter Tip Rack\nOpentrons 200\u00b5L Filter Tips\nCustom Opentrons 6x5 tube racks\n\n\n\nSlots 1 (if applicable): Tuberack A\nSlot 2 (if applicable): Tuberack B\nSlot 4: Opentrons 200\u00b5L Filter Tip Rack\n\n\nUsing the customizations field (below), set up your protocol.\n Using Tuberack A (tubes T1, T2, T3): Specify if Tuberack A will be pooled.\n Tuberack A tubes: Specify whether you will be populating Tuberack A with T1-T3 tubes or T7 tubes. Leave as is if not using Tuberack A.\n Number of pooling rows for Tuberack A: Specify how many rows to pool (1-5).\n Using Tuberack B (tubes T4, T5, T6): Specify if Tuberack B will be pooled\n Tuberack B tubes: Specify whether you will be populating tuberack B with T4 & T5 tubes or T6 tubes. Leave as is if not using Tuberack B.\n Number of pooling rows for Tuberack B: Specify how many rows to pool (1-5).\n Delay Time after Aspirating (in seconds): Since saliva is being transferred, often times a delay after aspiration allows the pipette to achieve the full volume. Specify the amount of time in seconds after each aspiration (a value of 0 can be inputted).\n P300 Single GEN2 Mount: Specify the mount side for the P300 Single GEN2 pipette\n* P300 Single GEN2 Mount: Specify the mount side for the P300 Single GEN2 pipette",
"internal": "384b23-pooling",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Normalization\n\t* Normalization and Pooling\n\n\n",
"description": "This protocol pools 100\u00b5L from wells across each row to the recipient tube in column 6 of the respective row (total of 500\u00b5L in each recipient tube).\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 200\u00b5L Filter Tips](hhttps://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* Custom Opentrons 6x5 tube racks\n\n\n\n---\n\n\nSlots 1 (if applicable): Tuberack A\n\nSlot 2 (if applicable): Tuberack B\n\nSlot 4: Opentrons 200\u00b5L Filter Tip Rack\n\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Using Tuberack A (tubes T1, T2, T3)**: Specify if Tuberack A will be pooled.\n* **Tuberack A tubes**: Specify whether you will be populating Tuberack A with T1-T3 tubes or T7 tubes. Leave as is if not using Tuberack A.\n* **Number of pooling rows for Tuberack A**: Specify how many rows to pool (1-5).\n* **Using Tuberack B (tubes T4, T5, T6)**: Specify if Tuberack B will be pooled\n* **Tuberack B tubes**: Specify whether you will be populating tuberack B with T4 & T5 tubes or T6 tubes. Leave as is if not using Tuberack B.\n* **Number of pooling rows for Tuberack B**: Specify how many rows to pool (1-5).\n* **Delay Time after Aspirating (in seconds)**: Since saliva is being transferred, often times a delay after aspiration allows the pipette to achieve the full volume. Specify the amount of time in seconds after each aspiration (a value of 0 can be inputted).\n* **P300 Single GEN2 Mount**: Specify the mount side for the P300 Single GEN2 pipette\n* **P300 Single GEN2 Mount**: Specify the mount side for the P300 Single GEN2 pipette\n\n",
"internal": "384b23-pooling\n",
diff --git a/protoBuilds/384b23-tube-transfer/README.json b/protoBuilds/384b23-tube-transfer/README.json
index ca2ceba2f8..463efd7544 100644
--- a/protoBuilds/384b23-tube-transfer/README.json
+++ b/protoBuilds/384b23-tube-transfer/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol transfers 500\u00b5L from tubes in 24 tube tuberacks to a 6x5 tube rack. New tips are granted between each transfer. Transfers are completed by column in the source tube rack and dispensed by row to the recipient tube racks. A new row starts with each column.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P1000 Single-Channel Pipette\nOpentrons 1000\u00b5L Filter Tip Rack\nCustom Opentrons 24-tube tube rack\nCustom Opentrons 30-tube tube rack\n\n\n\nSlots 1: Source tube rack (6x4)\nSlot 2: Destination tube rack (6x5)\nSlot 3: Opentrons 1000\u00b5L Filter Tip Rack\n\n\nUsing the customizations field (below), set up your protocol.\n Number of Samples: Specify number of samples in the source tube rack to be transferred.\n Delay Time after Aspirating (in seconds): Since saliva is being transferred, often times a delay after aspiration allows the pipette to achieve the full volume. Specify the amount of time in seconds after each aspiration (a value of 0 can be inputted).\n Aspiration Height for Recipient tube: Specify the height (in mm) from the bottom of the tubes in the recipient tube rack the pipette will aspirate from. Change this value if switching between T4, T5, or T6 tubes.\n Dispense Height for Recipient tube: Specify the height (in mm) from the bottom of the tubes in the destination tube rack the pipette will dispense at.\n* P1000 Single GEN2 Mount: Specify the mount side for the P300 Single GEN2 pipette",
"internal": "384b23-tube-transfer",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol transfers 500\u00b5L from tubes in 24 tube tuberacks to a 6x5 tube rack. New tips are granted between each transfer. Transfers are completed by column in the source tube rack and dispensed by row to the recipient tube racks. A new row starts with each column.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P1000 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 1000\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips)\n* Custom Opentrons 24-tube tube rack\n* Custom Opentrons 30-tube tube rack\n\n\n\n---\n\n\nSlots 1: Source tube rack (6x4)\n\nSlot 2: Destination tube rack (6x5)\n\nSlot 3: Opentrons 1000\u00b5L Filter Tip Rack\n\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Samples**: Specify number of samples in the source tube rack to be transferred.\n* **Delay Time after Aspirating (in seconds)**: Since saliva is being transferred, often times a delay after aspiration allows the pipette to achieve the full volume. Specify the amount of time in seconds after each aspiration (a value of 0 can be inputted).\n* **Aspiration Height for Recipient tube**: Specify the height (in mm) from the bottom of the tubes in the recipient tube rack the pipette will aspirate from. Change this value if switching between T4, T5, or T6 tubes.\n* **Dispense Height for Recipient tube**: Specify the height (in mm) from the bottom of the tubes in the destination tube rack the pipette will dispense at. \n* **P1000 Single GEN2 Mount**: Specify the mount side for the P300 Single GEN2 pipette\n\n",
"internal": "384b23-tube-transfer\n",
diff --git a/protoBuilds/387961/README.json b/protoBuilds/387961/README.json
index 5a196e7871..106f992608 100644
--- a/protoBuilds/387961/README.json
+++ b/protoBuilds/387961/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol automates the filling of a specialized 384-well (rotated 90 degrees and takes up two slots). Using a 96-well plate containing the reagents, the p20 will aspirate 20\u00b5L from the 96-well plate and dispense 10\u00b5L into the 384-well plate, swapping tips each time the the p20 moves to another column (different reagents) until the 384-well plate is filled.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons p20 Multi-Channel Pipette (attached to right mount)\nOpentrons 20\u00b5L Tip Rack\nNEST 96-Well PCR Plate\n384-Well Plate with Custom Adapter\n\n\n\nDeck Layout\n\nSlot 1: Opentrons 20\u00b5L Tip Rack\n\nSlot 5: NEST 96-Well PCR Plate\nReagent Locations\n Reagent 1: B1 (40\u00b5L), H11 (160\u00b5L), and H12 (160\u00b5L)\n Reagent 2: B2 (40\u00b5L), G11 (160\u00b5L), and G12 (160\u00b5L)\n Reagent 3: A1 (40\u00b5L), F11 (160\u00b5L), and F12 (160\u00b5L)\n Reagent 4: B3 (40\u00b5L), E11 (160\u00b5L), and E12 (160\u00b5L)\n Reagent 5: B4 (40\u00b5L), D11 (160\u00b5L), and D12 (160\u00b5L)\n Reagent 6: A3 (40\u00b5L), C11 (160\u00b5L), and C12 (160\u00b5L)\n Reagent 7: B6 (40\u00b5L), B11 (160\u00b5L), and B12 (160\u00b5L)\n Reagent 8: B5 (40\u00b5L), A11 (160\u00b5L), and A12 (160\u00b5L)\n Reagent 9: D1-D8 + B8 (40\u00b5L)\n Reagent 10: C1-C8 + B7 (40\u00b5L)\n\nSlot 3/6: 384-Well Plate with Custom Adapter",
"internal": "387961",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol automates the filling of a specialized 384-well (rotated 90 degrees and takes up two slots). Using a 96-well plate containing the reagents, the p20 will aspirate 20\u00b5L from the 96-well plate and dispense 10\u00b5L into the 384-well plate, swapping tips each time the the p20 moves to another column (different reagents) until the 384-well plate is filled.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons p20 Multi-Channel Pipette (attached to right mount)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [Opentrons 20\u00b5L Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* 384-Well Plate with Custom Adapter\n\n\n---\n\n\n**Deck Layout**\n\nSlot 1: [Opentrons 20\u00b5L Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 5: [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n*Reagent Locations*\n* Reagent 1: B1 (40\u00b5L), H11 (160\u00b5L), and H12 (160\u00b5L)\n* Reagent 2: B2 (40\u00b5L), G11 (160\u00b5L), and G12 (160\u00b5L)\n* Reagent 3: A1 (40\u00b5L), F11 (160\u00b5L), and F12 (160\u00b5L)\n* Reagent 4: B3 (40\u00b5L), E11 (160\u00b5L), and E12 (160\u00b5L)\n* Reagent 5: B4 (40\u00b5L), D11 (160\u00b5L), and D12 (160\u00b5L)\n* Reagent 6: A3 (40\u00b5L), C11 (160\u00b5L), and C12 (160\u00b5L)\n* Reagent 7: B6 (40\u00b5L), B11 (160\u00b5L), and B12 (160\u00b5L)\n* Reagent 8: B5 (40\u00b5L), A11 (160\u00b5L), and A12 (160\u00b5L)\n* Reagent 9: D1-D8 + B8 (40\u00b5L)\n* Reagent 10: C1-C8 + B7 (40\u00b5L)\n\nSlot 3/6: 384-Well Plate with Custom Adapter\n\n",
"internal": "387961\n",
diff --git a/protoBuilds/38efd4/README.json b/protoBuilds/38efd4/README.json
index d047415a2e..fabd666a62 100644
--- a/protoBuilds/38efd4/README.json
+++ b/protoBuilds/38efd4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons, Ricardo Padua, Brandeis University",
+ "author": "Opentrons, Ricardo Padua, Brandeis University\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -9,7 +9,7 @@
"description": "This protocol uses p300 single channel and p1000 single channel pipettes to transfer and mix custom volumes (ranging from 20 ul - 2 mL specified in input csv file) of water and each of up to 8 stock reagents to the wells of a 96-deep-well plate followed by optional mixing of the well contents.\nLinks:\n example reagent csv\n example formulation csv\n* supplemental information spreadsheet\nThis protocol was developed to combine and mix custom volumes (specified in input csv file) of water and up to 8 stock reagents for the screening of protein crystallization conditions in support of protein structure studies.",
"internal": "38efd4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/), Ricardo Padua, Brandeis University\n\n",
+ "author": "[Opentrons](https://opentrons.com/), Ricardo Padua, Brandeis University\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Plate Filling\n\n",
"deck-setup": "\n\n* Opentrons p300 and p1000 tips (Deck Slots 10, 11)\n* Crystallization Plate usascientific_96_wellplate_2.4ml_deep (Deck Slot 9)\n* Deck Slots 1-8 will either be empty or contain any of the following labware (based on selections made from the pulldown list at the time of protocol download) opentrons_10_tuberack_falcon_4x50ml_6x15ml_conical, opentrons_15_tuberack_falcon_15ml_conical, opentrons_24_tuberack_eppendorf_1.5ml_safelock_snapcap, opentrons_24_tuberack_eppendorf_2ml_safelock_snapcap, opentrons_6_tuberack_falcon_50ml_conical.\n\n\n\n\n\n",
"description": "\nThis protocol uses p300 single channel and p1000 single channel pipettes to transfer and mix custom volumes (ranging from 20 ul - 2 mL specified in input csv file) of water and each of up to 8 stock reagents to the wells of a 96-deep-well plate followed by optional mixing of the well contents.\n\nLinks:\n* [example reagent csv](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/38efd4/Configuration_0725.csv)\n* [example formulation csv](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/38efd4/Formulation_0725.csv)\n* [supplemental information spreadsheet](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/38efd4/ScreenMaker.xlsx)\n\nThis protocol was developed to combine and mix custom volumes (specified in input csv file) of water and up to 8 stock reagents for the screening of protein crystallization conditions in support of protein structure studies.\n\n",
diff --git a/protoBuilds/3955db/README.json b/protoBuilds/3955db/README.json
index 564096fc46..e7c7db85b5 100644
--- a/protoBuilds/3955db/README.json
+++ b/protoBuilds/3955db/README.json
@@ -13,13 +13,13 @@
"description": "This flexible protocol automates the pooling of samples in 12mL tubes and the transfer of pooled samples to Visby device for rapid Covid testing.\n\n\n---\n\n**Protocol Steps**\n1. The protocol begins by pooling samples (1-5) from Tube Racks in slots 2 and 3 into tubes in slot 5.\n2. Once all samples are pooled appropriately, *total volume of pooled samples* will be added from pooled sample to Visby.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 4.0.0 or later)](https://opentrons.com/ot-app/)\n* [P1000 Single-Channel GEN2](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 1000\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips)\n* [Visby Device with Adapter](https://www.visbymedical.com/covid-19-test/)\n* Custom Tube Rack with 12mL Tubes\n\n---\n\n\n**Deck Layout**\n\n\n\n\nExplanation of adjustable parameters below:\n* `P1000 Single GEN2 Mount`: Select which mount (left, right) the pipette is attached to.\n* `Total Volume of Pooled Samples`: Specify the volume (in \u00b5L) that should be transferred to the Visby. This volume will be divided by the `Number of Samples per Pool` to determine how much volume from each individual sample when creating the pools. Example: if this is set to 500 and there are 5 samples per pool, 100\u00b5L will be transferred from each individual sample to the pooled sample tube, then 500\u00b5L of the pooled volume will be added to the Visby.\n* `Number of Visbys`: Select the number of Visbys (1-6).\n* `Number of Samples per Pool`: Select the number of samples to pool into one tube (2-5).\n\n\n",
"internal": "3955db\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[BasisDx](https://www.basisdx.org/)\n\n",
+ "partner": "[BasisDx](https://www.basisdx.org/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"robot": "* [OT-2](https://opentrons.com/ot-2)\n\n",
"title": "Visby Test with Pooling p1000"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "BasisDx",
+ "partner": "BasisDx\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"robot": [
"OT-2"
diff --git a/protoBuilds/39b4c7/README.json b/protoBuilds/39b4c7/README.json
index 2caa2166af..813ccf75aa 100644
--- a/protoBuilds/39b4c7/README.json
+++ b/protoBuilds/39b4c7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"qPCR setup"
@@ -8,7 +8,7 @@
"description": "Reformat up to four 96 well plates into a 384 well PCR plate for downstream qPCR. This protocol will also add mastermix to the necessary wells and then transfer patient samples to the corresponding wells.\n\n\n\n\nOpentrons 20uL Tips\nOpentrons 20uL Filter Tips\nKingFisher 96 Deep Well Plate 2mL\nMicroAmp\u2122 Optical 384-Well Reaction Plate with Barcode\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\nP20 Single Channel GEN2\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nDeck Setup\n\nNote: A full run with 4 plates with a master mix transfer volume of 10 uL will require 3,760 uL of master mix. This is divided into 3 tubes in the picture above. There is a parameter below that asks for the Master Mix Volume per Tube. By default this is set to 1400 uL (Recommendation: Always add more than 1400 uL to prevent aspirating air) in order to account for dead volume. You can adjust this as needed and the protocol will automatically determine how many tubes it needs. \nFor Example: If the Master Mix Volume per Tube is set to 1000 uL the protocol will determine 4 tubes are needed (A1, B1, C1, D1) with at least 1000 uL in each tube for a run with four full plates. The last tube does not require a full 1000 uL volume; it would need a bit more than the difference (~760 uL). \nProtocol Steps\n\nTransfer 10 uL of Master Mix from A1 of the Tube Rack to A1 of the 384 well plate.\nStep 1 will repeat for all samples into the corresponding wells using the same tip.\nTransfer 10 uL of patient sample from A1 of Plate 1 to A1 of the 384 Well PCR plate.\nStep 2 will repeat for all patient samples into the corresponding wells using a new tip each time.\n",
"internal": "39b4c7",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * qPCR setup\n\n",
"description": "Reformat up to four 96 well plates into a 384 well PCR plate for downstream qPCR. This protocol will also add mastermix to the necessary wells and then transfer patient samples to the corresponding wells.\n\n\n\n---\n\n\n* [Opentrons 20uL Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 20uL Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [KingFisher 96 Deep Well Plate 2mL](https://www.thermofisher.com/order/catalog/product/95040450#/95040450)\n* [MicroAmp\u2122 Optical 384-Well Reaction Plate with Barcode](https://www.thermofisher.com/order/catalog/product/4309849?SID=srch-srp-4309849#/4309849?SID=srch-srp-4309849)\n* [Opentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap](https://shop.opentrons.com/products/tube-rack-set-1?_ga=2.232823964.387689945.1619373905-1181961818.1604785212&_gl=1%2Akkqpyx%2A_ga%2AMTE4MTk2MTgxOC4xNjA0Nzg1MjEy%2A_ga_GNSMNLW4RY%2AMTYxOTQ1ODk2Mi4xOTguMS4xNjE5NDU5NTQzLjA.)\n* [P20 Single Channel GEN2](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=31059478970462)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n---\n\n\n**Deck Setup**\n\n\n**Note**: A full run with 4 plates with a master mix transfer volume of 10 uL will require 3,760 uL of master mix. This is divided into 3 tubes in the picture above. There is a parameter below that asks for the `Master Mix Volume per Tube`. By default this is set to 1400 uL (**Recommendation: Always add more than 1400 uL to prevent aspirating air**) in order to account for dead volume. You can adjust this as needed and the protocol will automatically determine how many tubes it needs. \n\n**For Example**: If the `Master Mix Volume per Tube` is set to 1000 uL the protocol will determine 4 tubes are needed (A1, B1, C1, D1) with at least 1000 uL in each tube for a run with four full plates. The last tube does not require a full 1000 uL volume; it would need a bit more than the difference (~760 uL). \n\n**Protocol Steps**\n\n1. Transfer 10 uL of Master Mix from A1 of the Tube Rack to A1 of the 384 well plate.\n`Step 1 will repeat for all samples into the corresponding wells using the same tip.`\n2. Transfer 10 uL of patient sample from A1 of Plate 1 to A1 of the 384 Well PCR plate.\n`Step 2 will repeat for all patient samples into the corresponding wells using a new tip each time.`\n\n",
"internal": "39b4c7",
diff --git a/protoBuilds/39bbbb/README.json b/protoBuilds/39bbbb/README.json
index f8fe6e8d23..cd2687c5fd 100644
--- a/protoBuilds/39bbbb/README.json
+++ b/protoBuilds/39bbbb/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -9,7 +9,7 @@
"description": "This protocol uses a p300 multi-channel and p300 single-channel pipette to transfer up to 57 samples plus serial dilutions of IgG, IgM and IgE standards to 384-well ELISA plates.\nLinks:\n* plate layout and process steps\nThis protocol was developed to transfer up to 57 samples from eppendorf tubes in 24 tube racks to 96-well sample prep plates, then transfer and serially dilute IgG, IgM and IgE standards in a sample prep plate, followed by transfer of samples and serial dilutions to three 384-well ELISA plates. The sample arrangment for the tube racks, sample prep plates and ELISA plates is shown in the attached excel spreadsheet.",
"internal": "39bbbb",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Plate Filling\n\n",
"deck-setup": "\n\n* Opentrons p300 tips (Deck Slots 10, 11)\n* ELISA plates thermo_384_wellplate_50ul (Deck Slots 3, 6, 9)\n* Sample prep plates 96-well plates (Deck Slots 2, 5)\n* opentrons_24_tuberack_eppendorf_1.5ml_safelock_snapcap (Deck Slots 1, 4, 7)\n\n",
"description": "\nThis protocol uses a p300 multi-channel and p300 single-channel pipette to transfer up to 57 samples plus serial dilutions of IgG, IgM and IgE standards to 384-well ELISA plates.\n\nLinks:\n* [plate layout and process steps](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-06-27/gp23j3n/OT-2%20APA%20ELISA%20Protocol.zip)\n\nThis protocol was developed to transfer up to 57 samples from eppendorf tubes in 24 tube racks to 96-well sample prep plates, then transfer and serially dilute IgG, IgM and IgE standards in a sample prep plate, followed by transfer of samples and serial dilutions to three 384-well ELISA plates. The sample arrangment for the tube racks, sample prep plates and ELISA plates is shown in the attached excel spreadsheet.\n\n",
diff --git a/protoBuilds/3a49b6/README.json b/protoBuilds/3a49b6/README.json
index f5aa887976..f51279e0b9 100644
--- a/protoBuilds/3a49b6/README.json
+++ b/protoBuilds/3a49b6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -9,7 +9,7 @@
"description": "Normalize RNA concentration across up to 96 samples by combining custom volumes of water and RNA sample (volumes specified in csv file).\nLinks:\nexample csv\ndeck layout\nThis protocol combines custom volumes of input RNA with custom volumes of water (volumes listed in csv file, example above) to achieve uniform RNA concentration across all samples. This protocol normalizes up to 96 input RNA samples.",
"internal": "3a49b6",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Normalization\n\n",
"deck-setup": "* Opentrons Temperature Module (Deck Slot 3)\n* Opentrons p20 filter tips (Deck Slots 8 , 5)\n* Opentrons \"opentrons_10_tuberack_falcon_4x50ml_6x15ml_conical\" (Deck Slot 11)\n* Opentrons \"nest_96_wellplate_100ul_pcr_full_skirt\" (Deck Slot 9)\n\n",
"description": "Normalize RNA concentration across up to 96 samples by combining custom volumes of water and RNA sample (volumes specified in csv file).\n\nLinks:\n\n[example csv](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-03-09/0e24lpo/RNA%20conc_sample.xlsx)\n[deck layout](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-03-09/aw34l9t/1.jpg)\n\nThis protocol combines custom volumes of input RNA with custom volumes of water (volumes listed in csv file, example above) to achieve uniform RNA concentration across all samples. This protocol normalizes up to 96 input RNA samples.\n\n",
diff --git a/protoBuilds/3b0db0/README.json b/protoBuilds/3b0db0/README.json
index 0d21bca976..ce5d166797 100644
--- a/protoBuilds/3b0db0/README.json
+++ b/protoBuilds/3b0db0/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "3b0db0",
"labware": "\nBiorad 96 well plate 200ul\nPCR tube strips\nOpentrons custom 4x6 3D printed tube racks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol distributes reliance One Step Mulitplex RT-qPCR supermix to each well on a cool (4C) 96 well plate. After, saliva samples from custom Opentrons tube racks are distributed to each well containing mastermix, with a mix step to follow.\n\nExplanation of complex parameters below:\n* `Number of columns`: Specify the number of samples (wholly divisible by 8) that the OT-2 will distribute mastermix to on the 96 well plate (1-96).\n* `Delay after aspiration`: Specify the number of seconds after aspirating for the pipette to pause to achieve full volumes.\n* `Mix repetitions`: Specify the number of times to mix the mastermix and saliva.\n* `Tube Aspiration Height`: Specify the aspiration height from the bottom of the tube (in mm) to aspirate from when transferring saliva in the final step.\n* `Tube-Strip Aspiration Height for Multi Channel`: Specify the aspiration height from the bottom of the tube strip that the multi-channel pipette visits.\n* `Aspiration/Dispense Rate`: Specify the aspiration/dispense rate for the single and multi-channel pipettes. A value of 1 returns the default speed, 0.5 would return half of the default speed, etc. \n* `P20 Multi GEN2 Mount`: Specify the mount (left or right) of the P20 Multi GEN2 Pipette.\n* `P20 Single GEN2 Mount`: Specify the mount (left or right) of the P20 single GEN2 Pipette.\n\n---\n\n",
diff --git a/protoBuilds/3b3d2f/README.json b/protoBuilds/3b3d2f/README.json
index f2dcb29d35..0553aab5d6 100644
--- a/protoBuilds/3b3d2f/README.json
+++ b/protoBuilds/3b3d2f/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -9,7 +9,7 @@
"description": "This protocol uses a p20 single channel pipette to load (in triplicate) up to four 96-well plates (loading of each plate will be finished before loading of the next plate is started) in preparation for ddPCR from 2 mL source tubes containing reagent mix, water, up to 27 template samples and a positive control template sample (CSV-formatted plate maps are uploaded at the time of protocol download to specify the location and number of sample and control tubes).\nLinks:\n SOP_SARS-CoV2 in sewage Rose Lab_MSU_v2021draft_06.18.21\n Sample Plate Map CSV File\n* Sample Plate Map CSV File\nThis protocol was developed to perform the ddPCR plate loading part of the attached SOP for up to four ddPCR plates according to uploaded plate maps formatted as seen in the attached examples (based on plate map examples from Appendix A of the attached SOP).",
"internal": "3b3d2f",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * PCR Prep\n\n",
"deck-setup": "\n\n* Opentrons p20 filter tips (Deck Slots 10, 11)\n* Opentrons Temperature Module (with opentrons_24_aluminumblock_nest_2ml_snapcap Deck Slot 3)\n* ddPCR Plate (biorad_96_wellplate_200ul_pcr Deck Slot 5)\n\n",
"description": "\nThis protocol uses a p20 single channel pipette to load (in triplicate) up to four 96-well plates (loading of each plate will be finished before loading of the next plate is started) in preparation for ddPCR from 2 mL source tubes containing reagent mix, water, up to 27 template samples and a positive control template sample (CSV-formatted plate maps are uploaded at the time of protocol download to specify the location and number of sample and control tubes).\n\nLinks:\n* [SOP_SARS-CoV2 in sewage Rose Lab_MSU_v2021draft_06.18.21](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-06-22/ux22lt2/SOP_SARS-CoV2%20in%20sewage%20Rose%20Lab_MSU_v2021draft_06.18.21.pdf)\n* [Sample Plate Map CSV File](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/3b3d2f/N1N2+duplex+plate+map.csv)\n* [Sample Plate Map CSV File](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/3b3d2f/Phi6+simplex+plate+map.csv)\n\nThis protocol was developed to perform the ddPCR plate loading part of the attached SOP for up to four ddPCR plates according to uploaded plate maps formatted as seen in the attached examples (based on plate map examples from Appendix A of the attached SOP).\n\n",
diff --git a/protoBuilds/3b3f1c/README.json b/protoBuilds/3b3f1c/README.json
index 67252baac5..19ffb07cfa 100644
--- a/protoBuilds/3b3f1c/README.json
+++ b/protoBuilds/3b3f1c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol performs a specific set of genealogy dilutions and transferred to and from 96 deepwell plates and 1.5ml, 15ml, and 50ml. Input for the protocol should be specified in a .csv file as shown in this template.\n\n\n\nP300 and P1000 single-channel GEN2 electronic pipettes\nOpentrons 300ul and 1000ul tiprack\nOpentrons 4-in-1 tuberack sets with NEST 1.5ml snapcap, 15ml, and 50ml tubes\nNEST 96 Deepwell Plate 2mL #503001\n",
"internal": "3b3f1c",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol performs a specific set of genealogy dilutions and transferred to and from 96 deepwell plates and 1.5ml, 15ml, and 50ml. Input for the protocol should be specified in a `.csv` file as shown in [this template](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/3b3f1c/Alturas+Dilution+Temp+and+OT2+Pipette+Settings+332+312+V4.csv).\n\n---\n\n\n* [P300 and P1000 single-channel GEN2 electronic pipettes](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 300ul and 1000ul tiprack](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons 4-in-1 tuberack sets](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with [NEST 1.5ml snapcap, 15ml, and 50ml tubes](https://shop.opentrons.com/collections/verified-consumables)\n* [NEST 96 Deepwell Plate 2mL #503001](http://www.cell-nest.com/page94?product_id=101&_l=en)\n\n",
"internal": "3b3f1c\n",
diff --git a/protoBuilds/3bc25d/README.json b/protoBuilds/3bc25d/README.json
index 67f51d6de5..9d83e3e02e 100644
--- a/protoBuilds/3bc25d/README.json
+++ b/protoBuilds/3bc25d/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Nanopore"
@@ -8,7 +8,7 @@
"description": "This protocol automates the entire Oxford Nanopore sequencing kit SQK-RBK004 workflow. Input a variable number of samples (up to 12), and get out a pooled library. \n\n\n\nOpentrons P300-single channel electronic pipettes (GEN2)\nOpentrons P20-single channel electronic pipettes (GEN2)\nOpentrons 20ul Filter Tips\nOpentrons 200ul Filter Tips\nRapid Barcoding Kit (SQK-RBK004)\nFresh 70% Ethanol\nH2O\n1.5mL centrifuge tubes\n",
"internal": "3bc25d",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Nanopore\n\n",
"description": "This protocol automates the entire Oxford Nanopore sequencing kit SQK-RBK004 workflow. Input a variable number of samples (up to 12), and get out a pooled library. \n\n---\n\n\n* [Opentrons P300-single channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=5984549109789)\n* [Opentrons P20-single channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=31059478970462)\n* [Opentrons 20ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Rapid Barcoding Kit (SQK-RBK004)](https://store.nanoporetech.com/us/rapid-barcoding-kit.html)\n* Fresh 70% Ethanol\n* H2O\n* [1.5mL centrifuge tubes](https://shop.opentrons.com/collections/tubes/products/nest-microcentrifuge-tubes)\n\n",
"internal": "3bc25d\n",
diff --git a/protoBuilds/3c9aec/README.json b/protoBuilds/3c9aec/README.json
index 28441204b3..59b05138b2 100644
--- a/protoBuilds/3c9aec/README.json
+++ b/protoBuilds/3c9aec/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sterile Workflows": [
"Cell Culture"
@@ -10,7 +10,7 @@
"internal": "3c9ec",
"labware": "\nCorning 360ul 96 well plate flat\nNEST 12 well Reseroir 195mL\nOpentrons 200ul Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sterile Workflows\n\t* Cell Culture\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "This protocol preps a 96 well plate with cell culture and a variety of reagents. First, the protocol parses the csv to check for wells which have a confluency value of greater than 85. These values will be referenced and added to for the duration of the protocol. If there are a group of confluency wells with a value of greater than 85 in a single column, the multi-channel pipette will be used, only picking up the number of tips necessary. The user may be asked to replace tips if the protocol runs out. Note that a volume of at least 3mL of any reagent should be in each reservoir well AFTER the protocol is completed (i.e. there should never be > 3mL of reagent at any point of the protocol). See protocol steps below for details.\n\nExplanation of complex parameters below:\n* `csv file`: Upload the csv file as so:\n```\nWell,Confluency\nA1, 90\nB1, 83\n```\n* `Temperature module temperature (C)`: Specify temperature module temperatre in celsius\n* `First step aspiration rate (moving to waste)`: Specify the aspiration rate in step 1 of the protocol when included wells are moved to waste, where 1.0 is default, 1.5 is 50% faster, and 0.2 is 20% default speed, for example.\n* `PBS Dispense Flow Rate`: The PBS will be dispesed into the plate 2mm from the side of the well to avoid separation of culture. Specify the flow rate, where 1.0 is default, 1.5 is 50% faster, and 0.2 is 20% default speed, for example.\n* `Incubation Time (minutes)`: Specify the incubation time i nminutes after trypsin is added to the plate.\n* `Media Aspirate X (ul)`: Specify volume of media to move from column 12 of the reservoir to the plate.\n* `Media Dispense Y (ul)`: Specify the volume of the second media to be added to the plate.\n* `Track tips?`: Yes for tracking tips, no for fresh tip boxes.\n* `P300 Single-Channel Mount`: Specify which mount (left or right) to host your P300 pipettes.\n\n---\n\n",
diff --git a/protoBuilds/3cf31f/README.json b/protoBuilds/3cf31f/README.json
index 0b49eee89d..813da60098 100644
--- a/protoBuilds/3cf31f/README.json
+++ b/protoBuilds/3cf31f/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Distribution"
@@ -8,7 +8,7 @@
"description": "This protocol is part 1 of 2-part protocol designed to transfer biofluids and add the necessary reagents for preparation. In this part, The biofluids are distributed from cryovials to centrifuge tubes and two solutions are added. The samples are then transferred to a Nanosep 3k Ultrafiltration tube in preparation of centrifugation. See below for deck and reagent setup.\n\n\n\nP300 Single-channel electronic pipette\nP50 Single-channel electronic pipette\n50/300uL Opentrons tipracks\nOpentrons Temperature Module\nOpentrons aluminum block set\nCryovials (containing samples)\n0.5mL centrifuge tubes (clean and empty)\nNanosep 3K Ultrafiltration tubes (clean and empty)\nCustom container for holding cryovials\nCustom container for holding centrifuge tubes (x2)\n\n\n\nSlot 1/4/7: Custom container for holding centrifuge tubes, filled with Nanosep 3k Ultrafiltration tubes\nSlot 2/5/8: Custom container for holding centrifuge tubes, filled with 0.5mL centrifuge tubes\nSlot 3/6/9: Custom container for holding cryovials, filled with cryovials containing samples\nSlot 10: Temperature Module with 24-Well Aluminum Block :\n A1: Solution 1\n A2: Solution 2\n A3: Solution 2\n A4: Solution 2\n B1: Solution 2\n B2: Solution 2\nSlot 11: 50/300ul Opentrons tiprack",
"internal": "3cf31f",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Distribution\n\n\n",
"description": "This protocol is part 1 of 2-part protocol designed to transfer biofluids and add the necessary reagents for preparation. In this part, The biofluids are distributed from cryovials to centrifuge tubes and two solutions are added. The samples are then transferred to a Nanosep 3k Ultrafiltration tube in preparation of centrifugation. See below for deck and reagent setup.\n\n---\n\n\n* [P300 Single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P50 Single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [50/300uL Opentrons tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Opentrons aluminum block set](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set)\n* Cryovials (containing samples)\n* 0.5mL centrifuge tubes (clean and empty)\n* Nanosep 3K Ultrafiltration tubes (clean and empty)\n* Custom container for holding cryovials\n* Custom container for holding centrifuge tubes (x2)\n\n---\n\n\nSlot 1/4/7: Custom container for holding centrifuge tubes, filled with Nanosep 3k Ultrafiltration tubes\n\nSlot 2/5/8: Custom container for holding centrifuge tubes, filled with 0.5mL centrifuge tubes\n\nSlot 3/6/9: Custom container for holding cryovials, filled with cryovials containing samples\n\nSlot 10: [Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [24-Well Aluminum Block ](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set):\n* A1: Solution 1\n* A2: Solution 2\n* A3: Solution 2\n* A4: Solution 2\n* B1: Solution 2\n* B2: Solution 2\n\nSlot 11: [50/300ul Opentrons tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n",
"internal": "3cf31f",
diff --git a/protoBuilds/3d02be/README.json b/protoBuilds/3d02be/README.json
index 99c826598b..093475d67b 100644
--- a/protoBuilds/3d02be/README.json
+++ b/protoBuilds/3d02be/README.json
@@ -18,7 +18,7 @@
"labware": "* [Bio-RAD 96 Well Plate 200ul PCR](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?_gl=1*10feo1p*_gcl_aw*R0NMLjE2MTcwNjQ4OTguQ2owS0NRanc5WVdEQmhEeUFSSXNBRHQ2c0dhUUYzZ19ranNKV2R6Z1R6UmdzdG5QeE1DRmNBWm9zNEk3NFJ3YUpDUTItTWI2UDg4Xy1qWWFBbThnRUFMd193Y0I.*_ga*ODQ1NDAxMzU2LjE2MTIxOTA0Nzc.*_ga_GNSMNLW4RY*MTYxOTIwMTM3Mi4xODcuMS4xNjE5MjAxNDIxLjA.&_ga=2.87335384.1162033746.1618926044-845401356.1612190477&_gac=1.122034937.1617064898.Cj0KCQjw9YWDBhDyARIsADt6sGaQF3g_kjsJWdzgTzRgstnPxMCFcAZos4I74RwaJCQ2-Mb6P88_-jYaAm8gEALw_wcB)\n* [NEST 96 2mL Deep Well Plate](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?_gl=1*10feo1p*_gcl_aw*R0NMLjE2MTcwNjQ4OTguQ2owS0NRanc5WVdEQmhEeUFSSXNBRHQ2c0dhUUYzZ19ranNKV2R6Z1R6UmdzdG5QeE1DRmNBWm9zNEk3NFJ3YUpDUTItTWI2UDg4Xy1qWWFBbThnRUFMd193Y0I.*_ga*ODQ1NDAxMzU2LjE2MTIxOTA0Nzc.*_ga_GNSMNLW4RY*MTYxOTIwMTM3Mi4xODcuMS4xNjE5MjAxNDIxLjA.&_ga=2.87335384.1162033746.1618926044-845401356.1612190477&_gac=1.122034937.1617064898.Cj0KCQjw9YWDBhDyARIsADt6sGaQF3g_kjsJWdzgTzRgstnPxMCFcAZos4I74RwaJCQ2-Mb6P88_-jYaAm8gEALw_wcB)\n* Thomas Sci LabForce 96 Well Plate 200ul\n* FisherSci 96 Well Plate 1mL\n\n\n\n",
"modules": "No modules are required for this protocol.\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Partner Name](partner website link)\n\n",
+ "partner": "[Partner Name](partner website link)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P300-Multi GEN 2 Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885m)\n\n\n---\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Aspirate user-specified volume (from csv) from Column 1 of Source Plate (8-Channel p300)\n2. Dispense user-specified volume (from csv) into Column 1 of Destination Plate A (8-Channel p300)\n3. Dispense user-specified volume (from csv) into Column 1 of Destination Plate B (8-Channel p300)\n4. Dispense user-specified volume (from csv) into Column 1 of Destination Plate C (8-Channel p300)\n5. Discard Tips\n6. Repeat steps 1-5 over the breadth of the CSV file.\n\n",
@@ -26,7 +26,7 @@
},
"modules": [],
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Partner Name",
+ "partner": "Partner Name\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP300-Multi GEN 2 Pipette\n\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nAspirate user-specified volume (from csv) from Column 1 of Source Plate (8-Channel p300)\nDispense user-specified volume (from csv) into Column 1 of Destination Plate A (8-Channel p300)\nDispense user-specified volume (from csv) into Column 1 of Destination Plate B (8-Channel p300)\nDispense user-specified volume (from csv) into Column 1 of Destination Plate C (8-Channel p300)\nDiscard Tips\nRepeat steps 1-5 over the breadth of the CSV file.\n",
diff --git a/protoBuilds/3d942d/README.json b/protoBuilds/3d942d/README.json
index 671e338acc..86c0ff5a12 100644
--- a/protoBuilds/3d942d/README.json
+++ b/protoBuilds/3d942d/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "3d942d",
"labware": "\nOpentrons 20\u00b5L Filter Tips\nNEST 96-Well Plate\nCorning 384-Well Plate\nOpentrons Tube Rack\n1.5mL Microcentrigue Tubes\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n**Slot 1**: Source Plate ([NEST 96-Well Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt))\n\n\n**Slot 2**: Preamp Plate ([NEST 96-Well Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt))\n\n\n**Slot 3**: Loading Plate ([Corning 384-Well Plate](https://labware.opentrons.com/corning_384_wellplate_112ul_flat?category=wellPlate))\n\n\n**Slot 4**: [Opentrons 20\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips) (Tiprack 1)\n\n\n**Slot 5**: [Opentrons Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 24-Tube Topper\n\n\n**Slot 7**: [Opentrons 20\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips) (Tiprack 2)\n\n\n**Slot 10**: [Opentrons 20\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips) (Tiprack 3)\n\n\n\n",
"description": "Theis protocol is used to set up DNA samples for analysis with [Life Technologies QuantStudio\u2122 12K Flex Real-Time PCR System, OpenArray\u2122 block, Accufill\u2122 System](https://www.thermofisher.com/order/catalog/product/4471090?SID=srch-srp-4471090#/4471090?SID=srch-srp-4471090). Using the [P20 Single-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette), two PCR preps are accomplished. During the first prep, samples are reformatted and transferred from a 96-well plate to another 96-well plate before amplification. Once amplification is complete, the user returns the amplified plate and the second prep occurs - this time samples are transferred from the 96-well plate to a 384-well plate.\n\n\nExplanation of complex parameters below:\n* **Transfer CSV**: CSV file that contains the transfer information. It should contain the following header: SourcePos,SampleID,PreampPos,LoadPos1,LoadPos2\n* **P20 Single Mount**: Select which mount the [P20 Single-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) is attached to.\n\n*Note*: In the current configuration, up to 95 samples can be completed in one run.\n\n---\n\n\n",
diff --git a/protoBuilds/3db190-manual/README.json b/protoBuilds/3db190-manual/README.json
index 5ac1f60cdc..4f1bc45d06 100644
--- a/protoBuilds/3db190-manual/README.json
+++ b/protoBuilds/3db190-manual/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol automates certain steps of the Lyra Direct SARS-CoV Assay. This protocol assumes buffer is being added manually. The automated steps include mixing of the deep well block, and transferring samples to the PCR plate. \n\n\n\nP20 single-channel GEN2 electronic pipette\nP1000 single-channel GEN2 electronic pipette\nOpentrons 20ul filter tiprack\nOpentrons 1000ul filter tiprack\nMicroAmp\u2122 EnduraPlate\u2122 Optical 96-Well PCR Plates\nEppendorf 96 Deep Well Block 1000ul\n\n\n\nDeck Setup\n Opentrons 1000ul filter tiprack (Slot 4)\n Opentrons 20ul filter tiprack (Slot 7)\n Eppendorf 96 Deep Well Block 1000ul (Slot 2)\n MicroAmp\u2122 EnduraPlate\u2122 Optical 96-Well PCR Plate (Slot 3, should be rested on top of the Eppendorf cooling block)",
"internal": "3db190",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol automates certain steps of the Lyra Direct SARS-CoV Assay. This protocol assumes buffer is being added manually. The automated steps include mixing of the deep well block, and transferring samples to the PCR plate. \n\n---\n\n\n* [P20 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [P1000 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 20ul filter tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-filter-tip)\n* [Opentrons 1000ul filter tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips)\n* [MicroAmp\u2122 EnduraPlate\u2122 Optical 96-Well PCR Plates](https://www.thermofisher.com/order/catalog/product/4483354#/4483354)\n* [Eppendorf 96 Deep Well Block 1000ul](https://online-shop.eppendorf.us/US-en/Laboratory-Consumables-44512/Plates-44516/Eppendorf-Deepwell-Plates-PF-55960.html)\n\n---\n\n\nDeck Setup\n* Opentrons 1000ul filter tiprack (Slot 4)\n* Opentrons 20ul filter tiprack (Slot 7)\n* Eppendorf 96 Deep Well Block 1000ul (Slot 2)\n* MicroAmp\u2122 EnduraPlate\u2122 Optical 96-Well PCR Plate (Slot 3, should be rested on top of the Eppendorf cooling block)\n\n",
"internal": "3db190",
diff --git a/protoBuilds/3db190/README.json b/protoBuilds/3db190/README.json
index 55c41ae049..dfb7cc37d1 100644
--- a/protoBuilds/3db190/README.json
+++ b/protoBuilds/3db190/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Featured": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol automates certain steps of the Lyra Direct SARS-CoV Assay. These steps include the addition of process buffer to the deep well block, mixing of the deep well block, and transferring samples to the PCR plate. \n\n\n\nP20 multi-channel GEN2 electronic pipette\nP300 multi-channel GEN2 electronic pipette\nOpentrons 20ul filter tiprack\nOpentrons 200ul filter tiprack\nMicroAmp\u2122 EnduraPlate\u2122 Optical 96-Well PCR Plates\nEppendorf 96 Deep Well Block 1000ul\nOpentrons 10 Tube Rack with Falcon 4x50 mL, 6x15 mL Conical\nNEST 12 Well Reservoir 15 mL\n\n\n\nSingle Channel Setup:\n Pipettes: P1000 Single GEN2 AND P20 Single GEN2\n Opentrons 1000ul filter/standard tipracks\n Opentrons 20ul filter/standard tipracks \n Opentrons 10 Tube Rack with Falcon 4x50 mL, 6x15 mL Conical (Slot 1, Process Buffer in A1)\n Eppendorf 96 Deep Well Block 1000ul (Slot 2)\n MicroAmp\u2122 EnduraPlate\u2122 Optical 96-Well PCR Plate (Slot 3, should be rested on top of the Eppendorf cooling block)\nMulti Channel Setup:\n Pipettes: P300 Multi GEN2 AND P20 Multi GEN2\n Opentrons 200ul/300ul filter/standard tiprack\n Opentrons 20ul filter/standard tiprack\n NEST 12 Well Reservoir 15 mL (10 mL of Process Buffer in Channels 1-4)\n Eppendorf 96 Deep Well Block 1000ul (Slot 2)\n MicroAmp\u2122 EnduraPlate\u2122 Optical 96-Well PCR Plate (Slot 3, should be rested on top of the Eppendorf cooling block)",
"internal": "3db190",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Featured\n\t* PCR Prep\n\n",
"description": "This protocol automates certain steps of the [Lyra Direct SARS-CoV Assay](https://www.quidel.com/molecular-diagnostics/lyra-direct-sars-cov-2-assay). These steps include the addition of process buffer to the deep well block, mixing of the deep well block, and transferring samples to the PCR plate. \n\n---\n\n\n* [P20 multi-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5978988707869)\n* [P300 multi-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 20ul filter tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-filter-tip)\n* [Opentrons 200ul filter tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips)\n* [MicroAmp\u2122 EnduraPlate\u2122 Optical 96-Well PCR Plates](https://www.thermofisher.com/order/catalog/product/4483354#/4483354)\n* [Eppendorf 96 Deep Well Block 1000ul](https://online-shop.eppendorf.us/US-en/Laboratory-Consumables-44512/Plates-44516/Eppendorf-Deepwell-Plates-PF-55960.html)\n* [Opentrons 10 Tube Rack with Falcon 4x50 mL, 6x15 mL Conical](https://shop.opentrons.com/products/tube-rack-set-1?_ga=2.93128221.1266032643.1606143320-1181961818.1604785212)\n* [NEST 12 Well Reservoir 15 mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n\n---\n\n\n**Single Channel Setup:**\n* Pipettes: P1000 Single GEN2 AND P20 Single GEN2\n* Opentrons 1000ul filter/standard tipracks\n* Opentrons 20ul filter/standard tipracks \n* Opentrons 10 Tube Rack with Falcon 4x50 mL, 6x15 mL Conical (Slot 1, Process Buffer in A1)\n* Eppendorf 96 Deep Well Block 1000ul (Slot 2)\n* MicroAmp\u2122 EnduraPlate\u2122 Optical 96-Well PCR Plate (Slot 3, should be rested on top of the Eppendorf cooling block)\n\n**Multi Channel Setup:**\n* Pipettes: P300 Multi GEN2 AND P20 Multi GEN2\n* Opentrons 200ul/300ul filter/standard tiprack\n* Opentrons 20ul filter/standard tiprack\n* NEST 12 Well Reservoir 15 mL (10 mL of Process Buffer in Channels 1-4)\n* Eppendorf 96 Deep Well Block 1000ul (Slot 2)\n* MicroAmp\u2122 EnduraPlate\u2122 Optical 96-Well PCR Plate (Slot 3, should be rested on top of the Eppendorf cooling block)\n\n",
"internal": "3db190",
diff --git a/protoBuilds/3e0bef/README.json b/protoBuilds/3e0bef/README.json
index aa4ac776f1..0cabe7089b 100644
--- a/protoBuilds/3e0bef/README.json
+++ b/protoBuilds/3e0bef/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Omega Bio-Tek Mag-Bind Plant DNA DS Kit"
@@ -10,7 +10,7 @@
"internal": "3e0b3f",
"labware": "\nSample Input Labware (specified in parameters)\nNEST 12-Well Reservoir, 15mL\nNEST 1-Well Reservoir, 195mL\nNEST 96-Well PCR Plate\nNEST 96-Well Deepwell Plate, 2mL\nOpentrons 200\u00b5L Filter Tip Racks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Omega Bio-Tek Mag-Bind Plant DNA DS Kit\n\n",
"deck-setup": "\n\n",
"description": "This protocol automates the [Omega Bio-Tek Mag-Bind\u00ae Plant DNA DS 96 Kit](https://www.omegabiotek.com/product/plant-extraction-mag-bind-plant-dna-ds-96/) on the OT-2.\n\nUsing the [P300 8-Channel Pipette, GEN2](https://shop.opentrons.com/8-channel-electronic-pipette/) all liquid handling steps outlined in [the protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-12-14/kf03tt7/Omega%20BioTek%20Mag-Bind%20Plant%20DNA%20extraction%20automation%20protocol.pdf) are handled on the OT-2, with minimal user intervention. To optimize liquid handling on the automated platform, some reagents should be combined prior to adding to the deck (more details below).\n\nExplanation of complex parameters below:\n* **Pipette Mount**: Select which mount (left or right) the [P300 8-Channel Pipette, GEN2](https://shop.opentrons.com/8-channel-electronic-pipette/) is attached to.\n* **Number of Samples**: Specify the number of samples (1-96). Please note ~ because the 8-channel pipette is used in this protocol, any number will be rounded up to 8.\n* **Binding Time (minutes)**: Specify the number of minutes for binding, prior to the first supernatant removal. Please note that this is an estimate, as there will be continuous mixing throughout this step.\n* **MagDeck Incubation (minutes)**: Specify the number of minutes to delay the robot, while the MagDeck is engaged.\n* **Air Dry Time (minutes)**: Specify the number of minutes for air drying after the final wash step.\n* **Add Water Before Air Dry**: Specify whether or not to add, then remove 100\u00b5L of water immediately before the air dry step.\n* **Labware for Samples Input**: Select the labware that will contain the samples from the drop down.\n* **[QIAGEN ONLY] Aspiration Height**: Specify the height (in mm) from the bead to aspirate from (3mm is the default)\n* **Final Wash Removal (in \u00b5L)**: Specify the volume for the supernatant removal step during the final wash.\n* **Elution Volume (in \u00b5L)**: The volume of Elution Buffer added to each well. Please note ~ [the written protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-12-14/kf03tt7/Omega%20BioTek%20Mag-Bind%20Plant%20DNA%20extraction%20automation%20protocol.pdf) states 100-200\u00b5L and this protocol has been designed accordingly.\n* **Perform Resuspension Off-Deck**: Specify whether to perform resuspension with manual intervention (Yes) or with the OT-2 Pipette (No). This will apply to the initial 10 minute incubation and the 4 wash steps.\n* **Play Custom Music at Pause**: Specify whether or not to play custom music during pause steps. Please note, selecting **'No'** will result in the pre-installed music playing. Please only select **'Yes'** if you've worked with the Opentrons team on this protocol prior to download\n\n\n*Note*: This protocol was last updated on October 27th, 2022\n\n---\n\n",
diff --git a/protoBuilds/3e3c9d-protocol-Adding-BTM/README.json b/protoBuilds/3e3c9d-protocol-Adding-BTM/README.json
index 6955edd6c1..0dd1663110 100644
--- a/protoBuilds/3e3c9d-protocol-Adding-BTM/README.json
+++ b/protoBuilds/3e3c9d-protocol-Adding-BTM/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol updates an existing protocol request from Opentrons API Version 1 to Opentrons API Version 2.\n\nUsing the customizable parameters below, you can specify which column to start picking up tips from and how many samples are in each run for each protocol. \n\nThis protocol uses custom labware definitions for the glass trough (Electron Microscopy Sciences), Omegaquant 96-Well Plate, Blood Tube Rack, and 200ul Tip Rack. When downloading the protocol, the labware definitions (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: Using labware in your protocols\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P50 Multi-Channel Pipette\nOpentrons Corning 96 Well Plate 360 \u00b5L Flat\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 300\u00b5L Tips\nElectron Microscopy Sciences Rectangular Staining Dish [https://www.emsdiasum.com/microscopy/products/histology/staining.aspx#70314]\nReagents\n\n\n\nSlot 1: Electron Microscopy Sciences Rectangular Staining Dish\nSlot 2: Corning 96 Well Plate 360 \u00b5L Flat\nSlot 4: Opentrons 96 Tip-Rack 300ul\nP300-Multi Mount: Left Mount\n\n\nUsing the customizations field (below), set up your protocol.\n Number of Samples: Specify number of samples in well plate.\n Starting Tip Column: Specify which column of tips to begin drawing from.",
"internal": "3e3c9d-protocol-Adding-BTM",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol updates an existing protocol request from Opentrons API Version 1 to Opentrons API Version 2.\n\nUsing the customizable parameters below, you can specify which column to start picking up tips from and how many samples are in each run for each protocol. \n\nThis protocol uses custom labware definitions for the glass trough (Electron Microscopy Sciences), Omegaquant 96-Well Plate, Blood Tube Rack, and 200ul Tip Rack. When downloading the protocol, the labware definitions (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: [Using labware in your protocols](https://support.opentrons.com/en/articles/3136506-using-labware-in-your-protocols)\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P50 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons Corning 96 Well Plate 360 \u00b5L Flat](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate)\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://labware.opentrons.com/opentrons_96_tiprack_300ul?category=tipRack)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* Electron Microscopy Sciences Rectangular Staining Dish [https://www.emsdiasum.com/microscopy/products/histology/staining.aspx#70314]\n* Reagents\n\n\n\n---\n\n\nSlot 1: Electron Microscopy Sciences Rectangular Staining Dish\n\nSlot 2: Corning 96 Well Plate 360 \u00b5L Flat\n\nSlot 4: Opentrons 96 Tip-Rack 300ul\n\nP300-Multi Mount: Left Mount\n\n\n\n\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Samples**: Specify number of samples in well plate.\n* **Starting Tip Column**: Specify which column of tips to begin drawing from.\n\n\n\n",
"internal": "3e3c9d-protocol-Adding-BTM\n",
diff --git a/protoBuilds/3e3c9d-protocol-Adding-Water-RBC/README.json b/protoBuilds/3e3c9d-protocol-Adding-Water-RBC/README.json
index a81bb2ac17..7d4dc82345 100644
--- a/protoBuilds/3e3c9d-protocol-Adding-Water-RBC/README.json
+++ b/protoBuilds/3e3c9d-protocol-Adding-Water-RBC/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol updates an existing protocol request from Opentrons API Version 1 to Opentrons API Version 2.\n\nUsing the customizable parameters below, you can specify which column to start picking up tips from and how many samples are in each run for each protocol. \n\nThis protocol uses custom labware definitions for the glass trough (Electron Microscopy Sciences), Omegaquant 96-Well Plate, Blood Tube Rack, and 200ul Tip Rack. When downloading the protocol, the labware definitions (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: Using labware in your protocols\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P50 Multi-Channel Pipette\nOpentrons Corning 96 Well Plate 360 \u00b5L Flat\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 300\u00b5L Tips\nElectron Microscopy Sciences Rectangular Staining Dish [https://www.emsdiasum.com/microscopy/products/histology/staining.aspx#70314]\nReagents\n\n\n\nSlot 1: Electron Microscopy Sciences Rectangular Staining Dish\nSlot 2: Corning 96 Well Plate 360 \u00b5L Flat\nSlot 4: Opentrons 96 Tip-Rack 300ul\nP300-Multi Mount: Left\n\n\nUsing the customizations field (below), set up your protocol.\n Number of Samples: Specify number of samples in well plate.\n Starting Tip Column: Specify which column of tips to begin drawing from.",
"internal": "3e3c9d-protocol-Adding-Water-RBC",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol updates an existing protocol request from Opentrons API Version 1 to Opentrons API Version 2.\n\nUsing the customizable parameters below, you can specify which column to start picking up tips from and how many samples are in each run for each protocol. \n\nThis protocol uses custom labware definitions for the glass trough (Electron Microscopy Sciences), Omegaquant 96-Well Plate, Blood Tube Rack, and 200ul Tip Rack. When downloading the protocol, the labware definitions (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: [Using labware in your protocols](https://support.opentrons.com/en/articles/3136506-using-labware-in-your-protocols)\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P50 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons Corning 96 Well Plate 360 \u00b5L Flat](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate)\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://labware.opentrons.com/opentrons_96_tiprack_300ul?category=tipRack)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* Electron Microscopy Sciences Rectangular Staining Dish [https://www.emsdiasum.com/microscopy/products/histology/staining.aspx#70314]\n* Reagents\n\n\n\n---\n\n\nSlot 1: Electron Microscopy Sciences Rectangular Staining Dish\n\nSlot 2: Corning 96 Well Plate 360 \u00b5L Flat\n\nSlot 4: Opentrons 96 Tip-Rack 300ul\n\nP300-Multi Mount: Left\n\n\n\n\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Samples**: Specify number of samples in well plate.\n* **Starting Tip Column**: Specify which column of tips to begin drawing from.\n\n\n\n",
"internal": "3e3c9d-protocol-Adding-Water-RBC\n",
diff --git a/protoBuilds/3e3c9d-protocol-master-rbc-transfer/README.json b/protoBuilds/3e3c9d-protocol-master-rbc-transfer/README.json
index e327689f32..5574ae7d17 100644
--- a/protoBuilds/3e3c9d-protocol-master-rbc-transfer/README.json
+++ b/protoBuilds/3e3c9d-protocol-master-rbc-transfer/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol updates an existing protocol request from Opentrons API Version 1 to Opentrons API Version 2.\n\nUsing the customizable parameters below, you can specify which column to start picking up tips from and how many samples are in each run for each protocol. \n\nThis protocol uses custom labware definitions for the glass trough (Electron Microscopy Sciences), Omegaquant 96-Well Plate, Blood Tube Rack, and 200ul Tip Rack. When downloading the protocol, the labware definitions (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: Using labware in your protocols\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P50 Single-Channel Pipette\nOpentrons P50 Multi-Channel Pipette\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 300\u00b5L Tips\nElectron Microscopy Sciences Rectangular Staining Dish [https://www.emsdiasum.com/microscopy/products/histology/staining.aspx#70314]\nOmegaquant 96-Well Plate\nBlood Tube Rack\n200ul Tip-Rack and Tips\nReagents\n\n\n\nSlot 2: Omegaquant 96-Well Plate\nSlot 3: Electron Microscopy Sciences Rectangular Staining Dish\nSlot 4: Opentrons 96 Tip-Rack 300ul\nSlot 5: Electron Microscopy Sciences Rectangular Staining Dish\nSlot 6: 200ul Tip-Rack\nSlot 7: Blood Tube Rack\nSlot 8: Blood Tube Rack\nSlot 10: Blood Tube Rack\nSlot 11: Blood Tube Rack\nP300-Multi Mount: Left\nP50-Single Mount: Right\n\n\nUsing the customizations field (below), set up your protocol.\n Number of Samples: Specify number of samples in well plate.\n Starting Tip Column: Specify which column of tips to begin drawing from.",
"internal": "3e3c9d-protocol-Master-RBC-Transfer",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n\n",
"description": "This protocol updates an existing protocol request from Opentrons API Version 1 to Opentrons API Version 2.\n\nUsing the customizable parameters below, you can specify which column to start picking up tips from and how many samples are in each run for each protocol. \n\nThis protocol uses custom labware definitions for the glass trough (Electron Microscopy Sciences), Omegaquant 96-Well Plate, Blood Tube Rack, and 200ul Tip Rack. When downloading the protocol, the labware definitions (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: [Using labware in your protocols](https://support.opentrons.com/en/articles/3136506-using-labware-in-your-protocols)\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P50 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons P50 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://labware.opentrons.com/opentrons_96_tiprack_300ul?category=tipRack)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* Electron Microscopy Sciences Rectangular Staining Dish [https://www.emsdiasum.com/microscopy/products/histology/staining.aspx#70314]\n* Omegaquant 96-Well Plate\n* Blood Tube Rack\n* 200ul Tip-Rack and Tips\n* Reagents\n\n\n\n---\n\n\nSlot 2: Omegaquant 96-Well Plate\n\nSlot 3: Electron Microscopy Sciences Rectangular Staining Dish\n\nSlot 4: Opentrons 96 Tip-Rack 300ul\n\nSlot 5: Electron Microscopy Sciences Rectangular Staining Dish\n\nSlot 6: 200ul Tip-Rack\n\nSlot 7: Blood Tube Rack\n\nSlot 8: Blood Tube Rack\n\nSlot 10: Blood Tube Rack\n\nSlot 11: Blood Tube Rack\n\nP300-Multi Mount: Left\n\nP50-Single Mount: Right\n\n\n\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Samples**: Specify number of samples in well plate.\n* **Starting Tip Column**: Specify which column of tips to begin drawing from.\n\n\n\n",
"internal": "3e3c9d-protocol-Master-RBC-Transfer\n",
diff --git a/protoBuilds/3e3c9d-protocols-adding-wistd-water/README.json b/protoBuilds/3e3c9d-protocols-adding-wistd-water/README.json
index 64cdd55a00..a559fd3b8b 100644
--- a/protoBuilds/3e3c9d-protocols-adding-wistd-water/README.json
+++ b/protoBuilds/3e3c9d-protocols-adding-wistd-water/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol updates an existing protocol request from Opentrons API Version 1 to Opentrons API Version 2.\n\nUsing the customizable parameters below, you can specify which column to start picking up tips from and how many samples are in each run for each protocol. \n\nThis protocol uses custom labware definitions for the glass trough (Electron Microscopy Sciences), Omegaquant 96-Well Plate, Blood Tube Rack, and 200ul Tip Rack. When downloading the protocol, the labware definitions (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: Using labware in your protocols\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P50 Multi-Channel Pipette\nOpentrons Corning 96 Well Plate 360 \u00b5L Flat\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 300\u00b5L Tips\nElectron Microscopy Sciences Rectangular Staining Dish [https://www.emsdiasum.com/microscopy/products/histology/staining.aspx#70314]\nReagents\n\n\n\nSlot 2: Corning 96 Well Plate 360 \u00b5L Flat\nSlot 4: Opentrons 96 Tip-Rack 300ul\nSlot 5: Electron Microscopy Sciences Rectangular Staining Dish\nSlot 9: Electron Microscopy Sciences Rectangular Staining Dish\nP300-Multi Mount: Left\n\n\nUsing the customizations field (below), set up your protocol.\n Number of Samples: Specify number of samples in well plate.\n Starting Tip Column: Specify which column of tips to begin drawing from.",
"internal": "3e3c9d-protocols-Adding-WISTD-Water",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "This protocol updates an existing protocol request from Opentrons API Version 1 to Opentrons API Version 2.\n\nUsing the customizable parameters below, you can specify which column to start picking up tips from and how many samples are in each run for each protocol. \n\nThis protocol uses custom labware definitions for the glass trough (Electron Microscopy Sciences), Omegaquant 96-Well Plate, Blood Tube Rack, and 200ul Tip Rack. When downloading the protocol, the labware definitions (a JSON file) will be included for use with this protocol. For more information on using custom labware on the OT-2, please see this article: [Using labware in your protocols](https://support.opentrons.com/en/articles/3136506-using-labware-in-your-protocols)\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P50 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons Corning 96 Well Plate 360 \u00b5L Flat](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate)\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://labware.opentrons.com/opentrons_96_tiprack_300ul?category=tipRack)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* Electron Microscopy Sciences Rectangular Staining Dish [https://www.emsdiasum.com/microscopy/products/histology/staining.aspx#70314]\n* Reagents\n\n\n\n---\n\n\nSlot 2: Corning 96 Well Plate 360 \u00b5L Flat\n\nSlot 4: Opentrons 96 Tip-Rack 300ul\n\nSlot 5: Electron Microscopy Sciences Rectangular Staining Dish\n\nSlot 9: Electron Microscopy Sciences Rectangular Staining Dish\n\nP300-Multi Mount: Left\n\n\n\n\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Samples**: Specify number of samples in well plate.\n* **Starting Tip Column**: Specify which column of tips to begin drawing from.\n\n\n\n",
"internal": "3e3c9d-protocols-Adding-WISTD-Water\n",
diff --git a/protoBuilds/3fad82-part-2/README.json b/protoBuilds/3fad82-part-2/README.json
index 6945776fc6..9ac4dea3f4 100644
--- a/protoBuilds/3fad82-part-2/README.json
+++ b/protoBuilds/3fad82-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II FS DNA Library Prep Kit for Illumina"
@@ -9,7 +9,7 @@
"description": "Part 2 of 3: Clean Up and PCR Enrichment.\nLinks:\n Part 1: End Prep and Adapter Ligation\n Part 2: Clean Up and PCR Enrichment\n* Part 3: Final Clean Up\nWith this protocol, your robot can perform a modified Quarter Volume NEBNext Ultra II FS DNA Library Prep Kit for Illumina protocol described by the Experimental Protocol. NEB Instruction Manual for NEBNext.\nThis is part 2 of the protocol: Clean up and PCR enrichment.\nThis protocol assumes up to 96 input samples and follows the attached experimental protocol.\nAfter the steps carried out in this protocol (part 2), proceed with part 3: Final Clean Up.",
"internal": "3fad82-part-2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * NEBNext Ultra II FS DNA Library Prep Kit for Illumina\n\n",
"deck-setup": "* Opentrons p20 tips (Deck Slots 2,3,5)\n* Opentrons p300 tips (Deck Slot 6)\n* Opentrons Thermocycler Module (Deck Slots 7,8,10,11)\n* Opentrons Magnetic Module with Sample Plate (Deck Slot 4)\n* Reagent Reservoir (Deck Slot 1)\n\n",
"description": "Part 2 of 3: Clean Up and PCR Enrichment.\n\nLinks:\n* [Part 1: End Prep and Adapter Ligation](http://protocols.opentrons.com/protocol/3fad82)\n* [Part 2: Clean Up and PCR Enrichment](http://protocols.opentrons.com/protocol/3fad82-part-2)\n* [Part 3: Final Clean Up](http://protocols.opentrons.com/protocol/3fad82-part-3)\n\nWith this protocol, your robot can perform a modified Quarter Volume NEBNext Ultra II FS DNA Library Prep Kit for Illumina protocol described by the [Experimental Protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-05-07/ia13rz0/Quarter%20volume%20NEB%20Next%20Ultra%20II%20DNA%20Library%20Prep%20Kit%20for%20Illumina.docx). [NEB Instruction Manual for NEBNext](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-05-07/ce23ruu/manualE7805.pdf).\n\nThis is part 2 of the protocol: Clean up and PCR enrichment.\n\nThis protocol assumes up to 96 input samples and follows the attached experimental protocol.\n\nAfter the steps carried out in this protocol (part 2), proceed with part 3: Final Clean Up.\n\n\n",
diff --git a/protoBuilds/3fad82-part-3/README.json b/protoBuilds/3fad82-part-3/README.json
index 2b54a6fa1c..f2622f4f85 100644
--- a/protoBuilds/3fad82-part-3/README.json
+++ b/protoBuilds/3fad82-part-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II FS DNA Library Prep Kit for Illumina"
@@ -9,7 +9,7 @@
"description": "Part 3 of 3: Final Clean Up.\nLinks:\n Part 1: End Prep and Adapter Ligation\n Part 2: Clean Up and PCR Enrichment\n* Part 3: Final Clean Up\nWith this protocol, your robot can perform a modified Quarter Volume NEBNext Ultra II FS DNA Library Prep Kit for Illumina protocol described by the Experimental Protocol. NEB Instruction Manual for NEBNext.\nThis is part 3 of the protocol: Final Clean Up.\nThis protocol assumes up to 96 input samples and follows the attached experimental protocol.",
"internal": "3fad82-part-3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * NEBNext Ultra II FS DNA Library Prep Kit for Illumina\n\n",
"deck-setup": "* Opentrons p20 tips (Deck Slots 2,3)\n* Opentrons p300 tips (Deck Slots 5,6)\n* Opentrons Thermocycler Module-deactivated (Deck Slots 7,8,10,11)\n* Opentrons Magnetic Module with Sample Plate (Deck Slot 4)\n* Reagent Reservoir (Deck Slot 1)\n\n",
"description": "Part 3 of 3: Final Clean Up.\n\nLinks:\n* [Part 1: End Prep and Adapter Ligation](http://protocols.opentrons.com/protocol/3fad82)\n* [Part 2: Clean Up and PCR Enrichment](http://protocols.opentrons.com/protocol/3fad82-part-2)\n* [Part 3: Final Clean Up](http://protocols.opentrons.com/protocol/3fad82-part-3)\n\nWith this protocol, your robot can perform a modified Quarter Volume NEBNext Ultra II FS DNA Library Prep Kit for Illumina protocol described by the [Experimental Protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-05-07/ia13rz0/Quarter%20volume%20NEB%20Next%20Ultra%20II%20DNA%20Library%20Prep%20Kit%20for%20Illumina.docx). [NEB Instruction Manual for NEBNext](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-05-07/ce23ruu/manualE7805.pdf).\n\nThis is part 3 of the protocol: Final Clean Up.\n\nThis protocol assumes up to 96 input samples and follows the attached experimental protocol.\n\n\n",
diff --git a/protoBuilds/3fad82/README.json b/protoBuilds/3fad82/README.json
index 5616d9dac1..585e6df3c7 100644
--- a/protoBuilds/3fad82/README.json
+++ b/protoBuilds/3fad82/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II FS DNA Library Prep Kit for Illumina"
@@ -9,7 +9,7 @@
"description": "Part 1 of 3: End Prep and Adapter Ligation.\nLinks:\n Part 1: End Prep and Adapter Ligation\n Part 2: Clean Up and PCR Enrichment\n* Part 3: Final Clean Up\nWith this protocol, your robot can perform a modified Quarter Volume NEBNext Ultra II FS DNA Library Prep Kit for Illumina protocol described by the Experimental Protocol. NEB Instruction Manual for NEBNext.\nThis is part 1 of the protocol: End prep and adapter ligation.\nThis protocol assumes up to 96 input samples and follows the attached experimental protocol.\nAfter the steps carried out in this protocol (part 1), proceed with part 2: Clean Up and PCR Enrichment.",
"internal": "3fad82",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * NEBNext Ultra II FS DNA Library Prep Kit for Illumina\n\n",
"deck-setup": "* Opentrons p20 tips (Deck Slots 2,3,5,6)\n* Opentrons Thermocycler Module (Deck Slots 7,8,10,11)\n* Master Mix Plate (Deck Slot 4)\n\n",
"description": "Part 1 of 3: End Prep and Adapter Ligation.\n\nLinks:\n* [Part 1: End Prep and Adapter Ligation](http://protocols.opentrons.com/protocol/3fad82)\n* [Part 2: Clean Up and PCR Enrichment](http://protocols.opentrons.com/protocol/3fad82-part-2)\n* [Part 3: Final Clean Up](http://protocols.opentrons.com/protocol/3fad82-part-3)\n\nWith this protocol, your robot can perform a modified Quarter Volume NEBNext Ultra II FS DNA Library Prep Kit for Illumina protocol described by the [Experimental Protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-05-07/ia13rz0/Quarter%20volume%20NEB%20Next%20Ultra%20II%20DNA%20Library%20Prep%20Kit%20for%20Illumina.docx). [NEB Instruction Manual for NEBNext](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-05-07/ce23ruu/manualE7805.pdf).\n\nThis is part 1 of the protocol: End prep and adapter ligation.\n\nThis protocol assumes up to 96 input samples and follows the attached experimental protocol.\n\nAfter the steps carried out in this protocol (part 1), proceed with part 2: Clean Up and PCR Enrichment.\n\n\n",
diff --git a/protoBuilds/3fb582/README.json b/protoBuilds/3fb582/README.json
index 85b2e5e9c2..51670cf804 100644
--- a/protoBuilds/3fb582/README.json
+++ b/protoBuilds/3fb582/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol automates PCR setups using a CSV file. Slots 2, 5, 8, and 11 are reserved for input tube racks. Slot 6 is reserved for a 384 well PCR plate and slot 9 is reserved for a 96 well PCR plate. The rest of the slots are reserved for tips.\n\nUpdate (Jan 13, 2021): A fix was added that was preventing use of slots 5, 8, and 11.\nCustomize the PCR setup CSV and upload it to get a custom protocol. Template CSV can be found here.\n\n\n\nOpentrons P20-single channel electronic pipettes (GEN2)\nOpentrons 20ul Filter Tips\nOpentrons 4-in-1 Tube Rack Set\n384 or 96 well PCR plate\n",
"internal": "3fb582",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol automates PCR setups using a CSV file. Slots 2, 5, 8, and 11 are reserved for input tube racks. Slot 6 is reserved for a 384 well PCR plate and slot 9 is reserved for a 96 well PCR plate. The rest of the slots are reserved for tips.\n\n**Update (Jan 13, 2021)**: A fix was added that was preventing use of slots 5, 8, and 11.\n\nCustomize the PCR setup CSV and upload it to get a custom protocol. Template CSV can be found [here](https://opentrons-protocol-library-website.s3.amazonaws.com/Technical+Notes/3fb582/PCR+Setup+Template+-+Sheet1.csv).\n\n---\n\n\n* [Opentrons P20-single channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=31059478970462)\n* [Opentrons 20ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/products/tube-rack-set-1)\n* 384 or 96 well PCR plate\n\n",
"internal": "3fb582\n",
diff --git a/protoBuilds/400b8e/README.json b/protoBuilds/400b8e/README.json
index d0d6a67ed1..7a7b9c6b75 100644
--- a/protoBuilds/400b8e/README.json
+++ b/protoBuilds/400b8e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -8,7 +8,7 @@
"description": "\nCherrypicking, or hit-picking, is a key component of many workflows from high-throughput screening to microbial transfections. With this protocol, you can easily select specific wells in any labware without worrying about missing or selecting the wrong well. Just upload your properly formatted CSV file (keep scrolling for an example), customize your parameters, and download your ready-to-run protocol. You will be prompted to interact with the deck during pauses if necessary throughout the protocol.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Single-Channel Pipette and corresponding Tips\nMicroplates (96-well or 384-well)\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nLabware will be loaded automatically by specifying the labware loadname and labware slot in the .csv file. All available empty slots will be filled with the necessary tipracks, and the user will be prompted to refill the tipracks if all are emptied in the middle of the protocol. To pause mid-protocol to replenish source plates on the deck, add a line in the .csv that reads pause (see example below for a deeper explanation).\nCSV Format\nYour cherrypicking transfers must be saved as a comma separated value (.csv) file type. Your CSV must contain values corresponding to volumes in microliters (\u03bcL). Note that the header line (first row of the .csv file) should also be included!\nHere's an example of how a short cherrypicking protocol should be properly formatted:\nSource Labware ID,Source Labware Type,Source Slot,Source Well,Source Aspiration Height Above Bottom (in mm),Dest Labware Type,Dest Slot,Dest Well,Volume (in ul)\nRNA1,agilent_1_reservoir_290ml,3,A1,1,nest_96_wellplate_100ul_pcr_full_skirt,4,A11,1\nRNA2,nest_12_reservoir_15ml,4,A1,1,nest_96_wellplate_2ml_deep,5,A5,3\npause,,,,,,,,\nRNA3,nest_1_reservoir_195ml,5,A1,1,nest_96_wellplate_2ml_deep,5,H12,7\nIn this example, 1\u03bcL will be transferred from 1mm above the bottom of well A1 in an Agilent 1-well 290ml reservoir (slot 1) to well A11 in the destination NEST 96-well plate 100\u00b5l (slot 4). After this, 3\u03bcL will be transferred from 1mm above the bottom of well A1 in a NEST 12-well 15ml reservoir (slot 2) to well A5 in the destination NEST 96-well plate 100\u00b5l (slot 5). Last, 7\u03bcL will be transferred from 1mm above the bottom of well A1 in a NEST 1-well 195ml reservoir (slot 3) to well H12 in the destination NEST 96-well plate 100\u00b5l (slot 5).\nIf you\u2019d like to follow our template, you can make a copy of this spreadsheet, fill out your values, and export as CSV for use with this protocol.\nUsing the customizations fields, below set up your protocol.\n Transfer .csv File: Upload the .csv file containing your well locations, volumes, and source plate (optional).\n Pipette Model: Select which pipette you will use for this protocol.\n Pipette Mount: Specify which mount your single-channel pipette is on (left or right)\n Tip Type: Specify whether you want to use filter tips.\n* Tip Usage Strategy: Specify whether you'd like to use a new tip for each transfer, or keep the same tip throughout the protocol.",
"internal": "400b8e",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"description": "\n\nCherrypicking, or hit-picking, is a key component of many workflows from high-throughput screening to microbial transfections. With this protocol, you can easily select specific wells in any labware without worrying about missing or selecting the wrong well. Just upload your properly formatted CSV file (keep scrolling for an example), customize your parameters, and download your ready-to-run protocol. You will be prompted to interact with the deck during pauses if necessary throughout the protocol.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes) and corresponding [Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Microplates (96-well or 384-well)](https://labware.opentrons.com/?category=wellPlate)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n\n---\n\n\nLabware will be loaded automatically by specifying the labware loadname and labware slot in the .csv file. All available empty slots will be filled with the necessary [tipracks](https://shop.opentrons.com/collections/opentrons-tips), and the user will be prompted to refill the tipracks if all are emptied in the middle of the protocol. To pause mid-protocol to replenish source plates on the deck, add a line in the .csv that reads `pause` (see example below for a deeper explanation).\n\n**CSV Format**\n\nYour cherrypicking transfers must be saved as a comma separated value (.csv) file type. Your CSV must contain values corresponding to volumes in microliters (\u03bcL). Note that the header line (first row of the .csv file) should also be included!\n\nHere's an example of how a short cherrypicking protocol should be properly formatted:\n\n```\nSource Labware ID,Source Labware Type,Source Slot,Source Well,Source Aspiration Height Above Bottom (in mm),Dest Labware Type,Dest Slot,Dest Well,Volume (in ul)\nRNA1,agilent_1_reservoir_290ml,3,A1,1,nest_96_wellplate_100ul_pcr_full_skirt,4,A11,1\nRNA2,nest_12_reservoir_15ml,4,A1,1,nest_96_wellplate_2ml_deep,5,A5,3\npause,,,,,,,,\nRNA3,nest_1_reservoir_195ml,5,A1,1,nest_96_wellplate_2ml_deep,5,H12,7\n```\n\nIn this example, 1\u03bcL will be transferred from 1mm above the bottom of well A1 in an Agilent 1-well 290ml reservoir (slot 1) to well A11 in the destination NEST 96-well plate 100\u00b5l (slot 4). After this, 3\u03bcL will be transferred from 1mm above the bottom of well A1 in a NEST 12-well 15ml reservoir (slot 2) to well A5 in the destination NEST 96-well plate 100\u00b5l (slot 5). Last, 7\u03bcL will be transferred from 1mm above the bottom of well A1 in a NEST 1-well 195ml reservoir (slot 3) to well H12 in the destination NEST 96-well plate 100\u00b5l (slot 5).\n\nIf you\u2019d like to follow our template, you can make a copy of [this spreadsheet](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/400b8e/cp_example.csv), fill out your values, and export as CSV for use with this protocol.\n\nUsing the customizations fields, below set up your protocol.\n* Transfer .csv File: Upload the .csv file containing your well locations, volumes, and source plate (optional).\n* Pipette Model: Select which pipette you will use for this protocol.\n* Pipette Mount: Specify which mount your single-channel pipette is on (left or right)\n* Tip Type: Specify whether you want to use filter tips.\n* Tip Usage Strategy: Specify whether you'd like to use a new tip for each transfer, or keep the same tip throughout the protocol.\n\n\n",
"internal": "400b8e\n",
diff --git a/protoBuilds/407d5e/README.json b/protoBuilds/407d5e/README.json
index fe9a59579f..925eee75b3 100644
--- a/protoBuilds/407d5e/README.json
+++ b/protoBuilds/407d5e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Assay"
@@ -8,7 +8,7 @@
"description": "This workflow is comprised of a protein labelling protocol that begins by adding several reagents to samples and then incubating at 37C for several days (up to 4) and aliquoting 20\u00b5L of the incubating samples. For more information about this protocol, including materials needed and customizable parameters, please see below before downloading.\n\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons Temperature Module with Aluminum Blocks\nP300 Single-Channel Pipette\nP20 Single-Channel Pipette\nOpentrons 300\u00b5l Pipette Tips\nOpentrons 20\u00b5L Pipette Tips\nNEST 96-Well PCR Plates\nNEST 1.5mL Microcentrifuge Tubes & Tubes for Samples\nReagents\nSamples\n\n\n\nUsing the customizations field (below), set up your protocol.\n Number of Samples (1-19): Specify the number of samples to run (1-19).\n Number of Incubations: Specify the number of incubations. The protocol will transfer 20\u00b5L to the destination plate and repeat the process, up to three times, at the 1, 2, and 4 day mark.\n Labware Containing Samples: Select the type of tube that will be used to contain the samples. This tube should be place in the 24-Well Aluminum Block.\n Temperature Module: Select which generation of the Temperature Module will be used.\n* Reset Tipracks?: This protocol can save the state of the tipracks after each run for the P300 and the P20. If this is set to \"Yes\" (or the protocol is run for the first time on the robot), the protocol will begin picking up tips from the A1 of Tiprack 1 for each pipette. If set to \"No\", the saved tip state from the previous run will be accessed and the pipette will begin using tips where the previous protocol run left off. The user will be prompted to replace the corresponding tip racks.\n\n\nDeck Layout\n\nSlot 1: Destination Plate (NEST 96-Well PCR Plates) - 20\u00b5L of sample aliquots will be transferred here. This should be replaced for transfer after incubation(s).\n\nSlot 2: 24-Well Aluminum Block containing Tubes (specified by user) with Samples\n\nSlot 3: Opentrons 20\u00b5L Tiprack (Tiprack 3)\n\nSlot 4: Opentrons Temperature Module with 96-Well Aluminum Block and NEST 96-Well PCR Plates\n\nSlot 5: NEST 96-Well PCR Plates (Empty, for mixing)\n\nSlot 6: Opentrons 20\u00b5L Tiprack (Tiprack 2)\n\nSlot 7: 24-Well Aluminum Block containing 1.5mL NEST Tubes with Reagents\n\nSlot 8: Opentrons 300\u00b5L Tiprack (Tiprack 1)\n\nSlot 9: Opentrons 20\u00b5L Tiprack (Tiprack 1)\n\nSlot 10: Opentrons 300\u00b5L Tiprack (Tiprack 3)\n\nSlot 11: Opentrons 300\u00b5L Tiprack (Tiprack 2)\n",
"internal": "407d5e",
"markdown": {
- "author": "[Opentrons](http://www.opentrons.com/)\n\n",
+ "author": "[Opentrons](http://www.opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n * Assay\n\n",
"description": "This workflow is comprised of a protein labelling protocol that begins by adding several reagents to samples and then incubating at 37C for several days (up to 4) and aliquoting 20\u00b5L of the incubating samples. For more information about this protocol, including materials needed and customizable parameters, please see below before downloading.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons Temperature Module with Aluminum Blocks](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [P300 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 300\u00b5l Pipette Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 20\u00b5L Pipette Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [NEST 96-Well PCR Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 1.5mL Microcentrifuge Tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes) & Tubes for Samples\n* Reagents\n* Samples\n\n\n---\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Samples (1-19)**: Specify the number of samples to run (1-19).\n* **Number of Incubations**: Specify the number of incubations. The protocol will transfer 20\u00b5L to the destination plate and repeat the process, up to three times, at the 1, 2, and 4 day mark.\n* **Labware Containing Samples**: Select the type of tube that will be used to contain the samples. This tube should be place in the 24-Well Aluminum Block.\n* **Temperature Module**: Select which generation of the Temperature Module will be used.\n* **Reset Tipracks?**: This protocol can save the state of the tipracks after each run for the P300 and the P20. If this is set to \"Yes\" (or the protocol is run for the first time on the robot), the protocol will begin picking up tips from the A1 of Tiprack 1 for each pipette. If set to \"No\", the saved tip state from the previous run will be accessed and the pipette will begin using tips where the previous protocol run left off. The user will be prompted to replace the corresponding tip racks.\n\n\n\n**Deck Layout**\n\n**Slot 1**: Destination Plate ([NEST 96-Well PCR Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)) - 20\u00b5L of sample aliquots will be transferred here. This should be replaced for transfer after incubation(s).\n\n**Slot 2**: [24-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) containing Tubes (specified by user) with Samples\n\n**Slot 3**: [Opentrons 20\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips) (Tiprack 3)\n\n**Slot 4**: [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with 96-Well Aluminum Block and [NEST 96-Well PCR Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n\n**Slot 5**: [NEST 96-Well PCR Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) (Empty, for mixing)\n\n**Slot 6**: [Opentrons 20\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips) (Tiprack 2)\n\n**Slot 7**: [24-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) containing 1.5mL NEST Tubes with Reagents\n\n**Slot 8**: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips) (Tiprack 1)\n\n**Slot 9**: [Opentrons 20\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips) (Tiprack 1)\n\n**Slot 10**: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips) (Tiprack 3)\n\n**Slot 11**: [Opentrons 300\u00b5L Tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips) (Tiprack 2)\n\n\n",
"internal": "407d5e\n",
diff --git a/protoBuilds/412ec7/README.json b/protoBuilds/412ec7/README.json
index a138f6624d..ed2d723d97 100644
--- a/protoBuilds/412ec7/README.json
+++ b/protoBuilds/412ec7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Serial Dilution": [
"Optional Temperature Module Serial Dilution"
@@ -9,7 +9,7 @@
"description": "With this protocol, you can do the requested serial dilution with or without a temperature module under the 96-well plate using an 8-channel P300 pipette. The dilution factor and choice of plate are also modifiable.\n\nExample Setup\nThis protocol uses the inputs you define for \"Dilution Factor\" and \"Total Mixing Volume\" to automatically infer the necessary transfer volume for each dilution across your plate. For a 1 in 3 dilution series across an entire plate, as seen above:\n-- Start with your samples/reagents in Column 1 of your plate. In this example, you would pre-add 150 uL of concentrated sample to the first column of your 96-well plate.\n-- Define a Total Mixing Volume of 150uL, a Dilution Factor of 3, and set Number of Dilutions = 11.\n-- Your OT-2 will add 100uL of diluent to each empty well in your plate. Then it will transfer 50uL from Column 1 between each well/column in the plate.\n-- \"Total mixing volume\" = transfer volume + diluent volume.\n-- \"Tip Use Strategy\" will set whether a new tip will be used for every transfer or the same tip reused\n-- \"Blank in Well Plate\" will set whether a blank will be added to column 12\n\n-- Opentrons OT-2\n-- Opentrons OT-2 Run App (Version 4.7 or later)\n-- Opentrons Tips for selected Opentrons Pipette\n-- 12-Row, Automation-Friendly Trough\n-- 96-Well Plate, Bio-Rad or NEST full skirted plates or equivalent aluminum block plates\n-- Diluent (Pre-loaded in row 1 of trough)\n-- Samples/Standards (Pre-loaded in Column 1 of a standard 96-well plate)",
"internal": "412ec7",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Serial Dilution\n\t* Optional Temperature Module Serial Dilution\n\n",
"deck-setup": "\nDECK SETUP IMAGE HERE\n\nSlot 1: Nest 12 Well 15ml Reservoir\n\nSlot 2 and 3: Opentrons 300ul Tiprack\n\nSlot 4: 96 Well Plate, with or without temperature module (specified in parameters)\n\n",
"description": "With this protocol, you can do the requested serial dilution with or without a temperature module under the 96-well plate using an 8-channel P300 pipette. The dilution factor and choice of plate are also modifiable.\n\n\n\n***Example Setup***\n\nThis protocol uses the inputs you define for \"***Dilution Factor***\" and \"***Total Mixing Volume***\" to automatically infer the necessary transfer volume for each dilution across your plate. For a 1 in 3 dilution series across an entire plate, as seen above:\n\n-- Start with your samples/reagents in Column 1 of your plate. In this example, you would pre-add 150 uL of concentrated sample to the first column of your 96-well plate.\n\n-- Define a ***Total Mixing Volume*** of 150uL, a ***Dilution Factor*** of 3, and set ***Number of Dilutions*** = 11.\n\n-- Your OT-2 will add 100uL of diluent to each empty well in your plate. Then it will transfer 50uL from Column 1 between each well/column in the plate.\n\n-- \"***Total mixing volume***\" = transfer volume + diluent volume.\n\n-- \"Tip Use Strategy\" will set whether a new tip will be used for every transfer or the same tip reused\n\n-- \"Blank in Well Plate\" will set whether a blank will be added to column 12\n\n\n\n-- [Opentrons OT-2](http://opentrons.com/ot-2)\n\n-- [Opentrons OT-2 Run App (Version 4.7 or later)](http://opentrons.com/ot-app)\n\n-- [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips-racks-9-600-tips) for selected Opentrons Pipette\n\n-- [12-Row, Automation-Friendly Trough](https://shop.opentrons.com/nest-12-well-reservoirs-15-ml/)\n\n-- 96-Well Plate, Bio-Rad or NEST full skirted plates or equivalent aluminum block plates\n\n-- Diluent (Pre-loaded in row 1 of trough)\n\n-- Samples/Standards (Pre-loaded in Column 1 of a standard 96-well plate)\n\n",
diff --git a/protoBuilds/414aaa/README.json b/protoBuilds/414aaa/README.json
index 08d6872feb..79da48b44c 100644
--- a/protoBuilds/414aaa/README.json
+++ b/protoBuilds/414aaa/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "g42lab",
"labware": "\nNEST 12 Well Reservoir 15 mL #360102\nNEST 1 Well Reservoir 195 mL #360103\nDeepwell Plates\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n",
"description": "This protocol prepares deepwell plates for extraction. In the case that the protocol runs out of tips, it will automatically pause to prompt the user to refill tips.\n\n\n",
diff --git a/protoBuilds/4175de/README.json b/protoBuilds/4175de/README.json
index c098595611..eb69bce994 100644
--- a/protoBuilds/4175de/README.json
+++ b/protoBuilds/4175de/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom cherrypicked PCR/qPRC prep. It requires a CSV input for the source and destination wells of the samples. It will automatically add mastermix based on the number of samples inputted to the PCR plate sequentially (A1-12 then B1-12, etc...).\n\n\nOpentrons Temperature Module\nP20 single-channel GEN2 electronic pipette\nOpentrons 20ul filter tiprack\nCorning 96-Well Nonbinding Surface Microplates (3641)\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOpentrons 24 Tube Rack with Generic 2 mL Screwcap\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\n\n\n\nDeck Setup\n Opentrons 20ul filter tiprack (Slot 1, Slot 4, Slot 7)\n Opentrons 24 Tube Rack with Generic 2 mL Screwcap (Slot 2, Mastermix in well A6)\n Opentrons Temperature Module (Slot 3)\n 96 well PCR Strip (Aluminum Block) OR 96 well NEST 100uL PCR Plate (Slot 3 on Temperature Module)\n* Corning 96-Well Nonbinding Surface Microplates OR Opentrons 24-slot Tube Rack (Slot 5, Slot 6, Slot 8, Slot 11)",
"internal": "4175de",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"description": "This protocol performs a custom cherrypicked PCR/qPRC prep. It requires a CSV input for the source and destination wells of the samples. It will automatically add mastermix based on the number of samples inputted to the PCR plate sequentially (A1-12 then B1-12, etc...).\n---\n\n\n\n* [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [P20 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 20ul filter tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-filter-tip)\n* [Corning 96-Well Nonbinding Surface Microplates (3641)](https://www.fishersci.com/shop/products/corning-96-well-nonbinding-surface-nbs-microplates-flat-wells-clear/07200777)\n* [NEST 0.1 mL 96-Well PCR Plate, Full Skirt](https://shop.opentrons.com/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons 24 Tube Rack with Generic 2 mL Screwcap](https://shop.opentrons.com/products/tube-rack-set-1?_ga=2.48408495.884537678.1605539831-1181961818.1604785212)\n* [Opentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set?_ga=2.124502019.884537678.1605539831-1181961818.1604785212)\n\n---\n\n\nDeck Setup\n* Opentrons 20ul filter tiprack (Slot 1, Slot 4, Slot 7)\n* Opentrons 24 Tube Rack with Generic 2 mL Screwcap (Slot 2, Mastermix in well A6)\n* Opentrons Temperature Module (Slot 3)\n* 96 well PCR Strip (Aluminum Block) OR 96 well NEST 100uL PCR Plate (Slot 3 on Temperature Module)\n* Corning 96-Well Nonbinding Surface Microplates OR Opentrons 24-slot Tube Rack (Slot 5, Slot 6, Slot 8, Slot 11)\n\n",
"internal": "4175de\n",
diff --git a/protoBuilds/41ece2/README.json b/protoBuilds/41ece2/README.json
index 0cb6c09a7e..3d274c5e10 100644
--- a/protoBuilds/41ece2/README.json
+++ b/protoBuilds/41ece2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Serial Dilution"
@@ -8,7 +8,7 @@
"description": "Note: This protocol is a modified version of this drug dilution protocol. For a note on the differences, please see the note on modifications.\n\nThis protocol is an updated version of a drug dilution protocol designed with Protocol Designer.\n\nIn this protocol, the p300-multi channel pipette (GEN1) is used to transfer 100\u00b5L media to all of the columns of the three dilution plates (10uM, 1uM, and 0.1uM). When transferring the media, the same tips (column 1) will be used for all transfers, but after every four transfers, the pipette will return the tips to the tiprack and pick up them up again to ensure a good seal with the tips.\nAfter the media is added, the p10-multi channel pipette is used for the dilutions, transferring 10\u00b5L from a stock plate in slot 10, to the first dilution plate and so on... (using the same set of tip for each column of dilutions).\nFinally, the p300-multi will be used again, this time transferring 100\u00b5L from the reservoir in slot 11 to the three dilution plates. These transfers will follow the same pattern for tip usage as the earlier transfer with the p300-multi, but instead use the tips from column 2.\n\nModifications: In this modified version of the protocol, 100\u00b5L of media is added to columns 2-12 of the first two plates and 90\u00b5L of media is added to columns 2-12 of the last plate. When performing the dilution, only columns 2-11 are accessed, leaving column 1 completely empty and column 12 without drug. Finally, there is an optional parameter, Add Cells + Media?, that if selected, will add 100\u00b5L from the reservoir to columns 2-12 of the three dilution plates, 2mm above the dispense height of the original protocol, otherwise, the protocol will end after the dilution.\n\n\nUpdate: A new parameter - Number of Dilution Plates - has been added that will give the user the ability to choose 3 or 4 total dilution plates. This change also includes a change to the location of the labware.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nP300 Multi-Channel Pipette (GEN1)\nP10 Multi-Channel Pipette\nOpentrons 300\u00b5l Pipette Tips\nOpentrons 10\u00b5L Pipette Tips\nAxygen 1-Well Reservoir, 90mL\nCorning 96-Well Plate, 360\u00b5L Flat\n\n\n\nDeck Layout\n\nSlot 1: Corning 96-Well Plate (Drug Dilution 0.01\u00b5M), if using\n\nSlot 2: Axygen 1-Well Reservoir, 90mL (containing media, added to columns 2-12 of dilution plates)\n\nSlot 3: Opentrons 300\u00b5l Pipette tips\n\nSlot 4: Corning 96-Well Plate (Drug Dilution 0.1\u00b5M)\n\nSlot 5: Axygen 1-Well Reservoir, 90mL (containing cells, added to columns 2-12 of dilution plate), if using\n\nSlot 6: Opentrons 10\u00b5L Pipette Tips\n\nSlot 7: Corning 96-Well Plate (Drug Dilution 1\u00b5M)\n\nSlot 10: Corning 96-Well Plate (Drug Dilution 10\u00b5M)\n\nSlot 11: Corning 96-Well Plate (Drug Stock)\n",
"internal": "41ece2",
"markdown": {
- "author": "[Opentrons](http://www.opentrons.com/)\n\n",
+ "author": "[Opentrons](http://www.opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Serial Dilution\n\n",
"description": "**Note**: This protocol is a modified version of this [drug dilution protocol](https://protocols.opentrons.com/protocol/3662e6). For a note on the differences, please see the note on *modifications*.\n\nThis protocol is an updated version of a drug dilution protocol designed with Protocol Designer.\n\nIn this protocol, the p300-multi channel pipette (GEN1) is used to transfer 100\u00b5L media to all of the columns of the three dilution plates (10uM, 1uM, and 0.1uM). When transferring the media, the same tips (column 1) will be used for all transfers, but after every four transfers, the pipette will return the tips to the tiprack and pick up them up again to ensure a good seal with the tips.\nAfter the media is added, the p10-multi channel pipette is used for the dilutions, transferring 10\u00b5L from a stock plate in slot 10, to the first dilution plate and so on... (using the same set of tip for each column of dilutions).\nFinally, the p300-multi will be used again, this time transferring 100\u00b5L from the reservoir in slot 11 to the three dilution plates. These transfers will follow the same pattern for tip usage as the earlier transfer with the p300-multi, but instead use the tips from column 2.\n\n**Modifications**: In this modified version of the protocol, 100\u00b5L of media is added to columns 2-12 of the first two plates and 90\u00b5L of media is added to columns 2-12 of the last plate. When performing the dilution, only columns 2-11 are accessed, leaving column 1 completely empty and column 12 without drug. Finally, there is an optional parameter, *Add Cells + Media?*, that if selected, will add 100\u00b5L from the reservoir to columns 2-12 of the three dilution plates, 2mm above the dispense height of the original protocol, otherwise, the protocol will end after the dilution.\n\n\n**Update**: A new parameter - *Number of Dilution Plates* - has been added that will give the user the ability to choose 3 or 4 total dilution plates. This change also includes a change to the location of the labware.\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* P300 Multi-Channel Pipette (GEN1)\n* P10 Multi-Channel Pipette\n* [Opentrons 300\u00b5l Pipette Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 10\u00b5L Pipette Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [Axygen 1-Well Reservoir, 90mL](https://labware.opentrons.com/axygen_1_reservoir_90ml/)\n* [Corning 96-Well Plate, 360\u00b5L Flat](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate)\n\n\n---\n\n\n**Deck Layout**\n\n**Slot 1**: [Corning 96-Well Plate](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (Drug Dilution 0.01\u00b5M), if using\n\n**Slot 2**: [Axygen 1-Well Reservoir, 90mL](https://labware.opentrons.com/axygen_1_reservoir_90ml/) (containing media, added to columns 2-12 of dilution plates)\n\n**Slot 3**: [Opentrons 300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n**Slot 4**: [Corning 96-Well Plate](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (Drug Dilution 0.1\u00b5M)\n\n**Slot 5**: [Axygen 1-Well Reservoir, 90mL](https://labware.opentrons.com/axygen_1_reservoir_90ml/) (containing cells, added to columns 2-12 of dilution plate), if using\n\n**Slot 6**: [Opentrons 10\u00b5L Pipette Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n\n**Slot 7**: [Corning 96-Well Plate](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (Drug Dilution 1\u00b5M)\n\n**Slot 10**: [Corning 96-Well Plate](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (Drug Dilution 10\u00b5M)\n\n**Slot 11**: [Corning 96-Well Plate](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (Drug Stock)\n\n\n",
"internal": "41ece2\n",
diff --git a/protoBuilds/422b1e/README.json b/protoBuilds/422b1e/README.json
index a55c35e569..f8739b70dd 100644
--- a/protoBuilds/422b1e/README.json
+++ b/protoBuilds/422b1e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Titration": [
"Custom Titration"
@@ -8,7 +8,7 @@
"description": "This titration protocol utilizes a custom labware definition (beaker + stir plate) for use on the OT-2.\nThe P1000 Pipette (attached to the right mount) will pick up a tip from the tip rack located in slot 11. For each tube in the Opentrons Tube Rack (located in slot 9), the pipette will transfer 2mL to the beaker in slot 4, wait a predetermined amount of time, then transfer 2mL from the beaker in slot 4 to the beaker in slot 8.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons P1000 Single-Channel Pipette\nOpentrons 1000\u00b5L Tips\nOpentrons 4-in-1 Tube Rack Set, 6x50mL Top\n50mL Conical Tubes\nCustom Beaker + Stir Plate\nReagents\n\n\n\nSlot 4: Custom Beaker + Stir Plate\nSlot 8: Custom Beaker + Stir Plate (Waste Container)\nSlot 9: Opentrons 4-in-1 Tube Rack Set, 6x50mL Top with 50mL Conical Tubes containing reagents\n The P1000 will aspirate liquid from the bottom of the tube, so do not overfill the tube to ensure the pipette does not submerge itself in liquid.\nSlot 11: Opentrons 1000\u00b5L Tips",
"internal": "422b1e",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Titration\n\t* Custom Titration\n\n\n",
"description": "This titration protocol utilizes a custom labware definition (beaker + stir plate) for use on the OT-2.\nThe P1000 Pipette (attached to the right mount) will pick up a tip from the tip rack located in slot 11. For each tube in the Opentrons Tube Rack (located in slot 9), the pipette will transfer 2mL to the beaker in slot 4, wait a predetermined amount of time, then transfer 2mL from the beaker in slot 4 to the beaker in slot 8.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P1000 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 1000\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* [Opentrons 4-in-1 Tube Rack Set, 6x50mL Top](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [50mL Conical Tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-50-ml-centrifuge-tube)\n* Custom Beaker + Stir Plate\n* Reagents\n\n\n\n---\n\n\nSlot 4: Custom Beaker + Stir Plate\n\nSlot 8: Custom Beaker + Stir Plate (Waste Container)\n\nSlot 9: [Opentrons 4-in-1 Tube Rack Set, 6x50mL Top](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with [50mL Conical Tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-50-ml-centrifuge-tube) containing reagents*\n* The P1000 will aspirate liquid from the bottom of the tube, so do not overfill the tube to ensure the pipette does not submerge itself in liquid.\n\nSlot 11: [Opentrons 1000\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n\n\n",
"internal": "422b1e\n",
diff --git a/protoBuilds/42d27b/README.json b/protoBuilds/42d27b/README.json
index f423e93f7d..3060b11811 100644
--- a/protoBuilds/42d27b/README.json
+++ b/protoBuilds/42d27b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Quant-iT dsDNA Kit"
@@ -8,7 +8,7 @@
"description": "This protocol automates the steps outlined in the Quant-iT dsDNA Broad-Range Assay Kit. Using a P10 Multi-Channel Pipette and a P300 Multi-Channel Pipette, this protocol adds reagent mix (buffer + dye) to 96-well plate, before adding 1\u00b5L of DNA from sample tubes.\nUsing the customizations fields, below set up your protocol.\n P300 Multi Mount: Select which mount (left or right) the P300 Multi is attached to.\n P10 Multi Mount: Select which mount (left or right) the P10 Multi is attached to.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons P10 Multi-Channel Pipette\nOpentrons P300 Multi-Channel Pipette\nOpentrons 10\u00b5L Tips\nOpentrons 300\u00b5L Tips\nNEST 12-Well Reservoir, 15mL\nAxygen 96-Well Plate (PCR-96-FS-C)\nMicronics 96-Tubes in Plate (MP52757S-Y20)\nReagents\nSamples\n\n\n\nSlot 1: NEST 12-Well Reservoir\n* A1: Reagent Mix\nSlot 2: Axygen 96-Well Plate (PCR-96-FS-C)\nSlot 3: Micronics 96-Tubes in Plate (MP52757S-Y20)\nSlot 4: Opentrons 300\u00b5L Tips\nSlot 5: Opentrons 10\u00b5L Tips",
"internal": "42d27b",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Quant-iT dsDNA Kit\n\n\n",
"description": "This protocol automates the steps outlined in the [Quant-iT dsDNA Broad-Range Assay Kit](https://assets.thermofisher.com/TFS-Assets/LSG/manuals/Quant_iT_dsDNA_BR_Assay_UG.pdf). Using a P10 Multi-Channel Pipette and a P300 Multi-Channel Pipette, this protocol adds reagent mix (buffer + dye) to 96-well plate, before adding 1\u00b5L of DNA from sample tubes.\n\n\n\n**Using the customizations fields, below set up your protocol.**\n* **P300 Multi Mount**: Select which mount (left or right) the P300 Multi is attached to.\n* **P10 Multi Mount**: Select which mount (left or right) the P10 Multi is attached to.\n\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P10 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [NEST 12-Well Reservoir, 15mL](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* Axygen 96-Well Plate (PCR-96-FS-C)\n* Micronics 96-Tubes in Plate (MP52757S-Y20)\n* Reagents\n* Samples\n\n\n---\n\n\nSlot 1: [NEST 12-Well Reservoir](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* A1: Reagent Mix\n\nSlot 2: Axygen 96-Well Plate (PCR-96-FS-C)\n\nSlot 3: Micronics 96-Tubes in Plate (MP52757S-Y20)\n\nSlot 4: [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 5: [Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n\n\n",
"internal": "42d27b\n",
diff --git a/protoBuilds/43163c/README.json b/protoBuilds/43163c/README.json
index e5fc1b8528..e1e4da8496 100644
--- a/protoBuilds/43163c/README.json
+++ b/protoBuilds/43163c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Plant DNA"
@@ -8,7 +8,7 @@
"description": "This is a protocol to run a nanomaterial toxicity test. The nanomaterials get added at different concentrations to a plate, are dried, and then cells are exposed to the dried nanosheets. \nThere are 3 steps to this protocol:\n\nNanomaterials are added at different concentrations to a 384 well plate. (drying done out of OT2 to make nanosheets)\nNanosheets are washed with PBS and bacteria are added to the sheets. (5hrs at 37.5c out of OT2)\nBacteria are removed from the nanosheets and the nanosheets are washed with PBS. (Plate ready for imaging on the Opera Phenix) \n\n\n\n\nOpentrons P300-multi channel electronic pipettes (GEN2)\nOpentrons 200ul Filter Tips\nNEST 1 Well Reservoir 195 mL\nNEST 12-Well Reservoirs, 15 mL\nNEST 96 Deepwell Plate 2mL\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOMNI Pre-filled 2 mL 96 deep well plates with 1.4 mm Ceramic beads\nMagMAX Plant DNA Isolation Kit\n",
"internal": "43163c",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Plant DNA\n\n",
"description": "This is a protocol to run a nanomaterial toxicity test. The nanomaterials get added at different concentrations to a plate, are dried, and then cells are exposed to the dried nanosheets. \n\nThere are 3 steps to this protocol:\n\n1. Nanomaterials are added at different concentrations to a 384 well plate. (drying done out of OT2 to make nanosheets)\n2. Nanosheets are washed with PBS and bacteria are added to the sheets. (5hrs at 37.5c out of OT2)\n3. Bacteria are removed from the nanosheets and the nanosheets are washed with PBS. (Plate ready for imaging on the Opera Phenix) \n\n---\n\n\n* [Opentrons P300-multi channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [NEST 1 Well Reservoir 195 mL](http://www.cell-nest.com/page94?_l=en&product_id=102)\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 96 Deepwell Plate 2mL](http://www.cell-nest.com/page94?product_id=101&_l=en)\n* [NEST 0.1 mL 96-Well PCR Plate, Full Skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [OMNI Pre-filled 2 mL 96 deep well plates with 1.4 mm Ceramic beads](https://www.omni-inc.com/consumables/well-plates/2-pack-96-well-plate-1-4mm-ceramic.html)\n* [MagMAX Plant DNA Isolation Kit](https://www.thermofisher.com/order/catalog/product/A32549#/A32549)\n\n\n",
"internal": "43163c\n",
diff --git a/protoBuilds/43d313-part-2/README.json b/protoBuilds/43d313-part-2/README.json
index 5c5431c8cd..4574f46b5a 100644
--- a/protoBuilds/43d313-part-2/README.json
+++ b/protoBuilds/43d313-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 4 part workflow, the OT-2 will follow ArcBio Continuous RNA Workflow - Pre-PCR Instrument experimental protocol to convert up to 96 input RNA samples into cDNA libraries. This is Part 2 of 4: This OT-2 protocol uses the RNA Continuous Workflow, Pre-PCR Instrument section of attached experimental protocol to perform cDNA synthesis.",
"internal": "43d313-part-2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul tips (deck slots 10, 11)\n* Opentrons 300ul tips (deck slots 7, 8, 9)\n* Opentrons Temperature Module (deck slot 3) with Sample Plate opentrons_96_aluminumblock_biorad_wellplate_200ul\n* Opentrons Magnetic Module (deck slot 1) with deep well plate\n* Reservoirs nest_1_reservoir_195ml (deck slots 4, 6)\n* Reagents opentrons_96_aluminumblock_generic_pcr_strip_200ul (deck slot 2)\n* Reservoir nest_12_reservoir_15ml (deck slot 5)\n\n\n\n\n\n",
"description": "\nWith this 4 part workflow, the OT-2 will follow [ArcBio Continuous RNA Workflow - Pre-PCR Instrument experimental protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2022-01-06/0h134y4/ArcBio_RNA_Workflow_Continuous.xlsx) to convert up to 96 input RNA samples into cDNA libraries. This is Part 2 of 4: This OT-2 protocol uses the RNA Continuous Workflow, Pre-PCR Instrument section of attached experimental protocol to perform cDNA synthesis.\n\n",
diff --git a/protoBuilds/43d313-part-3/README.json b/protoBuilds/43d313-part-3/README.json
index 570b19d7ca..7de2be529b 100644
--- a/protoBuilds/43d313-part-3/README.json
+++ b/protoBuilds/43d313-part-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 4 part workflow, the OT-2 will follow ArcBio Continuous RNA Workflow - Pre-PCR Instrument experimental protocol to convert up to 96 input RNA samples into cDNA libraries. This is Part 3 of 4: This OT-2 protocol uses the RNA Continuous Workflow, Pre-PCR Instrument section of attached experimental protocol to perform Library Purification, Library Prep.",
"internal": "43d313-part-3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul tips (deck slots 10, 11)\n* Opentrons 300ul tips (deck slots 7, 8, 9)\n* Opentrons Temperature Module (deck slot 3) with Sample Plate opentrons_96_aluminumblock_biorad_wellplate_200ul\n* Opentrons Magnetic Module (deck slot 1) with deep well plate\n* Reservoirs nest_1_reservoir_195ml (deck slots 4, 6)\n* Reagents opentrons_96_aluminumblock_generic_pcr_strip_200ul (deck slot 2)\n* Reservoir nest_12_reservoir_15ml (deck slot 5)\n\n\n\n\n\n",
"description": "\nWith this 4 part workflow, the OT-2 will follow [ArcBio Continuous RNA Workflow - Pre-PCR Instrument experimental protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2022-01-06/0h134y4/ArcBio_RNA_Workflow_Continuous.xlsx) to convert up to 96 input RNA samples into cDNA libraries. This is Part 3 of 4: This OT-2 protocol uses the RNA Continuous Workflow, Pre-PCR Instrument section of attached experimental protocol to perform Library Purification, Library Prep.\n\n",
diff --git a/protoBuilds/43d313-part-4/README.json b/protoBuilds/43d313-part-4/README.json
index 3ab9b50a7f..d5f1207e39 100644
--- a/protoBuilds/43d313-part-4/README.json
+++ b/protoBuilds/43d313-part-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 4 part workflow, the OT-2 will follow ArcBio Continuous RNA Workflow - Pre-PCR Instrument experimental protocol to convert up to 96 input RNA samples into cDNA libraries. This is Part 4 of 4: This OT-2 protocol uses the RNA Continuous Workflow, Pre-PCR Instrument section of attached experimental protocol to perform Library Purification, Ligation.",
"internal": "43d313-part-4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul tips (deck slots 10, 11)\n* Opentrons 300ul tips (deck slots 7, 8, 9)\n* Opentrons Temperature Module (deck slot 3) with Sample Plate opentrons_96_aluminumblock_biorad_wellplate_200ul\n* Opentrons Magnetic Module (deck slot 1) with deep well plate\n* Reservoirs nest_1_reservoir_195ml (deck slots 4, 6)\n* Reagents opentrons_96_aluminumblock_biorad_wellplate_200ul (deck slot 2)\n* Reservoir nest_12_reservoir_15ml (deck slot 5)\n\n\n\n",
"description": "\nWith this 4 part workflow, the OT-2 will follow [ArcBio Continuous RNA Workflow - Pre-PCR Instrument experimental protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2022-01-06/0h134y4/ArcBio_RNA_Workflow_Continuous.xlsx) to convert up to 96 input RNA samples into cDNA libraries. This is Part 4 of 4: This OT-2 protocol uses the RNA Continuous Workflow, Pre-PCR Instrument section of attached experimental protocol to perform Library Purification, Ligation.\n\n",
diff --git a/protoBuilds/43d313-part-5/README.json b/protoBuilds/43d313-part-5/README.json
index 0acf67bfa6..6658a17339 100644
--- a/protoBuilds/43d313-part-5/README.json
+++ b/protoBuilds/43d313-part-5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 9 part workflow, the OT-2 will follow ArcBio Continuous RNA Workflow - Pre-PCR Instrument and Post-PCR Instrument experimental protocol to convert up to 96 input RNA samples into cDNA libraries. This is Part 5 of 9: This OT-2 protocol uses the RNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform post-indexing workflow steps.",
"internal": "43d313-part-5",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul tips (deck slots 10, 11)\n* Opentrons 300ul tips (deck slots 7, 8, 9)\n* Opentrons Temperature Module (deck slot 3) with Sample Plate opentrons_96_aluminumblock_biorad_wellplate_200ul\n* Opentrons Magnetic Module (deck slot 1) with deep well plate\n* Reservoirs nest_1_reservoir_195ml (deck slots 4, 6)\n* Reagents opentrons_96_aluminumblock_generic_pcr_strip_200ul (deck slot 2)\n* Reservoir nest_12_reservoir_15ml (deck slot 5)\n\n\n\n\n\n",
"description": "\nWith this 9 part workflow, the OT-2 will follow [ArcBio Continuous RNA Workflow - Pre-PCR Instrument and Post-PCR Instrument experimental protocol](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/43d313/ArcBio_RNA_Workflow_020922.xlsx) to convert up to 96 input RNA samples into cDNA libraries. This is Part 5 of 9: This OT-2 protocol uses the RNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform post-indexing workflow steps.\n\n",
diff --git a/protoBuilds/43d313-part-6/README.json b/protoBuilds/43d313-part-6/README.json
index 22e8ce01de..a998aed97f 100644
--- a/protoBuilds/43d313-part-6/README.json
+++ b/protoBuilds/43d313-part-6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 9 part workflow, the OT-2 will follow ArcBio Continuous RNA Workflow - Pre-PCR Instrument and Post-PCR Instrument experimental protocol to convert up to 96 input RNA samples into cDNA libraries. This is Part 6 of 9: This OT-2 protocol uses the RNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform post-indexing purification workflow steps.",
"internal": "43d313-part-6",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul tips (deck slots 10, 11)\n* Opentrons Temperature Module (deck slot 3) with Sample Plate opentrons_96_aluminumblock_biorad_wellplate_200ul\n* Opentrons Magnetic Module (deck slot 1) with deep well plate\n* Reagents opentrons_96_aluminumblock_generic_pcr_strip_200ul (deck slot 2)\n\n\n\n",
"description": "\nWith this 9 part workflow, the OT-2 will follow [ArcBio Continuous RNA Workflow - Pre-PCR Instrument and Post-PCR Instrument experimental protocol](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/43d313/ArcBio_RNA_Workflow_020922.xlsx) to convert up to 96 input RNA samples into cDNA libraries. This is Part 6 of 9: This OT-2 protocol uses the RNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform post-indexing purification workflow steps.\n\n",
diff --git a/protoBuilds/43d313-part-7/README.json b/protoBuilds/43d313-part-7/README.json
index 32245a9c70..3691c605d1 100644
--- a/protoBuilds/43d313-part-7/README.json
+++ b/protoBuilds/43d313-part-7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 9 part workflow, the OT-2 will follow ArcBio Continuous RNA Workflow - Pre-PCR Instrument and Post-PCR Instrument experimental protocol to convert up to 96 input RNA samples into cDNA libraries. This is Part 7 of 9: This OT-2 protocol uses the RNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform post-indexing cleanup steps.",
"internal": "43d313-part-7",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul tips (deck slots 10, 11)\n* Opentrons 300ul tips (deck slots 7, 8, 9)\n* Opentrons Temperature Module (deck slot 3) with Sample Plate opentrons_96_aluminumblock_biorad_wellplate_200ul\n* Opentrons Magnetic Module (deck slot 1) with deep well plate\n* Reservoirs nest_1_reservoir_195ml (deck slots 4, 6)\n* Reagents opentrons_96_aluminumblock_generic_pcr_strip_200ul (deck slot 2)\n* Reservoir nest_12_reservoir_15ml (deck slot 5)\n\n\n\n",
"description": "\nWith this 9 part workflow, the OT-2 will follow [ArcBio Continuous RNA Workflow - Pre-PCR Instrument and Post-PCR Instrument experimental protocol](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/43d313/ArcBio_RNA_Workflow_020922.xlsx) to convert up to 96 input RNA samples into cDNA libraries. This is Part 7 of 9: This OT-2 protocol uses the RNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform post-indexing cleanup steps.\n\n",
diff --git a/protoBuilds/43d313-part-8/README.json b/protoBuilds/43d313-part-8/README.json
index 32a9d7f8c2..8870ab2f9f 100644
--- a/protoBuilds/43d313-part-8/README.json
+++ b/protoBuilds/43d313-part-8/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 9 part workflow, the OT-2 will follow ArcBio Continuous RNA Workflow - Pre-PCR Instrument and Post-PCR Instrument experimental protocol to convert up to 96 input RNA samples into cDNA libraries. This is Part 8 of 9: This OT-2 protocol uses the RNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform library QC setup steps.",
"internal": "43d313-part-8",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul filter tips (deck slots 10, 11)\n* Opentrons 200ul filter tips (deck slots 7, 8)\n* Opentrons Temperature Module at room temperature (deck slot 3) with libraries in 200 uL PCR plate\n* Opentrons Magnetic Module (deck slot 1) with Agilent Tapestation plate\n* Reagents opentrons_96_aluminumblock_generic_pcr_strip_200ul (deck slot 2)\n* Reservoir nest_12_reservoir_15ml (deck slot 5)\n\n\n\n\n\n",
"description": "\nWith this 9 part workflow, the OT-2 will follow [ArcBio Continuous RNA Workflow - Pre-PCR Instrument and Post-PCR Instrument experimental protocol](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/43d313/ArcBio_RNA_Workflow_020922.xlsx) to convert up to 96 input RNA samples into cDNA libraries. This is Part 8 of 9: This OT-2 protocol uses the RNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform library QC setup steps.\n\n",
diff --git a/protoBuilds/43d313-part-9/README.json b/protoBuilds/43d313-part-9/README.json
index cf7d8ab9e9..a313b6fa50 100644
--- a/protoBuilds/43d313-part-9/README.json
+++ b/protoBuilds/43d313-part-9/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 9 part workflow, the OT-2 will follow ArcBio Continuous RNA Workflow - Pre-PCR Instrument and Post-PCR Instrument experimental protocol to convert up to 96 input RNA samples into cDNA libraries. This is Part 9 of 9: This OT-2 protocol uses the RNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform pooling steps.",
"internal": "43d313-part-9",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul filter tips (deck slot 10)\n* Opentrons Temperature Module at 4 degrees C (deck slot 3) with libraries in 200 uL PCR plate\n* Rack for pool tubes Opentrons 24-tube rack with 1.5 mL eppendorf snap cap tubes (deck slot 2)\n\n\n\n",
"description": "\nWith this 9 part workflow, the OT-2 will follow [ArcBio Continuous RNA Workflow - Pre-PCR Instrument and Post-PCR Instrument experimental protocol](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/43d313/ArcBio_RNA_Workflow_020922.xlsx) to convert up to 96 input RNA samples into cDNA libraries. This is Part 9 of 9: This OT-2 protocol uses the RNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform pooling steps.\n\n",
diff --git a/protoBuilds/43d313/README.json b/protoBuilds/43d313/README.json
index 5abdfaef93..f2dbb1c64c 100644
--- a/protoBuilds/43d313/README.json
+++ b/protoBuilds/43d313/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 4 part workflow, the OT-2 will follow ArcBio Continuous RNA Workflow - Pre-PCR Instrument experimental protocol to convert up to 96 input RNA samples into cDNA libraries. This is Part 1 of 4: This OT-2 protocol uses the RNA Continuous Workflow, Pre-PCR Instrument section of attached experimental protocol to treat input RNA samples with DNase.",
"internal": "43d313",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul tips (deck slots 10, 11)\n* Opentrons 300ul tips (deck slots 7, 8, 9)\n* Opentrons Temperature Module (deck slot 3) with Sample Plate opentrons_96_aluminumblock_biorad_wellplate_200ul\n* Opentrons Magnetic Module (deck slot 1)\n* Reservoirs nest_1_reservoir_195ml (deck slots 4, 6)\n* Reagents opentrons_96_aluminumblock_generic_pcr_strip_200ul (deck slot 2)\n* Reservoir nest_12_reservoir_15ml (deck slot 5)\n\n\n\n",
"description": "\nWith this 4 part workflow, the OT-2 will follow [ArcBio Continuous RNA Workflow - Pre-PCR Instrument experimental protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2022-01-06/0h134y4/ArcBio_RNA_Workflow_Continuous.xlsx) to convert up to 96 input RNA samples into cDNA libraries. This is Part 1 of 4: This OT-2 protocol uses the RNA Continuous Workflow, Pre-PCR Instrument section of attached experimental protocol to treat input RNA samples with DNase.\n\n",
diff --git a/protoBuilds/441cc8/README.json b/protoBuilds/441cc8/README.json
index 94e521f1fa..ad09116a93 100644
--- a/protoBuilds/441cc8/README.json
+++ b/protoBuilds/441cc8/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol uses a p300 multi-channel pipette to suspend a column of indexing primers (contained in a 96-well PCR plate) in master mix (contained in a reagent reservoir) and then distributes 14 ul of this mixture to the corresponding column of each of 9 PCR plates (in order as shown in the attached deck map). This process is repeated for all columns of the primer plate to fill all columns of the 9 PCR plates.\nThe liquid handling method used for the master mix includes the following features:\nslow flow rate for aspiration and dispense\nwait for liquid to finish moving after aspiration and dispense\navoid introducing air into liquid (avoid complete dispenses)\ndispense to a surface\nwithdraw tip slowly from liquid\nLinks:\n Deck Layout\n Process Steps\nWith this protocol, your robot can prepare stock plates containing master mix and indexing primers to be used for the indexing step of an NGS library prep workflow.",
"internal": "441cc8",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * PCR Prep\n\n",
"description": "This protocol uses a p300 multi-channel pipette to suspend a column of indexing primers (contained in a 96-well PCR plate) in master mix (contained in a reagent reservoir) and then distributes 14 ul of this mixture to the corresponding column of each of 9 PCR plates (in order as shown in the attached deck map). This process is repeated for all columns of the primer plate to fill all columns of the 9 PCR plates.\n\nThe liquid handling method used for the master mix includes the following features:\n\nslow flow rate for aspiration and dispense\nwait for liquid to finish moving after aspiration and dispense\navoid introducing air into liquid (avoid complete dispenses)\ndispense to a surface\nwithdraw tip slowly from liquid\n\nLinks:\n* [Deck Layout](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-04-29/ic13rvh/deck_layout_v2.pdf)\n* [Process Steps](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-04-29/zs03rlp/Opentrons%20-%20protocol%20development%20Quiagen%20multiplex%20plus%201.pdf)\n\nWith this protocol, your robot can prepare stock plates containing master mix and indexing primers to be used for the indexing step of an NGS library prep workflow.\n\n",
"internal": "441cc8\n",
diff --git a/protoBuilds/4471ba/README.json b/protoBuilds/4471ba/README.json
index f6f1a77045..1c4adc60b4 100644
--- a/protoBuilds/4471ba/README.json
+++ b/protoBuilds/4471ba/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol automates filling plates that have been turned 90-degrees on the deck. Using custom adapters, the P300 Multi-Channel Pipette is used to transfer a set volume (selected below) from a Beckman Coulter 8-Channel Reservoir to a Simport 96-Well Plate (on an adapter that allows the plate to be turned 90-degrees). The P300 Multi-Channel pipette picks up tips from an Opentrons Tip Rack (also on an adapter), by picking up 8/12 tips from the column (liquid transfer happens to 8/12 wells in column on plates) and then the last 4 tips in the column (for last 4/12 wells in columns on plates).\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.21.0 or later)\nOpentrons P300 Multi-Channel Pipette (GEN2)\nOpentrons 300\u00b5L Tip Rack\n(2x) Simport 96-Well Plate, 1200\u00b5L\nBeckman Coulter 8-Channel Reservoir, 19mL\nReagents\n\n\n\nDeck Layout\n\nSlot 2/5: Simport 96-Well Plate, in adapter (Plate 1)\n\nSlot 8/11: Simport 96-Well Plate, in adapter (Plate 1)\n\nSlot 4/7: Opentrons 300\u00b5L Tip Rack (in adapter)\n\nSlot 10: Beckman Coulter 8-Channel Reservoir with Reagents\n\n\nUsing the customizations field (below), set up your protocol.\n P300-Multi Mount: Select which mount (left or right) the P300-Multi is attached to.\n Transfer Volume (\u00b5L): Specify the volume (in \u00b5L) that should be transferred to each well.\n* Number of Columns to Fill (1-8): Specify the number of columns to fill in each plate.",
"internal": "4471ba",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol automates filling plates that have been turned 90-degrees on the deck. Using custom adapters, the P300 Multi-Channel Pipette is used to transfer a set volume (selected below) from a Beckman Coulter 8-Channel Reservoir to a Simport 96-Well Plate (on an adapter that allows the plate to be turned 90-degrees). The P300 Multi-Channel pipette picks up tips from an Opentrons Tip Rack (also on an adapter), by picking up 8/12 tips from the column (liquid transfer happens to 8/12 wells in column on plates) and then the last 4 tips in the column (for last 4/12 wells in columns on plates).\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.21.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 Multi-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 300\u00b5L Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n* (2x) Simport 96-Well Plate, 1200\u00b5L\n* Beckman Coulter 8-Channel Reservoir, 19mL\n* Reagents\n\n---\n\n\n\n**Deck Layout**\n\nSlot 2/5: Simport 96-Well Plate, in adapter (Plate 1)\n\nSlot 8/11: Simport 96-Well Plate, in adapter (Plate 1)\n\nSlot 4/7: [Opentrons 300\u00b5L Tip Rack](https://shop.opentrons.com/collections/opentrons-tips) (in adapter)\n\nSlot 10: Beckman Coulter 8-Channel Reservoir with Reagents\n\n\n**Using the customizations field (below), set up your protocol.**\n* **P300-Multi Mount**: Select which mount (left or right) the P300-Multi is attached to.\n* **Transfer Volume (\u00b5L)**: Specify the volume (in \u00b5L) that should be transferred to each well.\n* **Number of Columns to Fill (1-8)**: Specify the number of columns to fill in each plate.\n\n\n",
"internal": "4471ba\n",
diff --git a/protoBuilds/44981c/README.json b/protoBuilds/44981c/README.json
index a1db5a3c9e..fbb3d0ae1d 100644
--- a/protoBuilds/44981c/README.json
+++ b/protoBuilds/44981c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -8,7 +8,7 @@
"description": "This protocol performs cherrypicking from source to target plates as specified in an input CSV file. The protocol parses the CSV for slots to load the source and target plates. Only non-zero volume transfers are carried out for efficiency, and the user is prompted to refill tipracks if necessary.\n\nThis protocol is an update of this APIv1 protocol.\n\nYou will need:\n P10 Single-channel electronic pipette\n P50 Single-channel electronic pipette\n P300 Single-channel electronic pipette\n 10\u00b5l Pipette tips\n 300\u00b5l Pipette tips\n Biorad Hard-Shell 96-Well PCR Plates # HSP9601\n* Opentrons 4-in-1 Tube Rack Set",
"internal": "44981c",
"markdown": {
- "author": "[Opentrons](http://www.opentrons.com/)\n\n",
+ "author": "[Opentrons](http://www.opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Cherrypicking\n\n",
"description": "This protocol performs cherrypicking from source to target plates as specified in an input CSV file. The protocol parses the CSV for slots to load the source and target plates. Only non-zero volume transfers are carried out for efficiency, and the user is prompted to refill tipracks if necessary.\n\n**This protocol is an update of this [APIv1 protocol](https://protocol-delivery.protocols.opentrons.com/protocol/1617).**\n\n---\n\nYou will need:\n* [P10 Single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [P50 Single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette?variant=5984549077021)\n* [P300 Single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette?variant=5984549109789)\n* [10\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Biorad Hard-Shell 96-Well PCR Plates # HSP9601](http://www.bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n\n",
"internal": "44981c\n",
diff --git a/protoBuilds/44b1ac/README.json b/protoBuilds/44b1ac/README.json
index 90e3be1bed..9efe6d921e 100644
--- a/protoBuilds/44b1ac/README.json
+++ b/protoBuilds/44b1ac/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"QIAseq Targeted RNAscan Panel for Illumina Instruments"
@@ -10,7 +10,7 @@
"internal": "44b1ac",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nNEST 2 mL 96-Well Deep Well Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* QIAseq Targeted RNAscan Panel for Illumina Instruments\n\n",
"deck-setup": "* If the deck layout of a particular protocol is more or less static, it is often helpful to attach a preview of the deck layout, most descriptively generated with Labware Creator. Example:\n\n\n",
"description": "This protocol automates the QIAseq Targeted RNAscan Panel for Illumina Instruments protocol in the [QIAseq Targeted RNAscan Panel](https://www.qiagen.com/pt/products/discovery-and-translational-research/next-generation-sequencing/rna-sequencing/rna-fusions/qiaseq-targeted-rnascan-panel/?clear=true#orderinginformation) on the OT-2. The QIAseq Targeted RNAscan Panels are a complete Sample to Insight solution that applies the molecular barcode-based digital RNA sequencing strategy to quantify known and new fusion genes.\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `Samples Labware Type`: The starting samples can be placed in either 1.5 mL tubes on the Opentrons Tube Rack OR in a 96 Well Plate.\n* `P300 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `IL-N7 Adapter Row`: Choose the row the adapters will be used from either Plates A, B, C, or D.\n* `Magnetic Module Engage Height`: The height the magnets will raise on the magnetic module.\n* `Wash 1 Bead Volume`: Choose the bead volume based on RNA quality.\n* `Wash 2 Bead Volume`: Choose the bead volume based on RNA quality (This value is used for all subsequent bead washes). \n\n---\n\n",
diff --git a/protoBuilds/44b43c/README.json b/protoBuilds/44b43c/README.json
index 289bca2afd..90b7afe2b2 100644
--- a/protoBuilds/44b43c/README.json
+++ b/protoBuilds/44b43c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs a PCR preparation by transferring mastermix and controls from custom deepwell plates and samples from PCR plates to corresponding deepwell plates. The user is prompted to\n\n\n\nThermoFisher MicroAmp\u2122 Fast Optical 96-Well Reaction Plate with Barcode, 0.1 mL Cat # 4366932\nThermoFisher KingFisher\u2122 Deepwell 96-Well Plate Cat # 95040450\nOpentrons P300 GEN2 multi-channel electronic pipette\nOpentrons P20 GEN2 multi-channel electronic pipette\nOpentrons 50/300\u00b5l tipracks\nOpentrons 20\u00b5l tipracks\n\n",
"internal": "44b43c",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol performs a PCR preparation by transferring mastermix and controls from custom deepwell plates and samples from PCR plates to corresponding deepwell plates. The user is prompted to\n\n---\n\n\n* ThermoFisher MicroAmp\u2122 Fast Optical 96-Well Reaction Plate with Barcode, 0.1 mL Cat # 4366932\n* ThermoFisher KingFisher\u2122 Deepwell 96-Well Plate Cat # 95040450\n* [Opentrons P300 GEN2 multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons P20 GEN2 multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons 50/300\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 20\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n---\n\n",
"internal": "44b43c\n",
diff --git a/protoBuilds/453b5a/README.json b/protoBuilds/453b5a/README.json
index 12fbac096f..f79b3fe043 100644
--- a/protoBuilds/453b5a/README.json
+++ b/protoBuilds/453b5a/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom nucleic acid purification and PCR preparation on a PCR plate mounted on an Opentrons temperature module with aluminum block insert. The final step of the protocol is to transfer various PCR mixes in 1.5ml tubes to corresponding tubes. The source mix tubes and destination wells in the PCR plate should be specified in .csv format as in the following example:\nmix tube,PCR plate start well,PCR plate end well\nA1,A1,H2\nB1,A3,H6\nC1,A7,H8\nD1,A9,H12\nYou can also download a template for this input here, edit in a spreadsheet editor like Google Sheets, MS Excel, or Apple Numbers, export as a .csv file, and upload below as the .csv file to specify PCR mix destinations parameter.\nThe user is prompted to refill tipracks when necessary for both the P20 single-channel and P300 multi-channel pipettes.\n\n\n\nOpentrons temperature module with 96-well aluminum block insert holding Biozym 96-well PCR plate #712547 200\u00b5l\nCustom 24-cuvette racks for Greiner Bio-One Vacuette tubes (1-4)\nOpentrons 4-in-1 tuberack with 4x6 insert for Eppendorf 1.5 mL Safe-Lock Snapcap tubes\nUSA Scientific 12-channel reservoir 22ml #1061-8150\nOpentrons P20 single-channel GEN2 electronic pipette\nOpentrons P300 multi-channel electronic pipette\nOpentrons 20\u00b5l and 50/300\u00b5l tipracks\n\n\n\nblood collection tube racks (slots 7, 8, 10, 11):\n tubes 1-24: slot 7\n tubes 25-48: slot 8\n tubes 49-72: slot 10\n tubes 73-96: slot 11\n12-channel reservoir (slot 1)\n channel 1: concentration dilution\n channel 2: buffer B\n channels 3-5: buffer C\n channels 9-12: liquid waste (loaded empty)",
"internal": "453b5a",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol performs a custom nucleic acid purification and PCR preparation on a PCR plate mounted on an Opentrons temperature module with aluminum block insert. The final step of the protocol is to transfer various PCR mixes in 1.5ml tubes to corresponding tubes. The source mix tubes and destination wells in the PCR plate should be specified in `.csv` format as in the following example:\n\n```\nmix tube,PCR plate start well,PCR plate end well\nA1,A1,H2\nB1,A3,H6\nC1,A7,H8\nD1,A9,H12\n```\n\nYou can also download a template for this input [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/453b5a/csv_temp.csv), edit in a spreadsheet editor like Google Sheets, MS Excel, or Apple Numbers, export as a `.csv` file, and upload below as the `.csv file to specify PCR mix destinations` parameter.\n\nThe user is prompted to refill tipracks when necessary for both the P20 single-channel and P300 multi-channel pipettes.\n\n---\n\n\n* [Opentrons temperature module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [96-well aluminum block insert](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) holding [Biozym 96-well PCR plate #712547 200\u00b5l](https://www.biozym.com/DesktopModules/WebShop/shopdisplayproducts.aspx?id=4652&cat=Low+Profile)\n* Custom 24-cuvette racks for Greiner Bio-One Vacuette tubes (1-4)\n* [Opentrons 4-in-1 tuberack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 4x6 insert for [Eppendorf 1.5 mL Safe-Lock Snapcap tubes](https://online-shop.eppendorf.us/US-en/Laboratory-Consumables-44512/Tubes-44515/Eppendorf-Safe-Lock-Tubes-PF-8863.html)\n* [USA Scientific 12-channel reservoir 22ml #1061-8150](https://www.usascientific.com/12-channel-automation-reservoir/p/1061-8150)\n* [Opentrons P20 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons P300 multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 20\u00b5l and 50/300\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n\n---\n\n\nblood collection tube racks (slots 7, 8, 10, 11):\n* tubes 1-24: slot 7\n* tubes 25-48: slot 8\n* tubes 49-72: slot 10\n* tubes 73-96: slot 11\n\n12-channel reservoir (slot 1)\n* channel 1: concentration dilution\n* channel 2: buffer B\n* channels 3-5: buffer C\n* channels 9-12: liquid waste (loaded empty)\n\n",
"internal": "453b5a\n",
diff --git a/protoBuilds/4568fa-2/README.json b/protoBuilds/4568fa-2/README.json
index e2dc3df9ce..b9911b448b 100644
--- a/protoBuilds/4568fa-2/README.json
+++ b/protoBuilds/4568fa-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "4568fa",
"labware": "\nNEST 12 Well Reservoir 15 mL\nMicroAmp EnduraPlate Optical 96-Well Clear Reaction Plates with Barcode\nOpentrons 24 Tube Rack with Eppendorf 2 mL Snapcap Tubes or equivalent\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Normalization\n\n",
"deck-setup": "This example starting deck state shows the layout for 24 samples: \n\n\n* green on tuberacks: starting samples at various concentrations (specified in .csv file)\n* blue on reservoir A1: water\n* pink on reservoir A2: blank solution\n* purple on tuberack A6-C6: HS diluent\n\n---\n\n",
"description": "\nThis is a flexible normalization protocol a 2-fold dilution to an input desired concentration followed by sample aliquoting in triplicate. Normalization parameters should be input as a .csv file below, and should be formatted as shown in the following template **including headers line**:\n\n```\nsample number,sample name,sample concentration (<0.25 mg/ml)\n1,007007009-403-1,1.569\n2,007007009-403-2,0.984\n3,007007009-403-3,2.128\n4,007007009-403-4,2.413\n5,007007009-403-5,2.303\n6,007007009-403-6,1.541\n7,007007009-403-7,2.404\n8,007007009-403-8,2.352\n9,007007009-403-9,2.763\n10,007007009-403-10,1.763\n11,007007009-403-11,2.094\n12,007007009-403-12,0.906\n13,007007009-403-13,0.715\n```\n\nYou can also access this template for download [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/4568fa/ex.csv)\n\n---\n\n",
diff --git a/protoBuilds/4568fa/README.json b/protoBuilds/4568fa/README.json
index ad0624490a..767d41989c 100644
--- a/protoBuilds/4568fa/README.json
+++ b/protoBuilds/4568fa/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "4568fa",
"labware": "\nNEST 12 Well Reservoir 15 mL\nMicroAmp EnduraPlate Optical 96-Well Clear Reaction Plates with Barcode\nOpentrons 24 Tube Rack with Eppendorf 2 mL Snapcap Tubes or equivalent\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Normalization\n\n",
"deck-setup": "This example starting deck state shows the layout for 24 samples: \n\n\n* green on tuberacks: starting samples at various concentrations (specified in .csv file)\n* blue on reservoir A1: water\n* pink on reservoir A2: blank solution\n* orange on reservoir A11-A12: buffer for final plate (optional)\n* purple on tuberack B6-D6: HS diluent\n\n---\n\n",
"description": "\nThis is a flexible normalization protocol a 2-fold dilution to an input desired concentration followed by sample aliquoting in triplicate. Normalization parameters should be input as a .csv file below, and should be formatted as shown in the following template **including headers line**:\n\n```\nsample number,sample name,sample concentration (>0.25 mg/ml)\n1,007007009-403-1,1.569\n2,007007009-403-2,0.984\n3,007007009-403-3,2.128\n4,007007009-403-4,2.413\n5,007007009-403-5,2.303\n6,007007009-403-6,1.541\n7,007007009-403-7,2.404\n8,007007009-403-8,2.352\n9,007007009-403-9,2.763\n10,007007009-403-10,1.763\n11,007007009-403-11,2.094\n12,007007009-403-12,0.906\n13,007007009-403-13,0.715\n```\n\nYou can also access this template for download [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/4568fa/ex.csv)\n\n---\n\n",
diff --git a/protoBuilds/459a55/README.json b/protoBuilds/459a55/README.json
index 28f4926356..2425c333f0 100644
--- a/protoBuilds/459a55/README.json
+++ b/protoBuilds/459a55/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Mass Spec"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom mass spec sample prep for up to 20 samples.\n\n\n\nOpentrons temperature module with NEST 96-well PCR plate 100\u00b5l full skirt\n4x6 aluminum block for NEST 1.5ml snapcap tubes\nOpentrons 20\u00b5l tiprack\nOpentrons 300\u00b5l tiprack\nOpentrons P20 and P300 GEN2 single-channel electronic pipette\n\n\n\n4x6 aluminum block for NEST 1.5ml snapcap tubes (slot 2)\n tube D1: denaturing solution\n tube D2: DTT\n* all other tubes: samples that will be transferred to their corresponding locations in sample plate",
"internal": "459a55",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Mass Spec\n\n",
"description": "This protocol performs a custom mass spec sample prep for up to 20 samples.\n\n---\n\n\n* [Opentrons temperature module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [NEST 96-well PCR plate 100\u00b5l full skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [4x6 aluminum block](https://shop.opentrons.com/collections/verified-labware/products/aluminum-block-set) for [NEST 1.5ml snapcap tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes)\n* [Opentrons 20\u00b5l tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 300\u00b5l tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons P20 and P300 GEN2 single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n\n---\n\n\n4x6 aluminum block for NEST 1.5ml snapcap tubes (slot 2)\n* tube D1: denaturing solution\n* tube D2: DTT\n* all other tubes: samples that will be transferred to their corresponding locations in sample plate\n\n",
"internal": "459a55\n",
diff --git a/protoBuilds/459cc2/README.json b/protoBuilds/459cc2/README.json
index 96513c3aeb..f6b4725ceb 100644
--- a/protoBuilds/459cc2/README.json
+++ b/protoBuilds/459cc2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Mass Spec"
@@ -10,7 +10,7 @@
"internal": "459cc2",
"labware": "\nTBD\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Mass Spec\n\n",
"deck-setup": "\n**One Plate Example:**\n\n\n\n",
"description": "This protocol performs the preparation of samples from tubes to plates. It takes in a CSV file and uses the it to load the labware and make the necessary liquid transfers. It can support up to 2 sample plates.\n\nExplanation of complex parameters below:\n* `Input CSV File`: Upload a CSV file with the formatting shown in the block below for either One Plate or Two Plates:\n\n**One Plate**\n```\nSample ID,Slot number ,Rack position,Wellplate A position,Wellplate A well position\nSample1,1,1,3,A1\nSample2,1,2,3,A2\nSample3,1,3,3,A3\n```\n\n**Two Plates**\n```\nSample ID,Slot number ,Rack position,Wellplate A position,Wellplate A well position,Wellplate B position,Wellplate B well position\nSample1,1,1,3,A1,6,A1\nSample2,1,2,3,A2,6,A2\nSample3,1,3,3,A3,6,A3\n```\n\n* `Sample Volume`: The amount of sample to transfer from the tubes to the plate.\n* `Acetonitrile Transfer`: Whether to transfer acetonitrile into the plates (Steps 6-8).\n* `P300 Single Channel GEN2 Mount Position`: Select the pipette mount position.\n* `P300 Multi Channel GEN2 Mount Position`: Select the pipette mount position.\n* `Sample Aspiration/dispense Flow rate`: Specify the flow rate of the sample liquid handling in uL/s.\n* `MeCN Aspiration Flow Rate (uL/s)`: Specify aspiration/dispense flow rate for the multi-channel pipette.\n* `Use Temperature Module`: Specify whether to use the temperature module on slots 3 and 6 with mounted deepwell plates. Note: if using the temperature modules, they should be placed in slots 3, and 6 respectively, in that order depending on running 1 or 2 plates.\n* `Dispense Height in Destination Wells`: In millimeters.\n* `PL Blowout Height`: In millimeters.\n* `Airgap`: In microliters.\n* `Reservoir Location`: Specify which reservoir column the MeCN will transfer to and from.\n* `MeCN Dilution Location`: Specify slot MeCN is held in.\n* `Multi-Channel Starting tip pick-up column (1-12)`: Specify which column on slot 10 tip rack to start picking up tips with the multi-channel pipette. \n\n\n---\n\n",
diff --git a/protoBuilds/45a320/README.json b/protoBuilds/45a320/README.json
index d980c3d563..b4015fa4f6 100644
--- a/protoBuilds/45a320/README.json
+++ b/protoBuilds/45a320/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"RNA Extraction"
@@ -10,7 +10,7 @@
"internal": "45a320",
"labware": "\nOpentrons 4-in-1 Tube Rack Set\nOpentrons 1000ul Tips\nOpentrons 20ul Tips\nVacutainer tubes\n1.5mL tubes\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* RNA Extraction\n\n",
"deck-setup": "\n* On slot 2, empty tubes should be placed in every other column starting from column 1 (e.g. columns 1, 3, 5), which the protocol refers to when it pauses and prompts the user for instruction. Columns 2, 4, and 6 will be the binding column columns of the tube rack. Please see protocol steps below.\n\n\n",
"description": "This protocol extracts DNA using the ReliaPrep\u2122 Viral TNA Miniprep System extraction kit. Proteinase K, patient sample, cell lysis buffer and isopropanol are combined with a ReliaPrep Binding Column. After 3 washes, water is added to the column to form the eluate which can then be plated. \n\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples for this run. Samples should always be placed by column (A1, B1, C1,..,etc.).\n* `P20/P1000 Dispense Flow Rate`: Global control of P20 and P1000 dispense flow rate. A value of 1.0 is default, 0.5 is 50% of the default flow rate, 1.2 is 20% faster the default flow rate, etc.\n* `P20 Mount`: Specify which side (left or right) to mount the P20 single channel pipette.\n* `P1000 Mount`: Specify which side (left or right) to mount the P1000 single channel pipette.\n\n\n---\n\n",
diff --git a/protoBuilds/470d8c/README.json b/protoBuilds/470d8c/README.json
index f3188373f2..621019d2a3 100644
--- a/protoBuilds/470d8c/README.json
+++ b/protoBuilds/470d8c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol automates the steps of transferring mastermix from source plates to destination plates. It also automates the transfer of template to destination plates.\n\n\n\nP300 multi-channel GEN2 electronic pipette\nP20 multi-channel GEN2 electronic pipette\nOpentrons 300ul tiprack\nOpentrons 20ul tiprack\nThermo Scientific\u2122 PCR Plate, 96-well, semi-skirted\nEppendorf PCR cooler\n\n\n\nDeck Setup\n Source Plate (PCR Plate on top of Eppendorf Cooler) (Slot 1)\n Opentrons 20 uL tip rack (Slot 7, 10, 11)\n Opentrons 300 uL tip rack (Slot 4)\n Template Plates (PCR Plate on top of Eppendorf Cooler) (Slots 2, 5, 8)\n* Destination Plates (PCR Plate on top of Eppendorf Cooler) (Slots 3, 6, 9)\nRobot map:\n| Tips P20 | Tips P20 | Waste |\n| Tips P20 | Template Plate #1 | Dest Plate #1 |\n| Tips P300 | Template Plate #2 | Dest Plate #2 |\n| Source Plate #1 | Template Plate #3 | Dest Plate #3 | ",
"internal": "470d8c",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol automates the steps of transferring mastermix from source plates to destination plates. It also automates the transfer of template to destination plates.\n\n---\n\n\n* [P300 multi-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [P20 multi-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 300ul tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 20ul tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Thermo Scientific\u2122 PCR Plate, 96-well, semi-skirted](https://www.fishersci.com/shop/products/thermo-scientific-96-well-semi-skirted-plates-flat-deck/ab1400l)\n* [Eppendorf PCR cooler](https://www.daigger.com/eppendorf-pcr-coolers-14616-group?gclid=CjwKCAiAz4b_BRBbEiwA5XlVVkYoJn1xfnsYoEzsrHijqNP-YRCcVBJtWxD9-ENFfB_Pc9RZJUaXYRoCWjQQAvD_BwE)\n\n---\n\n\nDeck Setup\n* Source Plate (PCR Plate on top of Eppendorf Cooler) (Slot 1)\n* Opentrons 20 uL tip rack (Slot 7, 10, 11)\n* Opentrons 300 uL tip rack (Slot 4)\n* Template Plates (PCR Plate on top of Eppendorf Cooler) (Slots 2, 5, 8)\n* Destination Plates (PCR Plate on top of Eppendorf Cooler) (Slots 3, 6, 9)\n\nRobot map: \n| Tips P20 | Tips P20 | Waste | \n| Tips P20 | Template Plate #1 | Dest Plate #1 | \n| Tips P300 | Template Plate #2 | Dest Plate #2 | \n| Source Plate #1 | Template Plate #3 | Dest Plate #3 | \n\n\n",
"internal": "470d8c",
diff --git a/protoBuilds/48648f/README.json b/protoBuilds/48648f/README.json
index b1291cc54c..b3e256fab5 100644
--- a/protoBuilds/48648f/README.json
+++ b/protoBuilds/48648f/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Purification"
@@ -8,7 +8,7 @@
"description": "This protocol automates the transfer of up to 90 patient samples to an extraction plate for the purification of nucleic acids using an Opentrons OT-2 robot. This is a pythonized version of a Protocol Designer protocol. Each of the six patient sample racks should contain a full set of 15 samples. Wells A1, B1, H1, A7, B7, and H7 in the extraction plate are left empty unless otherwise specified in the parameters below. The protocol transfers 100uL of each sample into a new well on the extraction plate using the custom tube rack and custom NEST well plate.\n\n\n\nOpentrons P1000-single channel electronic pipettes (GEN2)\nOpentrons 1000ul Filter Tips\nOpentrons Temperature Module (GEN2)\nMTCbio 10 ml tube\nNEST 96 2 mL\n",
"internal": "48648f",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Nucleic Acid Purification\n\n",
"description": "This protocol automates the transfer of up to 90 patient samples to an extraction plate for the purification of nucleic acids using an Opentrons OT-2 robot. This is a pythonized version of a [Protocol Designer protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-01-27/jd23jyu/Sample%20Plating%2090%20samples.json). Each of the six patient sample racks should contain a full set of 15 samples. Wells A1, B1, H1, A7, B7, and H7 in the extraction plate are left empty unless otherwise specified in the parameters below. The protocol transfers 100uL of each sample into a new well on the extraction plate using the custom tube rack and custom NEST well plate.\n\n---\n\n\n* [Opentrons P1000-single channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=5984549142557)\n* [Opentrons 1000ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips)\n* [Opentrons Temperature Module (GEN2)](https://shop.opentrons.com/products/tempdeck?_pos=1&_sid=5b80a3c66&_ss=r)\n* [MTCbio 10 ml tube](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-01-27/jj43jf5/mtcbio_15_tuberack_10000ul.json)\n* [NEST 96 2 mL](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-01-27/ox33jt1/nest_96_wellplate_2000ul.json)\n\n\n",
"internal": "48648f\n",
diff --git a/protoBuilds/4883fc/README.json b/protoBuilds/4883fc/README.json
index 5f009936f8..14e71e40e6 100644
--- a/protoBuilds/4883fc/README.json
+++ b/protoBuilds/4883fc/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "4883fc",
"labware": "\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt\nOpentrons 4-in-1 Tube Rack Set with NEST 1.5 mL Microcentrifuge Tube\nOpentrons pipette tips\nNEST 12 Well Reservoir 15 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"deck-setup": "* if sources are arranged in 1.5ml tubes: \n\n\n* if sources are arranged in a 96-well plate: \n\n\n",
"description": "\n\nConcentration normalization is a key component of many genomic and proteomic applications, such as NGS library prep. With this protocol, you can easily normalize the concentrations of samples in a 96-well plate from either 1.5ml snapcap tubes, or another 96-well plate. The transfers should be specified as an input .csv file formatted as shown here:\n\n```\nSample number,Slot no number,Source well,Destination well,Start Concentration (ug/uL) Source,Final Concentraion (ug/ml) Destination,Sample volume (uL),Diluent Volume(ul),Total Volume(ul) Destination\n1,4,A1,A1,5,2,40,60,100.0\n2,4,C1,B1,3,2,67,33,100.0\n3,4,D1,C1,10,2,20,80,100.0\n4,4,E3,D1,20,2,10,90,100.0\n```\n\nYou can also download and edit [this template](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/4883fc/example.xlsx). Be sure to export the `.xlsx` format to `.csv` format before inputting as the `input .csv file` below!\n\n---\n\n",
diff --git a/protoBuilds/4920bc/README.json b/protoBuilds/4920bc/README.json
index 5bdc43705c..7d58982e4f 100644
--- a/protoBuilds/4920bc/README.json
+++ b/protoBuilds/4920bc/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Omega Mag-Bind\u00ae Total RNA 96 Kit"
@@ -10,7 +10,7 @@
"internal": "4920bc",
"labware": "\nNEST 12 Well Reservoir 15 mL\nNEST 1 Well Reservoir 195 mL\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt\nNEST 96 Deepwell Plate 2mL\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Omega Mag-Bind\u00ae Total RNA 96 Kit\n\n",
"deck-setup": "\n\n",
"description": "\nThis protocol performs the [Omega Mag-Bind\u00ae Total RNA 96 Kit](https://www.omegabiotek.com/product/total-cellular-rna-mag-bind-total-rna-96//?utm_source=google&utm_medium=ppc&utm_campaign=128157293756&utm_term=total%20rna%20extraction%20kit&utm_content=b&gclid=Cj0KCQjw-daUBhCIARIsALbkjSb5Gx-Bz7K1R_Dw_ePhpzhX8NNtwe_9GP1LXR7SfOaeOHWgEnehFE4aAtRVEALw_wcB) protocol.\n\nSamples should be loaded on the magnetic module in a NEST deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 4) to the elution PCR plate (temperature module, slot 1). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\n\n---\n\n",
diff --git a/protoBuilds/49b828/README.json b/protoBuilds/49b828/README.json
index e556569f02..27c85e6ee5 100644
--- a/protoBuilds/49b828/README.json
+++ b/protoBuilds/49b828/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "49b828",
"labware": "\nOpentrons 20ul Filter Tips\nNEST 12 well 15mL Reservoir\nThermo Scientific KingFisher Flex 96 well plates\nThermo-Fisher 384 well microplate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "* Note: Samples will be dispensed per plate to the 384 well plate by column. Column 1 plate 1 --> A1 of 384, column 2 plate 1 --> B1, column 3 plate 1 --> A2 so on and so forth. Each plate is then 6 columns of the 384 well plate. If there is an unfilled column (i.e. 54 samples = 0.5 plates + 6 samples), then the protocol will use 8 tips up to column 6 of the 96 well plate, pick up 6 tips, then go to dispense into A7 of the 384 well plate. For a full run (382 samples), mastermix will be supplied to all wells. For all other sample numbers, mastermix will not be provided to O24 and P24, and will have to be put in manually. Do not run this protocol for 368 < sample number < 384 samples.\n\n\n---\n\n",
"description": "This protocol preps a 384 well plate with mastermix and sample. Plates should loaded in order of slots 7, 8, 1, and 2. See below for how plates are transferred to the 384 well plate. If running a full run (382 samples), mastermix will be provided to the wells O24 and P24. If not running a full run, mastermix and controls will have to be placed manually. H11 and H12 of the final plate on slot 2 should be left empty if running a full run.\n\n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify the number of samples in this run. Samples 1-96 will go in plate 1 slot 7, samples 97-192 will go in plate 2 slot 8, samples 193-288 will go in plate 3 slot 1, samples 289-384 will go in plate 4 slot 2. Samples will be put into the 384 well plate in the following manner:\n* `96 Well Plate Type`: Specify whether using the four 96 plates as the Kingfisher plate (depth 12.8mm) or the thermofisher plate (depth 42.3mm).\n* `384 Well Plate Type`: Specify whether using the 384 plate with the clear numbers, or the 384 plate with the black numbers. Plate should be mounted on aluminum block.\n* `Use Temperature Module?`: Specify whether using the temperature module or not on the 384 plate in slot 3 for this run. The temperature module will be set at 4C. If not using the temperature module, just place the 384 plate in slot 3 with no aluminum block.\n* `P20 Multi-Channel Mount`: Specify which mount (left or right) to host the P20 Single-Channel pipette.\n\n---\n\n\n",
diff --git a/protoBuilds/49de51-pt1/README.json b/protoBuilds/49de51-pt1/README.json
index 0498adf475..d5a56f9579 100644
--- a/protoBuilds/49de51-pt1/README.json
+++ b/protoBuilds/49de51-pt1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Plant DNA"
@@ -8,7 +8,7 @@
"description": "This protocol automates the ThermoFischer MagMAX Plant DNA Isolation Kit on up to 192 samples (2 plates of 96). The entire workflow is broken into 2 different protocols. This is part 1, disrupting the tissue.\n\nIn this protocol, 590\u00b5L of buffer (combination of 500\u00b5L lysis Buffer A, 70\u00b5L of Lysis Buffer B, and 20\u00b5L of RNase A) is added to each well. The user is then prompted to remove the plate(s) for off deck vortexing/shaking and incubation. Once incubation is complete, the user returns the plate(s) to the deck and the robot will add 130\u00b5L of the precipitation solution. After one final incubation off the robot and a spin down, 400\u00b5L of supernatant is transferred to a deep well plate. The 400\u00b5L sample can be stored or purified with part 2 of the protocol.\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.21.0 or later)\nOpentrons p300 Multi-Channel Pipette (attached to right mount)\nOpentrons 200\u00b5L Filter Tip Rack\nNEST 1-Well Reservoir, 195mL\nNEST 12-Well Reservoir, 15mL\n96-Deep Well Plate (such as the OMNI 96-Well Deep Well Plate with Ceramic Beads)\nMagMAX\u2122 Plant DNA Isolation Kit\nSamples\n\n\n\nDeck Layout\n\nSlot 1: NEST 12-Well Reservoir, 15mL\n A1: Precipitation Solution, 13.5mL (for Plate 1)\n A2: Precipitation Solution, 13.5mL (for Plate 2)\nSlot 2: OMNI 96-Well Deep Well Plate with Ceramic Beads containing sample (Plate 1)\n\nSlot 3: NEST 96-Well Deep Well Plate (empty, for Plate 1 supernatant)\n\nSlot 4: NEST 1-Well Reservoir, 195mL\nFor each plate, the following should be added:\n Lysis Buffer A, 52mL\n Lysis Buffer B, 4.160mL\n* RNase A, 2.080mL\nSlot 5: OMNI 96-Well Deep Well Plate with Ceramic Beads containing sample (Plate 2)\n\nSlot 6: NEST 96-Well Deep Well Plate (empty, for Plate 2 supernatant)\n\nSlot 7: Opentrons 200\u00b5L Filter Tip Rack\n\nSlot 10: Opentrons 200\u00b5L Filter Tip Rack\n\nSlot 11: Opentrons 200\u00b5L Filter Tip Rack\n",
"internal": "49de51-pt1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Plant DNA\n\n\n",
"description": "This protocol automates the ThermoFischer MagMAX Plant DNA Isolation Kit on up to 192 samples (2 plates of 96). The entire workflow is broken into 2 different protocols. This is part 1, disrupting the tissue.\n\nIn this protocol, 590\u00b5L of buffer (combination of 500\u00b5L lysis Buffer A, 70\u00b5L of Lysis Buffer B, and 20\u00b5L of RNase A) is added to each well. The user is then prompted to remove the plate(s) for off deck vortexing/shaking and incubation. Once incubation is complete, the user returns the plate(s) to the deck and the robot will add 130\u00b5L of the precipitation solution. After one final incubation off the robot and a spin down, 400\u00b5L of supernatant is transferred to a deep well plate. The 400\u00b5L sample can be stored or purified with part 2 of the protocol.\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.21.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons p300 Multi-Channel Pipette (attached to right mount)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* 96-Deep Well Plate (such as the [OMNI 96-Well Deep Well Plate with Ceramic Beads](https://www.omni-inc.com/consumables/well-plates/2-pack-96-well-plate-1-4mm-ceramic.html))\n* [MagMAX\u2122 Plant DNA Isolation Kit](https://www.thermofisher.com/order/catalog/product/A32549#/A32549)\n* Samples\n\n\n---\n\n\n**Deck Layout**\n\nSlot 1: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* A1: Precipitation Solution, 13.5mL (for Plate 1)\n* A2: Precipitation Solution, 13.5mL (for Plate 2)\n\n\nSlot 2: [OMNI 96-Well Deep Well Plate with Ceramic Beads](https://www.omni-inc.com/consumables/well-plates/2-pack-96-well-plate-1-4mm-ceramic.html) containing sample (Plate 1)\n\nSlot 3: [NEST 96-Well Deep Well Plate](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate) (empty, for Plate 1 supernatant)\n\nSlot 4: [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\nFor each plate, the following should be added:\n* Lysis Buffer A, 52mL\n* Lysis Buffer B, 4.160mL\n* RNase A, 2.080mL\n\n\nSlot 5: [OMNI 96-Well Deep Well Plate with Ceramic Beads](https://www.omni-inc.com/consumables/well-plates/2-pack-96-well-plate-1-4mm-ceramic.html) containing sample (Plate 2)\n\nSlot 6: [NEST 96-Well Deep Well Plate](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate) (empty, for Plate 2 supernatant)\n\nSlot 7: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 10: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n\nSlot 11: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n\n\n\n",
"internal": "49de51-pt1\n",
diff --git a/protoBuilds/49de51-pt2/README.json b/protoBuilds/49de51-pt2/README.json
index 4e5be1789b..3cdc597547 100644
--- a/protoBuilds/49de51-pt2/README.json
+++ b/protoBuilds/49de51-pt2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Plant DNA"
@@ -8,7 +8,7 @@
"description": "This protocol automates the ThermoFischer MagMAX Plant DNA Isolation Kit on up to 96 samples. The entire workflow is broken into 2 different protocols. This is part 2, DNA purification.\n\nIn this protocol, 25\u00b5L of magnetic beads plus 400\u00b5L of ethanol is added to 400\u00b5L of sample (from part 1). After off-deck mixing, the plate is returned to the OT-2 for supernatant removal. This process continues with wash buffer 1, wash buffer 2, and the elution buffer. However, the supernatant of the elution buffer is preserved in a 96-well PCR plate and can be used for further downstream application.\n\n\nUpdate April 26, 2021: The addition of magnetic beads has been made optional.\n\nUpdate May 3, 2021: A second wash step has been added and heights of aspiration adjusted.\n\nUpdate June 8, 2021: Added the ability to park tips. This will cut down on tip usage when using 48+ samples.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.21.0 or later)\nOpentrons Magnetic Module, GEN2\nOpentrons p300 Multi-Channel Pipette (attached to right mount)\nOpentrons 200\u00b5L Filter Tip Rack\nNEST 1-Well Reservoir, 195mL\nNEST 12-Well Reservoir, 15mL\nNEST 96-Well Deep Well Plate\nNEST 96-Well PCR Plate\nMagMAX\u2122 Plant DNA Isolation Kit\nSamples\n\n\n\nDeck Layout\n\nSlot 11: NEST 1-Well Reservoir, 195mL (for liquid waste)\n\nSlot 4: NEST 12-Well Reservoir, 15mL\n A1: Magnetic Beads (see note 1 below)\n A2: Ethanol (Samples 1-32)\n A3: Ethanol (Samples 33-64)\n A4: Ethanol (Samples 65-96)\n A5: empty\n A6: empty\n A7: empty\n A8: empty\n A9: empty\n A10: Wash Buffer 1 (Samples 1-32)\n A11: Wash Buffer 1 (Samples 33-64)\n A12: Wash Buffer 1 (Samples 65-96)\nSlot 5: NEST 12-Well Reservoir, 15mL\n A1: Wash Buffer 2 (Samples 1-32)\n A2: Wash Buffer 2 (Samples 33-64)\n A3: Wash Buffer 2 (Samples 65-96)\n A4: Wash Buffer 2 (Samples 1-32)\n A5: Wash Buffer 2 (Samples 33-64)\n A6: Wash Buffer 2 (Samples 65-96)\n A7: empty\n A8: empty\n A9: empty\n A10: empty\n A11: Elution Buffer (Samples 1-48)\n A12: Elution Buffer (Samples 49-96)\nSlot 10: Labware Containing Magnetic Beads (see note 1 below)\n\nSlot 7: Opentrons Magnetic Module, GEN2 with NEST 96-Well Deep Well Plate containing Samples\n\nSlot 1: NEST 96-Well PCR Plate\n\nSlot 8: Opentrons 200\u00b5L Filter Tip Rack (see note 2 below)\n\nSlot 9: Opentrons 200\u00b5L Filter Tip Rack (see note 2 below)\n\nSlot 6: Opentrons 200\u00b5L Filter Tip Rack (see note 2 below)\n\nSlot 3: Opentrons 200\u00b5L Filter Tip Rack (see note 2 below)\n\n\nSlot 2: Opentrons 200\u00b5L Filter Tip Rack (see note 2 below)\n\n\nNote 1: If using, the Magnetic Beads can be placed in well 1 of the NEST 12-Well Reservoir (in slot 4) or in column 1 of a different piece of labware (in slot 10)\nNote 2: Ten (10) columns of tips are needed per column of sample and will be accessed in the following order: slot 8, 9, 6, 3, 2. In the case that more than 48 samples are used, the user will be prompted midway through the protocol to replace the tips.\n\nUsing the customization fields below, set up your protocol.\n Number of Samples: specify the number of samples (1-96)\n Deep Well Plate Type: select the type of deep well plate used (holding samples)\n Automate Bead Addition: select whether or not to automate the addition of magnetic beads\n Location of Magnetic Beads: select the location of the magnetic beads (labware; deck slot: well)\n* Park Tips: Select Yes or No. If Yes is selected, one tiprack (in slot 2) will be used for all mixing and supernatant removals; the other tips should be placed in slot 3 and 6.\n\n",
"internal": "49de51-pt2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Plant DNA\n\n\n",
"description": "This protocol automates the ThermoFischer MagMAX Plant DNA Isolation Kit on up to 96 samples. The entire workflow is broken into 2 different protocols. This is part 2, DNA purification.\n\nIn this protocol, 25\u00b5L of magnetic beads plus 400\u00b5L of ethanol is added to 400\u00b5L of sample (from [part 1](https://develop.protocols.opentrons.com/protocol/49de51-pt1)). After off-deck mixing, the plate is returned to the OT-2 for supernatant removal. This process continues with wash buffer 1, wash buffer 2, and the elution buffer. However, the supernatant of the elution buffer is preserved in a 96-well PCR plate and can be used for further downstream application.\n\n\n**Update April 26, 2021**: The addition of magnetic beads has been made optional.\n\n**Update May 3, 2021**: A second wash step has been added and heights of aspiration adjusted.\n\n**Update June 8, 2021**: Added the ability to park tips. This will cut down on tip usage when using 48+ samples.\n\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.21.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons p300 Multi-Channel Pipette (attached to right mount)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 96-Well Deep Well Plate](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [MagMAX\u2122 Plant DNA Isolation Kit](https://www.thermofisher.com/order/catalog/product/A32549#/A32549)\n* Samples\n\n\n---\n\n\n**Deck Layout**\n\nSlot 11: [NEST 1-Well Reservoir, 195mL](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml) (for liquid waste)\n\nSlot 4: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* A1: Magnetic Beads (*see note 1 below*)\n* A2: Ethanol (Samples 1-32)\n* A3: Ethanol (Samples 33-64)\n* A4: Ethanol (Samples 65-96)\n* A5: *empty*\n* A6: *empty*\n* A7: *empty*\n* A8: *empty*\n* A9: *empty*\n* A10: Wash Buffer 1 (Samples 1-32)\n* A11: Wash Buffer 1 (Samples 33-64)\n* A12: Wash Buffer 1 (Samples 65-96)\n\nSlot 5: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* A1: Wash Buffer 2 (Samples 1-32)\n* A2: Wash Buffer 2 (Samples 33-64)\n* A3: Wash Buffer 2 (Samples 65-96)\n* A4: Wash Buffer 2 (Samples 1-32)\n* A5: Wash Buffer 2 (Samples 33-64)\n* A6: Wash Buffer 2 (Samples 65-96)\n* A7: *empty*\n* A8: *empty*\n* A9: *empty*\n* A10: *empty*\n* A11: Elution Buffer (Samples 1-48)\n* A12: Elution Buffer (Samples 49-96)\n\nSlot 10: Labware Containing Magnetic Beads (*see note 1 below*)\n\nSlot 7: [Opentrons Magnetic Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [NEST 96-Well Deep Well Plate](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate) containing Samples\n\nSlot 1: [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n\nSlot 8: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips) (*see note 2 below*)\n\nSlot 9: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips) (*see note 2 below*)\n\nSlot 6: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips) (*see note 2 below*)\n\nSlot 3: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips) (*see note 2 below*)\n\n\nSlot 2: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips) (*see note 2 below*)\n\n\n**Note 1**: If using, the Magnetic Beads can be placed in well 1 of the [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) (in slot 4) or in column 1 of a different piece of labware (in slot 10)\n**Note 2**: Ten (10) columns of tips are needed per column of sample and will be accessed in the following order: slot 8, 9, 6, 3, 2. In the case that more than 48 samples are used, the user will be prompted midway through the protocol to replace the tips.\n\n**Using the customization fields below, set up your protocol.**\n* Number of Samples: specify the number of samples (1-96)\n* Deep Well Plate Type: select the type of deep well plate used (holding samples)\n* Automate Bead Addition: select whether or not to automate the addition of magnetic beads\n* Location of Magnetic Beads: select the location of the magnetic beads (*labware*; *deck slot*: *well*)\n* Park Tips: Select **Yes** or **No**. If **Yes** is selected, one tiprack (in slot 2) will be used for all mixing and supernatant removals; the other tips should be placed in slot 3 and 6.\n\n\n\n",
"internal": "49de51-pt2\n",
diff --git a/protoBuilds/49de51/README.json b/protoBuilds/49de51/README.json
index 9bfad4da00..ccb339905a 100644
--- a/protoBuilds/49de51/README.json
+++ b/protoBuilds/49de51/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Plant DNA"
@@ -8,7 +8,7 @@
"description": "This protocol automates the ThermoFischer MagMAX Plant DNA Isolation Kit on up to 96 samples in 1 96 well plate. This protocol is broken into 5 different phases that require human intervention. Throughout the protocol, tips are replaced as needed by the user. Please replace all tip boxes when requested.\nEach reagent mL value in the Process section was calculated using the raw minimum amount of reagenet + 10%. More or less of each may be required.\n\n\n\nOpentrons P300-multi channel electronic pipettes (GEN2)\nOpentrons 200ul Filter Tips\nNEST 1 Well Reservoir 195 mL\nNEST 12-Well Reservoirs, 15 mL\nNEST 96 Deepwell Plate 2mL\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOMNI Pre-filled 2 mL 96 deep well plates with 1.4 mm Ceramic beads\nMagMAX Plant DNA Isolation Kit\n",
"internal": "49de51",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Plant DNA\n\n",
"description": "This protocol automates the ThermoFischer MagMAX Plant DNA Isolation Kit on up to 96 samples in 1 96 well plate. This protocol is broken into 5 different phases that require human intervention. Throughout the protocol, tips are replaced as needed by the user. Please replace all tip boxes when requested.\n\nEach reagent mL value in the Process section was calculated using the raw minimum amount of reagenet + 10%. More or less of each may be required.\n\n---\n\n\n* [Opentrons P300-multi channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [NEST 1 Well Reservoir 195 mL](http://www.cell-nest.com/page94?_l=en&product_id=102)\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 96 Deepwell Plate 2mL](http://www.cell-nest.com/page94?product_id=101&_l=en)\n* [NEST 0.1 mL 96-Well PCR Plate, Full Skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [OMNI Pre-filled 2 mL 96 deep well plates with 1.4 mm Ceramic beads](https://www.omni-inc.com/consumables/well-plates/2-pack-96-well-plate-1-4mm-ceramic.html)\n* [MagMAX Plant DNA Isolation Kit](https://www.thermofisher.com/order/catalog/product/A32549#/A32549)\n\n\n",
"internal": "49de51\n",
diff --git a/protoBuilds/4a0be6/README.json b/protoBuilds/4a0be6/README.json
index bd59319b25..878764972d 100644
--- a/protoBuilds/4a0be6/README.json
+++ b/protoBuilds/4a0be6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Tube Filling"
@@ -10,7 +10,7 @@
"internal": "4a0be6",
"labware": "\nOpentrons Tiprack\nSource Labware:\n15mL Falcon Tube in Opentrons Tube Rack with 15 Tube Topper,\n50mL Falcon Tube in Opentrons Tube Rack with 6 Tube Topper,\nor Beckman Coulter Reservoir\nCustom Tube Rack with Sarstedt Tubes\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Tube Filling\n\n",
"deck-setup": "**Slot 1**: Source Labware containing reagent\n**Slots 2-10**: Custom Tube Racks containing Sarstedt Tubes\n**Slot 11**: [Opentrons Tiprack](https://shop.opentrons.com/collections/opentrons-tips)\n\n\n\n",
"description": "This protocol is an updated version (APIv1 to APIv2) of this protocol: [Tube Filling into 48-well or 24-well Tube Racks](https://protocol-delivery.protocols.opentrons.com/protocol/4a0be6)\n\nThis protocol allows your robot to transfer solution from a single source tube (15 mL or 50 mL Falcon conical tube) or Beckman Coulter reservoir to up to nine 48-well, 24-well, or 12-well tube racks containing Sarstedt screw-cap tubes (either 0.5 mL, 1.5 mL or 2 mL). User must specify parameters (explained below) for each run.\n\n\nExplanation of complex parameters below:\n* **Transfer Volume (in \u00b5L)**: volume to be transferred to each Sarstedt tube\n* **Tube Rack Type**: tube rack format to be used to hold the Sarstedt tubes\n* **Tube Type**: Sarstedt tube format to be used in the protocol\n* **Number of Racks**: total number of Sarstedt tube racks to be filled (1-9)\n* **Source Container Type**: Labware containing reagent to be distributed\n* **Starting Stock Volume (in mL)**: starting volume inside source container\n* **Pipette Type**: Single-channel Pipette to be used\n* **Pipette Mount**: the side to which you wish to attach the above pipette\n* **Starting Tip**: the tip to be used in the tip rack, pick from A1-H12\n* **Dispense Mode**: choose to either *transfer* (1-to-1) or *distribute* (1-to-many)\n* **Touch Tip?**: this will add an additional touch tip after dispensing liquid\n\n\n---\n\n",
diff --git a/protoBuilds/4a5f32/README.json b/protoBuilds/4a5f32/README.json
index 36d75f5083..2619a37103 100644
--- a/protoBuilds/4a5f32/README.json
+++ b/protoBuilds/4a5f32/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Purification"
@@ -9,7 +9,7 @@
"description": "This protocol automates the purification of nucleic acids using magnetic beads. It can support up to 96 samples in multiples of 8 using the P300 Multichannel pipette. The purification process is performed using the magnetic module for the OT-2.\nExplanation of parameters below:\n\nP300 Multichannel GEN2 Mount Position: Specify which mount (left or right) to load the P300 single channel pipette.\nNumber of Samples: Total number of samples (multiples of 8).\nMagnet Engage Height (mm): The height the magnet should be raised in millimeters.\n\n",
"internal": "4a5f32",
"markdown": {
- "author": "\n[Opentrons](https://opentrons.com/)\n\n",
+ "author": "\n[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "\n* Nucleic Acid Extraction & Purification\n * Nucleic Acid Purification\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "\nThis protocol automates the purification of nucleic acids using magnetic beads. It can support up to 96 samples in multiples of 8 using the P300 Multichannel pipette. The purification process is performed using the magnetic module for the OT-2.\n\nExplanation of parameters below:\n\n- `P300 Multichannel GEN2 Mount Position`: Specify which mount (left or right) to load the P300 single channel pipette.\n- `Number of Samples`: Total number of samples (multiples of 8).\n- `Magnet Engage Height (mm)`: The height the magnet should be raised in millimeters.\n---\n\n",
diff --git a/protoBuilds/4b4a80-adapter_ligation/README.json b/protoBuilds/4b4a80-adapter_ligation/README.json
index b1d67940de..af406c56c4 100644
--- a/protoBuilds/4b4a80-adapter_ligation/README.json
+++ b/protoBuilds/4b4a80-adapter_ligation/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L"
@@ -9,7 +9,7 @@
"description": "Part 2 of 4: Adapter ligation and User enzyme treatment.\nLinks:\n Part 1: Fragmentation\n Part 2: Adapter Ligation\n Part 3: Size Selection\n Part 4: PCR Enrichment\nWith this protocol, your robot can perform the NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L protocol described by the NEBNext E6177S/L. NEB Instruction Manual for NEBNext E6177S/L.\nThis is part 2 of the protocol: Adapter Ligation.\nThis step performs the ligation with the NEBNext Adaptor followed by U excision.\nThis protocol assumes up to 24 input DNA samples of > 100 ng and follows Section 2 of the NEB Instruction Manual.\nAfter the ligation and user enzyme steps carried out in this protocol (part 2), samples may be held overnight at -20 C before proceeding with part 3: Size Selection. If you are stopping, seal the plate and store at -20 C.",
"internal": "4b4a80-adapter_ligation",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L\n\n",
"deck-setup": "* Opentrons Temperature Module (Deck Slot 2)\n* Opentrons p300 tips (Deck Slot 3)\n* Opentrons p20 tips (Deck Slot 6)\n* Opentrons Thermocycler Module (Deck Slots 7,8,10,11)\n\n",
"description": "Part 2 of 4: Adapter ligation and User enzyme treatment.\n\nLinks:\n* [Part 1: Fragmentation](http://protocols.opentrons.com/protocol/4b4a80-fragmentation)\n* [Part 2: Adapter Ligation](http://protocols.opentrons.com/protocol/4b4a80-adapter_ligation)\n* [Part 3: Size Selection](http://protocols.opentrons.com/protocol/4b4a80-size_selection)\n* [Part 4: PCR Enrichment](http://protocols.opentrons.com/protocol/4b4a80-pcr_enrichment)\n\nWith this protocol, your robot can perform the NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L protocol described by the [NEBNext E6177S/L](https://www.neb.com/products/e6177-nebnext-ultra-ii-fs-dna-library-prep-with-sample-purification-beads#Product%20Information). [NEB Instruction Manual for NEBNext E6177S/L](https://s3.amazonaws.com./pf-upload-01/u-4256/0/2021-02-16/8q531ae/manualE6177-E7805.pdf).\n\nThis is part 2 of the protocol: Adapter Ligation.\n\nThis step performs the ligation with the NEBNext Adaptor followed by U excision.\n\nThis protocol assumes up to 24 input DNA samples of > 100 ng and follows Section 2 of the NEB Instruction Manual.\n\nAfter the ligation and user enzyme steps carried out in this protocol (part 2), samples may be held overnight at -20 C before proceeding with part 3: Size Selection. If you are stopping, seal the plate and store at -20 C.\n\n\n",
diff --git a/protoBuilds/4b4a80-fragmentation/README.json b/protoBuilds/4b4a80-fragmentation/README.json
index c249e749a0..20888f422e 100644
--- a/protoBuilds/4b4a80-fragmentation/README.json
+++ b/protoBuilds/4b4a80-fragmentation/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L"
@@ -9,7 +9,7 @@
"description": "Part 1 of 4: Fragment genomic DNA and prepare ends for adapter ligation.\nLinks:\n Part 1: Fragmentation\n Part 2: Adapter Ligation\n Part 3: Size Selection\n Part 4: PCR Enrichment\nWith this protocol, your robot can perform the NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L protocol described by the NEBNext E6177S/L. NEB Instruction Manual for NEBNext E6177S/L.\nThis is part 1 of the protocol: Fragmentation.\nThis step fragments the input DNA into a tunable size range (choose optimal size range by setting the incubation time with the \"Incubation Time for Fragmentation (minutes)\" parameter below following guidelines in the instruction manual) suitable for sequencing and prepares the ends for adapter ligation. This protocol assumes up to 24 input DNA samples of > 100 ng and follows Section 2 of the NEB Instruction Manual.\nAfter the steps carried out in this protocol (part 1), it is best to immediately proceed with part 2: Adapter Ligation. If you are stopping (not recommended-a slight loss in yield may occur), seal the plate and store at -20 C.",
"internal": "4b4a80-fragmentation",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L\n\n",
"deck-setup": "* Opentrons Temperature Module (Deck Slot 2)\n* Opentrons p300 tips (Deck Slot 3)\n* Opentrons p20 tips (Deck Slot 6)\n* Opentrons Thermocycler Module (Deck Slots 7,8,10,11)\n\n",
"description": "Part 1 of 4: Fragment genomic DNA and prepare ends for adapter ligation.\n\nLinks:\n* [Part 1: Fragmentation](http://protocols.opentrons.com/protocol/4b4a80-fragmentation)\n* [Part 2: Adapter Ligation](http://protocols.opentrons.com/protocol/4b4a80-adapter_ligation)\n* [Part 3: Size Selection](http://protocols.opentrons.com/protocol/4b4a80-size_selection)\n* [Part 4: PCR Enrichment](http://protocols.opentrons.com/protocol/4b4a80-pcr_enrichment)\n\nWith this protocol, your robot can perform the NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L protocol described by the [NEBNext E6177S/L](https://www.neb.com/products/e6177-nebnext-ultra-ii-fs-dna-library-prep-with-sample-purification-beads#Product%20Information). [NEB Instruction Manual for NEBNext E6177S/L](https://s3.amazonaws.com./pf-upload-01/u-4256/0/2021-02-16/8q531ae/manualE6177-E7805.pdf).\n\nThis is part 1 of the protocol: Fragmentation.\n\nThis step fragments the input DNA into a tunable size range (choose optimal size range by setting the incubation time with the \"Incubation Time for Fragmentation (minutes)\" parameter below following guidelines in the instruction manual) suitable for sequencing and prepares the ends for adapter ligation. This protocol assumes up to 24 input DNA samples of > 100 ng and follows Section 2 of the NEB Instruction Manual.\n\nAfter the steps carried out in this protocol (part 1), it is best to immediately proceed with part 2: Adapter Ligation. If you are stopping (not recommended-a slight loss in yield may occur), seal the plate and store at -20 C.\n\n\n",
diff --git a/protoBuilds/4b4a80-pcr_enrichment/README.json b/protoBuilds/4b4a80-pcr_enrichment/README.json
index 5ec0a4747f..362343c7a7 100644
--- a/protoBuilds/4b4a80-pcr_enrichment/README.json
+++ b/protoBuilds/4b4a80-pcr_enrichment/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L"
@@ -9,7 +9,7 @@
"description": "Part 4 of 4: Enrichment, barcoding, and post-PCR clean up of size-selected DNA fragments\nLinks:\n Part 1: Fragmentation\n Part 2: Adapter Ligation\n Part 3: Size Selection\n Part 4: PCR Enrichment\nWith this protocol, your robot can perform the NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L protocol described by the NEBNext E6177S/L. NEB Instruction Manual for NEBNext E6177S/L.\nThis is part 4 of the protocol: PCR enrichment\nThis step amplifies and barcodes the size-selected output from part 3, followed by a post-PCR cleanup.\nThis protocol assumes up to 24 input DNA samples of > 100 ng and follows Section 2 of the NEB Instruction Manual.\nAfter the pcr enrichment and cleanup steps carried out in this protocol, store samples at -20 C.",
"internal": "4b4a80-pcr_enrichment",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L\n\n",
"deck-setup": "* Opentrons Temperature Module (Deck Slot 2)\n* Opentrons p300 tips (Deck Slots 3,6,9)\n* Opentrons Thermocycler Module (Deck Slots 7,8,10,11)\n* Opentrons Magnetic Module (Deck Slot 4)\n* Reagent reservoir (Deck Slot 5)\n* Eluate plate (Deck Slot 1)\n\n",
"description": "Part 4 of 4: Enrichment, barcoding, and post-PCR clean up of size-selected DNA fragments\n\nLinks:\n* [Part 1: Fragmentation](http://protocols.opentrons.com/protocol/4b4a80-fragmentation)\n* [Part 2: Adapter Ligation](http://protocols.opentrons.com/protocol/4b4a80-adapter_ligation)\n* [Part 3: Size Selection](http://protocols.opentrons.com/protocol/4b4a80-size_selection)\n* [Part 4: PCR Enrichment](http://protocols.opentrons.com/protocol/4b4a80-pcr_enrichment)\n\nWith this protocol, your robot can perform the NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L protocol described by the [NEBNext E6177S/L](https://www.neb.com/products/e6177-nebnext-ultra-ii-fs-dna-library-prep-with-sample-purification-beads#Product%20Information). [NEB Instruction Manual for NEBNext E6177S/L](https://s3.amazonaws.com./pf-upload-01/u-4256/0/2021-02-16/8q531ae/manualE6177-E7805.pdf).\n\nThis is part 4 of the protocol: PCR enrichment\n\nThis step amplifies and barcodes the size-selected output from part 3, followed by a post-PCR cleanup.\n\nThis protocol assumes up to 24 input DNA samples of > 100 ng and follows Section 2 of the NEB Instruction Manual.\n\nAfter the pcr enrichment and cleanup steps carried out in this protocol, store samples at -20 C.\n\n\n",
diff --git a/protoBuilds/4b4a80-size_selection/README.json b/protoBuilds/4b4a80-size_selection/README.json
index 99ddfa71bd..3ecd316627 100644
--- a/protoBuilds/4b4a80-size_selection/README.json
+++ b/protoBuilds/4b4a80-size_selection/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L"
@@ -9,7 +9,7 @@
"description": "Part 3 of 4: Size select and clean up the adapter-ligated and User-enzyme-treated DNA fragments (the output from Part 2).\nLinks:\n Part 1: Fragmentation\n Part 2: Adapter Ligation\n Part 3: Size Selection\n Part 4: PCR Enrichment\nWith this protocol, your robot can perform the NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L protocol described by the NEBNext E6177S/L. NEB Instruction Manual for NEBNext E6177S/L.\nThis is part 3 of the protocol: Size Selection\nThis step size-selects and cleans up the sample output from part 2 to obtain a specific insert size range of 350-600 bp. Choose pcr labware compatible with the Opentrons Magnetic Module such as \"(Magnetic Module compatible) Nest 96 wellplate 100ul pcr full skirt\" using the parameters below.\nThis protocol assumes up to 24 input DNA samples of > 100 ng and follows Section 2 of the NEB Instruction Manual.\nAfter the size selection and cleanup steps carried out in this protocol (part 3), proceed with part 4: PCR enrichment, or seal the plate and store at -20 C.",
"internal": "4b4a80-size_selection",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L\n\n",
"deck-setup": "* Opentrons Temperature Module (Deck Slot 2)\n* Opentrons p300 tips (Deck Slots 6,9)\n* Opentrons p20 tips (Deck Slot 3)\n* Opentrons Magnetic Module (Deck Slot 4)\n* Reagent Reservoir (Deck Slot 5)\n* Supernatant Plate (Deck Slot 7)\n* Eluate Plate (Deck Slot 1)\n\n",
"description": "Part 3 of 4: Size select and clean up the adapter-ligated and User-enzyme-treated DNA fragments (the output from Part 2).\n\nLinks:\n* [Part 1: Fragmentation](http://protocols.opentrons.com/protocol/4b4a80-fragmentation)\n* [Part 2: Adapter Ligation](http://protocols.opentrons.com/protocol/4b4a80-adapter_ligation)\n* [Part 3: Size Selection](http://protocols.opentrons.com/protocol/4b4a80-size_selection)\n* [Part 4: PCR Enrichment](http://protocols.opentrons.com/protocol/4b4a80-pcr_enrichment)\n\nWith this protocol, your robot can perform the NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L protocol described by the [NEBNext E6177S/L](https://www.neb.com/products/e6177-nebnext-ultra-ii-fs-dna-library-prep-with-sample-purification-beads#Product%20Information). [NEB Instruction Manual for NEBNext E6177S/L](https://s3.amazonaws.com./pf-upload-01/u-4256/0/2021-02-16/8q531ae/manualE6177-E7805.pdf).\n\nThis is part 3 of the protocol: Size Selection\n\nThis step size-selects and cleans up the sample output from part 2 to obtain a specific insert size range of 350-600 bp. Choose pcr labware compatible with the Opentrons Magnetic Module such as \"(Magnetic Module compatible) Nest 96 wellplate 100ul pcr full skirt\" using the parameters below.\n\nThis protocol assumes up to 24 input DNA samples of > 100 ng and follows Section 2 of the NEB Instruction Manual.\n\nAfter the size selection and cleanup steps carried out in this protocol (part 3), proceed with part 4: PCR enrichment, or seal the plate and store at -20 C.\n\n\n",
diff --git a/protoBuilds/4b7268/README.json b/protoBuilds/4b7268/README.json
index e34b5a9552..4b60828091 100644
--- a/protoBuilds/4b7268/README.json
+++ b/protoBuilds/4b7268/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Purification"
@@ -9,7 +9,7 @@
"description": "This protocol automates the bead-based purification of nucleic acids from up to 96 cellular samples using a custom workflow.\nLinks:\n* Invitrogen DNA Binding Beads for MagMAX-96 DNA Multi-Sample Kit from ThermoFisher catalog number 4489112\nThis protocol was developed to perform the following steps: Bead binding, two wash steps, allow beads to air dry, followed by elution and transfer of the eluate to a fresh PCR plate.",
"internal": "4b7268",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Nucleic Acid Purification\n\n",
"deck-setup": "\n\n* Opentrons 300 ul tips (Deck Slots 4,5,7 - depends on tip parking and sample count)\n* Opentrons 1000 ul tips (Deck Slots 8,9,10 - depends on tip parking and sample count)\n* Elution Plate (96-well PCR Plate) (Deck Slot 1)\n* Reagent Reservoir (12-well reservoir) (Deck Slot 2)\n* Opentrons 6-Tube Rack with 50 mL Conical Tubes opentrons_6_tuberack_falcon_50ml_conical (Deck Slot 3)\n* Opentrons Magnetic Module with deep well plate (Deck Slot 6)\n\n\n\n\n\n",
"description": "\nThis protocol automates the bead-based purification of nucleic acids from up to 96 cellular samples using a custom workflow.\n\nLinks:\n* [Invitrogen DNA Binding Beads for MagMAX-96 DNA Multi-Sample Kit from ThermoFisher catalog number 4489112](https://www.thermofisher.com/order/catalog/product/4489112?SID=srch-srp-4489112#/4489112?SID=srch-srp-4489112)\n\nThis protocol was developed to perform the following steps: Bead binding, two wash steps, allow beads to air dry, followed by elution and transfer of the eluate to a fresh PCR plate.\n\n",
diff --git a/protoBuilds/4b76c0/README.json b/protoBuilds/4b76c0/README.json
index 4b5c03af56..3486eb5c1f 100644
--- a/protoBuilds/4b76c0/README.json
+++ b/protoBuilds/4b76c0/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -8,7 +8,7 @@
"description": "\nConcentration normalization is a key component of many genomic and proteomic applications, such as NGS library prep. With this protocol, you can easily normalize the concentrations of samples in a 96-well Axygen plate without worrying about missing a well or adding the wrong volume. Just upload your properly formatted CSV file (keep scrolling for an example), customize your parameters, and download your ready-to-run protocol.\nUsing the customizations fields, below set up your protocol.\n Volumes CSV: Upload the CSV (.csv) containing your diluent volumes.\n P10 Multi Mount: Select which mount (left or right) the P10 Multi is attached to.\nNote about CSV: All values corresponding to wells in the CSV must have a value (zero (0) is a valid value and nothing will be transferred to the corresponding well(s)). Additionally, the CSV can be formatted in \"portrait\" orientation. In portrait orientation, the bottom left corner is treated as A1 and the top right corner would correspond to the furthest well from A1 (H12 in a 96-well plate).\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons P10 Single-Channel Pipette\nOpentrons 10\u00b5L Tips\nNEST 12-Well Reservoir, 15mL\nAxygen 96-Well Plate (PCR-96-FS-C)\nDiluent\nSamples\n\n\n\nSlot 1: NEST 12-Well Reservoir\n* A1: Diluent\nSlot 2: Axygen 96-Well Plate (PCR-96-FS-C)\nSlot 3: Opentrons 10\u00b5L Tips",
"internal": "4b76c0",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n\n",
"description": "\n\nConcentration normalization is a key component of many genomic and proteomic applications, such as NGS library prep. With this protocol, you can easily normalize the concentrations of samples in a 96-well Axygen plate without worrying about missing a well or adding the wrong volume. Just upload your properly formatted CSV file (keep scrolling for an example), customize your parameters, and download your ready-to-run protocol.\n\n\n\n**Using the customizations fields, below set up your protocol.**\n* **Volumes CSV**: Upload the CSV (.csv) containing your diluent volumes.\n* **P10 Multi Mount**: Select which mount (left or right) the P10 Multi is attached to.\n\n*Note about CSV*: All values corresponding to wells in the CSV must have a value (zero (0) is a valid value and nothing will be transferred to the corresponding well(s)). Additionally, the CSV can be formatted in \"portrait\" orientation. In portrait orientation, the bottom left corner is treated as A1 and the top right corner would correspond to the furthest well from A1 (H12 in a 96-well plate).\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P10 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [NEST 12-Well Reservoir, 15mL](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* Axygen 96-Well Plate (PCR-96-FS-C)\n* Diluent\n* Samples\n\n\n---\n\n\nSlot 1: [NEST 12-Well Reservoir](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* A1: Diluent\n\nSlot 2: Axygen 96-Well Plate (PCR-96-FS-C)\n\nSlot 3: [Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n\n\n",
"internal": "4b76c0\n",
diff --git a/protoBuilds/4bdd14/README.json b/protoBuilds/4bdd14/README.json
index ac1407f806..ed9a978433 100644
--- a/protoBuilds/4bdd14/README.json
+++ b/protoBuilds/4bdd14/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol creates a custom plate-filling protocol from 1 source plate to up to 9 destination plate using a P300-multi channel pipette. The transfer scheme is shown here.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP300 Multi Channel Pipette\nNEST 96 Deepwell Plate 2mL\nGreiner Microlon 96 Well Plate 340 \u00b5L\n",
"internal": "4bdd14",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "This protocol creates a custom plate-filling protocol from 1 source plate to up to 9 destination plate using a P300-multi channel pipette. The transfer scheme is shown [here](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2020-11-24/ek13tro/image.png).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P300 Multi Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [NEST 96 Deepwell Plate 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep)\n* Greiner Microlon 96 Well Plate 340 \u00b5L\n\n",
"internal": "4bdd14\n",
diff --git a/protoBuilds/4c8861/README.json b/protoBuilds/4c8861/README.json
index 29a0d41e67..a9da78a504 100644
--- a/protoBuilds/4c8861/README.json
+++ b/protoBuilds/4c8861/README.json
@@ -17,14 +17,14 @@
"internal": "4c8861\n",
"labware": "* [Bio-Rad 96 Well Plate 200 \u00b5L PCR](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate)\n* [Opentrons 4-in-1 Tuberack](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [Opentrons 20ul Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons 200ul Tips](https://shop.opentrons.com/collections/opentrons-tips)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Partner Name](partner website link)\n\n",
+ "partner": "[Partner Name](partner website link)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [Opentrons P300 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n\n\n---\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Transfer X(ul) of dilution buffer (slot 11) to output plate (slot 8) using single channel pipette (volume based on input concentration)\n2. Pause after all 96 wells are filled, load input plate (slot 9)\n3. Transfer Y(ul) input to left side of output plate (slot 8) wells and tip touch (to ensure total volume dispensed)\n4. Pause, remove output (slot 8)\n5 . transfer X(ul) of dilution buffer (slot 11) to output plate (slot 5) using single channel pipette (volume based on input concentration)\n6. Pause after all 96 wells are filled, load input plate (slot 6)\n7. Transfer Y(ul) input to left side of output plate (slot 5) wells and tip touch (to ensure total volume dispensed)\n8. Pause, remove output (slot 5)\n9. Transfer X(ul) of dilution buffer (slot 11) to output plate (slot 2) using single channel pipette (volume based on input concentration)\n10. Pause after all 96 wells are filled, load input plate (slot 3)\n11. Transfer Y(ul) input to left side of output plate (slot 2) wells and tip touch (to ensure total volume dispensed)\n12. Pause, remove output (slot 2)\n\n\n",
"title": "Normalization with Input .CSV File"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Partner Name",
+ "partner": "Partner Name\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nOpentrons P20 Single-Channel Pipette\nOpentrons P300 Single-Channel Pipette\n\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nTransfer X(ul) of dilution buffer (slot 11) to output plate (slot 8) using single channel pipette (volume based on input concentration)\nPause after all 96 wells are filled, load input plate (slot 9)\nTransfer Y(ul) input to left side of output plate (slot 8) wells and tip touch (to ensure total volume dispensed)\nPause, remove output (slot 8)\n5 . transfer X(ul) of dilution buffer (slot 11) to output plate (slot 5) using single channel pipette (volume based on input concentration)\nPause after all 96 wells are filled, load input plate (slot 6)\nTransfer Y(ul) input to left side of output plate (slot 5) wells and tip touch (to ensure total volume dispensed)\nPause, remove output (slot 5)\nTransfer X(ul) of dilution buffer (slot 11) to output plate (slot 2) using single channel pipette (volume based on input concentration)\nPause after all 96 wells are filled, load input plate (slot 3)\nTransfer Y(ul) input to left side of output plate (slot 2) wells and tip touch (to ensure total volume dispensed)\nPause, remove output (slot 2)\n",
diff --git a/protoBuilds/4d9b8b/README.json b/protoBuilds/4d9b8b/README.json
index 096a0ddddd..2262b9fb1e 100644
--- a/protoBuilds/4d9b8b/README.json
+++ b/protoBuilds/4d9b8b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "4d9b8b",
"labware": "\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nCostar 96 Well Plate\nSilicon Wafer with Opentrons Labware Adapter\nCustom 8-tube 20ml tube rack\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol drop casts a mixed solution of polymer to a silicon wafer. The protocol can be broken down into the following parts:\n\n* Mix 3 user-specified volumes (via CSV) of polymer in a mix plate.\n* Mix solutions, drop cast 1-1 transfer from mix plate onto silicon wafer.\n\n The protocol begins by mixing 3 solutions of polymer (volumes of each respective solution specified by the user via a CSV file) in a 96 well plate, up to the number of samples specified. After a mix step, solutions are transferred 1-to-1 from the 96 well plate to the silicon wafer (A1 to A1, B1 to B1, etc.). The silicon wafer is treated as a 24, 48, or 96 well grid for this step, although in reality the surface is uniform.\n\n The option to have various grid sizes is to vary spacing between drops. Depending on the solution volume, viscosity, dispense flow rate, and dispense height above the wafer, an appropriate grid can be chosen. \n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify how many samples will be transferred onto the silicon wafer (number of wells).\n* `CSV`: Upload a CSV which specifies transfer volumes of the three solutions to be mixed and added to the silicon wafer.\nThe CSV should be formatted like so:\n\n`Well` | `Transfer Volume Solution 1 (ul)` | `Transfer Volume Solution 2 (ul)` | `Transfer Volume Solution 3 (ul)` | `Transfer Volume to Silicon Plate (ul)`\n\nExample Row: `A1`, `5`, `10`, `3`, `18`\n\nThe first row should contain headers (like above). All following rows should just include necessary information. \n\nSamples will be referenced in this protocol, as well as referenced in the CSV in order by column (e.g. A1, B1, C1, etc.) up to the number of samples for the run. The first `Well` column in the CSV should thus be ordered, A1, B1, C1, etc. \n* `Dispense Height Above Wafer`: Specify the height above the wafer to dispense mixed solution. **Caution:** a value of 0 returns the top of the wafer, with a value of 1 returning 1mm above the top of the wafer, for example. Negative numbers can also be passed (distance from top of the wafer) - use discretion to avoid tip crashing.\n* `Dispense Flow Rate`: Specify the dispense flow rate of solution onto the silicon wafer.\n* `Mix Repetitions`: Specify the number of times each well will be mixed in the mix well plate before being transferred onto the wafer.\n* `Mix Volume`: Specify the volume at which each well be mixed.\n* `Initial Volume in Solution`: Specify the initial volumes in all 3 tubes. This parameter is used for liquid tracking and ensuring the pipette is not submerged in solution.\n* `Delay after dispense before returning tip`: Specify the amount of time (in seconds) before the tip is returned to the tip rack after dispense. Following the dispense, there will be a delay, then blow out and touch tip. \n* `P20 Single Mount`: Specify which mount the P20 is on (left or right).\n\n---\n\n",
diff --git a/protoBuilds/4e4c0c/README.json b/protoBuilds/4e4c0c/README.json
index bcf75902c8..cc9c562231 100644
--- a/protoBuilds/4e4c0c/README.json
+++ b/protoBuilds/4e4c0c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "4e4c0c",
"labware": "\nOpentrons Filter Tips for the p300 and p20 (https://shop.opentrons.com)\nNest 195 mL Reservoir\nNest 12-Well Reservoir\nNest 96-Deep Well Plate 2 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\n**Slots 1-5**: Processing Plates (Sample Plate, Wash 1 Plate, Wash 2 plate, Wash 3 Plate, Elution Plate) (Nest 2 96-Deep Well 2 mL) \n**Slot 7**: 12-Well Reagent Reservoir (Nest 12-Well Reservoir) \n\n\n\n**Slot 8**: 80 Percent EtOH Reservoir (Nest 195 mL Reservoir) \n**Slot 9**: Wash Buffer Reservoir (Nest 195 mL Reservoir) \n**Slot 10**: Opentrons 20 uL Filter Tips \n**Slot 11**: Opentrons 200 uL Filter Tips \n\n\n---\n\n",
"description": "This protocol uses 8-channel P20 and P300 pipettes to add a fixed volume (10, 1000, 1000, 500, and 100 uL) of a specific reagent (Proteinase K Solution, Wash Buffer, 80 Percent EtOH, 80 Percent EtOH, and Elution Solution) to the corresponding processing plate on the OT-2 deck (Sample Plate, Wash 1 Plate, Wash 2 Plate, Wash 3 Plate, and Elution Plate). User-determined parameters are available to specify the number of columns to be filled in the five processing plates and the labware to be used for the processing plate, the wash buffer reservoir, the EtOH reservoir, and the 12-well reservoir holding the Proteinase K Solution and Elution Solution.\n\nLinks:\n* [Experimental Protocol](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/4e4c0c/Opentrons+Protocol+to+code.docx)\n\n\n---\n\n\n\n",
diff --git a/protoBuilds/4ee4d6-part2/README.json b/protoBuilds/4ee4d6-part2/README.json
index ce90568074..91d218cc2c 100644
--- a/protoBuilds/4ee4d6-part2/README.json
+++ b/protoBuilds/4ee4d6-part2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina DNA Prep with Enrichment"
@@ -9,7 +9,7 @@
"description": "Part 2 of 2: Clean Up and Pool Libraries, Hybridize and Capture Probes, Amplify Enriched Library, Clean Up Enriched Library\nLinks:\n Part 1: Tagmentation, Clean Up, Amplify Tagmented DNA\n Part 2: Clean Up and Pool Libraries, Hybridize and Capture Probes, Amplify Enriched Library, Clean Up Enriched Library\n* Protocol\nWith this protocol, your robot can perform the Illumina DNA Prep with Enrichment protocol described by the Illumina DNA Prep with Enrichment Reference Guide.\nThis is part 2 of the protocol, which includes the steps (1) Clean Up and Pool Libraries (2) Hybridize and Capture Probes (3) Amplify Enriched Library and (4) Clean Up Enriched Library.\nThe first clean up uses a double-sided bead purification for consistently narrow fragment size distribution. Up to 12 libraries with unique indexes are combined into one pool. Streptavidin magnetic beads are used to capture probes hybridized to target sequences. The enriched library is then amplified by PCR.",
"internal": "4ee4d6-part2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Illumina DNA Prep with Enrichment\n\n",
"deck-setup": "* Opentrons Temperature Module (Deck Slot 9)\n* Opentrons Magnetic Module (Deck Slot 6)\n* Opentrons p300 tips (Deck Slot 5)\n* Opentrons p20 tips (Deck Slot 4)\n* Opentrons Thermocycler Module (Deck Slots 7,8,10,11)\n* reagents block (opentrons_24_aluminumblock_nest_2ml_snapcap) (Deck Slot 3)\n* reagents reservoir (nest_12_reservoir_15ml) (Deck Slot 2)\n\n",
"description": "Part 2 of 2: Clean Up and Pool Libraries, Hybridize and Capture Probes, Amplify Enriched Library, Clean Up Enriched Library\n\nLinks:\n* [Part 1: Tagmentation, Clean Up, Amplify Tagmented DNA](http://protocols.opentrons.com/protocol/4ee4d6)\n* [Part 2: Clean Up and Pool Libraries, Hybridize and Capture Probes, Amplify Enriched Library, Clean Up Enriched Library](http://protocols.opentrons.com/protocol/4ee4d6-part2)\n* [Protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-03-24/dw03wwr/Opentron%20protocol.docx)\n\nWith this protocol, your robot can perform the Illumina DNA Prep with Enrichment protocol described by the [Illumina DNA Prep with Enrichment Reference Guide](https://support.illumina.com/sequencing/sequencing_kits/illumina-dna-prep-with-enrichment/documentation.html).\n\nThis is part 2 of the protocol, which includes the steps (1) Clean Up and Pool Libraries (2) Hybridize and Capture Probes (3) Amplify Enriched Library and (4) Clean Up Enriched Library.\n\nThe first clean up uses a double-sided bead purification for consistently narrow fragment size distribution. Up to 12 libraries with unique indexes are combined into one pool. Streptavidin magnetic beads are used to capture probes hybridized to target sequences. The enriched library is then amplified by PCR.\n\n",
diff --git a/protoBuilds/4ee4d6/README.json b/protoBuilds/4ee4d6/README.json
index d3f75d7dd9..832ca102d7 100644
--- a/protoBuilds/4ee4d6/README.json
+++ b/protoBuilds/4ee4d6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina DNA Prep with Enrichment"
@@ -9,7 +9,7 @@
"description": "Part 1 of 2: Tagmentation, Clean Up, Amplify Tagmented DNA\nLinks:\n Part 1: Tagmentation, Clean Up, Amplify Tagmented DNA\n Part 2: Clean Up and Pool Libraries, Hybridize and Capture Probes, Amplify Enriched Library, Clean Up Enriched Library\n* Protocol\nWith this protocol, your robot can perform the Illumina DNA Prep with Enrichment protocol described by the Illumina DNA Prep with Enrichment Reference Guide.\nThis is part 1 of the protocol, which includes the steps (1) Tagmentation (2) Clean Up and (3) Amplify Tagmented DNA.\nThe tagmentation uses a bead-based transposome complex to fragment a set number (if saturated with input DNA, the process is self-normalizing) of genomic DNA molecules and tag them with adapter sequences in one step. The amplification step increases the yield, adds indexes, and enables capability across all Illumina sequencing platforms.\nAfter the steps carried out in this protocol, you can safely stop work and return to it at a later point. If you are stopping, store at -25\u00b0C to -15\u00b0C for up to 30 days.",
"internal": "4ee4d6",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Illumina DNA Prep with Enrichment\n\n",
"deck-setup": "* Opentrons Temperature Module (Deck Slot 9)\n* Opentrons Magnetic Module (Deck Slot 6)\n* Opentrons p300 tips (Deck Slot 5)\n* Opentrons p20 tips (Deck Slot 4)\n* Opentrons Thermocycler Module (Deck Slots 7,8,10,11)\n* initial samples (nest_96_wellplate_100ul_pcr_full_skirt) (Deck Slot 1)\n* reagents block (opentrons_24_aluminumblock_nest_2ml_snapcap) (Deck Slot 3)\n* reagents reservoir (nest_12_reservoir_15ml) (Deck Slot 2)\n\n",
"description": "Part 1 of 2: Tagmentation, Clean Up, Amplify Tagmented DNA\n\nLinks:\n* [Part 1: Tagmentation, Clean Up, Amplify Tagmented DNA](http://protocols.opentrons.com/protocol/4ee4d6)\n* [Part 2: Clean Up and Pool Libraries, Hybridize and Capture Probes, Amplify Enriched Library, Clean Up Enriched Library](http://protocols.opentrons.com/protocol/4ee4d6-part2)\n* [Protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-03-24/dw03wwr/Opentron%20protocol.docx)\n\nWith this protocol, your robot can perform the Illumina DNA Prep with Enrichment protocol described by the [Illumina DNA Prep with Enrichment Reference Guide](https://support.illumina.com/sequencing/sequencing_kits/illumina-dna-prep-with-enrichment/documentation.html).\n\nThis is part 1 of the protocol, which includes the steps (1) Tagmentation (2) Clean Up and (3) Amplify Tagmented DNA.\n\nThe tagmentation uses a bead-based transposome complex to fragment a set number (if saturated with input DNA, the process is self-normalizing) of genomic DNA molecules and tag them with adapter sequences in one step. The amplification step increases the yield, adds indexes, and enables capability across all Illumina sequencing platforms.\n\nAfter the steps carried out in this protocol, you can safely stop work and return to it at a later point. If you are stopping, store at -25\u00b0C to -15\u00b0C for up to 30 days.\n\n",
diff --git a/protoBuilds/4f7a6f/README.json b/protoBuilds/4f7a6f/README.json
index 81fedec292..4998d6edb7 100644
--- a/protoBuilds/4f7a6f/README.json
+++ b/protoBuilds/4f7a6f/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "4f7a64",
"labware": "\nNEST 1-Well Reservoirs, 195 mL\n*Opentrons 300\u00b5L Tips\nHTD Dialysis Plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "This protocol performs an equilibrium dialysis assay on the OT-2 using HTD plates. The HTD plate is mounted directly on the temperature module to achieve cooling, as well as the buffer. Backup buffer reservoirs are available on the deck, as well as waste reservoirs. A csv is uploaded to the protocol to gain information such as transfer volume, incubation time, and buffer volume. The OT-2 will only be accessing the bottom half of the wells for the duration of the protocol, so as to operate below the partition line. For detailed protocol steps, please see below.\n\nExplanation of complex parameters below:\n* `csv`: Here, you should upload a .csv file formatted in the following way, being sure to include the header line (note sample number should always be in multiples of 8 e.g. 8, 16, 24, 32, etc.):\n```\nN.\u00ba PROTEIN,VOLUME_EXCHANGE (ul) | VOLUME_RESERVOIR (ml) | TEMPERATURE_MODULE 1 (\u00baC) | TEMPERATURE_MODULE 2 (\u00baC) | TIME_PROTOCOL (min) | TIME_INCUBATE (min)\n96,300,195,4,4,840,30\n```\n* `Initial volume of buffer reservoir`: Specify the volume inside each of the reservoirs in mL.\n* `P300 Multi-Channel Mount`: Specify which mount (left or right) to host the P300 Multi-Channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/4fa62e/README.json b/protoBuilds/4fa62e/README.json
index 5e62c8c70b..16e6976a1e 100644
--- a/protoBuilds/4fa62e/README.json
+++ b/protoBuilds/4fa62e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom PCR preparation on DNA samples in triplicate using pre-created mastermix.\n",
"internal": "4fa62e",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol performs a custom PCR preparation on DNA samples in triplicate using pre-created mastermix.\n\n---\n\n",
"internal": "4fa62e",
diff --git a/protoBuilds/4fc750/README.json b/protoBuilds/4fc750/README.json
index 874bcf0e7a..2367cb6664 100644
--- a/protoBuilds/4fc750/README.json
+++ b/protoBuilds/4fc750/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Omega Mag-Bind\u00ae Blood & Tissue DNA 96 Kit"
@@ -8,7 +8,7 @@
"description": "This is a flexible protocol accommodating the framework given for the 250\u00b5l Blood Protocol in the Omega Mag-Bind\u00ae Blood & Tissue DNA 96 Kit,\nLysed samples should be loaded on the magnetic module in a NEST or USA Scientific 96-deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 1) to the elution PCR plate (slot 5). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\nExplanation of complex parameters below:\n park tips: If set to yes (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to no, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n track tips across protocol runs: If set to yes, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to no, tips will be picked up from column 1 of the 1st tiprack.\n* flash: If set to yes, the robot rail lights will flash during any automatic pauses in the protocol. If set to no, the lights will not flash.\n\n \nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons magnetic module\nNEST 12 Well Reservoir 15 mL or USA Scientific 12 Well Reservoir 22 mL\nNEST 1 Well Reservoir 195 mL or Agilent 1 Well Reservoir 290 mL\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt or Bio-Rad 96 Well Plate 200 \u00b5L PCR\nNEST 96 Deepwell Plate 2mL or USA Scientific 96 Deep Well Plate 2.4 mL\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n\n\n\n\nReservoir 1: slot 2\nReservoir 2: slot 3\n\n",
"internal": "4fc750",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Omega Mag-Bind\u00ae Blood & Tissue DNA 96 Kit\n\n",
"description": "This is a flexible protocol accommodating the framework given for the 250\u00b5l Blood Protocol in the [Omega Mag-Bind\u00ae Blood & Tissue DNA 96 Kit](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-01-08/fl13zq3/Mag-Bind%C2%AE%20Blood%20%26%20Tissue%20DNA%20HDQ%2096%20Kit%20QMF27.0118.M6399%20v8.0.pdf),\n\nLysed samples should be loaded on the magnetic module in a NEST or USA Scientific 96-deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 1) to the elution PCR plate (slot 5). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\nExplanation of complex parameters below:\n* `park tips`: If set to `yes` (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to `no`, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n* `flash`: If set to `yes`, the robot rail lights will flash during any automatic pauses in the protocol. If set to `no`, the lights will not flash.\n\n---\n\n \n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons magnetic module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [NEST 12 Well Reservoir 15 mL](https://labware.opentrons.com/nest_12_reservoir_15ml) or [USA Scientific 12 Well Reservoir 22 mL](https://labware.opentrons.com/usascientific_12_reservoir_22ml)\n* [NEST 1 Well Reservoir 195 mL](https://labware.opentrons.com/nest_1_reservoir_195ml) or [Agilent 1 Well Reservoir 290 mL](https://www.agilent.com/store/en_US/Prod-201252-100/201252-100)\n* [NEST 96 Well Plate 100 \u00b5L PCR Full Skirt](https://labware.opentrons.com/nest_96_wellplate_100ul_pcr_full_skirt) or [Bio-Rad 96 Well Plate 200 \u00b5L PCR](https://www.bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [NEST 96 Deepwell Plate 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep) or [USA Scientific 96 Deep Well Plate 2.4 mL](https://labware.opentrons.com/usascientific_96_wellplate_2.4ml_deep)\n* [Opentrons 96 Filter Tip Rack 200 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n\n---\n\n\n* Reservoir 1: slot 2\n* Reservoir 2: slot 3 \n\n\n",
"internal": "4fc750\n",
diff --git a/protoBuilds/50486f-v2-part1/README.json b/protoBuilds/50486f-v2-part1/README.json
index 67308254f5..e07605c229 100644
--- a/protoBuilds/50486f-v2-part1/README.json
+++ b/protoBuilds/50486f-v2-part1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs PCR prep - HYB (part 1/4) for 96 samples using the P20 Multi-channel pipette.\n\n\n\nP20 Multi-channel Pipette\n10/20uL Opentrons tipracks\nBio-Rad Hard Shell 96-well PCR plate 200ul #hsp9601\nLabcon 0.2mL PCR strips #3940-550\nLabcon Plate\n\n\n\nSlot 1: 96-well Labcon plate (clean and empty) on PCR cooler.\nSlot 2: Pre-Chilled PCR Thermal Block with PCR strip placed in column 1 and filled with master mix\nSlot 3: 96-well Bio-Rad PCR plate filled with DNA samples.\nSlots 4-11: 10/20ul Opentrons tiprack",
"internal": "50486f-v2-part1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"description": "This protocol performs PCR prep - HYB (part 1/4) for 96 samples using the P20 Multi-channel pipette.\n\n---\n\n\n* [P20 Multi-channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [10/20uL Opentrons tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Bio-Rad Hard Shell 96-well PCR plate 200ul #hsp9601](bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [Labcon 0.2mL PCR strips #3940-550](http://www.labcon.com/microstrips.html)\n* Labcon Plate\n\n\n---\n\n\nSlot 1: 96-well Labcon plate (clean and empty) on PCR cooler.\n\nSlot 2: Pre-Chilled [PCR Thermal Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) with PCR strip placed in column 1 and filled with master mix\n\nSlot 3: 96-well Bio-Rad PCR plate filled with DNA samples.\n\nSlots 4-11: [10/20ul Opentrons tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n",
"internal": "50486f-v2-part1\n",
diff --git a/protoBuilds/50486f-v2-part2/README.json b/protoBuilds/50486f-v2-part2/README.json
index e5129054b8..1e2e5ca5a9 100644
--- a/protoBuilds/50486f-v2-part2/README.json
+++ b/protoBuilds/50486f-v2-part2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs PCR prep - GAP (part 2/4) for 96 samples using the P20 Multi-channel pipette.\n\n\n\nP20 Multi-channel Pipette\n10/20uL Opentrons tipracks\nLabcon 0.2mL PCR strips #3940-550\nLabcon Plate\nPCR Cooler\n\n\n\nSlot 1: 96-well PCR plate on PCR Cooler (filled with sample from previous step).\nSlot 2: PCR strip on PCR Cooler placed in column 1 and filled with master mix\nSlots 4-11: 10/20ul Opentrons tiprack",
"internal": "50486f-v2-part2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"description": "This protocol performs PCR prep - GAP (part 2/4) for 96 samples using the P20 Multi-channel pipette.\n\n---\n\n\n* [P20 Multi-channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [10/20uL Opentrons tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Labcon 0.2mL PCR strips #3940-550](http://www.labcon.com/microstrips.html)\n* Labcon Plate\n* PCR Cooler\n\n\n---\n\n\nSlot 1: 96-well PCR plate on PCR Cooler (filled with sample from previous step).\n\nSlot 2: PCR strip on PCR Cooler placed in column 1 and filled with master mix\n\nSlots 4-11: [10/20ul Opentrons tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n\n",
"internal": "50486f-v2-part2\n",
diff --git a/protoBuilds/50486f-v2-part3/README.json b/protoBuilds/50486f-v2-part3/README.json
index 9ef1c18dc5..eed263dcf9 100644
--- a/protoBuilds/50486f-v2-part3/README.json
+++ b/protoBuilds/50486f-v2-part3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs PCR prep - EXO (part 3/4) for 96 samples using the P20 Multi-channel pipette.\n\n\n\nP20 Multi-channel Pipette\n10/20uL Opentrons tipracks\nLabcon 0.2mL PCR strips #3940-550\nLabcon Plate\nPCR Cooler\n\n\n\nSlot 1: 96-well Labcon plate on PCR cooler (filled with sample from previous step).\nSlot 2: Pre-Chilled PCR Cooler with PCR strip placed in column 1 and filled with master mix\nSlot 4-11: 10/20ul Opentrons tiprack",
"internal": "50486f-v2-part3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"description": "This protocol performs PCR prep - EXO (part 3/4) for 96 samples using the P20 Multi-channel pipette.\n\n---\n\n\n* [P20 Multi-channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [10/20uL Opentrons tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Labcon 0.2mL PCR strips #3940-550](http://www.labcon.com/microstrips.html)\n* Labcon Plate\n* PCR Cooler\n\n\n---\n\n\nSlot 1: 96-well Labcon plate on PCR cooler (filled with sample from previous step).\n\nSlot 2: Pre-Chilled PCR Cooler with PCR strip placed in column 1 and filled with master mix\n\nSlot 4-11: [10/20ul Opentrons tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n\n",
"internal": "50486f-v2-part3\n",
diff --git a/protoBuilds/50486f-v2-part4/README.json b/protoBuilds/50486f-v2-part4/README.json
index 4446f869f0..8fe084ac92 100644
--- a/protoBuilds/50486f-v2-part4/README.json
+++ b/protoBuilds/50486f-v2-part4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs PCR prep - PCR (part 4/4) for 96 samples using the P20 Multi-channel pipette.\n\n\n\nP20 Multi-channel Pipette\n10/20uL Opentrons tipracks\nBio-Rad Hard Shell 96-well PCR plate 200ul #hsp9601\nLabcon 0.2mL PCR strips #3940-550\nLabcon Plate\nPCR Cooler\n\n\n\nSlot 1: 96-well Labcon plate on PCR cooler\nSlot 2: Pre-Chilled PCR Cooler with PCR strip placed in column 1 and filled with master mix\nSlot 3: Aluminum Block Adapter with 96-well PCR plate filled with DNA samples (from EXO step 3/4)\nSlot 4: 96-well PCR plate filled with primers\nSlots 5-11: 10/20ul Opentrons tiprack",
"internal": "50486f-v2-part4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"description": "This protocol performs PCR prep - PCR (part 4/4) for 96 samples using the P20 Multi-channel pipette.\n\n---\n\n\n* [P20 Multi-channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [10/20uL Opentrons tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Bio-Rad Hard Shell 96-well PCR plate 200ul #hsp9601](bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [Labcon 0.2mL PCR strips #3940-550](http://www.labcon.com/microstrips.html)\n* Labcon Plate\n* PCR Cooler\n\n\n---\n\n\nSlot 1: 96-well Labcon plate on PCR cooler\n\nSlot 2: Pre-Chilled PCR Cooler with PCR strip placed in column 1 and filled with master mix\n\nSlot 3: [Aluminum Block Adapter](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) with 96-well PCR plate filled with DNA samples (from EXO step 3/4)\n\nSlot 4: 96-well PCR plate filled with primers\n\nSlots 5-11: [10/20ul Opentrons tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n\n",
"internal": "50486f-v2-part4\n",
diff --git a/protoBuilds/50486f-v2-part5/README.json b/protoBuilds/50486f-v2-part5/README.json
index 9eba878814..e52a92094e 100644
--- a/protoBuilds/50486f-v2-part5/README.json
+++ b/protoBuilds/50486f-v2-part5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol consolidates the sample in a 96-well PCR plate into a 8-well PCR strip with the option to use the same tips.\n\n\n\nP20 Multi-channel Pipette\n10/20uL Opentrons tipracks\nOpentrons aluminum block set\nBio-Rad Hard Shell 96-well PCR plate 200ul #hsp9601\nLabcon 0.2mL PCR strips #3940-550\n\n\n\nSlot 1: Pre-Chilled PCR Thermal Block with PCR strip placed in column 1 (empty)\nSlots 2-5: 96-well PCR plate with samples from previous steps.\nSlots 6-11: 10ul Opentrons tiprack",
"internal": "50486f-v2-part5",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"description": "This protocol consolidates the sample in a 96-well PCR plate into a 8-well PCR strip with the option to use the same tips.\n\n---\n\n\n* [P20 Multi-channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [10/20uL Opentrons tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons aluminum block set](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set)\n* [Bio-Rad Hard Shell 96-well PCR plate 200ul #hsp9601](bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [Labcon 0.2mL PCR strips #3940-550](http://www.labcon.com/microstrips.html)\n\n\n---\n\n\n\nSlot 1: Pre-Chilled [PCR Thermal Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) with PCR strip placed in column 1 (empty)\n\nSlots 2-5: 96-well PCR plate with samples from previous steps.\n\nSlots 6-11: [10ul Opentrons tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n\n",
"internal": "50486f-v2-part5\n",
diff --git a/protoBuilds/50c11b/README.json b/protoBuilds/50c11b/README.json
index 07a4c7ee36..fd5e619b49 100644
--- a/protoBuilds/50c11b/README.json
+++ b/protoBuilds/50c11b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "50c11b",
"labware": "\nOpentrons 20\u00b5L Filter Tips\nKingFisher 96 Deepwell Plate (containing samples)\nApplied Biosystems MicroAmp Optical 384-Well Reaction Plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "The deck should be setup as follows:\n\n\n**Tipracks**: These will be accessed in the numbered order (starting with slot 8 and ending with slot 7)\n**Sample Plates**: These can be loaded as needed and with as many samples as available, with the locations of plates being static. In the image above, Plates 1 and 2 are loaded completely while Plates 3 and 4 only have 16 samples.\n**Destination Plate**: This is the 384-well plate and should be pre-filled with the corresponding mastermix needed for the protocol.\n\n",
"description": "This protocol automates two different PCR setup protocols, transferring up to 96 samples from up to 4 different source plates to a single 384-well plate.\n\nUsing the [P20 8-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette), samples are transferred from KingFisher 96-Deepwell plates to an Applied Biosystems MicroAmp Optical 384-Well Reaction Plate (pre-filled with mastermix), mixing the samples slightly before transfer and again after transfer.\n\nTo optimize waste bin storage and reduce the need for human interference, tips used after the first plate of transfers are replaced in the empty tiprack slots from the earlier transfers (ex. tips used for Plate 2 transfers are replaced in the tiprack used for Plate 1 transfers).\n\nExplanation of complex parameters below:\n* **Protocol Type**: If set to *Covid*, 5.3\u00b5l of sample will be transferred to the destination plate and 12\u00b5L will be mixed within the well. If set to *UTI & More*, 2\u00b5l of sample will be transferred to the destination plate and 4\u00b5L will be mixed within the well.\n* **Plate 1 Number of Samples**: Specify the number of samples in Plate 1 (up to 96). The plate can be skipped by setting this value to 0 (zero).\n* **Plate 2 Number of Samples**: Specify the number of samples in Plate 2 (up to 96). The plate can be skipped by setting this value to 0 (zero).\n* **Plate 3 Number of Samples**: Specify the number of samples in Plate 3 (up to 96). The plate can be skipped by setting this value to 0 (zero).\n* **Plate 4 Number of Samples**: Specify the number of samples in Plate 4 (up to 96). The plate can be skipped by setting this value to 0 (zero).\n* **P20-Multi Mount**: Select which mount the P20-Multi Pipette is attached to.\n\n---\n\n",
diff --git a/protoBuilds/516336-part-2/README.json b/protoBuilds/516336-part-2/README.json
index 658c9bff54..fa2ea4cc53 100644
--- a/protoBuilds/516336-part-2/README.json
+++ b/protoBuilds/516336-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Oxford Nanopore Ligation Sequencing Kit"
@@ -10,7 +10,7 @@
"internal": "516336-part-2",
"labware": "\nThermoFisher 96 Well 0.8mL Polypropylene Deepwell Storage Plate\nBio-Rad 96 Well Plate 200 \u00b5L PCR\nNEST 12-Well Reservoirs, 15 mL\nOpentrons 300uL Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Oxford Nanopore Ligation Sequencing Kit\n\n",
"deck-setup": "\n\n",
"description": "This protocol automates the Adapter Ligation and Clean-Up portion for preparing sequencing libraries in the Oxford Nanopore Ligation Sequencing Kit. There is a first part of this protocol for the [DNA Repair and End-Prep](https://protocols.opentrons.com/protocol/516336).\n\nExplanation of parameters below:\n* `Number of Samples`: Specify the number of samples in multiples of 8 (Max: 96).\n* `P300 Multichannel GEN2 Mount Position`: Specify the mount position of the P300 Multichannel.\n* `P300 Single GEN2 Mount Position`: Specify the mount position of the P300 Multichannel.\n* `Magnetic Module Engage Height from Well Bottom (mm)`: Specify the height of the magnets from the bottom of the well.\n\n---\n\n",
diff --git a/protoBuilds/516336/README.json b/protoBuilds/516336/README.json
index 763b20097b..f2f7109485 100644
--- a/protoBuilds/516336/README.json
+++ b/protoBuilds/516336/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Oxford Nanopore Ligation Sequencing Kit"
@@ -10,7 +10,7 @@
"internal": "516336",
"labware": "\nThermoFisher 96 Well 0.8mL Polypropylene Deepwell Storage Plate\nBio-Rad 96 Well Plate 200 \u00b5L PCR\nNEST 12-Well Reservoirs, 15 mL\nOpentrons 300uL Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Oxford Nanopore Ligation Sequencing Kit\n\n",
"deck-setup": "\n\n",
"description": "This protocol automates the DNA Repair and End Prep portion for preparing sequencing libraries in the Oxford Nanopore Ligation Sequencing Kit. The second part of this protocol can be found here: [Adapter Ligation and Clean-Up](https://protocols.opentrons.com/protocol/516336-part-2)\n\n**Note**: The protocol was updated on September 22nd, 2022\n\n\nExplanation of parameters below:\n* `Number of Samples`: Specify the number of samples in multiples of 8 (Max: 96).\n* `P300 Multichannel GEN2 Mount Position`: Specify the mount position of the P300 Multichannel.\n* `P300 Single GEN2 Mount Position`: Specify the mount position of the P300 Multichannel.\n* `Magnetic Module Generation`: Specify whether using Gen 1 or Gen 2 magnetic module.\n* `Magnetic Module Engage Height from Well Bottom (mm)`: Specify the height of the magnets from the bottom of the well.\n\n---\n\n",
diff --git a/protoBuilds/5191b6/README.json b/protoBuilds/5191b6/README.json
index e96d2e481d..76cae8b2eb 100644
--- a/protoBuilds/5191b6/README.json
+++ b/protoBuilds/5191b6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom cherrypicked high-throughput synthesis. It requires a CSV input for the mapping of the wells and vials on the tube racks. It also requires a CSV for high-throughput synthesis which contains the dispending volumes from each reservoir.\n\n\n\nP1000 single-channel GEN2 electronic pipette\nP300 single-channel GEN2 electronic pipette\nOpentrons Tube Racks\n\n\n\n\nVial Map CSV Example\nHigh-Throughput CSV Example\n\nDeck Setup\n P1000 Tips (Slot 1)\n P300 Tips (Slot 2)\n Reagent Reservoirs (Use High-Throughput CSV)\n Opentrons Tube Racks (Use Vial Map CSV)\nExample of the Vial Map CSV Download Example:\nWell Number,Labware,Slot,Well Position\n1,opentrons_24_tuberack_nest_2ml_screwcap,4,A1\n2,opentrons_24_tuberack_nest_2ml_screwcap,4,A2\n3,opentrons_24_tuberack_nest_2ml_screwcap,4,A3\n...\n96,opentrons_24_tuberack_nest_2ml_screwcap,7,D6\nThe Vial Map CSV can be used to place custom labware and different tube racks from the labware library. It also allows mapping of the well positions of each vial on a tube rack corresponding to the well number in the high-throughput synthesis protocol.\nExample of the High-Throughput Synthesis Protocol CSV Download Example\n*Note numbering starts from top left to top right,Reservoir 1,Reservoir 2,Reservoir 3,Reservoir 4,Reservoir 5,Reservoir 6,Reservoir 7,Reservoir 8,Reservoir 9,Reservoir 10,Reservoir 11,Reservoir 12,Reservoir 13,Reservoir 14,Reservoir 15,\nLabware,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,\nSlot,3,3,3,3,3,3,8,8,8,8,8,9,9,9,9,\nWell Position,A1,B1,C1,A2,B2,C2,A3,B3,C3,A4,B4,C4,A5,B5,C5,\nWell Number,Metal Salt Stock Soln 1 (1 M) (uL),Metal Salt Stock Soln 2 (1 M) (uL),Metal Salt Stock Soln 3 (1 M) (uL),Metal Salt Stock Soln 4 (1 M) (uL),Organic Linker Stock Soln 1 (1 M) (uL),Organic Linker Stock Soln 2 (1 M) (uL),Organic Linker Stock Soln 3 (1 M) (uL),Organic Linker Stock Soln 4 (1M) (uL),Extra Solvent 1 (uL),Extra Solvent 2 (uL),Extra Solvent 3 (uL),Extra Solvent 4 (uL),,,,Total Volume (uL)\n1,270,270,270,150,150,150,150,150,64,64,150,150,270,150,270,1088\n2,27,270,270,150,150,150,150,150,48,48,150,150,27,150,27,813\n3,34,270,270,150,150,150,150,150,48,48,150,150,34,150,34,821",
"internal": "5191b6",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n\n",
"description": "This protocol performs a custom cherrypicked high-throughput synthesis. It requires a CSV input for the mapping of the wells and vials on the tube racks. It also requires a CSV for high-throughput synthesis which contains the dispending volumes from each reservoir.\n\n---\n\n\n\n* [P1000 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [P300 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons Tube Racks](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n\n---\n\n\n* [Vial Map CSV Example](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/5191b6/vial_map.csv)\n* [High-Throughput CSV Example](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/5191b6/htp_multi_rack.csv)\n\nDeck Setup\n* P1000 Tips (Slot 1)\n* P300 Tips (Slot 2)\n* Reagent Reservoirs (Use High-Throughput CSV)\n* Opentrons Tube Racks (Use Vial Map CSV)\n\nExample of the Vial Map CSV [Download Example](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/5191b6/vial_map.csv):\n\n```\nWell Number,Labware,Slot,Well Position\n1,opentrons_24_tuberack_nest_2ml_screwcap,4,A1\n2,opentrons_24_tuberack_nest_2ml_screwcap,4,A2\n3,opentrons_24_tuberack_nest_2ml_screwcap,4,A3\n...\n96,opentrons_24_tuberack_nest_2ml_screwcap,7,D6\n```\n\nThe Vial Map CSV can be used to place custom labware and different tube racks from the labware library. It also allows mapping of the well positions of each vial on a tube rack corresponding to the well number in the high-throughput synthesis protocol.\n\nExample of the High-Throughput Synthesis Protocol CSV [Download Example](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/5191b6/htp_multi_rack.csv)\n\n```\n*Note numbering starts from top left to top right,Reservoir 1,Reservoir 2,Reservoir 3,Reservoir 4,Reservoir 5,Reservoir 6,Reservoir 7,Reservoir 8,Reservoir 9,Reservoir 10,Reservoir 11,Reservoir 12,Reservoir 13,Reservoir 14,Reservoir 15,\nLabware,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,rssrack_15_tuberack_2500ul,\nSlot,3,3,3,3,3,3,8,8,8,8,8,9,9,9,9,\nWell Position,A1,B1,C1,A2,B2,C2,A3,B3,C3,A4,B4,C4,A5,B5,C5,\nWell Number,Metal Salt Stock Soln 1 (1 M) (uL),Metal Salt Stock Soln 2 (1 M) (uL),Metal Salt Stock Soln 3 (1 M) (uL),Metal Salt Stock Soln 4 (1 M) (uL),Organic Linker Stock Soln 1 (1 M) (uL),Organic Linker Stock Soln 2 (1 M) (uL),Organic Linker Stock Soln 3 (1 M) (uL),Organic Linker Stock Soln 4 (1M) (uL),Extra Solvent 1 (uL),Extra Solvent 2 (uL),Extra Solvent 3 (uL),Extra Solvent 4 (uL),,,,Total Volume (uL)\n1,270,270,270,150,150,150,150,150,64,64,150,150,270,150,270,1088\n2,27,270,270,150,150,150,150,150,48,48,150,150,27,150,27,813\n3,34,270,270,150,150,150,150,150,48,48,150,150,34,150,34,821\n```\n\n",
"internal": "5191b6\n",
diff --git a/protoBuilds/51b9a5/README.json b/protoBuilds/51b9a5/README.json
index 340a6f8c12..5ed9b6bfc4 100644
--- a/protoBuilds/51b9a5/README.json
+++ b/protoBuilds/51b9a5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Agar Plating"
@@ -10,7 +10,7 @@
"internal": "51b9a5",
"labware": "\nOpentrons 20\u00b5L Tips\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nNunc rectangular agar plates\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Agar Plating\n\n",
"deck-setup": "* slot 1: 20\u00b5l tiprack\n* slot 2: source plate\n* slots 3-11: agar plates\n\n---\n\n",
"description": "\nThis protocol transfers up to 96 samples to up to 9 different source plates. Samples are transferred to their corresponding positions in the agar plates. Tips are changed between transfers of different samples to avoid contamination. The user has the option to puncture the agar plate before dispensing the sample on the agar.\n\n---\n\n",
diff --git a/protoBuilds/51df08/README.json b/protoBuilds/51df08/README.json
index 360dc8c095..74b3bb77c1 100644
--- a/protoBuilds/51df08/README.json
+++ b/protoBuilds/51df08/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Assay"
@@ -8,7 +8,7 @@
"description": "This protocol performs a mass spec sample prep on up to 10 samples with up to 5 antibodies on up to 2 custom 72-sample slide mounts (the top row of 9 wells are left blank). Samples and antibodies should be loaded in their respective tuberacks down the columns and then across the rows. Samples and antibodies are filled in the slides down the first column of each slide, then up the second, and finally down the third column. For more detailed sample and reagent setup, please see below.\n\n\n\ncustom 3-grid 3x8 slide mount\nOpentrons 4x6 tuberack insert with 1.5ml Eppendorf snapcap tubes\nOpentrons 2x3 tuberack insert with 50ml Falcon tubes\nOpentrons 20ul pipette tips\nOpentrons 300ul pipette tips\nOpentrons P20- and P300-single GEN2 electronic pipettes\n\n\n\n2x3 50ml tuberack (slot 1)\n detergent wash buffer: tube A1\n BSA blocking buffer: tube B1\n PBS: tubes A2-B2\n water: tubes A3-B3",
"internal": "51df08",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n * Assay\n\n",
"description": "This protocol performs a mass spec sample prep on up to 10 samples with up to 5 antibodies on up to 2 custom 72-sample slide mounts (the top row of 9 wells are left blank). Samples and antibodies should be loaded in their respective tuberacks down the columns and then across the rows. Samples and antibodies are filled in the slides down the first column of each slide, then up the second, and finally down the third column. For more detailed sample and reagent setup, please see below.\n\n---\n\n\n* custom 3-grid 3x8 slide mount\n* [Opentrons 4x6 tuberack insert](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 1.5ml Eppendorf snapcap tubes\n* [Opentrons 2x3 tuberack insert](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 50ml Falcon tubes\n* [Opentrons 20ul pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 300ul pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips?variant=15954632802398)\n* [Opentrons P20- and P300-single GEN2 electronic pipettes](https://shop.opentrons.com/collections/ot-2-pipettes)\n\n---\n\n\n2x3 50ml tuberack (slot 1)\n* detergent wash buffer: tube A1\n* BSA blocking buffer: tube B1\n* PBS: tubes A2-B2\n* water: tubes A3-B3\n\n",
"internal": "51df08\n",
diff --git a/protoBuilds/528c16/README.json b/protoBuilds/528c16/README.json
index 0168eec795..432f982dd5 100644
--- a/protoBuilds/528c16/README.json
+++ b/protoBuilds/528c16/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "528c16",
"labware": "\nOpentrons 4-in-1 tube rack with 50mL & 15mL Falcon tubes\nOpentrons 4-in-1 tube rack with 1.5mL Eppendorf snap cap tubes\nOpentrons 1000ul tips\nOpentrons 20ul tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This protocol preps 1.5mL and 2mL tubes from stock solutions including but not limited to acetonitrile, water, and methanol. Sources and destinations are determined via a csv uploaded by the user, as well as transfer volumes.\n\n\nExplanation of complex parameters below:\n* `csv file`: The csv file should be formatted like so. Note - for no mix steps, input \"0\" for the mix repetition. Also specify whether to mix at the source, or destination tube for that row:\n\n\n* Note: for aspiration height percent (column J), a value of 10 means that we will be aspirating from 10% of the tube depth, 50 will be 50% of the tube depth, etc.\n* `P20/P1000 Mount`: Specify which mount (left or right) for each single channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/52d238/README.json b/protoBuilds/52d238/README.json
index 25a28a0e05..69bbb1a43b 100644
--- a/protoBuilds/52d238/README.json
+++ b/protoBuilds/52d238/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEB Ultra II FS DNA Library Prep"
@@ -8,7 +8,7 @@
"description": "This protocol automates the PCR, Bead Clean Up and Elution steps of the NEB Ultra II FS DNA Library Prep\n\n\n\nP20-multi channel GEN2 electronic pipette\nP300-multi channel GEN2 electronic pipette\nOpentrons Thermocycler Module\nOpentrons Temperature Module\nOpentrons Magnetic Module\nNEST 100uL PCR Plate\nOpentrons 96 Well Aluminum Block with 200 uL PCR Strips\n\n\n\n\nThermocycler (Slot 7)\nTemperature Module (Slot 4)\nMagnetic Module (Slot 9)\nPlate with Index-Primers (Slot 5)\nOpentrons 200 uL Filter Tips (Slot 2, 3)\nOpentrons 20 uL Filter Tips (Slots 1)\n\nInitial Starting State:\n- Samples with beads in a 96 well PCR plate in the thermocycler.\n- EB Tween is in column 1 on the 96 Well Aluminum block on the temperature module.\n- Reservoir channel A1 contains ethanol\n- Plate on the magnetic module contains pre-aliquoted magnetic beads\n- Plate with Index-Primers starts in a 96 well plate on Position 5 of deck",
"internal": "52d238",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEB Ultra II FS DNA Library Prep\n\n\n",
"description": "This protocol automates the PCR, Bead Clean Up and Elution steps of the [NEB Ultra II FS DNA Library Prep](https://www.neb.com/products/e7805-nebnext-ultra-ii-fs-dna-library-prep-kit-for-illumina#Protocols,%20Manuals%20&%20Usage)\n\n\n---\n\n\n* [P20-multi channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [P300-multi channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons Thermocycler Module](https://opentrons.com/modules/thermocycler-module/)\n* [Opentrons Temperature Module](https://opentrons.com/modules/temperature-module/)\n* [Opentrons Magnetic Module](https://opentrons.com/modules/temperature-module/)\n* [NEST 100uL PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons 96 Well Aluminum Block with 200 uL PCR Strips](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n\n---\n\n\n* Thermocycler (Slot 7)\n* Temperature Module (Slot 4)\n* Magnetic Module (Slot 9)\n* Plate with Index-Primers (Slot 5)\n* Opentrons 200 uL Filter Tips (Slot 2, 3)\n* Opentrons 20 uL Filter Tips (Slots 1)\n\nInitial Starting State:\n- Samples with beads in a 96 well PCR plate in the thermocycler.\n- EB Tween is in column 1 on the 96 Well Aluminum block on the temperature module.\n- Reservoir channel A1 contains ethanol\n- Plate on the magnetic module contains pre-aliquoted magnetic beads\n- Plate with Index-Primers starts in a 96 well plate on Position 5 of deck\n\n\n\n",
"internal": "52d238",
diff --git a/protoBuilds/53134e/README.json b/protoBuilds/53134e/README.json
index 2816b27e15..632870902f 100644
--- a/protoBuilds/53134e/README.json
+++ b/protoBuilds/53134e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Aliquoting"
@@ -8,7 +8,7 @@
"description": "This protocol performs sample aliquoting from a single-channel reservoir to up to 24 custom cryotubes. The sample order is down columns and then across rows (A1, B1, C1, D1, A2, B2, etc.)\n\n\n\nNEST 1 Well Reservoir 195 mL\nOpentrons P1000 GEN2 single-channel electronic pipette\nOpentrons 96 Tip Rack 1000 \u00b5L\n",
"internal": "53134e",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Aliquoting\n\n",
"description": "This protocol performs sample aliquoting from a single-channel reservoir to up to 24 custom cryotubes. The sample order is down columns and then across rows (A1, B1, C1, D1, A2, B2, etc.)\n\n---\n\n\n* [NEST 1 Well Reservoir 195 mL](https://labware.opentrons.com/nest_1_reservoir_195ml)\n* [Opentrons P1000 GEN2 single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette?variant=5984549142557)\n* [Opentrons 96 Tip Rack 1000 \u00b5L](https://labware.opentrons.com/opentrons_96_tiprack_1000ul)\n\n",
"internal": "53134e\n",
diff --git a/protoBuilds/53e6bc_mastermix_creation/README.json b/protoBuilds/53e6bc_mastermix_creation/README.json
index f28c0c6311..ddcef5caf6 100644
--- a/protoBuilds/53e6bc_mastermix_creation/README.json
+++ b/protoBuilds/53e6bc_mastermix_creation/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Assay"
@@ -8,7 +8,7 @@
"description": "This protocol performs custom mastermix creation from antibodies. The volumes of each antibody per sample to be transferred from a custom deepwell plate to the mastermix tube are specfied in a .csv input file, formatted as follows:\nVolume per antibody(in ul),Antibody,Well\n2.2,CD3 BUV395,A1\n1.1,CD45 BUV496,A3\n1.1,CD15 BUV563,A5\n1.1,CD45RA BUV615,A7\n2.2,CD14 BUV661,A9\n0.55,CD8 BUV737,A11\n2.2,CD11c BUV805,B2\n1.1,CD25 BV421,B4\n0.55,CD4 BV480,B6\n2.2,CD16 BV605,B8\n2.2,CD123 BV650,B10\n2.2,CD127 BV711,B12\n2.2,IgD BV750,C1\n2.2,CD304 BV786,C3\n2.2,CD141 BB515,C5\nNote that the headers line should be included, and empty lines are ignored.\nLinks:\n* Flow Cytometry Staining\n\n\n\nVWR 96-deepwell plate 2.2ml #10755-250\nVWR conical self-standing tubes 5ml #89497-740 (or equivalent) seated in Opentrons 4-in-1 tuberack with 3x5 insert\nVWR screwcap microcentrifuge tubes 1.5ml # (or equivalent) seated in Opentrons 4-in-1 tuberack with 4x6 insert\nOpentrons P300 GEN1 single-channel pipette\nOpentrons P10 GEN1 single-channel pipette\nOpentrons 300ul tiprack\nOpentrons 10ul tiprack\n\n\n\n3x5 tuberack for 5ml screwcap tubes (slot 2)\n tube A1: mastermix (loaded empty)\n tube B1: PBS\n4x6 tuberack for 1.5ml screwcap tubes (slot 4)\n* tube A1: Brilliant Stain Buffer",
"internal": "53e6bc",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Assay\n\n",
"description": "This protocol performs custom mastermix creation from antibodies. The volumes of each antibody per sample to be transferred from a custom deepwell plate to the mastermix tube are specfied in a `.csv` input file, formatted as follows:\n\n```\nVolume per antibody(in ul),Antibody,Well\n2.2,CD3 BUV395,A1\n1.1,CD45 BUV496,A3\n1.1,CD15 BUV563,A5\n1.1,CD45RA BUV615,A7\n2.2,CD14 BUV661,A9\n0.55,CD8 BUV737,A11\n2.2,CD11c BUV805,B2\n1.1,CD25 BV421,B4\n0.55,CD4 BV480,B6\n2.2,CD16 BV605,B8\n2.2,CD123 BV650,B10\n2.2,CD127 BV711,B12\n2.2,IgD BV750,C1\n2.2,CD304 BV786,C3\n2.2,CD141 BB515,C5\n```\n\n**Note that the headers line should be included, and empty lines are ignored.**\n\nLinks:\n* [Flow Cytometry Staining](./53e6bc_flow_cytometry_staining)\n\n---\n\n\n* [VWR 96-deepwell plate 2.2ml #10755-250](https://us.vwr.com/store/product/4693284/vwr-96-well-deep-well-plates)\n* [VWR conical self-standing tubes 5ml #89497-740](https://us.vwr.com/store/product/11707931/self-standing-sample-tubes-5-and-10-ml-globe-scientific) (or equivalent) seated in [Opentrons 4-in-1 tuberack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 3x5 insert\n* [VWR screwcap microcentrifuge tubes 1.5ml #](https://us.vwr.com/store/product/4674084/vwr-microcentrifuge-tubes-with-socket-screw-cap) (or equivalent) seated in [Opentrons 4-in-1 tuberack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 4x6 insert\n* [Opentrons P300 GEN1 single-channel pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette?variant=5984549109789)\n* [Opentrons P10 GEN1 single-channel pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette?variant=31059478970462)\n* [Opentrons 300ul tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 10ul tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\n---\n\n\n3x5 tuberack for 5ml screwcap tubes (slot 2)\n* tube A1: mastermix (loaded empty)\n* tube B1: PBS\n\n4x6 tuberack for 1.5ml screwcap tubes (slot 4)\n* tube A1: Brilliant Stain Buffer\n\n",
"internal": "53e6bc\n",
diff --git a/protoBuilds/53fec2/README.json b/protoBuilds/53fec2/README.json
index d6592ea9b0..e538cbac64 100644
--- a/protoBuilds/53fec2/README.json
+++ b/protoBuilds/53fec2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"Cleanup"
@@ -10,7 +10,7 @@
"internal": "53fec2",
"labware": "\nEppendorf 96-deepwell plate 500\u00b5l\nEppendorf 96-well PCR plate 200\u00b5l, skirted\nEppendorf 1-channel automation reservoirs 30ml/100ml\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * Cleanup\n\n",
"deck-setup": "\n\n\n",
"description": "This is a flexible protocol accommodating Beckman Coulter AMPure XP PCR Cleanup and Size Selection. It consists of the following major sections:\n* binding beads pre-addition\n* sample pooling and mixing with binding beads\n* 2x ethanol wash\n* final elution in H2O\n\nSamples to be pooled should be loaded in 2 PCR plates on the deck. For reagent layout used in this protocol, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 7) to the elution PCR plate (slot 8). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\nExplanation of complex parameters below:\n* `park tips`: If set to `yes` (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to `no`, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n\n---\n\n",
diff --git a/protoBuilds/543bf9/README.json b/protoBuilds/543bf9/README.json
index 4de7e6b01e..64a616d0e2 100644
--- a/protoBuilds/543bf9/README.json
+++ b/protoBuilds/543bf9/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Serial Dilution"
@@ -8,7 +8,7 @@
"description": "This protocol performs standard curve serial dilutions using data from a CSV file. It has the option to perform up to 2 standard curves per run and the ability to modify stock concentrations.\n\nNote: The layout above depicts a full plate with 2 standard curves.\n\n\n\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt\nOpentrons 300uL Tips\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nDeck Setup\n\nCSV File Information\nThe protocol will require a CSV file to be uploaded in order to properly function. Please use the following Serial Dilutions Excel Spreadsheet Template (.XLSX Format) to update your values. Once the values are updated click Save As and then save it in .CSV format. You can then upload that CSV to the protocol for your run.\nProtocol Steps\n\nTransfer 44.5 uL of Diluent into Int1 (Well A1).\nTransfer 5.5 uL from Stock1 into Int1\nMix Int1 3 times, perform blow out, touch tip, then discard tip.\n\nNote: This process will repeat as it iterates through the rows in the CSV and add diluent first, then the sample from the previous well or specified well.",
"internal": "543bf9",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Serial Dilution\n\n",
"description": "This protocol performs standard curve serial dilutions using data from a CSV file. It has the option to perform up to 2 standard curves per run and the ability to modify stock concentrations.\n\n\n**Note**: The layout above depicts a full plate with 2 standard curves.\n\n---\n\n\n* [Opentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [NEST 96 Well Plate 100 \u00b5L PCR Full Skirt](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons 300uL Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n---\n\n\n**Deck Setup**\n\n\n\n**CSV File Information**\n\nThe protocol will require a CSV file to be uploaded in order to properly function. Please use the following [Serial Dilutions Excel Spreadsheet Template (.XLSX Format)](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/543bf9/serial_dilutions_template.xlsx) to update your values. Once the values are updated click `Save As` and then save it in `.CSV` format. You can then upload that CSV to the protocol for your run.\n\n**Protocol Steps**\n\n1. Transfer 44.5 uL of Diluent into Int1 (Well A1).\n2. Transfer 5.5 uL from Stock1 into Int1\n3. Mix Int1 3 times, perform blow out, touch tip, then discard tip.\n\n**Note**: This process will repeat as it iterates through the rows in the CSV and add diluent first, then the sample from the previous well or specified well.\n\n",
"internal": "543bf9",
diff --git a/protoBuilds/5520f0/README.json b/protoBuilds/5520f0/README.json
index 9b699dff4f..bcd35b8bb2 100644
--- a/protoBuilds/5520f0/README.json
+++ b/protoBuilds/5520f0/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Staining"
@@ -10,7 +10,7 @@
"internal": "5520f0",
"labware": "\nPerkin Elmer Cell Carrier Ultra 384 well plates\nNEST 12-Well Reservoirs, 15 mL\nOpentrons 300uL Tips\n",
"markdown": {
- "author": "\n[Opentrons](https://opentrons.com/)\n\n",
+ "author": "\n[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "\n* Sample Prep\n * Staining\n\n",
"deck-setup": "\n\n\n",
"description": "\nThis protocol automates the staining of cell-based 384 well assay plates. It performs various washes with PBS, 4% PFA, Perm Buffer, including Primary and Secondary Antibody solutions. The protocol utilizes various reservoirs and keeps track of reagent and waste volumes.\n\nExplanation of parameters below:\n\n- `P300 Multi GEN2 Mount`: Specify which mount (left or right) to load the P300 multi channel pipette.\n- `Number of Plates`: Total number of plates being run (Max: 6). Please follow the placement order in the deck layout below.\n- `New Tip per Plate`: Everytime a plate is completed, you can either use a new tip (Always) or continue using the same tip (Never).\n- `Dispense Flow Rate`: The flow rate in uL/s as liquid is dispensed throughout the protocol. The default dispense rate of a P300 Multichannel is 94 uL/s.\n- `Aspiration Height from Well Bottom`: The dispensing height from the bottom of the well in (mm). A value of 0.5 means, 0.5 mm from the very bottom of the well.\n\n---\n\n",
diff --git a/protoBuilds/558d02/README.json b/protoBuilds/558d02/README.json
index 67215e99be..8ca067c76e 100644
--- a/protoBuilds/558d02/README.json
+++ b/protoBuilds/558d02/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Purification"
@@ -8,7 +8,7 @@
"description": "This protocol performs a magbead-based peptide enrichment using the Opentrons magnetic module.\n\n\n\nUSA Scientific 96-deepwell plate 2.4ml #1896-2000\nThermo Scientific 96-well plate 300ul #AB0600 mounted in empty Opentrons tip rack\nNest 12-channel reservoir 15ml #360102\nNest 1-channel reservoir 195ml #360103\nOpentrons magnetic module\nOpentrons P300 GEN2 single-channel pipette\nOpentrons 300ul tipracks\n",
"internal": "558d02",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Purification\n\n",
"description": "This protocol performs a magbead-based peptide enrichment using the Opentrons magnetic module.\n\n---\n\n\n* [USA Scientific 96-deepwell plate 2.4ml #1896-2000](https://labware.opentrons.com/usascientific_96_wellplate_2.4ml_deep?category=wellPlate)\n* [Thermo Scientific 96-well plate 300ul #AB0600](https://www.fishersci.com/shop/products/thermo-scientific-96-well-non-skirted-plates-17/ab0600?crossRef=410088#?keyword=410088) mounted in empty Opentrons tip rack\n* [Nest 12-channel reservoir 15ml #360102](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* [Nest 1-channel reservoir 195ml #360103](https://labware.opentrons.com/nest_1_reservoir_195ml?category=reservoir)\n* [Opentrons magnetic module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P300 GEN2 single-channel pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons 300ul tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n",
"internal": "558d02\n",
diff --git a/protoBuilds/559aa0/README.json b/protoBuilds/559aa0/README.json
index d222399645..852496fe0a 100644
--- a/protoBuilds/559aa0/README.json
+++ b/protoBuilds/559aa0/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"qPCR setup"
@@ -8,7 +8,7 @@
"description": "This protocol automates the distribution of 4 different primers to the wells of up to nine 384-well plates in a specific arrangement shown in the attached plate map. The nine plates can be fully or partially filled (each plate filled to a different extent, as specified by choice of parameter values below prior to download of the protocol).\nLinks:\n* PCR/qPCR prep: distribution of patient samples to 384-well plates\n\nplate map\n\n\n\n\nOpentrons P20-multi channel electronic pipettes (GEN2)\n\n\nOpentrons 20ul Filter Tips\n\n",
"internal": "559aa0",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * qPCR setup\n\n",
"description": "This protocol automates the distribution of 4 different primers to the wells of up to nine 384-well plates in a specific arrangement shown in the attached plate map. The nine plates can be fully or partially filled (each plate filled to a different extent, as specified by choice of parameter values below prior to download of the protocol).\n\nLinks:\n* [PCR/qPCR prep: distribution of patient samples to 384-well plates](http://protocols.opentrons.com/protocol/165a77)\n\n* [plate map](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-03-08/e373l2s/384%20plate%20map.png)\n\n\n\n* [Opentrons P20-multi channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n\n* [Opentrons 20ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n\n\n",
"internal": "559aa0\n",
diff --git a/protoBuilds/563624/README.json b/protoBuilds/563624/README.json
index e5c23bf9c3..5f88ce8341 100644
--- a/protoBuilds/563624/README.json
+++ b/protoBuilds/563624/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Purification"
@@ -9,7 +9,7 @@
"internal": "563624",
"labware": "\nPhytips 300ul resin tipracks #93-05-01\nVWR 96-deepwell plate 500ul #76210-518\nEppendorf twin.tec 96-well PCR plate 150ul #951020401\nNEST 96 Deepwell Plate 2mL\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n\t* Purification\n\n",
"description": "This protocol performs resin Phytip preparation with 5 different buffers and samples for up to 12 samples.\n\n---\n\n",
"internal": "563624\n",
diff --git a/protoBuilds/5654c0/README.json b/protoBuilds/5654c0/README.json
index adfe593253..502a40091e 100644
--- a/protoBuilds/5654c0/README.json
+++ b/protoBuilds/5654c0/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -9,7 +9,7 @@
"description": "This protocol performs a custom sample normalization from a source PCR plate to a second PCR plate and a third norm plate. Transfer take place from multiple locations specified below based on the volumes described in the example CSV below:\nPlate LC (Source),Source Well #,ID ,concentration (ug/ml),yield (ug),Destination 1 volume (ul),Destination 1 (Dil),Destination 1 (Dil Plate) Well,concentration dil (ug/ml),yield (ug),Destination 2 volume (ul),Destination 2 (final norm plate),Destination 2 (well #)\nLC 96-well plate,A1,pET4286,3000.00,6.00,10,LC Dil Plate,A1,300.00,6.00,20.00,Norm Plate,A1\nLC 96-well plate,G3,pET4016,3000,6.00,10,LC Dil Plate,G3,300.00,6.00,20.00,Norm Plate,G3\nLC 96-well plate,H3,pET4017,3300,6.00,10,LC Dil Plate,H3,330.00,6.00,18.18,Norm Plate,H3\nHC 96-well plate,F3,pET4308,3000,6.00,10,HC Dil Plate,F3,300,6.00,20.00,Norm Plate,F3\nHC 96-well plate,G3,pET4308,3000,6.00,10,HC Dil Plate,G3,300,6.00,20.00,Norm Plate,G3\nHC 96-well plate,H3,pET4308,3000,6.00,10,HC Dil Plate,H3,300,6.00,20.00,Norm Plate,H3\nwater,F3,,,,,,,,,81.82,Norm Plate,F3\nwater,G3,,,,,,,,,80.00,Norm Plate,G3\nwater,H3,,,,,,,,,81.82,Norm Plate,H3\nNote: Save any .xlsx files as a .csv",
"internal": "5654c0",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"deck-setup": "* Opentrons 300 uL Tip Rack (Slot 1, 2)\n* Opentrons 20 uL Tip Rack (Slot 3, 4)\n* LC 96-well plate (Slot 5)\n* HC 96-well plate (Slot 6)\n* Water (Slot 7)\n* LC Dil Plate (Slot 8)\n* HC Dil Plate (Slot 9)\n* Norm Plate (Slot 10)\n\n",
"description": "This protocol performs a custom sample normalization from a source PCR plate to a second PCR plate and a third norm plate. Transfer take place from multiple locations specified below based on the volumes described in the example CSV below:\n\n```\nPlate LC (Source),Source Well #,ID ,concentration (ug/ml),yield (ug),Destination 1 volume (ul),Destination 1 (Dil),Destination 1 (Dil Plate) Well,concentration dil (ug/ml),yield (ug),Destination 2 volume (ul),Destination 2 (final norm plate),Destination 2 (well #)\nLC 96-well plate,A1,pET4286,3000.00,6.00,10,LC Dil Plate,A1,300.00,6.00,20.00,Norm Plate,A1\nLC 96-well plate,G3,pET4016,3000,6.00,10,LC Dil Plate,G3,300.00,6.00,20.00,Norm Plate,G3\nLC 96-well plate,H3,pET4017,3300,6.00,10,LC Dil Plate,H3,330.00,6.00,18.18,Norm Plate,H3\nHC 96-well plate,F3,pET4308,3000,6.00,10,HC Dil Plate,F3,300,6.00,20.00,Norm Plate,F3\nHC 96-well plate,G3,pET4308,3000,6.00,10,HC Dil Plate,G3,300,6.00,20.00,Norm Plate,G3\nHC 96-well plate,H3,pET4308,3000,6.00,10,HC Dil Plate,H3,300,6.00,20.00,Norm Plate,H3\nwater,F3,,,,,,,,,81.82,Norm Plate,F3\nwater,G3,,,,,,,,,80.00,Norm Plate,G3\nwater,H3,,,,,,,,,81.82,Norm Plate,H3\n```\n\n**Note: Save any .xlsx files as a .csv**\n\n",
diff --git a/protoBuilds/5689f5/README.json b/protoBuilds/5689f5/README.json
index a9351f1cbc..c6ffb215cc 100644
--- a/protoBuilds/5689f5/README.json
+++ b/protoBuilds/5689f5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Clean Up"
@@ -10,7 +10,7 @@
"internal": "5689f5",
"labware": "\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Clean Up\n\n\n",
"deck-setup": " \n* green on magnetic plate (slot 4): starting samples\n* blue on reagent plate (slot 2, column 1): DNA binding buffer\n* pink on reagent plate (slot 2, column 2): Ampure beads\n* orange on reagent plate (slot 2, column 3): elution buffer\n* purple on reagent reservoir (slot 5, column 1): 80% ethanol\n* dark blue on reagent reservoir (slot 5, column 12): waste (loaded empty)\n\n\n",
"description": "This protocol performs an NGS clean up protocol for up to 96 samples. The user is prompted to manually centrifuge the plate and replace on the magnetic module after 2x ethanol washes.\n\n\n",
diff --git a/protoBuilds/56a6a1/README.json b/protoBuilds/56a6a1/README.json
index 02bb2e62a2..7074188a70 100644
--- a/protoBuilds/56a6a1/README.json
+++ b/protoBuilds/56a6a1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins & Proteomics": [
"Assay"
@@ -8,7 +8,7 @@
"description": "This protocol performs a protein array on ONCYTE SuperNOVA slides mounted on an Inheco Teleshake 1536 using a P300 multi-channel pipette. For reagent setup in the 12-channel reservoir, see 'Additional Notes' below. Note that the user is prompted three times to engage and disengage the shaker (mounted on the deck). The Teleshake with slides will occupy slots 1, 2, 4, and 5, and will be calibrated from slot 4.\n\n\n\nGrace ONCYTE SuperNOVA 16-well slides #705116\nCustom 4-slide adapter for Inheco Teleshake 1536 (each slide with 8x2 square wells)\nBio-Rad Hard Shell 96-Well PCR plate 200ul #hsp9601 (if automatically adding samples)\nAgilent 1-Well Reservoir 290mL #201252-100\nUSA Scientific 12-Well Reservoir 22mL #1061-8150\nOpentrons P300 multi-channel electronic pipette\nOpentrons 300ul pipette tips\nInheco Teleshake 1536\n\n\n\n12-channel reservoir\n Super G plus blocking buffer: channel 1\n Secondary antibody: channel 2\n PBST: channels 3-6\n Super G: channel 7",
"internal": "56a6a1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins & Proteomics\n * Assay\n\n",
"description": "This protocol performs a protein array on ONCYTE SuperNOVA slides mounted on an Inheco Teleshake 1536 using a P300 multi-channel pipette. For reagent setup in the 12-channel reservoir, see 'Additional Notes' below. Note that the user is prompted three times to engage and disengage the shaker (mounted on the deck). The Teleshake with slides will occupy slots 1, 2, 4, and 5, and will be calibrated from slot 4.\n\n---\n\n\n* [Grace ONCYTE SuperNOVA 16-well slides #705116](https://gracebio.com/product/oncyte-supernova-705116/)\n* Custom 4-slide adapter for Inheco Teleshake 1536 (each slide with 8x2 square wells)\n* [Bio-Rad Hard Shell 96-Well PCR plate 200ul #hsp9601 (if automatically adding samples)](http://www.bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [Agilent 1-Well Reservoir 290mL #201252-100](https://www.agilent.com/store/en_US/Prod-201252-100/201252-100)\n* [USA Scientific 12-Well Reservoir 22mL #1061-8150](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n* [Opentrons P300 multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 300ul pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips?variant=15954632802398)\n* [Inheco Teleshake 1536](https://www.inheco.com/fileadmin/web_data/Downloads/Data-Sheets/Shaker.pdf)\n\n---\n\n\n12-channel reservoir\n* Super G plus blocking buffer: channel 1\n* Secondary antibody: channel 2\n* PBST: channels 3-6\n* Super G: channel 7\n\n",
"internal": "56a6a1\n",
diff --git a/protoBuilds/56b4ec/README.json b/protoBuilds/56b4ec/README.json
index bbd51de9c5..5e819194dc 100644
--- a/protoBuilds/56b4ec/README.json
+++ b/protoBuilds/56b4ec/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol performs all of the steps as outlined in the MethodA_map_labware_protocol file. This protocol requires the use of custom labware (Charles River Tube Rack) and custom tips (200\u00b5L, extended length).\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons P50 Single Pipette\nOpentrons P1000 Single Pipette\nOpentrons 1000\u00b5L Filter Tips\nOpentrons 4-in-1 Tube Rack\nCorning 96-Well Plate, 360\u00b5L\nCustom 200\u00b5L Tips\nCustom Tube Rack for 10mL Charles River Tubes\nFalcon 50mL Conical Tube\nEppendorf 2mL Tubes\n\n\n\nSlot 1: Custom 200\u00b5L Tips\nSlot 2: Opentrons 1000\u00b5L Filter Tips\nSlot 3: Corning 96-Well Plate, 360\u00b5L\nSlot 4: Empty\nSlot 5: Empty\nSlot 6: Empty\nSlot 7: Opentrons 6 Tube Rack with 50mL Tube (Rack for water)\n* A1: Water, 30mL\nSlot 8: Custom Charles River Tube Rack (Rack for sample 1 dilution)\n A1: Sample 1, 4mL\n A2: Charles River Tube, empty\n A3: Charles River Tube, empty\n A4: Charles River Tube, empty\n B1: Charles River Tube, empty\n B2: Charles River Tube, empty\n B3: Charles River Tube, empty\n B4: Charles River Tube, empty\nSlot 9: Empty\nSlot 10: Opentrons 24 Tube Rack with 2mL Tubes (Rack for RSE & NEP)\n A1: RSE, 1mL\n A2: HKSA, 500\u00b5L\nSlot 11: Custom Charles River Tube Rack (Rack for RSE & HKSA dilution)\n A1: Charles River Tube, empty\n A2: Charles River Tube, empty\n A3: Charles River Tube, empty\n A4: Charles River Tube, empty\n A5: Charles River Tube, empty\n B1: Charles River Tube, empty\n B2: Charles River Tube, empty\n B3: Charles River Tube, empty\n C1: Charles River Tube, empty\n C2: Charles River Tube, empty",
"internal": "56b4ec",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol performs all of the steps as outlined in the `MethodA_map_labware_protocol` file. This protocol requires the use of custom labware (Charles River Tube Rack) and custom tips (200\u00b5L, extended length).\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P50 Single Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons P1000 Single Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 1000\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips)\n* [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [Corning 96-Well Plate, 360\u00b5L](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate)\n* Custom 200\u00b5L Tips\n* Custom Tube Rack for 10mL Charles River Tubes\n* Falcon 50mL Conical Tube\n* Eppendorf 2mL Tubes\n\n\n---\n\n\nSlot 1: Custom 200\u00b5L Tips\n\nSlot 2: Opentrons 1000\u00b5L Filter Tips\n\nSlot 3: [Corning 96-Well Plate, 360\u00b5L](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate)\n\nSlot 4: *Empty*\n\nSlot 5: *Empty*\n\nSlot 6: *Empty*\n\nSlot 7: [Opentrons 6 Tube Rack](https://labware.opentrons.com/opentrons_6_tuberack_falcon_50ml_conical?category=tubeRack) with 50mL Tube (Rack for water)\n* A1: Water, 30mL\n\nSlot 8: Custom Charles River Tube Rack (Rack for sample 1 dilution)\n* A1: Sample 1, 4mL\n* A2: Charles River Tube, empty\n* A3: Charles River Tube, empty\n* A4: Charles River Tube, empty\n* B1: Charles River Tube, empty\n* B2: Charles River Tube, empty\n* B3: Charles River Tube, empty\n* B4: Charles River Tube, empty\n\nSlot 9: *Empty*\n\nSlot 10: [Opentrons 24 Tube Rack](https://labware.opentrons.com/opentrons_24_tuberack_eppendorf_2ml_safelock_snapcap?category=tubeRack) with 2mL Tubes (Rack for RSE & NEP)\n* A1: RSE, 1mL\n* A2: HKSA, 500\u00b5L\n\nSlot 11: Custom Charles River Tube Rack (Rack for RSE & HKSA dilution)\n* A1: Charles River Tube, empty\n* A2: Charles River Tube, empty\n* A3: Charles River Tube, empty\n* A4: Charles River Tube, empty\n* A5: Charles River Tube, empty\n* B1: Charles River Tube, empty\n* B2: Charles River Tube, empty\n* B3: Charles River Tube, empty\n* C1: Charles River Tube, empty\n* C2: Charles River Tube, empty\n\n\n",
"internal": "56b4ec\n",
diff --git a/protoBuilds/581011-pt2/README.json b/protoBuilds/581011-pt2/README.json
index b548e9d1b3..2b430342d9 100644
--- a/protoBuilds/581011-pt2/README.json
+++ b/protoBuilds/581011-pt2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -10,7 +10,7 @@
"internal": "581011",
"labware": "\nCorning 384 Well Plate 112 \u00b5L Flat\nNEST 12-Well Reservoirs, 15 mL\nOpentrons 200uL Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"deck-setup": "* Deck layout - example 384 plate populated with target cells, random column chosen for effector cells. Note: only one column of effector cells should be loaded, but any column can be loaded. Effector cells should always be put in the 96 well plate from row A down (i.e. effector cell 1 should always be A1, A2, A3, A4,..., or A12).\n\n\n\n\n---\n\n",
"description": "This protocol parses an uploaded .csv file (see below) and determines the number of tips to pick up with a multi-channel pipette. The pipette transfers the specified volume of effector cell to a 384 well-plate to the parsed wells by multi-dispensing as much as the pipette tip can accommodate before returning to the reservoir. The pipette keeps the same tip for the duration of the run.\n\nNote: Part 1 and Part 2 should receive the exact same .csv file.\n\nFind part one of the protocol (target cell loading) below:\n\n[Part 1 - Target Cell Loading](https://protocols.opentrons.com/protocol/581011)\n\nExplanation of complex parameters below:\n* `.CSV File`: Upload the .csv file according to the template shown [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/581011/co-culture_template_protocol.numbers). Any well to the left of a forward slash (`/`) will correspond to the well in the reservoir containing the desired target cell. Any well to the right of a forward slash (`/`) will correspond to the effector cell in the 96 well plate for part 2 of this protocol. If a cell is only to receive one of the effector or target cells, place a `0` (e.g. A1/0 would mean target cell from A1 of the reservoir with no effector cell). All cells that are to be skipped (receive neither effector nor target cell) should have a lowercase `'x'` in the resepective cell in the .csv file. For the parameters below, make sure there are NO commas inside of the wells, and just the values. For the mount, put nothing else but \"left\" or \"right\". For the pre-mix variable, you must start the well with \"Yes\" or \"No\", but can put anything else after that (e.g. \"Yes premix\", or \"No premix\").\n* `Transfer Volume Target Cell`: Specify the volume (in ul) to transfer between target cells in reservoir to well plate.\n* `Pre-mix before aspirating in source reservoir?`: Specify whether or not to pre-mix before each transfer of target cell in reservoir to plate.\n* `Pre-mix height in reservoir`: If `Pre-mix` variable is set to `Yes`, specify the height in the reservoir wells in which to pre-mix.\n* `Pre-mix repetitions`: If `Pre-mix` variable is set to `Yes`, specify the number of repetitions to mix before each transfer.\n* `Pre-mix Volume`: If `Pre-mix` variable is set to `Yes`, specify the pre-mix volume (in ul).\n* `Pre-mix Rate`: If `Pre-mix` variable is set to `Yes`, specify the mix rate. A value of `1` is default mix speed, whereas a value of `0.5` is half of the default mix speed, etc.\n* `Dispense Reservoir Height`: Specify the height above the reservoir bottom to dispense residual solution.\n* `Transfer Aspiration Height`: Specify the height in which to aspirate from in the reservoir. Default is 1mm from the bottom of the reservoir.\n* `Transfer Dispense Height`: Specify the height in which to dispense from in the well plate. Default is 1mm from the bottom of the reservoir.\n* `Aspiration Rate`: Specify the aspiration rate for transfers. A value of `1` is default aspiration speed, whereas a value of `0.5` is half of the default aspiration speed, etc.\n* `Dispense Rate`: Specify the dispense rate for transfers. A value of `1` is default dispense speed, whereas a value of `0.5` is half of the default dispense speed, etc.\n* `P300 Multi-Channel Mount`: Specify whether the P300 Multi-Channel pipette is on the left or right mount.\n---\n\n",
diff --git a/protoBuilds/581011/README.json b/protoBuilds/581011/README.json
index 77be64fb74..008db6ccec 100644
--- a/protoBuilds/581011/README.json
+++ b/protoBuilds/581011/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -10,7 +10,7 @@
"internal": "581011",
"labware": "\nCorning 384 Well Plate 112 \u00b5L Flat\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nOpentrons 200uL Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"deck-setup": "* Deck layout with 6 target cells in the reservoir on Slot 5.\n\n\n\n---\n\n",
"description": "This protocol parses an uploaded .csv file (see below) and determines the number of tips to pick up with a multi-channel pipette. The pipette transfers the specified volume of target cell to a 384 well-plate to the parsed wells by multi-dispensing as much as the pipette tip can accommodate before returning to the reservoir. New tips are awarded between each target cell.\n\nNote: Part 1 and Part 2 should receive the exact same .csv file.\n\nFind part two of the protocol (effector cell loading) below:\n\n[Part 2 - Effector Cell Loading](https://protocols.opentrons.com/protocol/581011-pt2)\n\nExplanation of complex parameters below:\n* `.CSV File`: Upload the .csv file according to the template shown [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/581011/co-culture_template_protocol.numbers). Any well to the left of a forward slash (`/`) will correspond to the well in the reservoir containing the desired target cell. Any well to the right of a forward slash (`/`) will correspond to the effector cell in the 96 well plate for part 2 of this protocol. If a cell is only to receive one of the effector or target cells, place a `0` (e.g. A1/0 would mean target cell from A1 of the reservoir with no effector cell). All cells that are to be skipped (receive neither effector nor target cell) should have a lowercase `'x'` in the resepective cell in the .csv file. For the parameters below, make sure there are NO commas inside of the wells, and just the values. For the mount, put nothing else but \"left\" or \"right\". For the pre-mix variable, you must start the well with \"Yes\" or \"No\", but can put anything else after that (e.g. \"Yes premix\", or \"No premix\").\n* `Transfer Volume Effector Cell`: Specify the volume (in ul) to transfer between effector cells in reservoir to well plate.\n* `Pre-mix before aspirating in source well plate?`: Specify whether or not to pre-mix before each transfer of target cell in reservoir to plate.\n* `Pre-mix height in well plate`: If `Pre-mix` variable is set to `Yes`, specify the height in the reservoir wells in which to pre-mix.\n* `Pre-mix repetitions`: If `Pre-mix` variable is set to `Yes`, specify the number of repetitions to mix before each transfer.\n* `Pre-mix Volume`: If `Pre-mix` variable is set to `Yes`, specify the pre-mix volume (in ul).\n* `Pre-mix Rate`: If `Pre-mix` variable is set to `Yes`, specify the mix rate. A value of `1` is default mix speed, whereas a value of `0.5` is half of the default mix speed, etc.\n* `Dispense Reservoir Height`: Specify the height above the reservoir bottom to dispense residual solution.\n* `Transfer Aspiration/Dispense Height`: Specify the height in which to aspirate/dispense from in the well plate. Default is 1mm from the bottom of the reservoir.\n* `Transfer Dispense Height`: Specify the height in which to dispense from in the well plate. Default is 1mm from the bottom of the reservoir.\n* `Aspiration Rate`: Specify the aspiration rate for transfers. A value of `1` is default aspiration speed, whereas a value of `0.5` is half of the default aspiration speed, etc.\n* `Dispense Rate`: Specify the dispense rate for transfers. A value of `1` is default dispense speed, whereas a value of `0.5` is half of the default dispense speed, etc.\n* `P300 Multi-Channel Mount`: Specify whether the P300 Multi-Channel pipette is on the left or right mount.\n\n\n---\n\n",
diff --git a/protoBuilds/5829d7/README.json b/protoBuilds/5829d7/README.json
index 33108d00f5..9e2a976ba0 100644
--- a/protoBuilds/5829d7/README.json
+++ b/protoBuilds/5829d7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "5829d7",
"labware": "\nOpentrons 1000uL Filter Tips\nQiagen 96 Well plate\n(3) Custom Opentrons 32-tube tube racks.\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "* Deck layout with example 90 tubes. Note: tubes should be placed by row in the tube racks, starting from the tube rack in slot 1. Tubes are transferred 1-1 with well plate wells by row.\n\n\n---\n\n",
"description": "200ul of viral transport media (VTM) is reformatted from (up to) 96 tubes split amongst 3 tube racks to a 96 well plate. Tubes are reformatted by row in tube racks 1 (slot 1), 2 (slot 4), and then 3 (slot 7).\n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify the number of samples for this run.\n* `Tube Aspiration Height`: Specify the aspiration height in the tubes. Default is 1mm.\n* `Well Dispense Height`: Specify the dispense height in the wells. Default is 1mm.\n* `Aspirate/Dispense Flow rate`: Specify the aspirate/dispense flow rate in the wells. Default is 274ul/sec.\n* `P1000 Single-Channel Mount`: Specify (left) or (right) mount for the P1000 single channel pipette.\n\n---\n\n\n",
diff --git a/protoBuilds/58d57d/README.json b/protoBuilds/58d57d/README.json
index c7d1ed4582..79b1190571 100644
--- a/protoBuilds/58d57d/README.json
+++ b/protoBuilds/58d57d/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol performs transfers of reagents for the Promega ADP-Glo Kinase Assay. It will transfer four different liquids into the test wells with pauses for centrifugation and incubations in between transfers.\n\n\n\nP20 single-channel GEN2 electronic pipette\nGreiner Microplate 384 Wells\n\n\n\nDeck Setup\n Greiner 384 Well Plate (Slot 1)\n Opentrons 20uL Tips (Slot 2)\n* Opentrons 24 Tube Rack 1.5mL Tubes (Slot 4)\nReagent Setup: Opentrons 24 Tube Rack 1.5mL Tubes (Slot 4):\n Liquid A (A1)\n Liquid B (B1)\n Liquid C (C1)\n Liquid D (D1)",
"internal": "58d57d",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol performs transfers of reagents for the Promega ADP-Glo Kinase Assay. It will transfer four different liquids into the test wells with pauses for centrifugation and incubations in between transfers.\n\n---\n\n\n\n* [P20 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Greiner Microplate 384 Wells](https://shop.gbo.com/en/usa/products/bioscience/covid-19/covid-19-non-binding-microplates/781904.html)\n\n---\n\n\nDeck Setup\n* Greiner 384 Well Plate (Slot 1)\n* Opentrons 20uL Tips (Slot 2)\n* Opentrons 24 Tube Rack 1.5mL Tubes (Slot 4)\n\nReagent Setup: Opentrons 24 Tube Rack 1.5mL Tubes (Slot 4):\n* Liquid A (A1)\n* Liquid B (B1)\n* Liquid C (C1)\n* Liquid D (D1)\n\n",
"internal": "58d57d\n",
diff --git a/protoBuilds/5a2a35/README.json b/protoBuilds/5a2a35/README.json
index d0bbe8a2b0..c5888392bb 100644
--- a/protoBuilds/5a2a35/README.json
+++ b/protoBuilds/5a2a35/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -9,7 +9,7 @@
"description": "This protocol uses a p300 single channel pipette to transfer custom volumes of DNA from a source plate to a destination plate (starting tip, volume, deck slots and well locations specified in a csv file which has been made available by the user on the OT-2 using the OT-2 jupyter notebook).\nLinks:\n* example input csv\nThis protocol was developed to transfer custom volumes of DNA from one of possibly several source plates to one of possibly several destination plates according to volumes, deck slot numbers, and well locations in a csv file made available by the user on the OT-2 using the OT-2 jupyter notebook.",
"internal": "5a2a35",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Cherrypicking\n\n",
"deck-setup": "\n\n* Opentrons 200ul filter tips (Deck Slots 7, 8, and 9)\n* Selected Plate Labware for Source and Destination Plates (Deck Slots 1-6)\n\n\n\n",
"description": "\nThis protocol uses a p300 single channel pipette to transfer custom volumes of DNA from a source plate to a destination plate (starting tip, volume, deck slots and well locations specified in a csv file which has been made available by the user on the OT-2 using the OT-2 jupyter notebook).\n\nLinks:\n* [example input csv](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/5a2a35/Cherrypicking+CSV.csv)\n\nThis protocol was developed to transfer custom volumes of DNA from one of possibly several source plates to one of possibly several destination plates according to volumes, deck slot numbers, and well locations in a csv file made available by the user on the OT-2 using the OT-2 jupyter notebook.\n\n",
diff --git a/protoBuilds/5a3797-protocol-1/README.json b/protoBuilds/5a3797-protocol-1/README.json
index 936c6e6769..47059c3529 100644
--- a/protoBuilds/5a3797-protocol-1/README.json
+++ b/protoBuilds/5a3797-protocol-1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -9,7 +9,7 @@
"description": "This protocol performs aliquoting of multiple buffers from different reservoirs into multiple 2 mL KingFisher deep well plates.",
"internal": "5a3797-protocol-1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "* Opentrons 200 uL Filter Tip Rack (Slot 1)\n* Wash 1 Deep Well Plate (Slot 4)\n* Wash 2 Deep Well Plate (Slot 5)\n* Elution Buffer Deep Well Plate (Slot 6)\n* 12-well reservoir (Wash 1 Solution) (Slot 7)\n* 12-well reservoir (Wash 2 Solution) (Slot 8)\n* 12-well reservoir (Elution Buffer Solution) (Slot 9)\n\n",
"description": "This protocol performs aliquoting of multiple buffers from different reservoirs into multiple 2 mL KingFisher deep well plates.\n\n",
diff --git a/protoBuilds/5a3797-protocol-2/README.json b/protoBuilds/5a3797-protocol-2/README.json
index be3b1c6098..b07f1216cf 100644
--- a/protoBuilds/5a3797-protocol-2/README.json
+++ b/protoBuilds/5a3797-protocol-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -9,7 +9,7 @@
"description": "This protocol performs the transfer of mastermix and samples from a source to a destination PCR plate.",
"internal": "5a3797-protocol-2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "* Opentrons 200 uL Filter Tip Rack (Slot 1, Slot 2)\n* Sample Deep Well Plate (Slot 4)\n* PCR Plate (Slot 5)\n* 12-well reservoir (Master Mix) (Slot 7)\n\n",
"description": "This protocol performs the transfer of mastermix and samples from a source to a destination PCR plate.\n\n",
diff --git a/protoBuilds/5a3797-protocol-3/README.json b/protoBuilds/5a3797-protocol-3/README.json
index 72b5afc9c2..0c8c255567 100644
--- a/protoBuilds/5a3797-protocol-3/README.json
+++ b/protoBuilds/5a3797-protocol-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -9,7 +9,7 @@
"description": "This protocol performs aliquoting from 7 different tube racks (first 96 tubes) into a 96 well plate. It then aliquots from a 12-well reservoir into the deep well plate.",
"internal": "5a3797-protocol-3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "* Opentrons 200 uL Filter Tip Rack (Slot 1, Slot 2)\n* Sample Deep Well Plate (Slot 3)\n* 12-well reservoir (Slot 4)\n* Tube Racks 1-7 (Slots 5-11, sequentially)\n\n",
"description": "This protocol performs aliquoting from 7 different tube racks (first 96 tubes) into a 96 well plate. It then aliquots from a 12-well reservoir into the deep well plate.\n\n",
diff --git a/protoBuilds/5a5767/README.json b/protoBuilds/5a5767/README.json
index 72a562a2c0..cd340a4c62 100644
--- a/protoBuilds/5a5767/README.json
+++ b/protoBuilds/5a5767/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol moves up to 384 samples from 1 source plate to 1 destination plate using a P50-multichannel GEN1 pipette. The source plate is a Greiner Bio-One 384-well plate, and the destination plate can be either a Corning 384-well plate or a Bio-Rad 384-well plate. Samples are transferred across row A (A1, C1, E1, G1, I1, K1, M1, O1; A2, C2, E2, G2, I2, K2, M2, O2; etc.), then across row B (B1, D1, F1, H1, J1, L1, N1, P1; B2, D2, F2, H2, J2, L2, N2, P2; etc.).\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 4.0.0 or later)\nOpentrons P50 Multi-Channel Pipette GEN1\nOpentrons 96 Tiprack 300ul\nGreiner Bio-One 384 Well Plate 100 \u00b5L #781720\nCorning 384 Well Plate 20 \u00b5L #4511\nBio-Rad 384 Well Plate 50 \u00b5L #HSR4801\n\nFor more detailed information on compatible labware, please visit our Labware Library.",
"internal": "5a5767",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "This protocol moves up to 384 samples from 1 source plate to 1 destination plate using a P50-multichannel GEN1 pipette. The source plate is a Greiner Bio-One 384-well plate, and the destination plate can be either a Corning 384-well plate or a Bio-Rad 384-well plate. Samples are transferred across row A (A1, C1, E1, G1, I1, K1, M1, O1; A2, C2, E2, G2, I2, K2, M2, O2; etc.), then across row B (B1, D1, F1, H1, J1, L1, N1, P1; B2, D2, F2, H2, J2, L2, N2, P2; etc.).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 4.0.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P50 Multi-Channel Pipette GEN1](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 96 Tiprack 300ul](https://shop.opentrons.com/collections/opentrons-tips)\n* [Greiner Bio-One 384 Well Plate 100 \u00b5L #781720](https://www.gbo.com/en_US.html)\n* [Corning 384 Well Plate 20 \u00b5L #4511](https://ecatalog.corning.com/life-sciences/b2c/EUOther/en/Microplates/Assay-Microplates/384-Well-Microplates/Corning\u00ae-384-well-Solid-Black-and-White-Polystyrene-Microplates/p/4511)\n* [Bio-Rad 384 Well Plate 50 \u00b5L #HSR4801](https://www.bio-rad.com/en-us/sku/hsr4801-hard-shell-384-well-pcr-plates-clear-clear-barcoded?ID=HSR4801)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n",
"internal": "5a5767\n",
diff --git a/protoBuilds/5b16ef-pt2/README.json b/protoBuilds/5b16ef-pt2/README.json
index 7d7ca08688..eabcfc905e 100644
--- a/protoBuilds/5b16ef-pt2/README.json
+++ b/protoBuilds/5b16ef-pt2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina Nextera Flex"
@@ -10,7 +10,7 @@
"internal": "5b16ef-pt2",
"labware": "\nNEST 12 well reservoir 15mL\nNEST 1 well reservoir 195mL\nBio-Rad 96 Well Plate 200 \u00b5L PCR\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nOpentrons 20ul Tips\nOpentrons 300ul Tips\nNEST 12 well reservoir 195mL\nIndex Plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina Nextera Flex\n\n",
"deck-setup": "Note: Split the ethanol evenly in columns 11 and 12 of the reservoir if running 5 or 6 columns of samples. If running 4 or fewer columns of samples, place all the ethanol in column 11 of the reservoir exclusively.\n\n\n",
"description": "This protocol is a semi-automated workflow which performs 8.1-8.4.6 of the SARS-CoV-2 Using Illumina Nextera Flex & MiSeqNEB kit. For detailed information on protocol steps, please see below. You can find part 1 of the protocol here:\n\n* [SARS-CoV-2 Using Illumina Nextera Flex & MiSeqNEB - Part 1](https://protocols.opentrons.com/protocol/5b16ef)\n\nExplanation of complex parameters below:\n* `Number of Sample Columns (1-6)`: Specify how many sample columns (1-6) this run will process. Samples will be placed in every other column, starting from column 1 (i.e. 1, 3, 5, 7, 9, 11 for 6 columns).\n* `csv`: csv should be formatted in the following order, including the header line (also be sure to skip columns):\n```\nWell, DNA Volume (ul), Pool Volume\nA1, 5, 5\nB1, 3, 5\nC1, 5, 5\nD1, 5, 5\nE1, 2, 5\nF1, 5, 5\nG1, 5, 5\nH1, 4, 5\nA3, 5, 5\n```\n* `Index Start Column (1-12)`: Specify which column in the index plate to begin aspirating from. Note: the number of index columns left should be greater or equal to the number of sample columns. If not, the protocol will throw an error at the beginning of the run.\n* `P20 Multi-Channel Mount`: Specify which mount (left or right) to host the P20 and P300 multi-channel pipettes.\n\n\n---\n\n",
diff --git a/protoBuilds/5b16ef/README.json b/protoBuilds/5b16ef/README.json
index d4321a687c..7875445527 100644
--- a/protoBuilds/5b16ef/README.json
+++ b/protoBuilds/5b16ef/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina Nextera Flex"
@@ -10,7 +10,7 @@
"internal": "5b16ef",
"labware": "\nBio-Rad 96 Well Plate 200 \u00b5L PCR\nOpentrons 20ul Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina Nextera Flex\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "This protocol is a semi-automated workflow which performs 8.1-8.4.6 of the SARS-CoV-2 Using Illumina Nextera Flex & MiSeqNEB kit. For detailed information on protocol steps, please see below. You can find part 2 of the protocol here:\n\n* [SARS-CoV-2 Using Illumina Nextera Flex & MiSeqNEB - Part 2](https://protocols.opentrons.com/protocol/5b16ef)\n\nExplanation of complex parameters below:\n* `Number of Sample Columns (1-6)`: Specify how many sample columns (1-6) this run will process. Samples will be placed in every other column, starting from column 1 (i.e. 1, 3, 5, 7, 9, 11 for 6 columns).\n* `P20 Multi-Channel Mount`: Specify which mount (left or right) to host the P20 pipette.\n\n\n---\n\n",
diff --git a/protoBuilds/5b8174/README.json b/protoBuilds/5b8174/README.json
index d880e2eb56..07c0e8a347 100644
--- a/protoBuilds/5b8174/README.json
+++ b/protoBuilds/5b8174/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -10,7 +10,7 @@
"internal": "5b8174",
"labware": "\nOpentrons 20 uL Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"deck-setup": "* If the deck layout of a particular protocol is more or less static, it is often helpful to attach a preview of the deck layout, most descriptively generated with Labware Creator. Example:\n\n\n---\n\n",
"description": "This protocol performs transfers of positive controls and patient samples to a 384 well plate loaded onto a temperature module. All labware and transfers can be controlled through the CSV files that are used as input for this protocol.\n\nExplanation of complex parameters below:\n* `P20 Single Channel Pipette Mount Position`: Choose the mount position of your pipette.\n* `Control CSV File`: Upload the CSV file for controls here. You can download an [example CSV file here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/5b8174/Control_updated_07302021.csv) OR follow the format directly below. **Note: The control source slot and destination slot are converted from the Eppendorf Rack format to the Opentrons 24 Tube Rack Format. Please refer to the Control Conversion Table at the end of this section.**\n```\nRack,Sr.Barcode,,Dest.Barcode,Dest.List Name,\n1,,PCRCTR15000004,,,\n2,,,,,\n3,,,,,\n4,,,,,\n,,,,,\nSource Labware,Source Slot,Source Well,Dest Labware,Dest Slot,Dest Well\neppendorf_24_tuberack_generic_2.0ml_screwcap,1,D2,appliedbiosystems_384_wellplate_20ul,9,B1\neppendorf_24_tuberack_generic_2.0ml_screwcap,1,D3,appliedbiosystems_384_wellplate_20ul,9,A4\neppendorf_24_tuberack_generic_2.0ml_screwcap,1,D4,appliedbiosystems_384_wellplate_20ul,9,D2\n```\n* `Assembly CSV File`: Upload the CSV file for assembly/patient samples here. You can download an [example CSV file here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/5b8174/Assembly.csv) OR follow the format directly below.\n```\nRack,Sr.Barcode, ,Dest.Barcode,Dest.List Name,\n1,,,,,\n2,,,,,\n3,,,,,\n4,,,,,\n,,,,,\nSource Labware,Source Slot,Source Well,Dest Labware,Dest Slot,Dest Well\nappliedbiosystems_96_wellplate_200ul_on_eppendorf_cooling_block,2,C11,appliedbiosystems_384_wellplate_20ul,6,B1\nappliedbiosystems_96_wellplate_200ul_on_eppendorf_cooling_block,2,E11,appliedbiosystems_384_wellplate_20ul,6,A4\nappliedbiosystems_96_wellplate_200ul_on_eppendorf_cooling_block,2,E11,appliedbiosystems_384_wellplate_20ul,6,D2\nappliedbiosystems_96_wellplate_200ul_on_eppendorf_cooling_block,2,E11,appliedbiosystems_384_wellplate_20ul,6,E2\n```\n* `96 Well Plates Aspiration Height (mm)`: The aspiration height from the bottom of the well when aspirating from the assembly 96 well plates.\n\n**Control Conversion Table**\n```\nEppendorf Source Well, Destination Slot, Destination Well\nA1,10,A1\nA2,10,A2\nA3,10,A3\nA4,10,A4\nA5,10,A5\nA6,10,A6\nB1,10,B1\nB2,10,B2\nB3,10,B3\nB4,10,B4\nB5,10,B5\nB6,10,B6\nC1,10,C1\nC2,10,C2\nC3,10,C3\nC4,10,C4\nC5,10,C5\nC6,10,C6\nD1,10,D1\nD2,10,D2\nD3,10,D3\nD4,10,D4\nD5,10,D5\nD6,10,D6\nE1,7,A1\nE2,7,A2\nE3,7,A3\nE4,7,A4\nE5,7,A5\nE6,7,A6\nF1,7,B1\nF2,7,B2\nF3,7,B3\nF4,7,B4\nF5,7,B5\nF6,7,B6\nG1,7,C1\nG2,7,C2\nG3,7,C3\nG4,7,C4\nG5,7,C5\nG6,7,C6\nH1,7,D1\nH2,7,D2\nH3,7,D3\nH4,7,D4\nH5,7,D5\nH6,7,D6\nI1,4,A1\nI2,4,A2\nI3,4,A3\nI4,4,A4\nI5,4,A5\nI6,4,A6\nJ1,4,B1\nJ2,4,B2\nJ3,4,B3\nJ4,4,B4\nJ5,4,B5\nJ6,4,B6\nK1,4,C1\nK2,4,C2\nK3,4,C3\nK4,4,C4\nK5,4,C5\nK6,4,C6\nL1,4,D1\nL2,4,D2\nL3,4,D3\nL4,4,D4\nL5,4,D5\nL6,4,D6\n```\n---\n\n",
diff --git a/protoBuilds/5c14ad/README.json b/protoBuilds/5c14ad/README.json
index 6d65473a95..0dab0db878 100644
--- a/protoBuilds/5c14ad/README.json
+++ b/protoBuilds/5c14ad/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol allows the user to fill custom adapters according to their specifications (found below):\nThe user can choose between: Salmonella or Listeria Fill (90\u00b5L, 70\u00b5L respectively), how many plates to fill (1-9), single channel or multi-channel pipette (P300), and which column to pick up a tip from (row A, to avoid cross contamination).\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP300 Single Channel Pipette, or\nP300 Multi Channel Pipette\nFisherbrand 20-200\u00b5L Tips\nSimport Scientific 500\u00b5L Tubes in Custom Holder\nNest 195mL Reservoir (or similar)\n\n\n\nFor this protocol, be sure that the correct Lysis Type is selected for correct volume transfers, as well as Pipette Type and Mount.\nLabware Setup\nSlot 1: Fisherbrand 20-200\u00b5L Tips\nSlot 2: Nest Reservoir\nSlot 3 - 11 (loaded sequentially): Simport Scientific 500\u00b5L Tubes in Custom Holder\nUsing the customization fields below, set up your protocol.\n Lysis Type (Salmonella/Listeria): Select either Salmonella or Listeria and the corresponding volume (90\u00b5L or 70\u00b5L, respectively) will be added to each well.\n Pipette Type: Select either Single Channel or Multi Channel. This will also modify the protocol behavior to the corresponding protocol.\n Pipette Mount: Specify which mount the P300 is on (left or right).\n Number of Plates: Specify how many plates to fill (1-9).\n* Tip Pick-Up at (Row A): Specify which column to pick up tip (1-12).\n",
"internal": "5c14ad",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol allows the user to fill custom adapters according to their specifications (found below):\n\nThe user can choose between: Salmonella or Listeria Fill (90\u00b5L, 70\u00b5L respectively), how many plates to fill (1-9), single channel or multi-channel pipette (P300), and which column to pick up a tip from (row A, to avoid cross contamination).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P300 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette), or\n* [P300 Multi Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Fisherbrand 20-200\u00b5L Tips](https://www.fishersci.com/shop/products/fisherbrand-sureone-aerosol-barrier-pipette-tips-20-200-l-beveled-tip-graduated-at-10-50-100-l/02707430)\n* [Simport Scientific 500\u00b5L Tubes in Custom Holder](http://www.simport.com/en/products/173-t100-1.html)\n* [Nest 195mL Reservoir (or similar)](https://labware.opentrons.com/nest_1_reservoir_195ml?category=reservoir)\n\n\n\n---\n\n\n\nFor this protocol, be sure that the correct Lysis Type is selected for correct volume transfers, as well as Pipette Type and Mount.\n\n**Labware Setup**\n\nSlot 1: [Fisherbrand 20-200\u00b5L Tips](https://www.fishersci.com/shop/products/fisherbrand-sureone-aerosol-barrier-pipette-tips-20-200-l-beveled-tip-graduated-at-10-50-100-l/02707430)\n\nSlot 2: [Nest Reservoir](https://labware.opentrons.com/nest_1_reservoir_195ml?category=reservoir)\n\nSlot 3 - 11 (loaded sequentially): [Simport Scientific 500\u00b5L Tubes in Custom Holder](http://www.simport.com/en/products/173-t100-1.html)\n\n**Using the customization fields below, set up your protocol.**\n* Lysis Type (Salmonella/Listeria): Select either Salmonella or Listeria and the corresponding volume (90\u00b5L or 70\u00b5L, respectively) will be added to each well.\n* Pipette Type: Select either `Single Channel` or `Multi Channel`. This will also modify the protocol behavior to the corresponding protocol.\n* Pipette Mount: Specify which mount the P300 is on (left or right).\n* Number of Plates: Specify how many plates to fill (1-9).\n* Tip Pick-Up at (Row A): Specify which column to pick up tip (1-12).\n\n---\n",
"internal": "5c14ad\n",
diff --git a/protoBuilds/5c24e2/README.json b/protoBuilds/5c24e2/README.json
index 080a8fe354..fb8229d27e 100644
--- a/protoBuilds/5c24e2/README.json
+++ b/protoBuilds/5c24e2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol transfers sample from NEST 96-Well Plates to Applied Biosystems MicroAmp 384-well plates with mastermix. Update: this protocol has been updated to accommodate the SSI 384-well plate as well.\n\nThe protocol can fill one or two, 384-well plates. The P20 Single-Channel Pipette will aspirate 5\u00b5L from each well of the NEST 96-Well Plates and dispense 2.5\u00b5L into two wells of the (A1 --> A1/B1; B1 --> C1/D1; etc.)\n\nUpdate (10/22/20): The protocol will now aspirate 10\u00b5L and dispense 5\u00b5L into two wells.\n\nUpdate (11/2/20): The protocol will now aspirate 20\u00b5L and dispense 10\u00b5L into two wells. Additionally, the option to use the p300-multi pipette has been added.\nUpdated (11/12/20): A new parameter has been added that allows the user to input the volume (in \u00b5L) of sample that should be transferred. The pipette will aspirate 2x the volume and dispense into two wells.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P20 Single-Channel Pipette\nOpentrons 20\u00b5L Tips\nApplied Biosystems MicroAmp 384-Well Plate or SSI 384-Well Plate with mastermix\nNEST 96-Well Plates with samples\n\n\n\nSlots 1: Opentrons 20\u00b5L Tips\nSlot 2: NEST 96-Well Plates with samples\nSlot 3: Applied Biosystems MicroAmp 384-Well Plate or SSI 384-Well Plate with mastermix\nSlot 4: Opentrons 20\u00b5L Tips\nSlot 5: NEST 96-Well Plates with samples\nSlot 6: Applied Biosystems MicroAmp 384-Well Plate or SSI 384-Well Plate with mastermix (if filling two plates)\nSlot 7: Opentrons 20\u00b5L Tips (if filling two plates)\nSlot 8: NEST 96-Well Plates with samples (if filling two plates)\nSlot 9: empty\nSlot 10: Opentrons 20\u00b5L Tips (if filling two plates)\nSlot 11: NEST 96-Well Plates with samples (if filling two plates)\n\n\nUsing the customizations field (below), set up your protocol.\n Pipette Type: Select which pipette will be used\n Pipette Mount: Select which mount (left or right) the pipette is attached to.\n Sample Volume (uL): Input the sample volume\n How many 384-well plates: Specify the number of 384-well plates to fill\n* Plate (384-Well) Type: Specify the type of 384-well plates that are being used",
"internal": "5c24e2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol transfers sample from [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) to [Applied Biosystems MicroAmp 384-well plates](https://www.thermofisher.com/order/catalog/product/4343370#/4343370) with mastermix. **Update**: this protocol has been updated to accommodate the SSI 384-well plate as well.\n\nThe protocol can fill one or two, 384-well plates. The [P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes) will aspirate 5\u00b5L from each well of the [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) and dispense 2.5\u00b5L into two wells of the (A1 --> A1/B1; B1 --> C1/D1; etc.)\n\n**Update (10/22/20):** The protocol will now aspirate 10\u00b5L and dispense 5\u00b5L into two wells.\n\n**Update (11/2/20):** The protocol will now aspirate 20\u00b5L and dispense 10\u00b5L into two wells. Additionally, the option to use the p300-multi pipette has been added.\n**Updated (11/12/20):** A new parameter has been added that allows the user to input the volume (in \u00b5L) of sample that should be transferred. The pipette will aspirate 2x the volume and dispense into two wells.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Applied Biosystems MicroAmp 384-Well Plate](https://www.thermofisher.com/order/catalog/product/4343370#/4343370) or SSI 384-Well Plate with mastermix\n* [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) with samples\n\n\n---\n\n\nSlots 1: [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\nSlot 2: [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) with samples\n\nSlot 3: [Applied Biosystems MicroAmp 384-Well Plate](https://www.thermofisher.com/order/catalog/product/4343370#/4343370) or SSI 384-Well Plate with mastermix\n\nSlot 4: [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\nSlot 5: [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) with samples\n\nSlot 6: [Applied Biosystems MicroAmp 384-Well Plate](https://www.thermofisher.com/order/catalog/product/4343370#/4343370) or SSI 384-Well Plate with mastermix (if filling two plates)\n\nSlot 7: [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips) (if filling two plates)\n\nSlot 8: [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) with samples (if filling two plates)\n\nSlot 9: *empty*\n\nSlot 10: [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips) (if filling two plates)\n\nSlot 11: [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) with samples (if filling two plates)\n\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Pipette Type**: Select which pipette will be used\n* **Pipette Mount**: Select which mount (left or right) the pipette is attached to.\n* **Sample Volume (uL)**: Input the sample volume\n* **How many 384-well plates**: Specify the number of 384-well plates to fill\n* **Plate (384-Well) Type**: Specify the type of 384-well plates that are being used\n\n\n\n",
"internal": "5c24e2\n",
diff --git a/protoBuilds/5c50d9/README.json b/protoBuilds/5c50d9/README.json
index bb692af8e4..43e77d1585 100644
--- a/protoBuilds/5c50d9/README.json
+++ b/protoBuilds/5c50d9/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "5c50d9",
"labware": "\nCustom 384 well plate\nOpentrons 4-in-1 tube rack with 15 tube insert\nOpentrons 200ul filter tips\nOpentrons 20ul tips\nNest 12 Well Reservoir, 195mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "* Deck map with lysis buffer in A1 (first well) of the Nest 12 well reservoir.\n\n\n\n---\n\n",
"description": "This protocol first preps one 384 well plate with lysis buffer and sample to undergo a thermocycler. In this step, samples are transferred from tube rack to the 384 well plate. If the number of samples selected exceeds the number of samples that can fit onto the deck, the protocol pauses and prompts the user to refill the tube racks with new samples up to the number of samples selected. Afterwards, 4ul of the resulting solution is transferred to the final 384 well plate.\n\nExplanation of complex parameters below:\n* `Number of Samples`: Select the number of samples (1-384) for this run. If larger than 90 samples are selected, the protocol will pause after the last sample is transferred on the deck and will prompt the user to refill samples up to the number specified. Samples should always be placed in the tuberack down by column, and in the order of the deck slots (i.e. 4, 5, 6 ..., etc.)\n* `P20/P300 Dispense Flow Rate`: Global control of P20 and P300 dispense flow rate. A value of 1.0 is default, 0.5 is 50% of the default flow rate, 1.2 is 20% faster the default flow rate, etc. \n* `P20 Single Mount`: Specify whether the P20 single channel pipette will be mounted on the left or right.\n* `P300 Single Mount`: Specify whether the P300 single channel pipette will be mounted on the left or right.\n\n\n---\n\n",
diff --git a/protoBuilds/5c7384-hhu/README.json b/protoBuilds/5c7384-hhu/README.json
index d3378c35fa..cc1207d8f4 100644
--- a/protoBuilds/5c7384-hhu/README.json
+++ b/protoBuilds/5c7384-hhu/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Mass Spec"
@@ -8,7 +8,7 @@
"description": "This protocol performs mass spec sample prep per this SP3 Proteomics manual (Appendix Protocol B). The following steps are performed throughout the protocol with the use of the Opentrons OT-2, Thermocycler Module, and Magnetic Module (GEN2):\n\nReduction and Alkylation\nProtein Binding\nEthanol Wash\nAcetonitrile Wash\nOn-Bead Digestion\n\nDue to limited deck space, the user is prompted to change tipracks on the deck when needed, and to move labware when needed.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 4.0 or later)\nOpentrons P20 GEN2 Single-Channel Pipette\nOpentrons 20\u00b5L Tips\nOpentrons P300 GEN2 Single-Channel Pipette\nOpentrons 300\u00b5L Tips\nNEST 96-Well Plates or comparable PCR plates\n\n\n\nReagent plate (slot 5):\n column 1: DTT\n column 2: CAA\n column 3: magnetic bead stock\n columns 4-5: ABC\n* column 6: trypsin\nEthanol plate (slot 2)\nAcetonitrile plate (slot 3)",
"internal": "5c7384",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Mass Spec\n\n\n",
"description": "This protocol performs mass spec sample prep per this [SP3 Proteomics manual (Appendix Protocol B)](https://www.embopress.org/action/downloadSupplement?doi=10.15252%2Fmsb.20199111&file=msb199111-sup-0001-Appendix.pdf). The following steps are performed throughout the protocol with the use of the Opentrons OT-2, Thermocycler Module, and Magnetic Module (GEN2):\n\n* Reduction and Alkylation\n* Protein Binding\n* Ethanol Wash\n* Acetonitrile Wash\n* On-Bead Digestion\n\nDue to limited deck space, the user is prompted to change tipracks on the deck when needed, and to move labware when needed.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 4.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P20 GEN2 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons P300 GEN2 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-30ul-tips)\n* [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) or comparable PCR plates\n\n\n---\n\n\nReagent plate (slot 5):\n* column 1: DTT\n* column 2: CAA\n* column 3: magnetic bead stock\n* columns 4-5: ABC\n* column 6: trypsin\n\nEthanol plate (slot 2)\n\nAcetonitrile plate (slot 3)\n\n",
"internal": "5c7384\n",
diff --git a/protoBuilds/5c7384/README.json b/protoBuilds/5c7384/README.json
index d3378c35fa..cc1207d8f4 100644
--- a/protoBuilds/5c7384/README.json
+++ b/protoBuilds/5c7384/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Mass Spec"
@@ -8,7 +8,7 @@
"description": "This protocol performs mass spec sample prep per this SP3 Proteomics manual (Appendix Protocol B). The following steps are performed throughout the protocol with the use of the Opentrons OT-2, Thermocycler Module, and Magnetic Module (GEN2):\n\nReduction and Alkylation\nProtein Binding\nEthanol Wash\nAcetonitrile Wash\nOn-Bead Digestion\n\nDue to limited deck space, the user is prompted to change tipracks on the deck when needed, and to move labware when needed.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 4.0 or later)\nOpentrons P20 GEN2 Single-Channel Pipette\nOpentrons 20\u00b5L Tips\nOpentrons P300 GEN2 Single-Channel Pipette\nOpentrons 300\u00b5L Tips\nNEST 96-Well Plates or comparable PCR plates\n\n\n\nReagent plate (slot 5):\n column 1: DTT\n column 2: CAA\n column 3: magnetic bead stock\n columns 4-5: ABC\n* column 6: trypsin\nEthanol plate (slot 2)\nAcetonitrile plate (slot 3)",
"internal": "5c7384",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Mass Spec\n\n\n",
"description": "This protocol performs mass spec sample prep per this [SP3 Proteomics manual (Appendix Protocol B)](https://www.embopress.org/action/downloadSupplement?doi=10.15252%2Fmsb.20199111&file=msb199111-sup-0001-Appendix.pdf). The following steps are performed throughout the protocol with the use of the Opentrons OT-2, Thermocycler Module, and Magnetic Module (GEN2):\n\n* Reduction and Alkylation\n* Protein Binding\n* Ethanol Wash\n* Acetonitrile Wash\n* On-Bead Digestion\n\nDue to limited deck space, the user is prompted to change tipracks on the deck when needed, and to move labware when needed.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 4.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P20 GEN2 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons P300 GEN2 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-30ul-tips)\n* [NEST 96-Well Plates](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) or comparable PCR plates\n\n\n---\n\n\nReagent plate (slot 5):\n* column 1: DTT\n* column 2: CAA\n* column 3: magnetic bead stock\n* columns 4-5: ABC\n* column 6: trypsin\n\nEthanol plate (slot 2)\n\nAcetonitrile plate (slot 3)\n\n",
"internal": "5c7384\n",
diff --git a/protoBuilds/5dcd88/README.json b/protoBuilds/5dcd88/README.json
index aac7ed8353..af7d0e5d76 100644
--- a/protoBuilds/5dcd88/README.json
+++ b/protoBuilds/5dcd88/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "5dcd88",
"labware": "\nNEST 100ul PCR Plate Full Skirt\nOpentrons 20\u00b5L Filter Tips\nOpentrons 200\u00b5L Filter Tips\nNEST 12 Well Reservoir 195mL\nOpentrons 4-in-1 Tube Rack Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n\n\n---\n\n",
"description": "This protocol preps a 96 well plate with barcoded, amplified sample. Ultimately, barcoded samples are pooled into a final 1.5mL tube.\n\nWater and mastermix are added to samples, after which an incubation period follows with subsequent temperature changes. User will load a new PCR plate onto the temperature module for the final sample collecting. User will also pull the sample plate from the deck after the second barcode mastermix is added to barcode samples. After another incubation period with subsequent temperature changes, samples are pooled. The protocol will automatically pause for all steps requiring user intervention.\n\n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify the number of samples for this run\n* `P20 Multi-Channel Mount`: Specify which mount (left or right) to host the P20 Multi-Channel Pipette.\n* `Park Tips?`: Specify whether or not you would like to park tips for this run\n* `P300 Multi-Channel Mount`: Specify which mount (left or right) to host the P300 Multi-Channel\n\n---\n\n",
diff --git a/protoBuilds/5eee17/README.json b/protoBuilds/5eee17/README.json
index a960c57aab..e71c64edeb 100644
--- a/protoBuilds/5eee17/README.json
+++ b/protoBuilds/5eee17/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Dilution"
@@ -10,7 +10,7 @@
"internal": "5eee17",
"labware": "\nCustom 15ml, 50ml, and HPLC tuberacks\nOpentrons 1000\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Dilution\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nThis protocol performs a dilution on custom tuberacks with 2x P1000 pipettes in tandem. The deck layout is shown below: \n\n\n---\n\n",
diff --git a/protoBuilds/5f37a2/README.json b/protoBuilds/5f37a2/README.json
index 2c19eb3a3f..ae259e3cc3 100644
--- a/protoBuilds/5f37a2/README.json
+++ b/protoBuilds/5f37a2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"MagneSil"
@@ -8,7 +8,7 @@
"description": "This protocol performs a nucleic acid purification protocol based on the MagneSil Plasmid Purification System from Promega.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Magnetic Module\nOpentrons P300 Multi-Channel Pipette\nOpentrons 50/300\u00b5L Tips\nEppendorf Deep Well Plate 500\u00b5L\nEppendorf Deep Well Plate 2000\u00b5L\nCorning 96-Well Plate, 360\u00b5L\nUSA Scientific 12-Well Reservoir, 22mL\nReagents\nSamples\n\n\n\nSlot 1: Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\nSlot 2: Opentrons 50/300\u00b5L Tips, if needed\nSlot 3: Opentrons 50/300\u00b5L Tips, if needed\nSlot 4: Opentrons 50/300\u00b5L Tips, if needed\nSlot 5: Opentrons 50/300\u00b5L Tips, if needed\nSlot 6: Opentrons 50/300\u00b5L Tips, if needed\nSlot 7: Opentrons 50/300\u00b5L Tips, if needed\nSlot 8: Corning 96-Well Plate\nSlot 9: Eppendorf Deep Well Plate 500\u00b5L, with samples\nSlot 10: Opentrons Magnetic Module with Eppendorf Deep Well Plate 2000\u00b5L\nSlot 11: USA Scientific 12-Well Reservoir\n A1: Resuspension Buffer\n A2: Lysis Solution\n A3: Neutralization Buffer\n A4: MagneSil Blue\n A5: MagneSil Red\n A6: 80% Ethanol Solution\n* A7: Elution Buffer",
"internal": "5fe7a2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* MagneSil\n\n\n",
"description": "This protocol performs a nucleic acid purification protocol based on the MagneSil Plasmid Purification System from Promega.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Eppendorf Deep Well Plate 500\u00b5L](https://online-shop.eppendorf.us/US-en/Laboratory-Consumables-44512/Plates-44516/Eppendorf-Deepwell-Plates-PF-55960.html)\n* [Eppendorf Deep Well Plate 2000\u00b5L](https://online-shop.eppendorf.us/US-en/Laboratory-Consumables-44512/Plates-44516/Eppendorf-Deepwell-Plates-PF-55960.html)\n* [Corning 96-Well Plate, 360\u00b5L](https://labware.opentrons.com/corning_96_wellplate_360ul_flat)\n* [USA Scientific 12-Well Reservoir, 22mL](https://labware.opentrons.com/usascientific_12_reservoir_22ml/)\n* Reagents\n* Samples\n\n\n---\n\n\nSlot 1: Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 2: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips), if needed\n\nSlot 3: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips), if needed\n\nSlot 4: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips), if needed\n\nSlot 5: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips), if needed\n\nSlot 6: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips), if needed\n\nSlot 7: [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips), if needed\n\nSlot 8: [Corning 96-Well Plate](https://labware.opentrons.com/corning_96_wellplate_360ul_flat)\n\nSlot 9: [Eppendorf Deep Well Plate 500\u00b5L](https://online-shop.eppendorf.us/US-en/Laboratory-Consumables-44512/Plates-44516/Eppendorf-Deepwell-Plates-PF-55960.html), with samples\n\nSlot 10: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [Eppendorf Deep Well Plate 2000\u00b5L](https://online-shop.eppendorf.us/US-en/Laboratory-Consumables-44512/Plates-44516/Eppendorf-Deepwell-Plates-PF-55960.html)\n\nSlot 11: [USA Scientific 12-Well Reservoir](https://labware.opentrons.com/usascientific_12_reservoir_22ml/)\n* A1: Resuspension Buffer\n* A2: Lysis Solution\n* A3: Neutralization Buffer\n* A4: MagneSil Blue\n* A5: MagneSil Red\n* A6: 80% Ethanol Solution\n* A7: Elution Buffer\n\n\n",
"internal": "5fe7a2\n",
diff --git a/protoBuilds/5f791f/README.json b/protoBuilds/5f791f/README.json
index 48cd918876..10e94b7899 100644
--- a/protoBuilds/5f791f/README.json
+++ b/protoBuilds/5f791f/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep and Magnetic Bead Cleanup"
@@ -10,7 +10,7 @@
"internal": "5f791f",
"labware": "\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nNEST 12-Well Reservoirs, 15 mL\nOpentrons 200\u00b5L Filter Tips\nOpentrons 20\u00b5L Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep and Magnetic Bead Cleanup\n\n",
"deck-setup": " \n\n",
"description": "\nThis protocol performs a full custom PCR prep and post-PCR magnetic bead cleanup protocol for up to 96 samples. If both steps are run, the user will be prompted to run PCR off of the robot deck, and to replace the reaction plate onto the magnetic module for cleanup.\n\n---\n\n",
diff --git a/protoBuilds/5f7d18/README.json b/protoBuilds/5f7d18/README.json
index 0615cde700..9f504414ae 100644
--- a/protoBuilds/5f7d18/README.json
+++ b/protoBuilds/5f7d18/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"RNA Extraction"
@@ -8,7 +8,7 @@
"description": "This protocol isolates nucleic acid using a MagBio CTL lysis buffer and MAG-S1 particle beads suspended in 100% isopropanol. 200\u00b5L of sample is pre-loaded into each well within a Kingfisher deep-well plate on the Opentrons magnetic module. The sample undergoes extraction with 2 ethanol washes. The eluate is then transferred to a full-skirted PCR plate along with PCR master-mix for further processing.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Magnetic Module\nP20 Single Pipette\nP300 Multi-Channel Pipette\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons NEST 96 well plate 100ul PCR full skirt\nOpentrons NEST 12-Well Reservoir\nOpentrons 4-in-1 Tube Rack Set (24-tube insert)\n\n\n\nFor this protocol, be sure that the pipettes (P20 and P300) are attached.\nUsing the customization fields below, set up your protocol.\n Number of columns: Specify the number of columns of samples loaded onto the NEST deep well plate. Note that runs should only be completed in multiples of full columns (8 samples).\n Sample and lysis buffer mix repetitions: Specify the number of repetitions to mix the sample and lysis buffer as mentioned in Step 1 of the MAGBIO protocol\n Mix repetitions before first magnetic engage: Specify the number of mix repetitions to re-suspend the MAG-S1 particles before use (i.e. \"shake well\" in Step 3 of the MAGBIO protocol).\n Incubation time: Specify the amount of incubation time to allow RNA to bind to the beads\n Magnetic bead engagement time: Specify the amount of time to engage the magnet for every engagement in protocol\n Aspiration height from bottom of well (removing supernatant): Specify the height from the bottom of the well in which the pipette will aspirate from when removing supernatant.\n Multi_channel pipette aspiration flow rate (ul/s): Specify the aspiration flow rate\n Multi-channel pippette dispense flow rate (ul/s): Specify the dispense flow rate\n Distance from side of well opposite beads (1mm - 4.15mm): Specify the distance from the side of the well opposite the engaged magnetic beads to aspirate from. A value of 4.15mm returns the center of the well, with 1mm returning 1mm from the side of the well opposite the engaged beads.\n Bead drying time: Specify the amount of incubation time for the beads to dry at room temperature while magnetically engaged.\n Bead drying time (with nuclease free water): Specify the amount of time to to dry the beads at room temperature after adding nuclease free water.\n Nuclease water volume: Specify the amount of nuclease free water to add to the beads\n P20 single GEN2 mount: Specify which mount to load the P20 single GEN2 pipette.\n P300 multi GEN2 mount: Specify which mount to load the P300 multi GEN2 pipette.\nNote about 20\u00b5L tip racks\nWhen prompted to replace the 300ul tip racks, be sure to re-load all 3 tip racks as in the original configuration of the deck.\nLabware Setup\nSlots 1: Opentrons Magnetic Module with loaded NEST 96 2mL deep well plate. 200ul of viral sample pre-loaded into each well.\nSlot 2, 5: Opentrons 96 Filter Tip Rack 20 \u00b5L\nSlot 3, 6, 9: Opentrons 96 Filter Tip Rack 200 \u00b5L\nSlot 4: Opentrons NEST 12-Well Reservoir (empty)\nSlot 7: Opentrons 4-in-1 Tube Rack Set. Mastermix loaded into tubes A1, B1.\nSlot 8: Opentrons NEST 96 well plate 100ul PCR full skirt\nSlot 10: Opentrons NEST 12-Well Reservoir with ethanol loaded\nSlot 11: Opentrons NEST 12-Well Reservoir with CTL medium, isopropanol + beads, and nuclease free water loaded\nNote NEST 12-Well Reservoir with Reagents on Slot 11\nThe minimum volume of reagents in the NEST 12-Well Reservoir will be calculated dependent on the number of columns specified and displayed in the Opentrons app prior to beginning the run. \nEthanol is loaded into all 12 wells on the Nest 12-well reservoir on Slot 10.\n\nCTL medium is loaded onto the first two wells, isopropanol + beads onto wells 4-7, and nuclease free water loaded onto well 12 of the Nest 12-Well reservoir on Slot 11.\n",
"internal": "5f7d18",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* RNA Extraction\n\n",
"description": "This protocol isolates nucleic acid using a MagBio CTL lysis buffer and MAG-S1 particle beads suspended in 100% isopropanol. 200\u00b5L of sample is pre-loaded into each well within a Kingfisher deep-well plate on the Opentrons magnetic module. The sample undergoes extraction with 2 ethanol washes. The eluate is then transferred to a full-skirted PCR plate along with PCR master-mix for further processing.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module](https://opentrons.com/modules/)\n* [P20 Single Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 96 Filter Tip Rack 20 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_20ul?category=tipRack)\n* [Opentrons 96 Filter Tip Rack 200 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_200ul?category=tipRack)\n* [Opentrons NEST 96 well plate 100ul PCR full skirt](https://labware.opentrons.com/?category=wellPlate)\n* [Opentrons NEST 12-Well Reservoir](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [Opentrons 4-in-1 Tube Rack Set (24-tube insert)](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n\n\n\n---\n\n\nFor this protocol, be sure that the pipettes (P20 and P300) are attached.\n\nUsing the customization fields below, set up your protocol.\n* Number of columns: Specify the number of columns of samples loaded onto the NEST deep well plate. Note that runs should only be completed in multiples of full columns (8 samples).\n* Sample and lysis buffer mix repetitions: Specify the number of repetitions to mix the sample and lysis buffer as mentioned in Step 1 of the MAGBIO protocol\n* Mix repetitions before first magnetic engage: Specify the number of mix repetitions to re-suspend the MAG-S1 particles before use (i.e. \"shake well\" in Step 3 of the MAGBIO protocol).\n* Incubation time: Specify the amount of incubation time to allow RNA to bind to the beads\n* Magnetic bead engagement time: Specify the amount of time to engage the magnet for every engagement in protocol\n* Aspiration height from bottom of well (removing supernatant): Specify the height from the bottom of the well in which the pipette will aspirate from when removing supernatant.\n* Multi_channel pipette aspiration flow rate (ul/s): Specify the aspiration flow rate\n* Multi-channel pippette dispense flow rate (ul/s): Specify the dispense flow rate\n* Distance from side of well opposite beads (1mm - 4.15mm): Specify the distance from the side of the well opposite the engaged magnetic beads to aspirate from. A value of 4.15mm returns the center of the well, with 1mm returning 1mm from the side of the well opposite the engaged beads.\n* Bead drying time: Specify the amount of incubation time for the beads to dry at room temperature while magnetically engaged.\n* Bead drying time (with nuclease free water): Specify the amount of time to to dry the beads at room temperature after adding nuclease free water.\n* Nuclease water volume: Specify the amount of nuclease free water to add to the beads\n* P20 single GEN2 mount: Specify which mount to load the P20 single GEN2 pipette.\n* P300 multi GEN2 mount: Specify which mount to load the P300 multi GEN2 pipette.\n\n**Note about 20\u00b5L tip racks**\n\nWhen prompted to replace the 300ul tip racks, be sure to re-load all 3 tip racks as in the original configuration of the deck.\n\n**Labware Setup**\n\nSlots 1: Opentrons Magnetic Module with loaded NEST 96 2mL deep well plate. 200ul of viral sample pre-loaded into each well.\n\nSlot 2, 5: Opentrons 96 Filter Tip Rack 20 \u00b5L\n\nSlot 3, 6, 9: Opentrons 96 Filter Tip Rack 200 \u00b5L\n\nSlot 4: Opentrons NEST 12-Well Reservoir (empty)\n\nSlot 7: Opentrons 4-in-1 Tube Rack Set. Mastermix loaded into tubes A1, B1.\n\nSlot 8: Opentrons NEST 96 well plate 100ul PCR full skirt\n\nSlot 10: Opentrons NEST 12-Well Reservoir with ethanol loaded\n\nSlot 11: Opentrons NEST 12-Well Reservoir with CTL medium, isopropanol + beads, and nuclease free water loaded\n\n**Note NEST 12-Well Reservoir with Reagents on Slot 11**\nThe minimum volume of reagents in the NEST 12-Well Reservoir will be calculated dependent on the number of columns specified and displayed in the Opentrons app prior to beginning the run. \n\nEthanol is loaded into all 12 wells on the Nest 12-well reservoir on Slot 10.\n\n\n\nCTL medium is loaded onto the first two wells, isopropanol + beads onto wells 4-7, and nuclease free water loaded onto well 12 of the Nest 12-Well reservoir on Slot 11.\n\n\n\n\n",
"internal": "5f7d18\n",
diff --git a/protoBuilds/5fa647/README.json b/protoBuilds/5fa647/README.json
index e12d35d20d..56bfd19ead 100644
--- a/protoBuilds/5fa647/README.json
+++ b/protoBuilds/5fa647/README.json
@@ -18,7 +18,7 @@
"labware": "* [Opentrons 96 Filter Tip Rack 20 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_20ul?category=tipRack)\n* [Opentrons 96 Filter Tip Rack 200 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_200ul?category=tipRack)\n* [Opentrons 24 well Aluminum tube rack](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n* [NEST 100ul Full Skirted PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n\n",
"modules": "* [Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module)\n\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Invitrogen](https://www.thermofisher.com/order/catalog/product/11752250#/11752250)\n\n",
+ "partner": "[Invitrogen](https://www.thermofisher.com/order/catalog/product/11752250#/11752250)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P20 Single Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P300 Single Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Mix 10ul (per sample) of 2x RT Rxn Mix + 2ul (per sample) of RT Enzyme Mix in a tube using a single channel pipette.\n2. Pipette 12ul of the mix from step 1 into PCR Plate. (# of tubes = # samples per run). Use same tip.\n3. Using a single channel pipette, pipette designated amount of RNA (per CSV) from the sample tube rack into the PCR Plate containing the mix from step 1, and pipette mix. Change tips between each sample.\n4. Heat at 25C for 10 minutes\n5. Heat at 50C for 30 minutes\n6. Heat at 85C for 5 minutes, then chill on ice\n7. Add 1ul of RNase H to each sample (new tip for each sample)\n8. Heat at 37C for 20 minutes.\n9. Add 60ul of water to each sample (new tip each sample)\n10. Store on ice\n\n",
@@ -30,7 +30,7 @@
"Thermocycler Module"
],
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Invitrogen",
+ "partner": "Invitrogen\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP20 Single Pipette\nP300 Single Pipette\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nMix 10ul (per sample) of 2x RT Rxn Mix + 2ul (per sample) of RT Enzyme Mix in a tube using a single channel pipette.\nPipette 12ul of the mix from step 1 into PCR Plate. (# of tubes = # samples per run). Use same tip.\nUsing a single channel pipette, pipette designated amount of RNA (per CSV) from the sample tube rack into the PCR Plate containing the mix from step 1, and pipette mix. Change tips between each sample.\nHeat at 25C for 10 minutes\nHeat at 50C for 30 minutes\nHeat at 85C for 5 minutes, then chill on ice\nAdd 1ul of RNase H to each sample (new tip for each sample)\nHeat at 37C for 20 minutes.\nAdd 60ul of water to each sample (new tip each sample)\nStore on ice\n",
diff --git a/protoBuilds/6030b7/README.json b/protoBuilds/6030b7/README.json
index 5fd2473c4d..2abe4741c8 100644
--- a/protoBuilds/6030b7/README.json
+++ b/protoBuilds/6030b7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Lexogen QuantSeq"
@@ -8,7 +8,7 @@
"description": "This protocol performs the Lexogen QuantSeq 3'mRNA FWD NGS library prep for up to 96 samples.\n\n\n\nNEST 0.1mL 96-well PCR plates, full skirt\nOpentrons 4-in-1 tuberack set with 4x6 insert holding NEST 1.5mL microcentrifuge tubes\nOpentrons magnetic module\nOpentrons temperature module with 4x6 aluminum block holding NEST 1.5mL microcentrifuge tubes\nOpentrons P20 GEN2 single-channel electronic pipette\nOpentrons P300 multi-channel electronic pipette\nOpentrons 10/20\u00b5l tipracks\nOpentrons 50/300\u00b5l tipracks\n\n\n\n4x6 tuberack (slot 2):\n tube A1: RS\n tube A2: SS1\n* tube A3: SS2\n4x6 aluminum block on temperature module (slot 7):\n tube A1: FS1\n tube A2: FS2/E1\n tube A3: PB\n tube A4: EB\n tube A5: PS\n tube A6: PCR/E3",
"internal": "6030b7",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Lexogen QuantSeq\n\n",
"description": "This protocol performs the [Lexogen QuantSeq 3'mRNA FWD NGS library prep](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2019-12-09/0313rnd/015UG009V0251_QuantSeq_Illumina.pdf) for up to 96 samples.\n\n---\n\n\n* [NEST 0.1mL 96-well PCR plates, full skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons 4-in-1 tuberack set](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 4x6 insert holding [NEST 1.5mL microcentrifuge tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes)\n* [Opentrons magnetic module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons temperature module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [4x6 aluminum block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) holding [NEST 1.5mL microcentrifuge tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes)\n* [Opentrons P20 GEN2 single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons P300 multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 10/20\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 50/300\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n---\n\n\n4x6 tuberack (slot 2):\n* tube A1: RS\n* tube A2: SS1\n* tube A3: SS2\n\n4x6 aluminum block on temperature module (slot 7):\n* tube A1: FS1\n* tube A2: FS2/E1\n* tube A3: PB\n* tube A4: EB\n* tube A5: PS\n* tube A6: PCR/E3\n\n",
"internal": "6030b7\n",
diff --git a/protoBuilds/60d861-2/README.json b/protoBuilds/60d861-2/README.json
index 456000b4fa..21d5cf147e 100644
--- a/protoBuilds/60d861-2/README.json
+++ b/protoBuilds/60d861-2/README.json
@@ -17,14 +17,14 @@
"internal": "60d861\n",
"labware": "* [ThermoFisher MicroAmp\u2122 Fast Optical 96-Well Reaction Plate, 0.1 mL](https://www.thermofisher.com/order/catalog/product/4346907)\n* [NEST 1-Well Reservoirs, 195 mL](https://shop.opentrons.com/nest-1-well-reservoirs-195-ml/)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/opentrons-20-l-tips-160-racks-800-refills/)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Partner Name](partner website link)\n\n",
+ "partner": "[Partner Name](partner website link)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P20 8 Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"reagents": "* [ThermoFisher Scientific mRNA Catcher\u2122 PLUS Purification Kit #K157003](https://www.thermofisher.com/order/catalog/product/K157003)\n* [ThermoFisher Scientific SuperScript\u2122 IV VILO\u2122 Master Mix #11756500](https://www.thermofisher.com/order/catalog/product/11756500)\n\n---\n\n",
"title": "CYP Induction, Part 2/3"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Partner Name",
+ "partner": "Partner Name\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP20 8 Channel Electronic Pipette (GEN2)\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"reagents": [
diff --git a/protoBuilds/60d861-3/README.json b/protoBuilds/60d861-3/README.json
index d7d1c4ab66..1e052f0a87 100644
--- a/protoBuilds/60d861-3/README.json
+++ b/protoBuilds/60d861-3/README.json
@@ -17,14 +17,14 @@
"internal": "60d861\n",
"labware": "* [ThermoFisher MicroAmp\u2122 Fast Optical 96-Well Reaction Plate, 0.1 mL](https://www.thermofisher.com/order/catalog/product/4346907)\n* [NEST 1-Well Reservoirs, 195 mL](https://shop.opentrons.com/nest-1-well-reservoirs-195-ml/)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/opentrons-20-l-tips-160-racks-800-refills/)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Partner Name](partner website link)\n\n",
+ "partner": "[Partner Name](partner website link)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P20 8 Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"reagents": "* [ThermoFisher Scientific mRNA Catcher\u2122 PLUS Purification Kit #K157003](https://www.thermofisher.com/order/catalog/product/K157003)\n* [ThermoFisher Scientific SuperScript\u2122 IV VILO\u2122 Master Mix #11756500](https://www.thermofisher.com/order/catalog/product/11756500)\n\n---\n\n",
"title": "CYP Induction, Part 3/3"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Partner Name",
+ "partner": "Partner Name\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP20 8 Channel Electronic Pipette (GEN2)\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"reagents": [
diff --git a/protoBuilds/60d861/README.json b/protoBuilds/60d861/README.json
index 53efb182e7..c710943f0d 100644
--- a/protoBuilds/60d861/README.json
+++ b/protoBuilds/60d861/README.json
@@ -17,14 +17,14 @@
"internal": "60d861\n",
"labware": "* [Labware name](link to labware on shop.opentrons.com when applicable)\n* Nick is working on auto-filling these sections from the protocol (3/28/2021)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Partner Name](partner website link)\n\n",
+ "partner": "[Partner Name](partner website link)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P300 8 Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"reagents": "* [ThermoFisher Scientific mRNA Catcher\u2122 PLUS Purification Kit #K157003](https://www.thermofisher.com/order/catalog/product/K157003)\n* [ThermoFisher Scientific SuperScript\u2122 IV VILO\u2122 Master Mix #11756500](https://www.thermofisher.com/order/catalog/product/11756500)\n\n---\n\n",
"title": "CYP Induction, Part 1/3: Wash"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Partner Name",
+ "partner": "Partner Name\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP300 8 Channel Electronic Pipette (GEN2)\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"reagents": [
diff --git a/protoBuilds/6151af/README.json b/protoBuilds/6151af/README.json
index f73b61b5bd..2ca64898dd 100644
--- a/protoBuilds/6151af/README.json
+++ b/protoBuilds/6151af/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol automates sample plating from collection tubes to a 96-deepwell plate. \nUsing a Single-Channel Pipette, this protocol will transfer the user-specified volume from sample tubes placed in the Opentrons Tube Rack into a 96-well plate. Samples are transferred top to bottom, and then left to right (rack 1 tube A1 to plate well A1, rack 1 tube B1 to plate well B1, rack 1 tube C1 to plate well C1, rack 1 tube A2 to plate well D1, etc).\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 4.1.0 or later)\nOpentrons P300 GEN2 Single-Channel Pipette\nOpentrons 200\u00b5l Filter/300\u00b5l Standard Tips\nOpentrons 4-in-1 Tube Rack Set with 3x5 15ml tube insert for Avantik 3ml Viral Transport Medium Tubes #GL4692\nNEST 96 Deepwell Plate 2mL\n",
"internal": "6151af",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol automates sample plating from collection tubes to a 96-deepwell plate. \n\nUsing a [Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette), this protocol will transfer the user-specified volume from sample tubes placed in the [Opentrons Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) into a 96-well plate. Samples are transferred top to bottom, and then left to right (rack 1 tube A1 to plate well A1, rack 1 tube B1 to plate well B1, rack 1 tube C1 to plate well C1, rack 1 tube A2 to plate well D1, etc).\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 4.1.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 GEN2 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 200\u00b5l Filter/300\u00b5l Standard Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 3x5 15ml tube insert for [Avantik 3ml Viral Transport Medium Tubes #GL4692](https://www.avantik-us.com/specimen-handling/vtm-3ml-50-pack.asp)\n* [NEST 96 Deepwell Plate 2mL](http://www.cell-nest.com/page94?product_id=101&_l=en)\n\n\n",
"internal": "6151af\n",
diff --git a/protoBuilds/62679a/README.json b/protoBuilds/62679a/README.json
index 7afb7cb0ab..17df076f02 100644
--- a/protoBuilds/62679a/README.json
+++ b/protoBuilds/62679a/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Serial Dilution"
@@ -10,7 +10,7 @@
"internal": "62679a",
"labware": "\nGreiner Bio-One 96-Wellplate V-Bottom 340\u00b5L, #651201\nPerkinElmer 96-Well StorPlate, U-Bottom 450\u00b5L, #6008190\nBeckman Coulter 8-Channel Reservoir 19mL or NEST 12-Well Reservoirs 15 mL #999-00076\nOpentrons 200\u00b5L Filter Tips\nOpentrons 20\u00b5L Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Serial Dilution\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This protocol performs a compound plating and subsequent serial dilution for up to 36 stock compounds. Each compound's dilution will occupy a column of a 96-well plate, requiring up to 4x 96-well plates for a full 48-compound dilution protocol. Stock compound will be transferred to row A of the dilution plates, and a 6-step dilution will be carried down the plate through rows B-G. Well H of that column will contain pure DMSO. A P300 or P20 pipette is automatically selected for each compound's dilution, depending on the dilution factor, and thus, the transfer volume.\n\nThe input file for the workflow should be specified in a format similar to [this template](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/62679a/ex.csv).\n\n---\n\n",
diff --git a/protoBuilds/629f38/README.json b/protoBuilds/629f38/README.json
index d0f414284b..d43f88cb4b 100644
--- a/protoBuilds/629f38/README.json
+++ b/protoBuilds/629f38/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Zymo Quick-DNA HMW MagBead Kit"
@@ -10,7 +10,7 @@
"internal": "629f38",
"labware": "\nNEST 12 Well Reservoir 15 mL\nNEST 1 Well Reservoir 195 mL\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt\nNEST 96 Deepwell Plate 2mL\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Zymo Quick-DNA HMW MagBead Kit\n\n",
"deck-setup": "\n\n",
"description": "\nThis protocol performs the [Zymo Quick-DNA HMW MagBead Kit](https://www.zymoresearch.com/products/quick-dna-hmw-magbead-kit) protocol.\n\nSamples should be loaded on the magnetic module in a NEST deepwell plate. For reagent layout in the 12-channel and 1-channel reservoirs used in this protocol, please see \"Setup\" below.\n\n---\n\n",
diff --git a/protoBuilds/6401f1/README.json b/protoBuilds/6401f1/README.json
index aaf0c957c2..fd3a528149 100644
--- a/protoBuilds/6401f1/README.json
+++ b/protoBuilds/6401f1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -9,7 +9,7 @@
"description": "This protocol uses a p300 single channel and p20 single channel pipette to transfer custom volumes of master mixes to a 96-well PCR plate followed by addition of custom volumes of sample, positive control, or no-template control.\nLinks:\n* example input csv\nThis protocol was developed to combine custom volumes of master mixes with custom volumes of sample, positive control, or no-template control for PCR/qPCR setup.",
"internal": "6401f1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * PCR Prep\n\n",
"deck-setup": "\n\n* Opentrons p20 and p300 tips (Deck Slots 4, 5)\n* PCR Plate hardshell_96_wellplate_200ul (Deck Slot 1)\n* Sample Plate lvl_96_wellplate_1317.97ul (Deck Slot 3)\n* Positive and No Template Control Plate lvl_96_wellplate_1317.97ul (Deck Slot 6)\n* 24-Tube Rack (Master Mixes) beckman_24_tuberack_1000ul (Deck Slot 2)\n\n\n\n",
"description": "\nThis protocol uses a p300 single channel and p20 single channel pipette to transfer custom volumes of master mixes to a 96-well PCR plate followed by addition of custom volumes of sample, positive control, or no-template control.\n\nLinks:\n* [example input csv](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/6401f1/Sample%2Badd%2Bexample%2Bfile_2329C_updated.csv)\n\nThis protocol was developed to combine custom volumes of master mixes with custom volumes of sample, positive control, or no-template control for PCR/qPCR setup.\n\n",
diff --git a/protoBuilds/640a85/README.json b/protoBuilds/640a85/README.json
index a07e5890f3..c239d96c2f 100644
--- a/protoBuilds/640a85/README.json
+++ b/protoBuilds/640a85/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom PCR preparation on DNA samples in triplicate using pre-created mastermix.\nDNA samples should be arranged down columns before moving across rows (sample 1 in plate well A1, sample 2 in plate well B1, ..., sample 9 in plate well A2, etc.).\n\n\n\nThermo Scientific semi-skirted 96-well PCR plate 300ul #AB1400150\nNEST 12-channel reservoir 15ml #360102\nOpentrons P300 multi-channel electronic pipette\nOpentrons P20 single-channel GEN2 electronic pipette\nOpentrons 10/20ul and 50/300ul tipracks\n\n",
"internal": "640a85",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol performs a custom PCR preparation on DNA samples in triplicate using pre-created mastermix.\n\nDNA samples should be arranged down columns before moving across rows (sample 1 in plate well A1, sample 2 in plate well B1, ..., sample 9 in plate well A2, etc.).\n\n---\n\n\n* [Thermo Scientific semi-skirted 96-well PCR plate 300ul #AB1400150](https://www.fishersci.com/shop/products/thermo-scientific-96-well-semi-skirted-plates-flat-deck-11/ab1400150?searchHijack=true&searchTerm=AB1400150&searchType=RAPID&matchedCatNo=AB1400150)\n* [NEST 12-channel reservoir 15ml #360102](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* [Opentrons P300 multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons P20 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 10/20ul and 50/300ul tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n\n---\n\n",
"internal": "640a85\n",
diff --git a/protoBuilds/642c73/README.json b/protoBuilds/642c73/README.json
index 320dba2a6b..67d9139876 100644
--- a/protoBuilds/642c73/README.json
+++ b/protoBuilds/642c73/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol automates the creation of a PCR Plate with a variable number of samples.\n\nUsing the Opentrons 4-in-1 Tube Rack with 2mL Eppendorf tubes, 20\u00b5L of pre-made master mix is added to PCR Plate. Following the addition of master mix to all necessary wells, 5\u00b5L of water is added for the negative control and 5\u00b5L of each sample is added to the other well(s).\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.21.0 or later)\nOpentrons P20 Single-Channel Pipette\nOpentrons 20\u00b5L Filter Tip Rack\nOpentrons 4-in-1 Tube Rack\nBio-Rad 96-Well PCR Plate, 200\u00b5L\nEppendorf 2mL Tubes\nMaster Mix\nNegative Control (Water)\nSamples\n\n\n\nDeck Layout\n\nSlot 1: Opentrons 4-in-1 Tube Rack containing Samples (1-24)\n\nSlot 2: Bio-Rad 96-Well PCR Plate, 200\u00b5L (Final PCR Plate)\n\nSlot 3: Opentrons 4-in-1 Tube Rack\nA1: Master Mix\nD6: Negative Control (Water)\n\nSlot 4: Opentrons 4-in-1 Tube Rack containing Samples (25-48)\n\nSlot 7: Opentrons 4-in-1 Tube Rack containing Samples (49-72)\n\nSlot 10: Opentrons 4-in-1 Tube Rack containing Samples (73-95)\n\n\nUsing the customizations field (below), set up your protocol.\n* Number of Samples: Specify the number of samples to run (1-95).",
"internal": "642c73",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"description": "This protocol automates the creation of a PCR Plate with a variable number of samples.\n\nUsing the [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with [2mL Eppendorf tubes](https://labware.opentrons.com/opentrons_24_tuberack_eppendorf_2ml_safelock_snapcap?category=tubeRack), 20\u00b5L of pre-made master mix is added to PCR Plate. Following the addition of master mix to all necessary wells, 5\u00b5L of water is added for the negative control and 5\u00b5L of each sample is added to the other well(s).\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.21.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [Opentrons 20\u00b5L Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [Bio-Rad 96-Well PCR Plate, 200\u00b5L](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate)\n* Eppendorf 2mL Tubes\n* Master Mix\n* Negative Control (Water)\n* Samples\n\n\n\n---\n\n\n**Deck Layout**\n\n**Slot 1**: [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) containing Samples (1-24)\n\n**Slot 2**: [Bio-Rad 96-Well PCR Plate, 200\u00b5L](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate) (Final PCR Plate)\n\n**Slot 3**: [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\nA1: Master Mix\nD6: Negative Control (Water)\n\n**Slot 4**: [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) containing Samples (25-48)\n\n**Slot 7**: [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) containing Samples (49-72)\n\n**Slot 10**: [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) containing Samples (73-95)\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Samples**: Specify the number of samples to run (1-95).\n\n\n\n\n\n",
"internal": "642c73\n",
diff --git a/protoBuilds/649b67/README.json b/protoBuilds/649b67/README.json
index 98997d0cbd..9d8b812163 100644
--- a/protoBuilds/649b67/README.json
+++ b/protoBuilds/649b67/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Advanced cherrypicking"
@@ -10,7 +10,7 @@
"internal": "649b67",
"labware": "\nAny verified labware found in our Labware Library. This includes aluminum block labware for use with temperature modules, well plates, tuberacks and reservoirs.\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Advanced cherrypicking\n\n",
"deck-setup": "* Example deck setup\n\n\n---\n\n",
"description": "\nThis is an advanced cherrypicking protocol that allows for liquid transfers, aspirating and parking tips, pauses, and dispensing parked tips. The protocol allows for fine tuning how the instructions are carried out by specifying air gap volumes, tip touching and blow outs in the target wells.\n\n\n\nExplanation of complex parameters below:\n\n* `Transfer .csv File`: Here, you should upload a .CSV file formatted as shown below, making sure to include headers in your csv file.\n* `Pipette type in the left mount`: Select which pipette you will use in the left mount for this protocol.\n* `Pipette type in the right mount`: Select which pipette you will use in the right mount for this protocol.\n* `Tip type for the left pipette`: Specify whether you want to use filter tips.\n* `Tip type for the right pipette`: Specify whether you want to use filter tips.\n* `Tip Usage Strategy`: Specify whether you'd like to use a new tip for each transfer, or keep the same tip throughout the protocol.\n\n\n**CSV Format**\n\nExample CSV input:\n\n*Part 1 of an example CSV*\n\n*Part 2 of an example CSV*\n\n\n**Explanation of CSV Headers:**\n\n*step_id:* Succesive natural number that gives each instruction an identity\n\n*instruction:* What type of operation that this row specifies. There are four different instructions. **transfer:** Transfer a volume from the source well to the destination well. **aspirate_and_park_tip:** Aspirates from the source well and parks the tip, the tip can later be retrieved through its step_id. **pause:** Pause for some specified amount of time. **dispense_parked_tip** Dispense a parked tip into a destination well, specify the tip by identifying the step id from a previous aspirate_and_park_tip instruction.\n\n*instruction_parameters:* Some instructions like pause have parameters, in the case of pause it is `time` (see below).\n\n*source_labware:* The source labware used in this instruction, should be a valid Opentrons API name. The API name only needs to be specified the first time a slot is specified in an instruction.\n\n*source_magnetic_module:* yes or no depending on whether a magnetic module is used with the source labware, only needs to be specified the first time and can be left blank after that.\n\n*source_temperature_module:* yes or no depending on whether a temperature module is used with the source labware, only needs to be specified the first time and can be left blank after that.\n\n*source_slot:* A valid slot on the Opentrons deck, i.e. it's a slot from 1 to 11 and it is not occupied by other labware or tipracks. Once a labware has been defined it is enough to specify a slot and a well for any subsequent operation.\n\n*source_well:* The well to aspirate from, if it is a 15 or 50 mL tube the liquid height will be calculated for aspirations.\n\n*Source_well_starting_volume:* The starting volume of the given well, only needs to be specified the first time.\n\n*transfer_volume:* The volume (in uL) to transfer from the source well to the target well.\n\n*air_gap_volume:* An air gap volume for protecting from dripping. Can be 0 or blank if not used.\n\n*dest_labware:* The destination labware used in this instruction, should be a valid Opentrons API name. The API name only needs to be specified the first time the slot is specified in an instruction.\n\n*dest_magnetic_module:* yes or no depending on whether a magnetic module is used with the destination labware, only needs to be specified the first time and can be left blank after that.\n\n*dest_temperature_module:* yes or no depending on whether a temperature module is used with the destination labware, only needs to be specified the first time and can be left blank after that.\n\n*dest_slot:* The slot where the destination labware resides. Once a labware has been defined it is enough to specify a slot and a well for any subsequent operation.\n\n*dest_well:* The destination well to dispense liquid into.\n\n*dest_well_starting_volume:* The initial volume of the target well, only needs to be specified for the first occurence of the well.\n\n*touch_tip:* yes/no depending on if you want tip touching after dispensing into a well.\n\n*blow_out:* yes/no depending on if you want blow out after dispensing into a well.\n\n**Explanation of Instructions**\n\nThe labware is loaded by looking for the first definition of a slot and labware combination that occurs in a CSV instruction. The user should make sure that the first instruction where a source and/or destination slot is mentioned is fully specified with the source and/or destination labware and which (if any) modules it is using. If the temperature module is used the labware should be of an aluminum block type.\n\n**Transfer instructions**\n\nThe transfer instruction can be limited to the step_id, and instruction, source slot, source well, transfer volume, target slot and target well if all the labware has been specified previously. If not, the labware source and target API load names and module usage must also be specified (by answering yes/no in the module fields). The transfer instruction tells the protocol how and where to transfer one liquid from a source well to a target well. Additionally an air gap volume, tip touching in the target well, and blow out in the target well can be specified. There are no instruction_parameters (3rd column field) associated with transfers.\n\n**Pause instructions**\n\nRequires three entries: A step id (1st field), an instruction named \"pause\" (2nd field) and a time parameter (3rd parameter). The time parameter should be formatted likes this: time=[\\h][\\m][\\s]. For example all of the following time parameters are valid:\n`time=1h10m30s`, `time=25s`, `time=5m`, `time=3m10s`, etc.\n\n**Aspirate and park tip instructions**\n\nAspirates a given volume from the source well and parks the tip, can be used for binding an analyte or contaminants in a liquid to a resin filled tip for example.\n\n**Dispense parked tip instructions**\n\nPicks up a parked tip identified by an `aspirate_and_park_tip` instruction step_id and dispenses it to a target well. e.g. the parameter_instruction could be `step_id=3` if that step_id belongs to an earlier `aspirate_and_park_tip` instruction.\n\n---\n\n",
diff --git a/protoBuilds/64c54a/README.json b/protoBuilds/64c54a/README.json
index 5eeed8447d..4cdd7dd51f 100644
--- a/protoBuilds/64c54a/README.json
+++ b/protoBuilds/64c54a/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Mass Spec"
@@ -10,7 +10,7 @@
"internal": "64c54a",
"labware": "\nOpentrons 4-in-1 Tuberack Set with 4x6 adapter for Eppendorf 1.5 mL Safe-Lock Snapcap Tubes\nOpentrons 1000\u00b5L Tips\nNEST 12-Well Reservoirs, 15 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Mass Spec\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nThis custom protocol performs 2 parts of a mass spec sample prep:\n1. Precipitate supernatant transfer\n2. Mobile phase A reconstitution\n\nThe user is prompted to refill the tuberacks for as many samples as specified. The protocol begins with samples 1-24 loaded in the tuberack on slot 1.\n\n---\n\n",
diff --git a/protoBuilds/657ee9-2/README.json b/protoBuilds/657ee9-2/README.json
index bc54943728..e4ea186a73 100644
--- a/protoBuilds/657ee9-2/README.json
+++ b/protoBuilds/657ee9-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Oncomine Focus Assay"
@@ -10,7 +10,7 @@
"internal": "657ee9",
"labware": "\nNEST 96-Well PCR Plates\nOpentrons 20\u00b5L Filter Tip Rack\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Oncomine Focus Assay\n\n",
"deck-setup": "**Slot 7**: [Plate]((https://shop.opentrons.com/nest-0-1-ml-96-well-pcr-plate-full-skirt/)) containing samples (Destination Plate). Can be on [Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) or the [Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module).\n\n**Slot 4**: [Plate]((https://shop.opentrons.com/nest-0-1-ml-96-well-pcr-plate-full-skirt/)) containing FuPa in column 2. Can be on [Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck).\n\n**Slot 3**: [Opentrons 20\u00b5L Filter Tip Rack](https://shop.opentrons.com/opentrons-20ul-filter-tips/)\n\n---\n\n",
"description": "This is the second part in a multi-part protocol designed to automate the Oncomine Focus Assay. For a detailed description of the manual protocol, please see [this resource.](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2022-01-27/kk23ns4/MAN0015819_Part1_OFAv1S5FTLibraryPrep_UG.pdf)\n\nIn this protocol, the OT-2 automates the 'Partial Digestion' portion of the protocol. Using the complex parameters described below, the user can specify which, if any, modules they are using and select from different options pertaining to RNA/cDNA and DNA (**Columns with Samples**).\n\nExplanation of complex parameters below:\n* **Columns with Samples**: Specify which columns should we receive the FuPa reagent. Column numbers should be separated by a comma (,).\n* **Module for Destination Plate**: Select which module you are using for the destination plate (containing samples). If you are not using the [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module), you will be prompted to remove the plate after liquid handling to an off-deck thermal cycler.\n* **Module for Reagent Plate**: Select which module you will use for the plate containing mastermix.\n* **P20-Multi Mount**: Select which mount the [P20 Multi-Channel Pipette](https://shop.opentrons.com/8-channel-electronic-pipette/) is attached to. \n\n---\n\n",
diff --git a/protoBuilds/657ee9-3/README.json b/protoBuilds/657ee9-3/README.json
index d3c68f0da9..b00317c827 100644
--- a/protoBuilds/657ee9-3/README.json
+++ b/protoBuilds/657ee9-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Oncomine Focus Assay"
@@ -10,7 +10,7 @@
"internal": "657ee9-3",
"labware": "\nNEST 96-Well PCR Plates\nOpentrons 20\u00b5L Filter Tip Rack\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Oncomine Focus Assay\n\n",
"deck-setup": "**Slot 7**: [Plate]((https://shop.opentrons.com/nest-0-1-ml-96-well-pcr-plate-full-skirt/) containing samples (Destination Plate). Can be on [Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) or the [Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module).\n\n**Slot 4**: [Plate]((https://shop.opentrons.com/nest-0-1-ml-96-well-pcr-plate-full-skirt/) containing Switch Solution, DNA Ligase, and IonCode Adapters *or* diluted Ion Xpress barcode adapter mix. Can be on [Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck).\n\n**Slot 3**: [Opentrons 20\u00b5L Filter Tip Rack](https://shop.opentrons.com/opentrons-20ul-filter-tips/)\n\n",
"description": "This is the third part in a multi-part protocol designed to automate the Oncomine Focus Assay. For a detailed description of the manual protocol, please see [this resource.](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2022-01-27/kk23ns4/MAN0015819_Part1_OFAv1S5FTLibraryPrep_UG.pdf)\n\nIn this protocol, the OT-2 automates the 'Ligate Adapters' portion of the protocol. Using the complex parameters described below, the user can specify which, if any, modules they are using and select from different options pertaining to RNA/cDNA and DNA (**Columns with Samples**).\n\nExplanation of complex parameters below:\n* **Columns with Samples**: Specify which columns should we receive the FuPa reagent. Column numbers should be separated by a comma (,).\n* **Module for Destination Plate**: Select which module you are using for the destination plate (containing samples). If you are not using the [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module), you will be prompted to remove the plate after liquid handling to an off-deck thermal cycler.\n* **Module for Reagent Plate**: Select which module you will use for the plate containing mastermix.\n* **P20-Multi Mount**: Select which mount the [P20 Multi-Channel Pipette](https://shop.opentrons.com/8-channel-electronic-pipette/) is attached to. \n\n---\n\n",
diff --git a/protoBuilds/657ee9-4/README.json b/protoBuilds/657ee9-4/README.json
index 78431cf751..3cc345bcaa 100644
--- a/protoBuilds/657ee9-4/README.json
+++ b/protoBuilds/657ee9-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Oncomine Focus Assay"
@@ -10,7 +10,7 @@
"internal": "657ee9-4",
"labware": "\nNEST 96-Well PCR Plates\nNEST 12-Well Reservoir, 15mL\nOpentrons 200\u00b5L Filter Tip Rack\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Oncomine Focus Assay\n\n",
"deck-setup": "**Slot 1**: [[NEST 96-Well PCR Plates](https://shop.opentrons.com/nest-0-1-ml-96-well-pcr-plate-full-skirt/) containing samples on the [Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck).\n\n**Slot 2**: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/nest-12-well-reservoirs-15-ml/) containing reagents. \n\n**Slot 3**: [NEST 96-Well PCR Plates](https://shop.opentrons.com/nest-0-1-ml-96-well-pcr-plate-full-skirt/), empty for elutes.\n\n**Slot 4**: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/opentrons-200ul-filter-tips/)\n\n**Slot 5**: [Opentrons 200\u00b5L Filter Tip Rack](https://shop.opentrons.com/opentrons-200ul-filter-tips/)\n\n\n",
"description": "This is the fourth (and last) part in a multi-part protocol designed to automate the Oncomine Focus Assay. For a detailed description of the manual protocol, please see [this resource.](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2022-01-27/kk23ns4/MAN0015819_Part1_OFAv1S5FTLibraryPrep_UG.pdf)\n\nIn this protocol, the OT-2 automates the 'Purify The Library' and the 'Elute The Library' portions of the protocol. Using the complex parameters described below, the user can specify which, if any, modules they are using and select from different options pertaining to RNA/cDNA and DNA (**Columns with Samples**).\n\n**Update**: Protocol was updated to accommodate single column input.\n\nExplanation of complex parameters below:\n* **Columns with Samples**: Specify which columns should we receive the FuPa reagent. Column numbers should be separated by a comma (,).\n* **P300-Multi Mount**: Select which mount the [P300 Multi-Channel Pipette](https://shop.opentrons.com/8-channel-electronic-pipette/) is attached to. \n\n---\n\n",
diff --git a/protoBuilds/657ee9/README.json b/protoBuilds/657ee9/README.json
index bfde524631..1085b8e19b 100644
--- a/protoBuilds/657ee9/README.json
+++ b/protoBuilds/657ee9/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Oncomine Focus Assay"
@@ -10,7 +10,7 @@
"internal": "657ee9",
"labware": "\nNEST 96-Well PCR Plates\nOpentrons 20\u00b5L Filter Tip Rack\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Oncomine Focus Assay\n\n",
"deck-setup": "**Slot 7**: [Plate]((https://shop.opentrons.com/nest-0-1-ml-96-well-pcr-plate-full-skirt/)) containing samples (Destination Plate). Can be on [Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) or the [Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module).\n\n**Slot 4**: [Plate]((https://shop.opentrons.com/nest-0-1-ml-96-well-pcr-plate-full-skirt/)) containing master mix in column 1. Can be on [Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck).\n\n**Slot 3**: [Opentrons 20\u00b5L Filter Tip Rack](https://shop.opentrons.com/opentrons-20ul-filter-tips/)\n\n---\n\n",
"description": "This is the first part in a multi-part protocol designed to automate the Oncomine Focus Assay. For a detailed description of the manual protocol, please see [this resource.](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2022-01-27/kk23ns4/MAN0015819_Part1_OFAv1S5FTLibraryPrep_UG.pdf)\n\nIn this protocol, the OT-2 automates the 'Target Amplification' portion of the protocol. Using the complex parameters described below, the user can specify which, if any, modules they are using and select from different options pertaining to RNA/cDNA and DNA.\n\nExplanation of complex parameters below:\n* **Number of Samples**: Specify the number of samples.\n* **Master Mix Transfer Volume**: Select the volume of mastermix to transfer to the samples. 10\u00b5L is for cDNA synthesis and the other options are for DNA.\n* **Module for Destination Plate**: Select which module you are using for the destination plate (containing samples). If you are not using the [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module), you will be prompted to remove the plate after liquid handling to an off-deck thermal cycler.\n* **Number of Cycles**: If using the [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module), you can specify the number of cycles. For more information, please see [the manual](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2022-01-27/kk23ns4/MAN0015819_Part1_OFAv1S5FTLibraryPrep_UG.pdf).\n* **Module for Reagent Plate**: Select which module you will use for the plate containing mastermix.\n* **P20-Multi Mount**: Select which mount the [P20 Multi-Channel Pipette](https://shop.opentrons.com/8-channel-electronic-pipette/) is attached to. \n\n---\n\n",
diff --git a/protoBuilds/659888/README.json b/protoBuilds/659888/README.json
index f5ba03cb06..a50b62909b 100644
--- a/protoBuilds/659888/README.json
+++ b/protoBuilds/659888/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "659888",
"labware": "\nCorning 96 Well Plate 360 \u00b5L Flat\nOpentrons 20uL Filter Tips\nCustom Agar Plates\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This protocol dispenses diluted phage by column in a source 360ul corning plate to all specified agar plates. The tips are to not touch the agar medium but instead dispense right above it, saving up to 5 times the number of tips per run. Column 1 of the source plate is dispensed into into column 1 of all agar plates in a multi-dispense fashion to additionally save time.\n\nExplanation of complex parameters below:\n* `Number of plates`: Specify the number of agar plates (1-5) to fill for this run.\n* `Number of columns in each plate`: Specify the number of columns (1-12) in each agar plate to fill.\n* `Number of rows in each plate`: Specify the number of rows (1-8) in each agar plate to fill.\n* `P20 Multi-Channel Pipette Mount`: Specify which mount (left or right) to host the P20 multi-channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/65ed01/README.json b/protoBuilds/65ed01/README.json
index 35bfed4950..1c0bca7174 100644
--- a/protoBuilds/65ed01/README.json
+++ b/protoBuilds/65ed01/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Purification"
@@ -8,7 +8,7 @@
"description": "This protocol performs a nucleic acid purification with the Opentrons magnetic module.\nLysis buffer is added to sample and mixed. The subsequent lysate is then added to magnetic beads with isopropanol. After mixing, the Opentrons magnetic module is engaged and supernatant is removed and returned to reservoir. Two washes are then performed using wash and elution buffer, respectively. The final elute is transferred to an Opentrons 96-Well Aluminum Block plate. \n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P300 Multi-Channel Pipette\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 200\u00b5L Tips\nOpentrons Magnetic Module\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nNEST 1 Well Reservoir 195 mL\nYDP 96-Well Square Well-Plate 2200ul\n\n\n\nSlot 1: Opentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nSlot 2: Opentrons 96 Filter Tip Rack 200 \u00b5L\nSlot 3: Opentrons 96 Filter Tip Rack 200 \u00b5L\nSlot 6: YDP 96-Well Square Well-Plate 2200ul with Magnetic Module\nSlot 11: NEST 1 Well Reservoir 195 mL\n\n\nUsing the customizations field (below), set up your protocol.\n Distance From Side of well: Specify the distance from the well wall the pipette will aspirate from. The pipette will aspirate from this distance on the well wall opposite that of the magnetic beads. An input value of 4.05 returns the center of the well.\n Number of Plates: Specify the mount side for the P300 Multi Channel Pipette.",
"internal": "65ed01",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Nucleic Acid Purification\n\n\n",
"description": "This protocol performs a nucleic acid purification with the Opentrons magnetic module.\n\nLysis buffer is added to sample and mixed. The subsequent lysate is then added to magnetic beads with isopropanol. After mixing, the Opentrons magnetic module is engaged and supernatant is removed and returned to reservoir. Two washes are then performed using wash and elution buffer, respectively. The final elute is transferred to an Opentrons 96-Well Aluminum Block plate. \n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 96 Filter Tip Rack 200 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_200ul?category=tipRack)\n* [Opentrons 200\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons Magnetic Module](https://opentrons.com/modules/magnetic-module/)\n* [Opentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L](https://labware.opentrons.com/opentrons_96_aluminumblock_generic_pcr_strip_200ul?category=aluminumBlock)\n* [NEST 1 Well Reservoir 195 mL](https://labware.opentrons.com/nest_1_reservoir_195ml?category=reservoir)\n* YDP 96-Well Square Well-Plate 2200ul\n\n\n\n---\n\n\nSlot 1: Opentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\n\nSlot 2: Opentrons 96 Filter Tip Rack 200 \u00b5L\n\nSlot 3: Opentrons 96 Filter Tip Rack 200 \u00b5L\n\nSlot 6: YDP 96-Well Square Well-Plate 2200ul with Magnetic Module\n\nSlot 11: NEST 1 Well Reservoir 195 mL\n\n\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Distance From Side of well**: Specify the distance from the well wall the pipette will aspirate from. The pipette will aspirate from this distance on the well wall opposite that of the magnetic beads. An input value of 4.05 returns the center of the well.\n* **Number of Plates**: Specify the mount side for the P300 Multi Channel Pipette.\n\n",
"internal": "65ed01\n",
diff --git a/protoBuilds/66aa48/README.json b/protoBuilds/66aa48/README.json
index 19323b8562..4a338f3802 100644
--- a/protoBuilds/66aa48/README.json
+++ b/protoBuilds/66aa48/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -9,7 +9,7 @@
"internal": "66aa48",
"labware": "\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 24 Tube Rack with Eppendorf 2 mL Safe-Lock Snapcap\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n\n",
"description": "This protocol allows the robot to create up to 4 different pooling samples by consolidating cherrypicked wells from a 96 tall-well plates. The user will need upload one CSV each for increase, no change, decrease, and inactive property changes. Each CSV will contain information of the wells from which to be picked. See Additional Notes for the format of the CSVs, as well as the arrangement of pooling tubes in the Opentrons 4x6 2ml Eppendorf tuberack. If you would like to skip a pool as specified by 1 of the 4 CSV files, upload an empty CSV file corresponding to that pool.\n\nYour CSV files should contain source wells of the 96-well source plate each on its own line, as in the following: \n```\nA2\nA3\nA4\nA7\nA8\nB3\nB5\n```\n\nThe pooling tubes are seated in the Opentrons tube rack according to the following setup:\n* inactive: tube A1\n* decrease: tube B1\n* no change: tube C1\n* increase: tube D1\n\n\n",
"internal": "66aa48\n",
diff --git a/protoBuilds/66e60f/README.json b/protoBuilds/66e60f/README.json
index 52b99b6aba..e734b34da2 100644
--- a/protoBuilds/66e60f/README.json
+++ b/protoBuilds/66e60f/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "66e60f",
"labware": "\nNEST 12-Well Reservoirs, 15 mL\nAn Opentrons compatible 96-well sample source plates\nAn Opentrons compatible 96-well target plates\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"deck-setup": "Slots:\n1. 20 \u00b5L filter tip-rack (B)\n2. Empty\n3. 20 \u00b5L filter tip-rack (A)\n4. Final Plate B\n5. Tuberack\n6. Final Plate A\n7. DNA Plate B\n8. Empty\n9. DNA Plate A\n10. 200 \u00b5L filter tip-rack\n11. Water reservoir\n\n\n",
"description": "This protocol performs a custom sample normalization from one or two source plate(s) to one or two final plate(s), diluting with water from a 12 well reservoir on slot 11 based on an input csv file. After normalization the protocol samples 5 \u00b5L from each well from the final plate(s) and dispenses it into a DNA pool tube on the tuberack on slot 5 (the tube in position A2). Sample and diluent volumes are specified via the .csv file in the following format, including the header line (empty lines ignored, also note that this is just an example and that any number of lines less than and up to 2x96 may be specified):\n\n```\nPlate, Well, SampleID, Concentration, VolumeToDispense\nA, A1, Sample1, 3.28, 12.52,\n...\nA, H12, Sample96, 5.30, 9.70,\nB, A1, Sample97, 6.34, 8.60,\n...\nB, H12, Sample192, 7.34, 5.60,\n```\nThe plate is either A (to Final plate slot 6 from DNA plate slot 9) or B (to Final plate slot 4 from DNA plate slot 7)\n\n**Update (Jan 28, 2022)**: This protocol has been updated. The user can now select between the [Opentrons P300 GEN2 Single-Channel pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) or the [Opentrons P300 GEN2 8-Channel pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette).\n\nParameters:\n* `.csv input file`: Input file (see above)\n* `Source plate type`: Opentrons compatible 96 well plate containing DNA samples\n* `Destination plate type`: Opentrons compatible 96 well plates where DNA samples are diluted with water\n* `Tuberack/tubes`: Tuberack/tube combination for DNA and water bins and DNA pool.\n* `P300 type`: Single or multichannel: Only used for the initial water distribution\n* `Air gap volume`: Volume of air for all air gaps, default is 10 \u00b5L\n\n---\n\n\n\n",
diff --git a/protoBuilds/68ce98/README.json b/protoBuilds/68ce98/README.json
index 1a778a1ba8..834db01b69 100644
--- a/protoBuilds/68ce98/README.json
+++ b/protoBuilds/68ce98/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -9,7 +9,7 @@
"description": "This protocol performs the transfer of samples and reagent from one plate to another. It will pre-wet the tip and aspirate/dispense 300uL 3 times. Then it will transfer 750 uL to a second plate with a 30uL air gap, blow out and touch tip at the destination.\n\n\n\nP300 multi-channel GEN2 electronic pipette\nWaters 2mL 96 Well Plate\n\n\n",
"internal": "68ce98",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "* Waters 2mL 96 Well Plate (Slot 1)\n* Waters 2mL 96 Well Plate (Slot 2)\n* Opentrons 300uL Tip Rack (Slot 4)\n\n",
"description": "This protocol performs the transfer of samples and reagent from one plate to another. It will pre-wet the tip and aspirate/dispense 300uL 3 times. Then it will transfer 750 uL to a second plate with a 30uL air gap, blow out and touch tip at the destination.\n\n---\n\n\n\n* [P300 multi-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Waters 2mL 96 Well Plate](https://www.waters.com/nextgen/us/en/shop/vials-containers--collection-plates/186002482-96-well-sample-collection-plate-2-ml-square-well-50-pk.html)\n\n---\n\n\n",
diff --git a/protoBuilds/69486f/README.json b/protoBuilds/69486f/README.json
index d72f4a4a82..cbaa73ae2d 100644
--- a/protoBuilds/69486f/README.json
+++ b/protoBuilds/69486f/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"KAPA HiFi"
@@ -10,7 +10,7 @@
"internal": "69486f",
"labware": "\nAgilent AriaMx 96 Well PCR Plates 200ul #401490\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap Tubes\nOpentrons 20\u00b5L Filter Tips\nOpentrons 200\u00b5L Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* KAPA HiFi\n\n",
"deck-setup": "\nNote that these deck states depict a combination of the maximum number of PCR 1 forward primers.\n\nStarting state (showing normalization for <= 28 sample + subsample product): \n \n* green on slot 1: starting samples according to above example .csv\n* clear on slot 7 (tubes A1-D2): empty tubes for PCR 1 mix creation\n* orange on slot 7 (A3-D4): unique PCR 1 forward primers (in column order)\n* clear on slot 7 (tubes A5-D6): empty tubes for PCR 2 mix creation\n* blue on slot 8 (A1): water\n* pink on slot 8 (B1): PCR mastermix\n* purple on slot 8 (C1): PCR 1 reverse primer\n* purple on slot 8 (D1): PCR 2 reverse primer\n* dark blue on slot 8 (A2-D3): tubes for normalized pools\n* orange on slot 8 (A4-D5): unique PCR 1 forward primers (in column order)\n* light blue on slot 8 (A6): normalization binding buffer\n* light purple on slot 8 (B6-C6): normalization wash buffer\n* green on slot 8 (D6): normalization elution buffer\n\nStarting state (showing normalization for <= 28 sample + subsample product): \n\n* green on slot 1: starting samples according to above example .csv\n* clear on slot 7 (tubes A1-D2): empty tubes for PCR 1 mix creation\n* orange on slot 7 (A3-D4): unique PCR 1 forward primers (in column order)\n* clear on slot 7 (tubes A5-D6): empty tubes for PCR 2 mix creation\n* blue on slot 8 (A1): water\n* pink on slot 8 (B1): PCR mastermix\n* purple on slot 8 (C1): PCR 1 reverse primer\n* purple on slot 8 (D1): PCR 2 reverse primer\n* dark blue on slot 8 (A2-D3): tubes for normalized pools\n* orange on slot 8 (A4-D5): unique PCR 1 forward primers (in column order)\n* light blue on slot 11 (A1): normalization binding buffer\n* light purple on slot 11 (A2): normalization wash buffer\n* green on slot 11 (A3): normalization elution buffer\n\n---\n\n",
"description": "\nThis custom protocol performs part 2 of the [KAPA HiFi NGS Library Prep kit](https://sequencing.roche.com/en/products-solutions/products/sample-preparation/library-amplification/kapa-hifi-kits.html). The protocol can accommodate up to 8 samples. The operator is prompted to perform PCR and normalization off-deck.\n\nThe number of samples and subsamples should be specified in a .csv file formatted as follows:\n\n```\nsample index,# subsamples\n1,1\n2,2\n```\n\nYou can also access a template file [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/69486f/ex.csv).\n\n---\n\n",
diff --git a/protoBuilds/698b9e-part2/README.json b/protoBuilds/698b9e-part2/README.json
index 8aa7009c74..0009fb12ce 100644
--- a/protoBuilds/698b9e-part2/README.json
+++ b/protoBuilds/698b9e-part2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "698b9e-part2",
"labware": "\nOpentrons Filter Tip Racks 20ul\nOpentrons Filter Tip Racks 200ul\nNunc\u2122 96-Well Polypropylene Storage Microplates\nMicroAmp\u2122 Fast Optical 96-Well Reaction Plate with Barcode, 0.1 mL\nNEST 12-Well Reservoir, 15 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n\n\n\n\n\n\n---\n\n",
"description": "This protocol is part two of a two part series for performing a custom PCR prep with 1.5 mL Falcon conical tubes. Find Part 1 of the protocol here:\n\n* [PCR Prep with 1.5 mL Tubes Part 1 - Plate Filling](https://protocols.opentrons.com/protocol/698b9e)\n\nThis protocol makes a saliva and buffer solution, and then transfers this solution to a mastermix plate for further processing. \n\n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify the number of sample tubes you will use over this run. After samples from all tubes in the tube rack have been moved to the buffer plate, the protocol will pause and ask for the user to replace the tubes. For example, a value of `32` for `Number of Samples` would ask the user to replace the tube rack once (after 15 samples), and the OT-2 will then only aspirate from the first two tubes after replacement.\n* `Aspirate Saliva Delay Time (in seconds)`: Select the amount of time to delay after aspirating saliva or saliva/buffer solution to allow for full achievement of aspiration.\n* `P300 Multi Channel Mount`: Specify which side (left or right) to mount the P300 Multi Channel Pipette. \n* `P20 Single Channel Mount`: Specify which side (left or right) to mount the P20 Single Channel Pipette. \n\n\n---\n\n",
diff --git a/protoBuilds/698b9e/README.json b/protoBuilds/698b9e/README.json
index 451a90e4f2..06e5afd4f0 100644
--- a/protoBuilds/698b9e/README.json
+++ b/protoBuilds/698b9e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "698b9e",
"labware": "\nOpentrons Filter Tip Racks 20ul\nOpentrons Filter Tip Racks 200ul\nNunc\u2122 96-Well Polypropylene Storage Microplates\nMicroAmp\u2122 Fast Optical 96-Well Reaction Plate with Barcode, 0.1 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "* Deck setup if running 9 plates. If selecting less than 9 plates, fill the deck slots in numerical order starting from Deck Slot 3.\n\n\n",
"description": "This protocol is part one of a two part series for performing a custom PCR prep with 1.5 mL Falcon conical tubes. Find Part 2 of the protocol here:\n\n* [PCR Prep with 1.5 mL Tubes Part 2 - Adding Sample](https://protocols.opentrons.com/protocol/698b9e-part2)\n\nThis protocol fills one of two plates with either reaction buffer or mastermix as specified by the user. The user can also specify how many plates they would like to fill with buffer, or mastermix. Plates should then be kept in cold storage, ready for use as saliva samples come in.\n\n\nExplanation of complex parameters below:\n* `Number of Plates`: Specify the number of plates you want to populate with reagent.\n* `Reagent`: Specify whether you would like to fill the number of plates selected above with mastermix, or buffer. 80ul will be transferred if `Buffer` is selected, while 7.6ul will be transferred to each well in every plate if `Mastermix` is selected. The P300 Multi Channel pipette is used for `Buffer`, while the P20 Single Channel Pipette is used for `Mastermix`.\n* `P300 Multi Channel Mount`: Specify which side (left or right) to mount the P300 Multi Channel Pipette. \n* `P20 Single Channel Mount`: Specify which side (left or right) to mount the P20 Single Channel Pipette. \n\n\n---\n\n",
diff --git a/protoBuilds/69cf81-2/README.json b/protoBuilds/69cf81-2/README.json
index 1f0256bafc..a11b2a6801 100644
--- a/protoBuilds/69cf81-2/README.json
+++ b/protoBuilds/69cf81-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "69cf81",
"labware": "\nThermoFisher MicroAmp\u2122 Optical 96-Well Reaction Plate with Barcode #4306737\nNEST 15 Reservoir 12ml\nOpentrons 20\u00b5l Tipracks or Rainin 20\u00b5l Tipracks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "The following example shows a setup for 16 samples and 6 primers. Note that samples can continue across the plate, and primers can continue down and then across the tuberack. \n* green in reagent reservoir: unique primer, BigDye + water mix\n* blue in sample plate: PCR templates\n\n\n---\n\n",
"description": "\nThis protocol performs a custom PCR prep by transferring samples, primers, BigDye, and water to a clean PCR plate. New tips are used for each transfer, both to avoid contamination and to accurately dispense small volumes (1-6\u00b5l). See below for deck and reagent setups.\n\nNote that the product of the number of samples multiplied by the number of primers cannot exceed 96, or an exception will be thrown.\n\nAn example of the transfer scheme is shown in the following schematic: \n\n\n---\n\n",
diff --git a/protoBuilds/69cf81/README.json b/protoBuilds/69cf81/README.json
index 7f6af16660..9489bfba22 100644
--- a/protoBuilds/69cf81/README.json
+++ b/protoBuilds/69cf81/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "69cf81",
"labware": "\nThermoFisher MicroAmp\u2122 Optical 96-Well Reaction Plate with Barcode #4306737\nNEST 15 Reservoir 12ml\nOpentrons 24 Tube Rack with NEST 1.5 mL Snapcap (or equivalent)\nOpentrons 20\u00b5l Tipracks or Rainin 20\u00b5l Tipracks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "The following example shows a setup for 8 samples and 2 primers. Note that samples can continue across the plate, and primers can continue down and then across the tuberack. \n* green in reagent reservoir: BigDye + water mix\n* pink in primer rack: primer 1\n* orange in primer rack: primer 2\n\n\n---\n\n",
"description": "\nThis protocol performs a custom PCR prep by transferring samples, primers, BigDye, and water to a clean PCR plate. New tips are used for each transfer, both to avoid contamination and to accurately dispense small volumes (1-6\u00b5l). See below for deck and reagent setups.\n\nNote that the product of the number of samples multiplied by the number of primers cannot exceed 96, or an exception will be thrown.\n\nAn example of the transfer scheme is shown in the following schematic: \n\n\n---\n\n",
diff --git a/protoBuilds/69db93/README.json b/protoBuilds/69db93/README.json
index 1f995db029..7e1379b5eb 100644
--- a/protoBuilds/69db93/README.json
+++ b/protoBuilds/69db93/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"RNA Extraction"
@@ -8,7 +8,7 @@
"description": "This protocol isolates nucleic acid preloaded onto a NEST 96 deep well plate premixed with magnetic beads. The sample undergoes extraction with one wash step. The eluate is then transferred to a full-skirted PCR plate for further processing. Tips are parked throughout the protocol for potential reuse. No tips are discarded in the trash.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Magnetic Module\nP300 Multi-Channel Pipette\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons NEST 96 well plate 100ul PCR full skirt\nOpentrons NEST 96 deep well plate 2mL\nOpentrons NEST 12-Well Reservoir\nOpentrons NEST 1-Well Reservoir\n\n\n\nFor this protocol, be sure that the P300 pipette is attached.\nUsing the customization fields below, set up your protocol.\n Number of samples: Specify the number of samples loaded onto the NEST deep well plate. Note that runs should only be completed in multiples of full columns (8 samples).\n Aspiration height from bottom of well (removing supernatant): Specify the height from the bottom of the well in which the pipette will aspirate from when removing supernatant.\n Multi_channel pipette aspiration flow rate for mixing (ul/s): Specify the aspiration flow rate for all mix steps.\n Multi_channel pipette dispense flow rate for mixing (ul/s): Specify the dispense flow rate for all mix steps.\n Distance from side of well opposite beads (1mm - 4.15mm): Specify the distance from the side of the well opposite the engaged magnetic beads to aspirate from. A value of 4.15mm returns the center of the well, with 1mm returning 1mm from the side of the well opposite the engaged beads.\n P300 multi GEN2 mount: Specify which mount to load the P300 multi GEN2 pipette.\nNote About Reagents\nBind buffer + Etoh, wash buffer, and TE in the Nest 12 well reservoir should be loaded into their respective wells prior to the protocol. The minimum volume of which (dependent on how many samples are being run) can be found below.\n\n\n\nLabware Setup\nSlots 1: Opentrons Magnetic Module with loaded NEST 96 2mL deep well plate. 400ul sample and pre-mix loaded in each well.\nSlot 2: Nest 12 well reservoir with binding buffer + Etoh, wash buffer, and TE loaded.\nSlot 3: Opentrons NEST 96 well plate 100ul PCR full skirt.\nSlot 4, 7, 10: Opentrons 96 Filter Tip Rack 200 \u00b5L (empty for parking).\nSlot 5, 6, 8, 9: Opentrons 96 Filter Tip Rack 200 \u00b5L.\nSlot 11: Nest 1-well reservoir",
"internal": "69db93",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* RNA Extraction\n\n",
"description": "This protocol isolates nucleic acid preloaded onto a NEST 96 deep well plate premixed with magnetic beads. The sample undergoes extraction with one wash step. The eluate is then transferred to a full-skirted PCR plate for further processing. Tips are parked throughout the protocol for potential reuse. No tips are discarded in the trash.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module](https://opentrons.com/modules/)\n* [P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 96 Filter Tip Rack 200 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_200ul?category=tipRack)\n* [Opentrons NEST 96 well plate 100ul PCR full skirt](https://labware.opentrons.com/?category=wellPlate)\n* [Opentrons NEST 96 deep well plate 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate)\n* [Opentrons NEST 12-Well Reservoir](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [Opentrons NEST 1-Well Reservoir](https://shop.opentrons.com/collections/reservoirs/products/nest-1-well-reservoir-195-ml)\n\n\n\n\n---\n\n\nFor this protocol, be sure that the P300 pipette is attached.\n\nUsing the customization fields below, set up your protocol.\n* Number of samples: Specify the number of samples loaded onto the NEST deep well plate. Note that runs should only be completed in multiples of full columns (8 samples).\n* Aspiration height from bottom of well (removing supernatant): Specify the height from the bottom of the well in which the pipette will aspirate from when removing supernatant.\n* Multi_channel pipette aspiration flow rate for mixing (ul/s): Specify the aspiration flow rate for all mix steps.\n* Multi_channel pipette dispense flow rate for mixing (ul/s): Specify the dispense flow rate for all mix steps.\n* Distance from side of well opposite beads (1mm - 4.15mm): Specify the distance from the side of the well opposite the engaged magnetic beads to aspirate from. A value of 4.15mm returns the center of the well, with 1mm returning 1mm from the side of the well opposite the engaged beads.\n* P300 multi GEN2 mount: Specify which mount to load the P300 multi GEN2 pipette.\n\n**Note About Reagents**\nBind buffer + Etoh, wash buffer, and TE in the Nest 12 well reservoir should be loaded into their respective wells prior to the protocol. The minimum volume of which (dependent on how many samples are being run) can be found below.\n\n\n\n\n\n\n\n\n**Labware Setup**\n\nSlots 1: Opentrons Magnetic Module with loaded NEST 96 2mL deep well plate. 400ul sample and pre-mix loaded in each well.\n\nSlot 2: Nest 12 well reservoir with binding buffer + Etoh, wash buffer, and TE loaded.\n\nSlot 3: Opentrons NEST 96 well plate 100ul PCR full skirt.\n\nSlot 4, 7, 10: Opentrons 96 Filter Tip Rack 200 \u00b5L (empty for parking).\n\nSlot 5, 6, 8, 9: Opentrons 96 Filter Tip Rack 200 \u00b5L.\n\nSlot 11: Nest 1-well reservoir\n\n\n",
"internal": "69db93\n",
diff --git a/protoBuilds/6a77d9/README.json b/protoBuilds/6a77d9/README.json
index 354c6e9de9..e68fbfdedd 100644
--- a/protoBuilds/6a77d9/README.json
+++ b/protoBuilds/6a77d9/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Slide Spotting"
@@ -8,7 +8,7 @@
"description": "This protocol performs simple spotting transfers of variable volume to a custom plate occupying slots 1, 2, 3, 4, 5, 6, 7, 8, & 9 of the OT-2 deck.\nCalibration\nThe user should calibrate to the top left-most spot of the top left-most grid on the shaker (A). The pipette tip should be calibrated to just above the center of this spot during calibration.\nOffsets\nOffsets are used to define the location of each additional grid. The offset of the top left-most spot of each additional grid (B) relative to the calibration point (A) will be specified in a pair of x- and y-coordinates via .csv file input, as in the following example (be sure to include the header line):\nx offset (in mm),y offset (in mm)\n50,0\n0, 70\n50, 70\nThe above example will transfer to 3 grids with specified offsets in addition to the starting grid to which the protocol is calibrated.\nYou can also download and modify this template.\n\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons P20 Single-Channel Pipette\nOpentrons 20\u00b5l Tiprack\nUSA Scientific 96 Deep Well Plate 2.4 mL\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\n\nIf using a multi-channel pipette, liquids A-D should be in reservoir wells A1-D1, and liquids E-H should be in reservoir wells E2-H2. \nIf using a single-channel pipette, liquids A-H should be in reservoir wells A1-H1.\n",
"internal": "6a77a9",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Slide Spotting\n\n",
"description": "\nThis protocol performs simple spotting transfers of variable volume to a custom plate occupying slots 1, 2, 3, 4, 5, 6, 7, 8, & 9 of the OT-2 deck.\n\n**Calibration** \nThe user should calibrate to the top left-most spot of the top left-most grid on the shaker (A). The pipette tip should be calibrated to just above the center of this spot during calibration.\n\n**Offsets** \nOffsets are used to define the location of each additional grid. The offset of the top left-most spot of each additional grid (B) relative to the calibration point (A) will be specified in a pair of x- and y-coordinates via \t`.csv` file input, as in the following example (be sure to include the header line):\n```\nx offset (in mm),y offset (in mm)\n50,0\n0, 70\n50, 70\n```\n\nThe above example will transfer to 3 grids with specified offsets in addition to the starting grid to which the protocol is calibrated.\n\nYou can also download and modify [this template](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/6a77d9/ex.csv).\n\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 20\u00b5l Tiprack](https://shop.opentrons.com/collections/opentrons-tips)\n* [USA Scientific 96 Deep Well Plate 2.4 mL](https://labware.opentrons.com/usascientific_96_wellplate_2.4ml_deep)\n\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n---\n\n\n* If using a multi-channel pipette, liquids A-D should be in reservoir wells A1-D1, and liquids E-H should be in reservoir wells E2-H2. \n* If using a single-channel pipette, liquids A-H should be in reservoir wells A1-H1.\n\n",
"internal": "6a77a9\n",
diff --git a/protoBuilds/6a93a2-part2/README.json b/protoBuilds/6a93a2-part2/README.json
index c032e17c4d..1cb6951c3c 100644
--- a/protoBuilds/6a93a2-part2/README.json
+++ b/protoBuilds/6a93a2-part2/README.json
@@ -18,7 +18,7 @@
"labware": "* [NEST 0.1 mL 96-Well PCR Plate, Full Skirt](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 200uL Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 24-Tube Aluminum Block](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n\n",
"modules": "* [Temperature Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Swift Biosciences](https://swiftbiosci.com/protocols/)\n\n",
+ "partner": "[Swift Biosciences](https://swiftbiosci.com/protocols/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P20 GEN2 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P300 GEN2 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Adaptase mastermix is made on ice (4C)\n2. User vortexes mix tube\n3. Samples undergo thermocycler profile\n4. User moves samples to ice for 4 minutes\n5. Adaptase mastermix is added to samples\n6. Samples undergo thermocycler profile\n7. Samples are brought back down to 4C, ready for extension.\n\n\n\n\n",
@@ -31,7 +31,7 @@
"Thermocycler Module"
],
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Swift Biosciences",
+ "partner": "Swift Biosciences\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP20 GEN2 Single Channel Pipette\nP300 GEN2 Multi-Channel Pipette\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nAdaptase mastermix is made on ice (4C)\nUser vortexes mix tube\nSamples undergo thermocycler profile\nUser moves samples to ice for 4 minutes\nAdaptase mastermix is added to samples\nSamples undergo thermocycler profile\nSamples are brought back down to 4C, ready for extension.\n",
diff --git a/protoBuilds/6a93a2-part3/README.json b/protoBuilds/6a93a2-part3/README.json
index 2b1928f5cd..65b06634bb 100644
--- a/protoBuilds/6a93a2-part3/README.json
+++ b/protoBuilds/6a93a2-part3/README.json
@@ -18,7 +18,7 @@
"labware": "* [NEST 0.1 mL 96-Well PCR Plate, Full Skirt](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 200uL Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 24-Tube Aluminum Block](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n\n",
"modules": "* [Temperature Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Magnetic Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Swift Biosciences](https://swiftbiosci.com/protocols/)\n\n",
+ "partner": "[Swift Biosciences](https://swiftbiosci.com/protocols/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P20 GEN2 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P300 GEN2 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Extension Mastermix is made on the Opentrons Aluminum 24 tube rack at 4C.\n2. User is prompted to mix and pulse mastermix, place back in rack.\n3. 23ul of mastermix is added to each sample\n3. Samples undergo thermocycler profile.\n4. Samples are then moved to the magnetic module.\n5. 30ul of Low EDTA TE are added to samples.\n6. Magnetic beads are added to samples.\n7. Samples are incubated at room temperature.\n8. Protocol pauses - plate is removed from deck, sealed and spun down.\n9. Plate is placed back on magnetic module.\n10. Supernatant removed.\n11. Two ethanol washes of samples with 30 second incubation periods before supernatant is removed.\n12. Protocol pauses - plate is removed from deck, sealed and spun down.\n13. Low EDTA TE is added to beads, incubate. \n14. Protocol pauses - plate is removed from deck, sealed and spun down.\n15. Supernatant is moved to columns 5, 6, and 7.\n\n\n",
@@ -32,7 +32,7 @@
"Thermocycler Module"
],
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Swift Biosciences",
+ "partner": "Swift Biosciences\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP20 GEN2 Single Channel Pipette\nP300 GEN2 Multi-Channel Pipette\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nExtension Mastermix is made on the Opentrons Aluminum 24 tube rack at 4C.\nUser is prompted to mix and pulse mastermix, place back in rack.\n23ul of mastermix is added to each sample\nSamples undergo thermocycler profile.\nSamples are then moved to the magnetic module.\n30ul of Low EDTA TE are added to samples.\nMagnetic beads are added to samples.\nSamples are incubated at room temperature.\nProtocol pauses - plate is removed from deck, sealed and spun down.\nPlate is placed back on magnetic module.\nSupernatant removed.\nTwo ethanol washes of samples with 30 second incubation periods before supernatant is removed.\nProtocol pauses - plate is removed from deck, sealed and spun down.\nLow EDTA TE is added to beads, incubate. \nProtocol pauses - plate is removed from deck, sealed and spun down.\nSupernatant is moved to columns 5, 6, and 7.\n",
diff --git a/protoBuilds/6a93a2-part4/README.json b/protoBuilds/6a93a2-part4/README.json
index ee49f930d9..8bbc3d4dab 100644
--- a/protoBuilds/6a93a2-part4/README.json
+++ b/protoBuilds/6a93a2-part4/README.json
@@ -18,7 +18,7 @@
"labware": "* [NEST 0.1 mL 96-Well PCR Plate, Full Skirt](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 200uL Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 24-Tube Aluminum Block](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n\n",
"modules": "* [Magnetic Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Swift Biosciences](https://swiftbiosci.com/protocols/)\n\n",
+ "partner": "[Swift Biosciences](https://swiftbiosci.com/protocols/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P300 GEN2 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Magnetic beads are added to samples.\n2. Samples are incubated at room temperature.\n3. Protocol pauses - plate is removed from deck, sealed and spun down.\n4. Plate is placed back on magnetic module.\n5. Supernatant removed.\n6. Two ethanol washes of samples with 30 second incubation periods before supernatant is removed.\n7. Protocol pauses - plate is removed from deck, sealed and spun down.\n8. Low EDTA TE is added to beads, incubate. \n9. Protocol pauses - plate is removed from deck, sealed and spun down.\n10. Supernatant is moved to columns 5, 6, and 7 on the new PCR plate on Slot 9.\n\n\n\n",
@@ -30,7 +30,7 @@
"Magnetic Module (GEN2)"
],
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Swift Biosciences",
+ "partner": "Swift Biosciences\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP300 GEN2 Multi-Channel Pipette\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nMagnetic beads are added to samples.\nSamples are incubated at room temperature.\nProtocol pauses - plate is removed from deck, sealed and spun down.\nPlate is placed back on magnetic module.\nSupernatant removed.\nTwo ethanol washes of samples with 30 second incubation periods before supernatant is removed.\nProtocol pauses - plate is removed from deck, sealed and spun down.\nLow EDTA TE is added to beads, incubate. \nProtocol pauses - plate is removed from deck, sealed and spun down.\nSupernatant is moved to columns 5, 6, and 7 on the new PCR plate on Slot 9.\n",
diff --git a/protoBuilds/6a93a2-part5/README.json b/protoBuilds/6a93a2-part5/README.json
index f047416dbc..29a7fdf9ae 100644
--- a/protoBuilds/6a93a2-part5/README.json
+++ b/protoBuilds/6a93a2-part5/README.json
@@ -18,7 +18,7 @@
"labware": "* [NEST 0.1 mL 96-Well PCR Plate, Full Skirt](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 200uL Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 24-Tube Aluminum Block](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n* Thermofisher Armadillo 96 Wellplate 200ul\n\n\n",
"modules": "* [Temperature Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Magnetic Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Swift Biosciences](https://swiftbiosci.com/protocols/)\n\n",
+ "partner": "[Swift Biosciences](https://swiftbiosci.com/protocols/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P20 GEN2 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P300 GEN2 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Index primers are added to sample wells.\n2. PCR mastermix is added to each well.\n3. Samples undergo thermocycle profile.\n4. Magnetic beads are added to samples.\n5. Samples are incubated at room temperature.\n6. Protocol pauses - plate is removed from deck, sealed and spun down.\n7. Plate is placed back on magnetic module.\n8. Supernatant removed.\n9. Two ethanol washes of samples with 30 second incubation periods before supernatant is removed.\n10. Protocol pauses - plate is removed from deck, sealed and spun down.\n11. Low EDTA TE is added to beads, incubate. \n12. Protocol pauses - plate is removed from deck, sealed and spun down.\n13. Supernatant is moved to columns 9, 10, 11.\n\n\n\n",
@@ -32,7 +32,7 @@
"Thermocycler Module"
],
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Swift Biosciences",
+ "partner": "Swift Biosciences\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP20 GEN2 Single Channel Pipette\nP300 GEN2 Multi-Channel Pipette\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nIndex primers are added to sample wells.\nPCR mastermix is added to each well.\nSamples undergo thermocycle profile.\nMagnetic beads are added to samples.\nSamples are incubated at room temperature.\nProtocol pauses - plate is removed from deck, sealed and spun down.\nPlate is placed back on magnetic module.\nSupernatant removed.\nTwo ethanol washes of samples with 30 second incubation periods before supernatant is removed.\nProtocol pauses - plate is removed from deck, sealed and spun down.\nLow EDTA TE is added to beads, incubate. \nProtocol pauses - plate is removed from deck, sealed and spun down.\nSupernatant is moved to columns 9, 10, 11.\n",
diff --git a/protoBuilds/6a93a2/README.json b/protoBuilds/6a93a2/README.json
index c781bd65e0..f1fefa5bb9 100644
--- a/protoBuilds/6a93a2/README.json
+++ b/protoBuilds/6a93a2/README.json
@@ -18,7 +18,7 @@
"labware": "* [NEST 0.1 mL 96-Well PCR Plate, Full Skirt](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 200uL Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 24-Tube Aluminum Block](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n\n",
"modules": "* [Temperature Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Magnetic Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Swift Biosciences](https://swiftbiosci.com/protocols/)\n\n",
+ "partner": "[Swift Biosciences](https://swiftbiosci.com/protocols/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P20 GEN2 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P300 GEN2 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Reverse Transcription Mastermix is made on the Opentrons Aluminum 24 tube rack at 4C.\n2. Mastermix is distributed to all sample wells and mixed.\n3. Samples undergo thermocycler profile.\n4. Samples are then moved to the magnetic module.\n5. Magnetic beads are added to samples.\n6. Samples are incubated at room temperature.\n7. Protocol pauses - plate is removed from deck, sealed and spun down.\n8. Plate is placed back on magnetic module.\n9. Supernatant removed.\n10. Two ethanol washes of samples with 30 second incubation periods before supernatant is removed.\n11. Protocol pauses - plate is removed from deck, sealed and spun down.\n12. Low EDTA TE is added to beads, incubate. \n13. Protocol pauses - plate is removed from deck, sealed and spun down.\n14. Steps 5-13 are repeated.\n15. Supernatant is moved to columns 9, 10, and 11.\n16. Ligation Mastermix is made on the Opentrons Aluminum 24 tube rack at 4C.\n17. Mastermix added to samples, plate spun down.\n18. Samples undergo thermocycler profile.\n\n\n",
@@ -32,7 +32,7 @@
"Thermocycler Module"
],
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Swift Biosciences",
+ "partner": "Swift Biosciences\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP20 GEN2 Single Channel Pipette\nP300 GEN2 Multi-Channel Pipette\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nReverse Transcription Mastermix is made on the Opentrons Aluminum 24 tube rack at 4C.\nMastermix is distributed to all sample wells and mixed.\nSamples undergo thermocycler profile.\nSamples are then moved to the magnetic module.\nMagnetic beads are added to samples.\nSamples are incubated at room temperature.\nProtocol pauses - plate is removed from deck, sealed and spun down.\nPlate is placed back on magnetic module.\nSupernatant removed.\nTwo ethanol washes of samples with 30 second incubation periods before supernatant is removed.\nProtocol pauses - plate is removed from deck, sealed and spun down.\nLow EDTA TE is added to beads, incubate. \nProtocol pauses - plate is removed from deck, sealed and spun down.\nSteps 5-13 are repeated.\nSupernatant is moved to columns 9, 10, and 11.\nLigation Mastermix is made on the Opentrons Aluminum 24 tube rack at 4C.\nMastermix added to samples, plate spun down.\nSamples undergo thermocycler profile.\n",
diff --git a/protoBuilds/6aee6e/README.json b/protoBuilds/6aee6e/README.json
index 42778cf739..82864f97cf 100644
--- a/protoBuilds/6aee6e/README.json
+++ b/protoBuilds/6aee6e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol automates a request for a ammonia and ytrium dilution protocol. It uses 2 custom labwares, transferring 10ul of ammonia from a 50mL tube to a custom 99 well plate, then transferring 5ul of Ytrium from a custom 96 well plate on a shaker. This is repeated 96 times until all wells from the custom 96 well plate have been transferred.\n\n\n\nOpentrons P20-single channel electronic pipettes (GEN2)\nOpentrons 20ul Filter Tips\nNEST 50mL centrifuge tubes\n",
"internal": "6aee6e",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "This protocol automates a request for a ammonia and ytrium dilution protocol. It uses 2 custom labwares, transferring 10ul of ammonia from a 50mL tube to a custom 99 well plate, then transferring 5ul of Ytrium from a custom 96 well plate on a shaker. This is repeated 96 times until all wells from the custom 96 well plate have been transferred.\n\n---\n\n\n* [Opentrons P20-single channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=31059478970462)\n* [Opentrons 20ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [NEST 50mL centrifuge tubes](http://www.cell-nest.com/page94?product_id=110&_l=en)\n\n",
"internal": "6aee6e\n",
diff --git a/protoBuilds/6af807-amendment/README.json b/protoBuilds/6af807-amendment/README.json
index 66a61d4480..7ada2636da 100644
--- a/protoBuilds/6af807-amendment/README.json
+++ b/protoBuilds/6af807-amendment/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "6af807",
"labware": "\nThermofisher 384 well plate\nOpentrons 24 tube rack with 1.5mL Eppendorf tubes\nOpentrons 20ul Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "* Note: Mastermixes and cDNA tubes should be placed on their respective tube racks by column. Please look at the deck layout below for a full run (8 genes and 16 mastermixes).\n\n\n\n\n\n---\n\n",
"description": "This protocol preps a 384 well plate for up to 8 cDNA samples in triplicates. The protocol can be broken down into 2 main parts:\n\n* cDNA is added in triplicates down the column onto the plate.\n* Mastermix is added to the cDNA.\n\nExplanation of complex parameters below:\n* `Number of cDNA`: Specify the number of cDNA tubes that will be loaded onto the tube rack.\n* `Number of mastermix tubes`: Specify the number of mastermix tubes that will be added to the cDNA.\n* `cDNA/mastermix volume`: Specify the volume of both the cDNA and mastermix. Note, the mastermix volume will be calculated from this volume (total 20ul).\n* `P20 Mount`: Specify the mount (left or right) for the P20 single channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/6af807/README.json b/protoBuilds/6af807/README.json
index 7d1d18841d..721c6abb96 100644
--- a/protoBuilds/6af807/README.json
+++ b/protoBuilds/6af807/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "6af807",
"labware": "\nThermofisher 384 well plate\nOpentrons 24 tube rack with 1.5mL Eppendorf tubes\nOpentrons 20ul Tips\nCustom Tip Rack\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "* Note: Mastermixes and cDNA tubes should be placed on their respective tube racks by column. Please look at the deck layout below for a full run (5 genes and 24 mastermixes).\n\n\n\n\n\n---\n\n",
"description": "This protocol preps a 384 well plate for up to 5 cDNA samples in triplicates. The protocol can be broken down into 2 main parts:\n\n* cDNA is added in triplicates down the column onto the plate.\n* Mastermix is added to the cDNA\n\nExplanation of complex parameters below:\n* `Mastermix in triplicates across columns (new) or whole columns (original)`: Specify whether to run the new protocol (box method) or original protocol (which may break a box method).\n* `Number of cDNA`: Specify the number of cDNA tubes that will be loaded onto the tube rack.\n* `Number of mastermix tubes`: Specify the number of mastermix tubes that will be added to the cDNA.\n* `Dispense Flow Rate`: Specify the rate to dispense mastermix. A value of 1 is default, a value of 0.5 is half of the default value, and a value of 1.2 would be 20% higher than the default value, for example.\n* `Blowout Flow Rate`: Specify the rate to blowout mastermix. A value of 1 is default, a value of 0.5 is half of the default value, and a value of 1.2 would be 20% higher than the default value, for example.\n* `cDNA/mastermix volume`: Specify the volume of both the cDNA and mastermix. Note, the total volume of these two parameters should be equal to 20ul. \n* `Reset Tipracks?`: The OT-2 will track tips from run to run of the protocol (e.g. tip leaves off in H11 at the end of protocol 1; first tip pick up will be from H12 in Part 2). When tips run out for any particular run, the user will be prompted to replace all tip racks. The user will also have the option to \"Reset Tip Racks\" in the customized parameters section. Reset tip racks to pick up from A1 of the first tip rack of any one protocol.\n* `P20 Mount`: Specify the mount (left or right) for the P20 single channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/6b51bd/README.json b/protoBuilds/6b51bd/README.json
index c7ecdaa3a3..b59bc92695 100644
--- a/protoBuilds/6b51bd/README.json
+++ b/protoBuilds/6b51bd/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -9,7 +9,7 @@
"description": "This protocol uses a p300 single-channel pipette to distribute custom volumes of buffer from a 12-well reservoir to specified wells of up to three 96-well child plates followed by transfer of well contents from up to three 96-well parent plates to the corresponding well in a child plate (volumes, plate locations and well locations are specified in a csv file uploaded at the time of protocol download).\nLinks:\n* example input csv\nThis protocol was developed to distribute custom volumes of buffer from a 12-well reservoir to specific wells of a child plate and then transfer contents from specific wells of parent plates to corresponding wells of child plates.",
"internal": "6b51bd",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Normalization\n\n",
"deck-setup": "\n\n* Opentrons 200ul filter tips (Deck Slot 6)\n* Opentrons 20ul filter tips (Deck Slots 1, 2, 3)\n* 96-well Child Plates (Deck Slots 5, 8, 11)\n* 96-well Parent Plates (Deck Slots 4, 7, 10)\n* Reservoir (Deck Slot 9)\n\n\n\n",
"description": "\nThis protocol uses a p300 single-channel pipette to distribute custom volumes of buffer from a 12-well reservoir to specified wells of up to three 96-well child plates followed by transfer of well contents from up to three 96-well parent plates to the corresponding well in a child plate (volumes, plate locations and well locations are specified in a csv file uploaded at the time of protocol download).\n\nLinks:\n* [example input csv](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/6b51bd/Example+CSV.csv)\n\n\nThis protocol was developed to distribute custom volumes of buffer from a 12-well reservoir to specific wells of a child plate and then transfer contents from specific wells of parent plates to corresponding wells of child plates.\n\n",
diff --git a/protoBuilds/6b8c90-2/README.json b/protoBuilds/6b8c90-2/README.json
index 7cedcecb07..19693d66dd 100644
--- a/protoBuilds/6b8c90-2/README.json
+++ b/protoBuilds/6b8c90-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This is one of three protocols updated from protocols developed with Protocol Designer. The three protocols are:\n48 MCT to 1mL Vial\n48 Samples to MCT\nEtOH 48 Sample to 1mL Vial\n\nIn this protocol, the p300-single channel pipette is used to transfer 100\u00b5L between a custom 5mL tube rack and the acrylic Opentrons tuberack.\n\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nP300 Single-Channel Pipette (GEN1)\nOpentrons 300\u00b5l Pipette tips\nOpentrons Acrylic Tube Rack\nCustom 5mL Tube Rack + Tubes\n\n\n\nDeck Layout\n\nSlot 1: Custom 5mL Tube Rack + Tubes (for samples 1-24)\n\nSlot 2: Opentrons Acrylic Tube Rack (for samples 1-24)\n\nSlot 4: Custom 5mL Tube Rack + Tubes (for samples 25-48)\n\nSlot 5: Opentrons Acrylic Tube Rack (for samples 25-48)\n\nSlot 7: Opentrons 300\u00b5l Pipette tips\n",
"internal": "6b8c90-2",
"markdown": {
- "author": "[Opentrons](http://www.opentrons.com/)\n\n",
+ "author": "[Opentrons](http://www.opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Plate Filling\n\n",
"description": "This is one of three protocols updated from protocols developed with Protocol Designer. The three protocols are:\n[48 MCT to 1mL Vial](https://develop.protocols.opentrons.com/protocol/6b8c90)\n[48 Samples to MCT](https://develop.protocols.opentrons.com/protocol/6b8c90-2)\n[EtOH 48 Sample to 1mL Vial](https://develop.protocols.opentrons.com/protocol/6b8c90-3)\n\nIn this protocol, the p300-single channel pipette is used to transfer 100\u00b5L between a custom 5mL tube rack and the acrylic Opentrons tuberack.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [P300 Single-Channel Pipette (GEN1)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* Opentrons Acrylic Tube Rack\n* Custom 5mL Tube Rack + Tubes\n\n\n---\n\n\n**Deck Layout**\n\n**Slot 1**: Custom 5mL Tube Rack + Tubes (for samples 1-24)\n\n**Slot 2**: Opentrons Acrylic Tube Rack (for samples 1-24)\n\n**Slot 4**: Custom 5mL Tube Rack + Tubes (for samples 25-48)\n\n**Slot 5**: Opentrons Acrylic Tube Rack (for samples 25-48)\n\n**Slot 7**: [Opentrons 300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n",
"internal": "6b8c90-2\n",
diff --git a/protoBuilds/6b8c90-3/README.json b/protoBuilds/6b8c90-3/README.json
index 14dc673833..9043c17790 100644
--- a/protoBuilds/6b8c90-3/README.json
+++ b/protoBuilds/6b8c90-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This is one of three protocols updated from protocols developed with Protocol Designer. The three protocols are:\n48 MCT to 1mL Vial\n48 Samples to MCT\nEtOH 48 Sample to 1mL Vial\n\nIn this protocol, the p50-single channel pipette is used to transfer 50\u00b5L between a custom 5mL tube rack and a custom 1mL tube rack.\n\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nP300 Single-Channel Pipette (GEN1)\nOpentrons 300\u00b5l Pipette tips\nCustom 1mL Tube Rack + Tubes\nCustom 5mL Tube Rack + Tubes\n\n\n\nDeck Layout\n\nSlot 1: Custom 5mL Tube Rack + Tubes (for samples 1-24)\n\nSlot 2: Custom 1mL Tube Rack + Tubes (for samples 1-24)\n\nSlot 4: Custom 5mL Tube Rack + Tubes (for samples 25-48)\n\nSlot 5: Custom 1mL Tube Rack + Tubes (for samples 25-48)\n\nSlot 7: Opentrons 300\u00b5l Pipette tips\n",
"internal": "6b8c90-3",
"markdown": {
- "author": "[Opentrons](http://www.opentrons.com/)\n\n",
+ "author": "[Opentrons](http://www.opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Plate Filling\n\n",
"description": "This is one of three protocols updated from protocols developed with Protocol Designer. The three protocols are:\n[48 MCT to 1mL Vial](https://develop.protocols.opentrons.com/protocol/6b8c90)\n[48 Samples to MCT](https://develop.protocols.opentrons.com/protocol/6b8c90-2)\n[EtOH 48 Sample to 1mL Vial](https://develop.protocols.opentrons.com/protocol/6b8c90-3)\n\nIn this protocol, the p50-single channel pipette is used to transfer 50\u00b5L between a custom 5mL tube rack and a custom 1mL tube rack.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [P300 Single-Channel Pipette (GEN1)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* Custom 1mL Tube Rack + Tubes\n* Custom 5mL Tube Rack + Tubes\n\n\n---\n\n\n**Deck Layout**\n\n**Slot 1**: Custom 5mL Tube Rack + Tubes (for samples 1-24)\n\n**Slot 2**: Custom 1mL Tube Rack + Tubes (for samples 1-24)\n\n**Slot 4**: Custom 5mL Tube Rack + Tubes (for samples 25-48)\n\n**Slot 5**: Custom 1mL Tube Rack + Tubes (for samples 25-48)\n\n**Slot 7**: [Opentrons 300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n",
"internal": "6b8c90-3\n",
diff --git a/protoBuilds/6b8c90/README.json b/protoBuilds/6b8c90/README.json
index 70d30d6005..0fa2e2af0a 100644
--- a/protoBuilds/6b8c90/README.json
+++ b/protoBuilds/6b8c90/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This is one of three protocols updated from protocols developed with Protocol Designer. The three protocols are:\n48 MCT to 1mL Vial\n48 Samples to MCT\nEtOH 48 Sample to 1mL Vial\n\nIn this protocol, the p300-single channel pipette is used to transfer 300\u00b5L between the acrylic Opentrons tuberack and a custom 1mL tube rack.\n\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nP300 Single-Channel Pipette (GEN1)\nOpentrons 300\u00b5l Pipette tips\nOpentrons Acrylic Tube Rack\nCustom 1mL Tube Rack + Tubes\n\n\n\nDeck Layout\n\nSlot 2: Opentrons Acrylic Tube Rack (for samples 1-24)\n\nSlot 3: Custom 1mL Tube Rack + Tubes (for samples 1-24)\n\nSlot 5: Opentrons Acrylic Tube Rack (for samples 25-48)\n\nSlot 6: Custom 1mL Tube Rack + Tubes (for samples 25-48)\n\nSlot 7: Opentrons 300\u00b5l Pipette tips\n",
"internal": "6b8c90",
"markdown": {
- "author": "[Opentrons](http://www.opentrons.com/)\n\n",
+ "author": "[Opentrons](http://www.opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Plate Filling\n\n",
"description": "This is one of three protocols updated from protocols developed with Protocol Designer. The three protocols are:\n[48 MCT to 1mL Vial](https://develop.protocols.opentrons.com/protocol/6b8c90)\n[48 Samples to MCT](https://develop.protocols.opentrons.com/protocol/6b8c90-2)\n[EtOH 48 Sample to 1mL Vial](https://develop.protocols.opentrons.com/protocol/6b8c90-3)\n\nIn this protocol, the p300-single channel pipette is used to transfer 300\u00b5L between the acrylic Opentrons tuberack and a custom 1mL tube rack.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [P300 Single-Channel Pipette (GEN1)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* Opentrons Acrylic Tube Rack\n* Custom 1mL Tube Rack + Tubes\n\n\n---\n\n\n**Deck Layout**\n\n**Slot 2**: Opentrons Acrylic Tube Rack (for samples 1-24)\n\n**Slot 3**: Custom 1mL Tube Rack + Tubes (for samples 1-24)\n\n**Slot 5**: Opentrons Acrylic Tube Rack (for samples 25-48)\n\n**Slot 6**: Custom 1mL Tube Rack + Tubes (for samples 25-48)\n\n**Slot 7**: [Opentrons 300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n",
"internal": "6b8c90\n",
diff --git a/protoBuilds/6b9ee3/README.json b/protoBuilds/6b9ee3/README.json
index c630e4afa2..29e5fb8bb6 100644
--- a/protoBuilds/6b9ee3/README.json
+++ b/protoBuilds/6b9ee3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol automates the creation of a mastermix (containing OptiMEM, DNA samples, and Viafect) and then dispenses this mastermix in triplicate. Using a simple CSV and a couple key parameters, this protocol can be easily adjusted for diifferent scenarios.\n\nThe protocol begins by adding 5.3\u00b5L of OptiMEM to the number of wells as defined by the user (the last well can receive more). After this, 4.4\u00b5L of DNA will be added to the wells per the CSV, transferring from sample tubes in deck slot 4 to the mastermix tubes in deck slot 5 (with OptiMEM). The OT-2 will then add 1.3\u00b5L of Viafect to the mastermix wells and pause until the user is ready to resume. After resuming, the OT-2 will transfer 10\u00b5L of the mastermix, in triplicate, to a 96-well plate in deck slot 6.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P20 Single-Channel Pipette\nOpentrons P20 Multi-Channel Pipette\nOpentrons 20\u00b5L Tips\nOpentrons Aluminum Block Set\nOpentrons 4-in-1 Tube Rack Set\nCorning 96-Well Plate, 360\u00b5L Flat\n1.5mL Microcentrifuge Tube\nPCR Strips, 200\u00b5L\n\n\n\nDeck Setup\n\nSlot 1: Opentrons 20\u00b5L Tips\nSlot 2: Opentrons 20\u00b5L Tips\nSlot 3: Opentrons 20\u00b5L Tips\nSlot 4: Opentrons 96-Well Aluminum Block with PCR Strips containing samples/stock\nSlot 5: Opentrons 96-Well Aluminum Block with empty PCR Strips on the left for mastermix creation (columns 1-4) and a PCR Strip containing Viafect (column 12)\nSlot 6: Corning 96-Well Plate, 360\u00b5L Flat\nSlot 7: Opentrons 4-in-1 Tube Rack Set with a 1.5mL Microcentrifuge Tube containing OptiMEM in D6\n\nUsing the customizations field (below), set up your protocol.\n Transfer CSV: Upload a CSV that will dictate the transfer of DNA from deck slot 4 to deck slot 5 (see below for more detail)\n Number of Mastermix: Specify the number of mastermixes to be created (1-32).\n* Extra OptiMEM: Specify whether the last mastermix tube should receive 31.7\u00b5L of OptiMEM or the standard 5.3\u00b5L\n\n\nCSV Layout\n\n\n\nThe CSV should consist of two columns with headers, Source Well and Destination Well. Using capital letters, you can simply put well locations and the pipette will transfer 4.4\u00b5L from the source well (in deck slot 4) to the destination well (in deck slot 5).",
"internal": "6b9ee3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol automates the creation of a mastermix (containing OptiMEM, DNA samples, and Viafect) and then dispenses this mastermix in triplicate. Using a simple CSV and a couple key parameters, this protocol can be easily adjusted for diifferent scenarios.\n\nThe protocol begins by adding 5.3\u00b5L of OptiMEM to the number of wells as defined by the user (the last well can receive more). After this, 4.4\u00b5L of DNA will be added to the wells per the CSV, transferring from sample tubes in deck slot 4 to the mastermix tubes in deck slot 5 (with OptiMEM). The OT-2 will then add 1.3\u00b5L of Viafect to the mastermix wells and pause until the user is ready to resume. After resuming, the OT-2 will transfer 10\u00b5L of the mastermix, in triplicate, to a 96-well plate in deck slot 6.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons P20 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons Aluminum Block Set](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [Corning 96-Well Plate, 360\u00b5L Flat](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate)\n* [1.5mL Microcentrifuge Tube](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes)\n* PCR Strips, 200\u00b5L\n\n\n\n---\n\n\n\n**Deck Setup**\n\nSlot 1: [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips)\nSlot 2: [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips)\nSlot 3: [Opentrons 20\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips)\nSlot 4: [Opentrons 96-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) with PCR Strips containing samples/stock\nSlot 5: [Opentrons 96-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) with empty PCR Strips on the left for mastermix creation (columns 1-4) and a PCR Strip containing Viafect (column 12)\nSlot 6: [Corning 96-Well Plate, 360\u00b5L Flat](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate)\nSlot 7: [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with a [1.5mL Microcentrifuge Tube](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes) containing OptiMEM in D6\n\n**Using the customizations field (below), set up your protocol.**\n* **Transfer CSV**: Upload a CSV that will dictate the transfer of DNA from deck slot 4 to deck slot 5 (see below for more detail)\n* **Number of Mastermix**: Specify the number of mastermixes to be created (1-32).\n* **Extra OptiMEM**: Specify whether the last mastermix tube should receive 31.7\u00b5L of OptiMEM or the standard 5.3\u00b5L\n\n\n**CSV Layout**\n\n\n\nThe CSV should consist of two columns with headers, **Source Well** and **Destination Well**. Using capital letters, you can simply put well locations and the pipette will transfer 4.4\u00b5L from the source well (in deck slot 4) to the destination well (in deck slot 5).\n\n",
"internal": "6b9ee3\n",
diff --git a/protoBuilds/6bc986-part-2/README.json b/protoBuilds/6bc986-part-2/README.json
index 0c1178fbce..d2843ced6e 100644
--- a/protoBuilds/6bc986-part-2/README.json
+++ b/protoBuilds/6bc986-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "Part 2 of 6: Distribution of Prot K (Apostle)\nLinks:\n Distribution of Sample Lysis Buffer\n Distribution of Prot K (Apostle)\n Transfer of DNA Template\n Distribution of Master Mix\n Distribution of Elution Buffer\n Distribution of Prot K (Qiagen)\nThis protocol is a translation of the attached json protocol json protocol to python with a couple additional features for control of the blow out location in order to place it near the bottom of the source well: 4 ul Prot K is distributed from a reservoir to the columns of a 96 deep-well plate with a blow out of the disposal volume occuring in a location close to the bottom of the reservoir.\nThe deck layout can be displayed by uploading the attached json protocol to the Opentrons Protocol Designer web page.",
"internal": "6bc986-part-2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * PCR Prep\n\n",
"description": "Part 2 of 6: Distribution of Prot K (Apostle)\n\nLinks:\n* [Distribution of Sample Lysis Buffer](https://protocols.opentrons.com/protocol/6bc986)\n* [Distribution of Prot K (Apostle)](https://protocols.opentrons.com/protocol/6bc986-part-2)\n* [Transfer of DNA Template](https://protocols.opentrons.com/protocol/6bc986-part-3)\n* [Distribution of Master Mix](https://protocols.opentrons.com/protocol/6bc986-part-4)\n* [Distribution of Elution Buffer](https://protocols.opentrons.com/protocol/6bc986-part-5)\n* [Distribution of Prot K (Qiagen)](https://protocols.opentrons.com/protocol/6bc986-part-6)\n\nThis protocol is a translation of the attached json protocol [json protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-04-19/te53ia8/Apostle%20Prot.%20K.json) to python with a couple additional features for control of the blow out location in order to place it near the bottom of the source well: 4 ul Prot K is distributed from a reservoir to the columns of a 96 deep-well plate with a blow out of the disposal volume occuring in a location close to the bottom of the reservoir.\nThe deck layout can be displayed by uploading the attached json protocol to the [Opentrons Protocol Designer web page](https://opentrons.com/protocols/designer/).\n\n",
"internal": "6bc986-part-2\n",
diff --git a/protoBuilds/6bc986-part-3/README.json b/protoBuilds/6bc986-part-3/README.json
index 33d8e1357b..cd459d5368 100644
--- a/protoBuilds/6bc986-part-3/README.json
+++ b/protoBuilds/6bc986-part-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "Part 3 of 6: Transfer of DNA Template\nLinks:\n Distribution of Sample Lysis Buffer\n Distribution of Prot K (Apostle)\n Transfer of DNA Template\n Distribution of Master Mix\n Distribution of Elution Buffer\n Distribution of Prot K (Qiagen)\nThis protocol is a translation of the attached json protocol json protocol to python: 5 ul DNA template is transferred to the columns of a PCR plate.\nThe deck layout can be displayed by uploading the attached json protocol to the Opentrons Protocol Designer web page.",
"internal": "6bc986-part-3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * PCR Prep\n\n",
"description": "Part 3 of 6: Transfer of DNA Template\n\nLinks:\n* [Distribution of Sample Lysis Buffer](https://protocols.opentrons.com/protocol/6bc986)\n* [Distribution of Prot K (Apostle)](https://protocols.opentrons.com/protocol/6bc986-part-2)\n* [Transfer of DNA Template](https://protocols.opentrons.com/protocol/6bc986-part-3)\n* [Distribution of Master Mix](https://protocols.opentrons.com/protocol/6bc986-part-4)\n* [Distribution of Elution Buffer](https://protocols.opentrons.com/protocol/6bc986-part-5)\n* [Distribution of Prot K (Qiagen)](https://protocols.opentrons.com/protocol/6bc986-part-6)\n\nThis protocol is a translation of the attached json protocol [json protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-04-19/sy43ivh/4.%20DNA%20template-edit.json) to python: 5 ul DNA template is transferred to the columns of a PCR plate.\nThe deck layout can be displayed by uploading the attached json protocol to the [Opentrons Protocol Designer web page](https://opentrons.com/protocols/designer/).\n\n",
"internal": "6bc986-part-3\n",
diff --git a/protoBuilds/6bc986-part-4/README.json b/protoBuilds/6bc986-part-4/README.json
index 985c1e8e6f..1613cda6d3 100644
--- a/protoBuilds/6bc986-part-4/README.json
+++ b/protoBuilds/6bc986-part-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "Part 4 of 6: Distribution of Master Mix\nLinks:\n Distribution of Sample Lysis Buffer\n Distribution of Prot K (Apostle)\n Transfer of DNA Template\n Distribution of Master Mix\n Distribution of Elution Buffer\n Distribution of Prot K (Qiagen)\nThis protocol is a translation of the attached json protocol json protocol to python with a couple additional features for control of the blow out location in order to place it near the bottom of the source reservoir well: 20 ul Master Mix is distributed from a reservoir with a blow out of the disposal volume occuring in a location close to the bottom of the source well.\nThe deck layout can be displayed by uploading the attached json protocol to the Opentrons Protocol Designer web page.",
"internal": "6bc986-part-4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * PCR Prep\n\n",
"description": "Part 4 of 6: Distribution of Master Mix\n\nLinks:\n* [Distribution of Sample Lysis Buffer](https://protocols.opentrons.com/protocol/6bc986)\n* [Distribution of Prot K (Apostle)](https://protocols.opentrons.com/protocol/6bc986-part-2)\n* [Transfer of DNA Template](https://protocols.opentrons.com/protocol/6bc986-part-3)\n* [Distribution of Master Mix](https://protocols.opentrons.com/protocol/6bc986-part-4)\n* [Distribution of Elution Buffer](https://protocols.opentrons.com/protocol/6bc986-part-5)\n* [Distribution of Prot K (Qiagen)](https://protocols.opentrons.com/protocol/6bc986-part-6)\n\nThis protocol is a translation of the attached json protocol [json protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-04-19/sc33ipe/3.%20Master%20Mix%20only-edit.json) to python with a couple additional features for control of the blow out location in order to place it near the bottom of the source reservoir well: 20 ul Master Mix is distributed from a reservoir with a blow out of the disposal volume occuring in a location close to the bottom of the source well.\nThe deck layout can be displayed by uploading the attached json protocol to the [Opentrons Protocol Designer web page](https://opentrons.com/protocols/designer/).\n\n",
"internal": "6bc986-part-4\n",
diff --git a/protoBuilds/6bc986-part-5/README.json b/protoBuilds/6bc986-part-5/README.json
index 745ffe5ac5..ecc8a74294 100644
--- a/protoBuilds/6bc986-part-5/README.json
+++ b/protoBuilds/6bc986-part-5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "Part 5 of 6: Distribution of Elution Buffer\nLinks:\n Distribution of Sample Lysis Buffer\n Distribution of Prot K (Apostle)\n Transfer of DNA Template\n Distribution of Master Mix\n Distribution of Elution Buffer\n Distribution of Prot K (Qiagen)\nThis protocol is a translation of the attached json protocol json protocol to python with a couple additional features for control of the blow out location in order to place it near the bottom of the source reservoir well: 20 ul elution buffer is distributed from a reservoir to the columns of a 96-well plate with a blow out of the disposal volume occuring in a location close to the bottom of the source well.\nThe deck layout can be displayed by uploading the attached json protocol to the Opentrons Protocol Designer web page.",
"internal": "6bc986-part-5",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * PCR Prep\n\n",
"description": "Part 5 of 6: Distribution of Elution Buffer\n\nLinks:\n* [Distribution of Sample Lysis Buffer](https://protocols.opentrons.com/protocol/6bc986)\n* [Distribution of Prot K (Apostle)](https://protocols.opentrons.com/protocol/6bc986-part-2)\n* [Transfer of DNA Template](https://protocols.opentrons.com/protocol/6bc986-part-3)\n* [Distribution of Master Mix](https://protocols.opentrons.com/protocol/6bc986-part-4)\n* [Distribution of Elution Buffer](https://protocols.opentrons.com/protocol/6bc986-part-5)\n* [Distribution of Prot K (Qiagen)](https://protocols.opentrons.com/protocol/6bc986-part-6)\n\nThis protocol is a translation of the attached json protocol [json protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-04-19/eg83ite/2.%20Elution%20Buffer-edit.json) to python with a couple additional features for control of the blow out location in order to place it near the bottom of the source reservoir well: 20 ul elution buffer is distributed from a reservoir to the columns of a 96-well plate with a blow out of the disposal volume occuring in a location close to the bottom of the source well.\nThe deck layout can be displayed by uploading the attached json protocol to the [Opentrons Protocol Designer web page](https://opentrons.com/protocols/designer/).\n\n",
"internal": "6bc986-part-5\n",
diff --git a/protoBuilds/6bc986-part-6/README.json b/protoBuilds/6bc986-part-6/README.json
index ed47388006..40d9bccb6e 100644
--- a/protoBuilds/6bc986-part-6/README.json
+++ b/protoBuilds/6bc986-part-6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "Part 6 of 6: Distribution of Prot K (Qiagen)\nLinks:\n Distribution of Sample Lysis Buffer\n Distribution of Prot K (Apostle)\n Transfer of DNA Template\n Distribution of Master Mix\n Distribution of Elution Buffer\n Distribution of Prot K (Qiagen)\nThis protocol is a translation of the attached json protocol json protocol to python with a couple additional features for control of the blow out location in order to place it near the bottom of the well: 10 ul sample lysis buffer is distributed from a reservoir to the columns of a 96 deep-well plate with a blow out occuring after each dispense in a location close to the bottom of the well.\nThe deck layout can be displayed by uploading the attached json protocol to the Opentrons Protocol Designer web page.",
"internal": "6bc986-part-6",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * PCR Prep\n\n",
"description": "Part 6 of 6: Distribution of Prot K (Qiagen)\n\nLinks:\n* [Distribution of Sample Lysis Buffer](https://protocols.opentrons.com/protocol/6bc986)\n* [Distribution of Prot K (Apostle)](https://protocols.opentrons.com/protocol/6bc986-part-2)\n* [Transfer of DNA Template](https://protocols.opentrons.com/protocol/6bc986-part-3)\n* [Distribution of Master Mix](https://protocols.opentrons.com/protocol/6bc986-part-4)\n* [Distribution of Elution Buffer](https://protocols.opentrons.com/protocol/6bc986-part-5)\n* [Distribution of Prot K (Qiagen)](https://protocols.opentrons.com/protocol/6bc986-part-6)\n\nThis protocol is a translation of the attached json protocol [json protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-04-19/dd73ib0/1.%20QIAGEN%20Prot.%20K-edit.json) to python with a couple additional features for control of the blow out location in order to place it near the bottom of the well: 10 ul sample lysis buffer is distributed from a reservoir to the columns of a 96 deep-well plate with a blow out occuring after each dispense in a location close to the bottom of the well.\nThe deck layout can be displayed by uploading the attached json protocol to the [Opentrons Protocol Designer web page](https://opentrons.com/protocols/designer/).\n\n",
"internal": "6bc986-part-6\n",
diff --git a/protoBuilds/6bc986/README.json b/protoBuilds/6bc986/README.json
index 7b7e1ade43..37c19a1519 100644
--- a/protoBuilds/6bc986/README.json
+++ b/protoBuilds/6bc986/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "Part 1 of 6: Distribution of Sample Lysis Buffer\nLinks:\n Distribution of Sample Lysis Buffer\n Distribution of Prot K (Apostle)\n Transfer of DNA Template\n Distribution of Master Mix\n Distribution of Elution Buffer\n Distribution of Prot K (Qiagen)\nThis protocol is a translation of the attached json protocol json protocol to python with a couple additional features for control of the blow out location in order to place it near the bottom of the well: 10 ul sample lysis buffer is distributed from a reservoir to the columns of a 96 deep-well plate with a blow out occurring after each dispense in a location close to the bottom of the well.\nThe deck layout can be displayed by uploading the attached json protocol to the Opentrons Protocol Designer web page.",
"internal": "6bc986",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * PCR Prep\n\n",
"description": "Part 1 of 6: Distribution of Sample Lysis Buffer\n\nLinks:\n* [Distribution of Sample Lysis Buffer](https://protocols.opentrons.com/protocol/6bc986)\n* [Distribution of Prot K (Apostle)](https://protocols.opentrons.com/protocol/6bc986-part-2)\n* [Transfer of DNA Template](https://protocols.opentrons.com/protocol/6bc986-part-3)\n* [Distribution of Master Mix](https://protocols.opentrons.com/protocol/6bc986-part-4)\n* [Distribution of Elution Buffer](https://protocols.opentrons.com/protocol/6bc986-part-5)\n* [Distribution of Prot K (Qiagen)](https://protocols.opentrons.com/protocol/6bc986-part-6)\n\nThis protocol is a translation of the attached json protocol [json protocol](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-04-19/ul63i18/Apostle%20Sample%20Lysis%204.json) to python with a couple additional features for control of the blow out location in order to place it near the bottom of the well: 10 ul sample lysis buffer is distributed from a reservoir to the columns of a 96 deep-well plate with a blow out occurring after each dispense in a location close to the bottom of the well.\nThe deck layout can be displayed by uploading the attached json protocol to the [Opentrons Protocol Designer web page](https://opentrons.com/protocols/designer/).\n\n",
"internal": "6bc986\n",
diff --git a/protoBuilds/6be1f8/README.json b/protoBuilds/6be1f8/README.json
index 2fee352a53..4ba0e938f2 100644
--- a/protoBuilds/6be1f8/README.json
+++ b/protoBuilds/6be1f8/README.json
@@ -17,7 +17,7 @@
"internal": "6be1f8\n",
"labware": "* Greiner 384 well plate 200ul\n* Opentrons 4-in-1 tube rack - 2x3 grid with Appleton 50mL tubes\n* Opentrons 4-in-1 tube rack - 4x6 grid with Axygen 1.7mL tubes\n* Greiner 96 well plate 2000ul\n* [Opentrons 20ul tip rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 1000ul tip rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Partner Name](partner website link)\n\n",
+ "partner": "[Partner Name](partner website link)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P1000 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P20 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Transfer diluent to mAb210 100ug/ml tube\n2. Transfer mAb210 stock to mAb210 100ug/ml tube\n3. Mix mAb210 100ug/ml tube x10\n4. Repeat steps (1-3) for mAb210 20ug/ml and 2ug/ml\n5. Transfer diluent to 'A1', 'A2', 'B1', 'B2', 'C1', 'G1', 'C2', 'E2', 'D1', 'D2', 'E1', 'F1', 'G1', 'G2', 'H1', 'H2'\n6. Transfer mAb207 to same wells\n7. Transfer diluent and mAb210 to column 3 of masterblock.\n8. Transfer 180ul A1, B1, C1, D1, E1, F1, G1 and H1 masterblock to A1, C1, E1, G1, I1, K1, M1, O1 control plate 1 and control plate 2\n9. Transfer 80ul A1, B1, C1, D1, E1, F1, G1 and H1 masterblock to A1, C1, E1, G1, I1, K1, M1, O1 control plate 3\n10. Transfer 180ul A2, B2, C2, D2, E2, F2, G2 and H2 masterblock to B1, D1, F1, H1, J1, L1, N1, P1 control plate 1 and control plate 2\n11. Transfer 80ul A2, B2, C2, D2, E2, F2, G2 and H2 masterblock to B1, D1, F1, H1, J1, L1, N1, P1 control plate 3\n12. Transfer 180ul A3, B3, C3, D3, E3, F3, G3 and H3 masterblock to A2, C2, E2, G2, I2, K2, M2, O2 control plate 1 and control plate 2\n13. Transfer 80ul A3, B3, C3, D3, E3, F3, G3 and H3 masterblock to A2, C2, E2, G2, I2, K2, M2, O2 control plate 3\n14. Transfer 180ul A3, B3, C3, D3, E3, F3, G3 and H3 masterblock to B2, D2, F2, H2, J2, L2, N2, P2 control plate 1 and control plate 2\n15. Transfer 80ul A3, B3, C3, D3, E3, F3, G3 and H3 masterblock to B2, D2, F2, H2, J2, L2, N2, P2 control plate 3\n\n\n",
@@ -26,7 +26,7 @@
"title": "Sample Prep for ELISA Test of Monoclonal Antibodies"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Partner Name",
+ "partner": "Partner Name\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP1000 Single Channel Pipette\nP20 Single Channel Pipette\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nTransfer diluent to mAb210 100ug/ml tube\nTransfer mAb210 stock to mAb210 100ug/ml tube\nMix mAb210 100ug/ml tube x10\nRepeat steps (1-3) for mAb210 20ug/ml and 2ug/ml\nTransfer diluent to 'A1', 'A2', 'B1', 'B2', 'C1', 'G1', 'C2', 'E2', 'D1', 'D2', 'E1', 'F1', 'G1', 'G2', 'H1', 'H2'\nTransfer mAb207 to same wells\nTransfer diluent and mAb210 to column 3 of masterblock.\nTransfer 180ul A1, B1, C1, D1, E1, F1, G1 and H1 masterblock to A1, C1, E1, G1, I1, K1, M1, O1 control plate 1 and control plate 2\nTransfer 80ul A1, B1, C1, D1, E1, F1, G1 and H1 masterblock to A1, C1, E1, G1, I1, K1, M1, O1 control plate 3\nTransfer 180ul A2, B2, C2, D2, E2, F2, G2 and H2 masterblock to B1, D1, F1, H1, J1, L1, N1, P1 control plate 1 and control plate 2\nTransfer 80ul A2, B2, C2, D2, E2, F2, G2 and H2 masterblock to B1, D1, F1, H1, J1, L1, N1, P1 control plate 3\nTransfer 180ul A3, B3, C3, D3, E3, F3, G3 and H3 masterblock to A2, C2, E2, G2, I2, K2, M2, O2 control plate 1 and control plate 2\nTransfer 80ul A3, B3, C3, D3, E3, F3, G3 and H3 masterblock to A2, C2, E2, G2, I2, K2, M2, O2 control plate 3\nTransfer 180ul A3, B3, C3, D3, E3, F3, G3 and H3 masterblock to B2, D2, F2, H2, J2, L2, N2, P2 control plate 1 and control plate 2\nTransfer 80ul A3, B3, C3, D3, E3, F3, G3 and H3 masterblock to B2, D2, F2, H2, J2, L2, N2, P2 control plate 3\n",
diff --git a/protoBuilds/6c27ee/README.json b/protoBuilds/6c27ee/README.json
index dfb105a888..11f25ff5a2 100644
--- a/protoBuilds/6c27ee/README.json
+++ b/protoBuilds/6c27ee/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -8,7 +8,7 @@
"description": "This protocol performs cherrypicking from source to target plates as specified in an input CSV file. The protocol parses the CSV for slots to load the source and target plates. Only non-zero volume transfers are carried out for efficiency, and the user is prompted to refill tipracks if necessary.\n\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nP50 Single-channel electronic pipette\nP300 8-channel electronic pipette\nOpentrons 300\u00b5l Pipette tips\nCorning 96-Well Plate, Flat 360\u00b5L\nOpentrons 4-in-1 Tube Rack Set\nUSA Scientific 12-Well Reservoir, 22mL\n\n\n\nDeck Layout\n\nSlot 1: Opentrons 300\u00b5l Pipette tips (for P50 Single-Channel)\n\nSlot 2: Corning 96-Well Plate, Flat 360\u00b5L (ELISA plate A)\n\nSlot 3: USA Scientific 12-Well Reservoir, 22mL\n\nSlot 4: Opentrons 300\u00b5l Pipette tips (for P300 8-Channel)\n\nSlot 5: Corning 96-Well Plate, Flat 360\u00b5L (ELISA plate B)\n\nSlot 6: Corning 96-Well Plate, Flat 360\u00b5L (1st Dilution Plate)\n\nSlot 7: Opentrons 300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips) (for P300 8-Channel)\n\nSlot 8: Corning 96-Well Plate, Flat 360\u00b5L (ELISA plate C)\n\nSlot 9: Opentrons 4-in-1 Tube Rack containing 2mL Tubes\n\nSlot 10: Corning 96-Well Plate, Flat 360\u00b5L (2nd Dilution Plate)\n\nSlot 11: Corning 96-Well Plate, Flat 360\u00b5L (ELISA plate D)\n",
"internal": "6c27ee",
"markdown": {
- "author": "[Opentrons](http://www.opentrons.com/)\n\n",
+ "author": "[Opentrons](http://www.opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Cherrypicking\n\n",
"description": "This protocol performs cherrypicking from source to target plates as specified in an input CSV file. The protocol parses the CSV for slots to load the source and target plates. Only non-zero volume transfers are carried out for efficiency, and the user is prompted to refill tipracks if necessary.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [P50 Single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette?variant=5984549077021)\n* [P300 8-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette?variant=5984549109789)\n* [Opentrons 300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Corning 96-Well Plate, Flat 360\u00b5L](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [USA Scientific 12-Well Reservoir, 22mL](https://labware.opentrons.com/usascientific_12_reservoir_22ml?category=reservoir)\n\n---\n\n\n**Deck Layout**\n\n**Slot 1**: [Opentrons 300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips) (for P50 Single-Channel)\n\n**Slot 2**: [Corning 96-Well Plate, Flat 360\u00b5L](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (ELISA plate A)\n\n**Slot 3**: [USA Scientific 12-Well Reservoir, 22mL](https://labware.opentrons.com/usascientific_12_reservoir_22ml?category=reservoir)\n\n**Slot 4**: [Opentrons 300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips) (for P300 8-Channel)\n\n**Slot 5**: [Corning 96-Well Plate, Flat 360\u00b5L](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (ELISA plate B)\n\n**Slot 6**: [Corning 96-Well Plate, Flat 360\u00b5L](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (1st Dilution Plate)\n\n**Slot 7**: Opentrons 300\u00b5l Pipette tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips) (for P300 8-Channel)\n\n**Slot 8**: [Corning 96-Well Plate, Flat 360\u00b5L](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (ELISA plate C)\n\n**Slot 9**: [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) containing 2mL Tubes\n\n**Slot 10**: [Corning 96-Well Plate, Flat 360\u00b5L](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (2nd Dilution Plate)\n\n**Slot 11**: [Corning 96-Well Plate, Flat 360\u00b5L](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate) (ELISA plate D)\n\n\n",
"internal": "6c27ee\n",
diff --git a/protoBuilds/6c7447/README.json b/protoBuilds/6c7447/README.json
index 8435f27d8a..768a00f6ac 100644
--- a/protoBuilds/6c7447/README.json
+++ b/protoBuilds/6c7447/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol is an update to a qPCR prep protocol designed for the OT-1.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons P300 Single-Channel Pipette\nOpentrons P10 Multi-Channel Pipette\nOpentrons 300\u00b5L Tips\nOpentrons 10\u00b5L Tips\nOpentrons 4-in-1 Tube Rack\nBio-Rad 96-Well Plate, 200\u00b5L\nEppendorf 2mL Tubes\nReagents\nSamples\n\n\n\nSlot 1: Bio-Rad Plate (qPCR Plate)\nSlot 2: Bio-Rad Plate (DNA Plate)\nSlot 3: Opentrons 10\u00b5L Tips, Full Tip Rack\nSlot 4: Tube Rack with 2mL Tubes and Reagents\nSlot 5: Opentrons 10\u00b5L Tips, with only tips in row 'A'\nSlot 6: [Opentrons 10\u00b5L Tips, with only tips in 'A1' and 'A2'\nSlot 7: Opentrons 300\u00b5L Tips, Full Tip Rack\nUsing the customizations fields, below set up your protocol.\n P10 Multi Mount: Select which mount (left or right) the P10 Multi is attached to.\n P300 Single Mount: Select which mount (left or right) the P300 Single is attached to.\n* Pre-Wet Tips?: Specify whether or not you'd like a mix step prior to each transfer to pre-wet the tip.\nNote: The tip racks in slot 5 and 6 should be empty, with tips only in row 'A'. This will allow the P10-Multi to effectively operate as a single channel pipette.",
"internal": "6c7447",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"description": "This protocol is an update to a qPCR prep protocol designed for the OT-1.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons P10 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [Bio-Rad 96-Well Plate, 200\u00b5L](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr)\n* Eppendorf 2mL Tubes\n* Reagents\n* Samples\n\n\n---\n\n\nSlot 1: Bio-Rad Plate (qPCR Plate)\n\nSlot 2: Bio-Rad Plate (DNA Plate)\n\nSlot 3: [Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips), Full Tip Rack\n\nSlot 4: [Tube Rack](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1) with 2mL Tubes and Reagents\n\nSlot 5: [Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips), with only tips in row 'A'\n\nSlot 6: [[Opentrons 10\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips), with only tips in 'A1' and 'A2'\n\nSlot 7: [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips), Full Tip Rack\n\n\n\n\n**Using the customizations fields, below set up your protocol.**\n* **P10 Multi Mount**: Select which mount (left or right) the P10 Multi is attached to.\n* **P300 Single Mount**: Select which mount (left or right) the P300 Single is attached to.\n* **Pre-Wet Tips?**: Specify whether or not you'd like a mix step prior to each transfer to pre-wet the tip.\n\n\n\n**Note**: The tip racks in slot 5 and 6 should be empty, with tips only in row 'A'. This will allow the P10-Multi to effectively operate as a single channel pipette.\n\n",
"internal": "6c7447\n",
diff --git a/protoBuilds/6cb818/README.json b/protoBuilds/6cb818/README.json
index fa56dbbf47..d23f1fa538 100644
--- a/protoBuilds/6cb818/README.json
+++ b/protoBuilds/6cb818/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Staining"
@@ -9,7 +9,7 @@
"description": "This protocol uses a p20 single-channel pipette to transfer 5 uL anti-CD154 from a 1.5 mL screw-cap tube to wells A1 and A2 (already containing 200 uL of suspended cells) of a 96-well culture plate. The cells are then mixed and aliquoted into wells B1-E1 and B2-E2 of the culture plate. Wells B1-B2, C1-C2, D1-D2 and E1-E2 are then treated with either one of three peptides or PMA-Ca. Finally, well volumes are optionally adjusted to 100 uL total by addition of culture medium.\nLinks:\n* experimental protocol\nThis protocol was developed to mix anti-CD154 with cultured cells suspended in culture medium and then aliquot the cells to wells of a 96-well culture plate where the cells are treated either with one of three different peptides or PMA-Ca. Volumes are optionally adjusted to 100 uL total by addition of culture medium.",
"internal": "6cb818",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Staining\n\n",
"deck-setup": "\n\n\n\n\n* Opentrons 20 ul filter or regular tips (selected deck slot)\n* 96-Well Culture Plate with Cells in A1 and A2 (selected deck slot)\n* Opentrons 24-Tube Rack Containing 1.5 mL Snap Cap Tubes (containing peptides 1-3 and PMA-Ca) (selected deck slot)\n* Opentrons 24-Tube Rack Containing 1.5 mL Screw Cap Tubes (containing anti-CD154 and culture medium) (selected deck slot)\n\n",
"description": "\nThis protocol uses a p20 single-channel pipette to transfer 5 uL anti-CD154 from a 1.5 mL screw-cap tube to wells A1 and A2 (already containing 200 uL of suspended cells) of a 96-well culture plate. The cells are then mixed and aliquoted into wells B1-E1 and B2-E2 of the culture plate. Wells B1-B2, C1-C2, D1-D2 and E1-E2 are then treated with either one of three peptides or PMA-Ca. Finally, well volumes are optionally adjusted to 100 uL total by addition of culture medium.\n\nLinks:\n* [experimental protocol](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/6cb818/Protocol_ForCustom_Opentrons_9-21-21.docx)\n\n\nThis protocol was developed to mix anti-CD154 with cultured cells suspended in culture medium and then aliquot the cells to wells of a 96-well culture plate where the cells are treated either with one of three different peptides or PMA-Ca. Volumes are optionally adjusted to 100 uL total by addition of culture medium.\n\n",
diff --git a/protoBuilds/6cd9d6/README.json b/protoBuilds/6cd9d6/README.json
index a787312b11..045e807fd4 100644
--- a/protoBuilds/6cd9d6/README.json
+++ b/protoBuilds/6cd9d6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -10,7 +10,7 @@
"internal": "6cd9d6",
"labware": "\nBio-Rad 96 Well Plate 200 \u00b5L PCR\nOpentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nNEST 12 Well Reservoir 15 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Custom\n\n",
"deck-setup": "**Note: The deck setup changes throughout the course of the protocol. The initial calibration layout may differ from the final setup. Please follow the initial calibration setup and any instructions during the pauses.**\n\n* Slot 1: Temperature Module A + Bio Rad Plate (Sample Plate)\n* Slot 2: Bio Rad Plate (Reagent #2 Plate)\n* Slot 3: Temperature Module B + Bio Rad Plate (Reagent #1 Plate)\n* Slot 4: Magnetic Module\n* Slot 6: NEST 1 Well Reservoir (Ethanol Reservoir)\n* Slot 7: Opentrons 200 uL Filter Tips\n* Slot 8: Opentrons 200 uL Filter Tips\n* Slot 10: Opentrons 20 uL Filter Tips\n* Slot 11: Opentrons 20 uL Filter Tips\n\n---\n\n",
"description": "This protocol performs a custom NGS Library Prep. It utilizes two temperature modules in Slot 1 and Slot 3 for the cooling of reagents. It also utilizes the magnetic module for bead based purification of samples.\n\nExplanation of complex parameters below:\n* `P20 Multichannel GEN2 Mount Position`: Choose the mount position of the P20 Multichannel GEN2 pipette.\n* `P300 Multichannel GEN2 Mount Position`: Choose the mount position of the P300 Multichannel GEN2 pipette.\n* `Number of Samples`: Enter total number of samples in the protocol run. **Note: Because both pipettes are 8-channels, the number of samples should be a multiple of 8, otherwise it will use up additional tips for an entire column.**\n\n---\n\n",
diff --git a/protoBuilds/6d2e65/README.json b/protoBuilds/6d2e65/README.json
index 1490382a6b..ead48747ef 100644
--- a/protoBuilds/6d2e65/README.json
+++ b/protoBuilds/6d2e65/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Purification"
@@ -9,7 +9,7 @@
"description": "With this protocol, your robot can perform a bead clean-up of up to 96 input samples using Omega MagBind TotalPure NGS Beads.",
"internal": "6d2e65",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Nucleic Acid Purification\n\n",
"deck-setup": "* Opentrons Magnetic Module with NEST 100 ul PCR Plate (Deck Slot 3)\n* Opentrons 200 ul filter tips (Deck Slots 5, 6, 7, 8, 10)\n* NEST 12-Well Reservoir (Deck Slot 4)\n* Clean 96-Well PCR Plate for eluate (Deck Slot 1)\n\n",
"description": "With this protocol, your robot can perform a bead clean-up of up to 96 input samples using Omega MagBind TotalPure NGS Beads.\n\n",
diff --git a/protoBuilds/6d3eda/README.json b/protoBuilds/6d3eda/README.json
index b41f1eaca4..0566f03b04 100644
--- a/protoBuilds/6d3eda/README.json
+++ b/protoBuilds/6d3eda/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Plant DNA"
@@ -8,7 +8,7 @@
"description": "This protocol was built for Mass Spec sample prep for IPM doctors. It transfers a variable volume of reagent between 24 well input plate(s) and 24 well output plate(s). There are 3 custom labwares that this protocol normally uses - a 7ml tube rack (24 slot), a 1ml autosampler tube rack (24 slot), and a 24 MCT laser cut acrylic rack.\n\n\n\nOpentrons P300-multi channel electronic pipettes (GEN2)\nOpentrons 200ul Filter Tips\n24 well plate\n",
"internal": "6d3eda",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Plant DNA\n\n",
"description": "This protocol was built for Mass Spec sample prep for IPM doctors. It transfers a variable volume of reagent between 24 well input plate(s) and 24 well output plate(s). There are 3 custom labwares that this protocol normally uses - a 7ml tube rack (24 slot), a 1ml autosampler tube rack (24 slot), and a 24 MCT laser cut acrylic rack.\n\n---\n\n\n\n* [Opentrons P300-multi channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [24 well plate](example.com)\n\n\n",
"internal": "6d3eda\n",
diff --git a/protoBuilds/6d7eb7/README.json b/protoBuilds/6d7eb7/README.json
index e1307659bf..8e2edf26fc 100644
--- a/protoBuilds/6d7eb7/README.json
+++ b/protoBuilds/6d7eb7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -9,7 +9,7 @@
"description": "Our most robust cherrypicking protocol. Specify aspiration height, labware, pipette, as well as source and destination wells with this all inclusive cherrypicking protocol.\n\nYour Transfer .csv File should be a .csv file formatted in the following way:\nPipette Mount to use,Source Labware,Source Slot,Source Well,Source Aspiration Height Above Bottom (in mm),Destination Well,Volume (in ul)\nleft,4x6 2ml screw true,1,A1,1,A11,1\nleft,4x6 1.5ml snap,1,A1,1,A5,3\nright,8x12 0.5ml snap,2,A1,1.5,H12,7\n...\nYou can also download a template here. Please making sure to include headers in your .csv file.\nYour source labware selection in the .csv file must be one of the following options, which correspond to the different sample tubes you use in your workflow:\n 384 wellplate\n 96 wellplate\n 4x6 2ml screw true\n 4x6 2ml screw false\n 4x6 1.5ml snap\n 4x6 1.5ml screw\n 8x12 strip\n 8x12 1ml plug\n* 8x12 0.5ml snap\nThe height offsets of different tubes seated in the same tuberack will be automatically calculated in the Python protocol logic. However, please ensure to calibrate your tuberacks with the following tube types in spot A1:\n 4x6 Opentrons tuberack: 2ml screwcap false bottom in spot A1\n 8x12 ThermoScientific Matrix tuberack: 1ml plugcap in spot A1\nCalibration to these tubes and tuberacks will allow for offsets to be properly taken into account during the protocol run.\n",
"internal": "6d7eb7",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"deck-setup": "* Example deck setup - tip racks loaded onto remining slots.\n\n\n---\n\n",
"description": "\nOur most robust cherrypicking protocol. Specify aspiration height, labware, pipette, as well as source and destination wells with this all inclusive cherrypicking protocol.\n\n\n\nYour `Transfer .csv File` should be a .csv file formatted in the following way:\n\n```\nPipette Mount to use,Source Labware,Source Slot,Source Well,Source Aspiration Height Above Bottom (in mm),Destination Well,Volume (in ul)\nleft,4x6 2ml screw true,1,A1,1,A11,1\nleft,4x6 1.5ml snap,1,A1,1,A5,3\nright,8x12 0.5ml snap,2,A1,1.5,H12,7\n...\n```\nYou can also download a template [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/6d7eb7/ex.csv). **Please making sure to include headers in your .csv file.**\n\nYour source labware selection in the .csv file must be one of the following options, which correspond to the different sample tubes you use in your workflow:\n* `384 wellplate`\n* `96 wellplate`\n* `4x6 2ml screw true`\n* `4x6 2ml screw false`\n* `4x6 1.5ml snap`\n* `4x6 1.5ml screw`\n* `8x12 strip`\n* `8x12 1ml plug`\n* `8x12 0.5ml snap`\n\nThe height offsets of different tubes seated in the same tuberack will be automatically calculated in the Python protocol logic. However, **please ensure to calibrate your tuberacks with the following tube types in spot A1**:\n* 4x6 Opentrons tuberack: 2ml screwcap false bottom in spot A1\n* 8x12 ThermoScientific Matrix tuberack: 1ml plugcap in spot A1\n\nCalibration to these tubes and tuberacks will allow for offsets to be properly taken into account during the protocol run.\n\n---\n\n",
diff --git a/protoBuilds/6d7fc3-part-2/README.json b/protoBuilds/6d7fc3-part-2/README.json
index 80a163182d..1057eec55f 100644
--- a/protoBuilds/6d7fc3-part-2/README.json
+++ b/protoBuilds/6d7fc3-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung DNA UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "6d7fc3-part-2",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung DNA UMI Panel Kit\n\n",
"deck-setup": "\n* The example below illustrates the starting deck layout for Part 2 (Adapter Ligation).\n \n\n",
"description": "This protocol automates the second part of a seven part protocol for the [GeneRead QIAact Lung DNA UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Adapter Ligation part described in the [GeneRead QIAact Lung DNA UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=94ab92d2-1918-4388-989b-4cefa8eed203&lang=en).\n\n* Part 1: [Fragmentation, End-repair and A-addition](https://protocols.opentrons.com/protocol/6d7fc3)\n* Part 2: [Adapter Ligation](https://protocols.opentrons.com/protocol/6d7fc3-part-2)\n* Part 3: [Cleanup of Adapter-ligated DNA with QIAact Beads.](https://protocols.opentrons.com/protocol/6d7fc3-part-3)\n* Part 4: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/6d7fc3-part-4)\n* Part 5: [Cleanup of Target Enrichment PCR with QIAact Beads](https://protocols.opentrons.com/protocol/6d7fc3-part-5)\n* Part 6: [Universal PCR Amplification.](https://protocols.opentrons.com/protocol/6d7fc3-part-6)\n* Part 7: [Cleanup of Universal PCR with QIAact Beads](https://protocols.opentrons.com/protocol/6d7fc3-part-7)\n\nExplanation of complex parameters below:\n\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P300 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n\n---\n\n",
diff --git a/protoBuilds/6d7fc3-part-3/README.json b/protoBuilds/6d7fc3-part-3/README.json
index 9d0f9910ed..5de51b0e9c 100644
--- a/protoBuilds/6d7fc3-part-3/README.json
+++ b/protoBuilds/6d7fc3-part-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung DNA UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "6d7fc3-part-3",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung DNA UMI Panel Kit\n\n",
"deck-setup": "* The example below illustrates the starting deck layout for Part 3 (Cleanup of Adapter-ligated DNA with QIAact Beads).\n\n\n",
"description": "This protocol automates the third part of a seven part protocol for the [GeneRead QIAact Lung DNA UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Cleanup of Adapter-ligated DNA with QIAact Beads part described in the [GeneRead QIAact Lung DNA UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=94ab92d2-1918-4388-989b-4cefa8eed203&lang=en).\n\n* Part 1: [Fragmentation, End-repair and A-addition](https://protocols.opentrons.com/protocol/6d7fc3)\n* Part 2: [Adapter Ligation](https://protocols.opentrons.com/protocol/6d7fc3-part-2)\n* Part 3: [Cleanup of Adapter-ligated DNA with QIAact Beads.](https://protocols.opentrons.com/protocol/6d7fc3-part-3)\n* Part 4: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/6d7fc3-part-4)\n* Part 5: [Cleanup of Target Enrichment PCR with QIAact Beads](https://protocols.opentrons.com/protocol/6d7fc3-part-5)\n* Part 6: [Universal PCR Amplification.](https://protocols.opentrons.com/protocol/6d7fc3-part-6)\n* Part 7: [Cleanup of Universal PCR with QIAact Beads](https://protocols.opentrons.com/protocol/6d7fc3-part-7)\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P300 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `Magnetic Module Engage Height (mm)`: The height the magnets should raise.\n\n---\n\n",
diff --git a/protoBuilds/6d7fc3-part-4/README.json b/protoBuilds/6d7fc3-part-4/README.json
index c9d6760a2a..2ee9645d41 100644
--- a/protoBuilds/6d7fc3-part-4/README.json
+++ b/protoBuilds/6d7fc3-part-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung DNA UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "6d7fc3-part-4",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "\n[Opentrons](https://opentrons.com/)\n\n",
+ "author": "\n[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung DNA UMI Panel Kit\n\n",
"deck-setup": "\n* The example below illustrates the starting deck layout for Part 4 (Target Enrichment PCR).\n\n\n\n",
"description": "\nThis protocol automates the fourth part of a seven part protocol for the [GeneRead QIAact Lung DNA UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Target Enrichment PCR part described in the [GeneRead QIAact Lung DNA UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=94ab92d2-1918-4388-989b-4cefa8eed203&lang=en).\n\n* Part 1: [Fragmentation, End-repair and A-addition](https://protocols.opentrons.com/protocol/6d7fc3)\n* Part 2: [Adapter Ligation](https://protocols.opentrons.com/protocol/6d7fc3-part-2)\n* Part 3: [Cleanup of Adapter-ligated DNA with QIAact Beads.](https://protocols.opentrons.com/protocol/6d7fc3-part-3)\n* Part 4: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/6d7fc3-part-4)\n* Part 5: [Cleanup of Target Enrichment PCR with QIAact Beads](https://protocols.opentrons.com/protocol/6d7fc3-part-5)\n* Part 6: [Universal PCR Amplification.](https://protocols.opentrons.com/protocol/6d7fc3-part-6)\n* Part 7: [Cleanup of Universal PCR with QIAact Beads](https://protocols.opentrons.com/protocol/6d7fc3-part-7)\n\nExplanation of complex parameters below:\n\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P300 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n\n---\n\n",
diff --git a/protoBuilds/6d7fc3-part-5/README.json b/protoBuilds/6d7fc3-part-5/README.json
index edd3ce302c..2aea81a3cd 100644
--- a/protoBuilds/6d7fc3-part-5/README.json
+++ b/protoBuilds/6d7fc3-part-5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung DNA UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "6d7fc3-part-5",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung DNA UMI Panel Kit\n\n",
"deck-setup": "* The example below illustrates the starting deck layout for Part 5 (Cleanup of Target Enrichment PCR with QIAact Beads).\n\n\n",
"description": "This protocol automates the fifth part of a seven part protocol for the [GeneRead QIAact Lung DNA UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Cleanup of Target Enrichment PCR with QIAact Beads part described in the [GeneRead QIAact Lung DNA UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=94ab92d2-1918-4388-989b-4cefa8eed203&lang=en).\n\n* Part 1: [Fragmentation, End-repair and A-addition](https://protocols.opentrons.com/protocol/6d7fc3)\n* Part 2: [Adapter Ligation](https://protocols.opentrons.com/protocol/6d7fc3-part-2)\n* Part 3: [Cleanup of Adapter-ligated DNA with QIAact Beads.](https://protocols.opentrons.com/protocol/6d7fc3-part-3)\n* Part 4: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/6d7fc3-part-4)\n* Part 5: [Cleanup of Target Enrichment PCR with QIAact Beads](https://protocols.opentrons.com/protocol/6d7fc3-part-5)\n* Part 6: [Universal PCR Amplification.](https://protocols.opentrons.com/protocol/6d7fc3-part-6)\n* Part 7: [Cleanup of Universal PCR with QIAact Beads](https://protocols.opentrons.com/protocol/6d7fc3-part-7)\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P300 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `Magnetic Module Engage Height (mm)`: The height the magnets should raise.\n\n---\n\n",
diff --git a/protoBuilds/6d7fc3-part-6/README.json b/protoBuilds/6d7fc3-part-6/README.json
index d6f8a71e14..6af57a27bd 100644
--- a/protoBuilds/6d7fc3-part-6/README.json
+++ b/protoBuilds/6d7fc3-part-6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung DNA UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "6d7fc3-part-6",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "\n[Opentrons](https://opentrons.com/)\n\n",
+ "author": "\n[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung DNA UMI Panel Kit\n\n",
"deck-setup": "\n* The example below illustrates the starting deck layout for Part 6 (Universal PCR Amplification).\n\n\n\n",
"description": "\nThis protocol automates the sixth part of a seven part protocol for the [GeneRead QIAact Lung DNA UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Universal PCR Amplification part described in the [GeneRead QIAact Lung DNA UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=94ab92d2-1918-4388-989b-4cefa8eed203&lang=en).\n\n* Part 1: [Fragmentation, End-repair and A-addition](https://protocols.opentrons.com/protocol/6d7fc3)\n* Part 2: [Adapter Ligation](https://protocols.opentrons.com/protocol/6d7fc3-part-2)\n* Part 3: [Cleanup of Adapter-ligated DNA with QIAact Beads.](https://protocols.opentrons.com/protocol/6d7fc3-part-3)\n* Part 4: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/6d7fc3-part-4)\n* Part 5: [Cleanup of Target Enrichment PCR with QIAact Beads](https://protocols.opentrons.com/protocol/6d7fc3-part-5)\n* Part 6: [Universal PCR Amplification.](https://protocols.opentrons.com/protocol/6d7fc3-part-6)\n* Part 7: [Cleanup of Universal PCR with QIAact Beads](https://protocols.opentrons.com/protocol/6d7fc3-part-7)\n\nExplanation of complex parameters below:\n\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P300 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n\n---\n\n",
diff --git a/protoBuilds/6d7fc3-part-7/README.json b/protoBuilds/6d7fc3-part-7/README.json
index 499213137b..17efea546d 100644
--- a/protoBuilds/6d7fc3-part-7/README.json
+++ b/protoBuilds/6d7fc3-part-7/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung DNA UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "6d7fc3-part-7",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung DNA UMI Panel Kit\n\n",
"deck-setup": "* The example below illustrates the starting deck layout for Part 6 (Cleanup of Universal PCR with QIAact Beads).\n\n\n",
"description": "This protocol automates the seventh part of a seven part protocol for the [GeneRead QIAact Lung DNA UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Cleanup of Universal PCR with QIAact Beads part described in the [GeneRead QIAact Lung DNA UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=94ab92d2-1918-4388-989b-4cefa8eed203&lang=en).\n\n* Part 1: [Fragmentation, End-repair and A-addition](https://protocols.opentrons.com/protocol/6d7fc3)\n* Part 2: [Adapter Ligation](https://protocols.opentrons.com/protocol/6d7fc3-part-2)\n* Part 3: [Cleanup of Adapter-ligated DNA with QIAact Beads.](https://protocols.opentrons.com/protocol/6d7fc3-part-3)\n* Part 4: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/6d7fc3-part-4)\n* Part 5: [Cleanup of Target Enrichment PCR with QIAact Beads](https://protocols.opentrons.com/protocol/6d7fc3-part-5)\n* Part 6: [Universal PCR Amplification.](https://protocols.opentrons.com/protocol/6d7fc3-part-6)\n* Part 7: [Cleanup of Universal PCR with QIAact Beads](https://protocols.opentrons.com/protocol/6d7fc3-part-7)\n\nExplanation of complex parameters below:\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `P300 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `Magnetic Module Engage Height (mm)`: The height the magnets should raise.\n\n---\n\n",
diff --git a/protoBuilds/6d7fc3/README.json b/protoBuilds/6d7fc3/README.json
index 5f5c6d58bd..8e6a1f59f5 100644
--- a/protoBuilds/6d7fc3/README.json
+++ b/protoBuilds/6d7fc3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"GeneRead QIAact Lung DNA UMI Panel Kit"
@@ -10,7 +10,7 @@
"internal": "6d7fc3",
"labware": "\nOpentrons Filter Tips\nNEST 96 Well 100 uL PCR Plate\nOpentrons Aluminum Block Set\n",
"markdown": {
- "author": "\n[Opentrons](https://opentrons.com/)\n\n",
+ "author": "\n[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* GeneRead QIAact Lung DNA UMI Panel Kit\n\n",
"deck-setup": "\n* The example below illustrates the deck layout when the samples are placed in 1.5 mL Screwcap tubes in an Opentrons Tube Rack.\n\n\n\n",
"description": "\nThis protocol automates the first part of a seven part protocol for the [GeneRead QIAact Lung DNA UMI Panel Kit](https://www.qiagen.com/us/products/instruments-and-automation/genereader-system/generead-qiaact-lung-panels-ww/) which constructs molecularly bar-coded DNA libraries for digital sequencing. This protocol automates the Fragmentation, End-repair and A-addition part described in the [GeneRead QIAact Lung DNA UMI Panel Handbook](https://www.qiagen.com/us/resources/download.aspx?id=94ab92d2-1918-4388-989b-4cefa8eed203&lang=en).\n\n* Part 1: [Fragmentation, End-repair and A-addition](https://protocols.opentrons.com/protocol/6d7fc3)\n* Part 2: [Adapter Ligation](https://protocols.opentrons.com/protocol/6d7fc3-part-2)\n* Part 3: [Cleanup of Adapter-ligated DNA with QIAact Beads.](https://protocols.opentrons.com/protocol/6d7fc3-part-3)\n* Part 4: [Target Enrichment PCR](https://protocols.opentrons.com/protocol/6d7fc3-part-4)\n* Part 5: [Cleanup of Target Enrichment PCR with QIAact Beads](https://protocols.opentrons.com/protocol/6d7fc3-part-5)\n* Part 6: [Universal PCR Amplification.](https://protocols.opentrons.com/protocol/6d7fc3-part-6)\n* Part 7: [Cleanup of Universal PCR with QIAact Beads](https://protocols.opentrons.com/protocol/6d7fc3-part-7)\n\nExplanation of complex parameters below:\n\n* `Number of Samples`: The total number of DNA samples. Samples must range between 1 (minimum) and 12 (maximum).\n* `Samples Labware Type`: The starting samples can be placed in either NEST 2.0 mL tubes on the Opentrons Tube Rack OR in a 96 Well Plate.\n* `P300 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n* `P20 Single GEN2 Pipette Mount Position`: The position of the pipette, either left or right.\n\n---\n\n",
diff --git a/protoBuilds/6d901d-2/README.json b/protoBuilds/6d901d-2/README.json
index 72771e59a3..a564d6702b 100644
--- a/protoBuilds/6d901d-2/README.json
+++ b/protoBuilds/6d901d-2/README.json
@@ -17,14 +17,14 @@
"internal": "6d901d-2\n",
"labware": "* Any verified labware found in our [Labware Library](https://labware.opentrons.com/?category=wellPlate) and some additional microplates (see plate options for source and destination plates parameters below, e.g. Greiner Bio-One plates)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[AstraZeneca](https://www.astrazeneca.com/)\n\n",
+ "partner": "[AstraZeneca](https://www.astrazeneca.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P20 Multi GEN2 Pipette](https://shop.opentrons.com/8-channel-electronic-pipette/)\n* [P300 Multi GEN2 Pipette](https://shop.opentrons.com/8-channel-electronic-pipette/)\n\n---\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. The pipette will aspirate a user-specified volume from the well on the (optionally normalized, see [Protocol Part 1](https://protocols.opentrons.com/protocol/6d901d)) source plate according to the imported csv file. The CSV also defines the aspiration height from the bottom of the source well.\n2. The pipette will dispense this volume into a user-specified target well according to the csv file.\n3. Steps 1 and 2 repeated for each line the CSV.\n\n",
"title": "Cherrypicking with multi-channel pipette substituting for a single channel pipette"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "AstraZeneca",
+ "partner": "AstraZeneca\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP20 Multi GEN2 Pipette\nP300 Multi GEN2 Pipette\n\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nThe pipette will aspirate a user-specified volume from the well on the (optionally normalized, see Protocol Part 1) source plate according to the imported csv file. The CSV also defines the aspiration height from the bottom of the source well.\nThe pipette will dispense this volume into a user-specified target well according to the csv file.\nSteps 1 and 2 repeated for each line the CSV.\n",
diff --git a/protoBuilds/6d901d/README.json b/protoBuilds/6d901d/README.json
index f6934fe5a7..1b274f8620 100644
--- a/protoBuilds/6d901d/README.json
+++ b/protoBuilds/6d901d/README.json
@@ -15,14 +15,14 @@
"description": "\n\nConcentration normalization is a key component of many genomic and proteomic applications, such as NGS library preparation. With this protocol, you can easily normalize the concentrations of samples in a 96 or 384 microwell plate without worrying about missing a well or adding the wrong volume. Just upload your properly formatted CSV file (keep scrolling for an example), customize your parameters, and download your ready-to-run protocol. This protocol is a modified version of our [Normalization Protocol](https://protocols.opentrons.com/protocol/normalization), that instead uses a multi-channel pipette as single channel pipette by picking up one tip at a time.\n\nThere is an optional [Part 2: Cherrypicking Protocol](https://protocols.opentrons.com/protocol/6d901d-2) to this protocol which performs cherrypicking using a multi-channel pipette in the same way.\n\n**Note**: This protocol was updated for a change in our software stack and will require app 6.0 or greater.\n\nUsing the customization fields below, set up your protocol.\n* `Volumes CSV`: Your input CSV specifying the normalization volumes. See the Setup section below for details.\n* `P300 Multi Mount`: Select mount for P300 Multi-Channel Pipette\n* `P20 Multi Mount`: Select mount for P20 Multi-Channel Pipette\n* `Plate Type`: Select the model of microwell plate to normalize samples on.\n* `Reservoir Type`: The type of the diluent reservoir. If you are selecting a multi-well reservoir you should place your diluent in well A1.\n* `Use Filter Tips?`: Select whether your pipette will use regular or filter tips.\n* `Tip Usage Strategy`: You can select whether your pipette will reuse the same tip throughout the entire normalization procedure or change tips after every diluent transfer.\n\n---\n\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 4.4.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Multi-Channel Pipette](https://shop.opentrons.com/8-channel-electronic-pipette/) and corresponding [Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Samples in a compatible plate (96-well or 384-well)](https://labware.opentrons.com/?category=wellPlate)\n* [Automation-friendly reservoir](https://labware.opentrons.com/?category=reservoir)\n* Diluent\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n---\n\n\n\n**CSV Format**\n\nYour file must be saved as a comma separated value (.csv) file type. Your CSV must contain values corresponding to volumes in microliters (\u03bcL). It should be formatted in \u201clandscape\u201d orientation, with the value corresponding to well A1 in the upper left-hand corner of the value list.\n\n\n\nIn this example, 40\u03bcL will be added to A1, 41\u03bcL will be added to well B1, etc.\n\nIf you would like to follow our template, you can make a copy of [this spreadsheet](https://opentrons-protocol-library-website.s3.amazonaws.com/Technical+Notes/normalization/Opentrons+Normalization+Template.xlsx), fill out your values, and export as CSV from there.\n\n*Note about CSV*: All values corresponding to wells in the CSV must have a value (zero (0) is a valid value and nothing will be transferred to the corresponding well(s)). Additionally, the CSV can be formatted in \"portrait\" orientation. In portrait orientation, the bottom left corner is treated as A1 and the top right corner would correspond to the furthest well from A1 (H12 in a 96-well plate).\n\n\n",
"internal": "6d901d\n",
"notes": "\nIf you\u2019d like to request a protocol supporting multiple plates or require any other changes to this script, please fill out our [Protocol Request Form](https://opentrons-protocol-dev.paperform.co/). You can also modify the Python file directly by following our [API Documentation](https://docs.opentrons.com/v2/). If you\u2019d like to chat with an applications engineer about changes, please contact us at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n",
- "partner": "[AstraZeneca](https://www.astrazeneca.com/)\n\n",
+ "partner": "[AstraZeneca](https://www.astrazeneca.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"process": "\n1. Create your CSV file according to our instructions.\n2. Upload your CSV and select all desired settings according to the \u201cSetup\u201d section above to customize your protocol run.\n3. Download your customized OT-2 protocol using the blue \u201cDownload\u201d button, located above the deckmap.\n4. Upload your protocol into the Opentrons App and follow the instructions there to set up your deck, calibrate your labware, and proceed to run.\n5. Make sure to add reagents to your labware before placing it on the deck! Your diluent should be in your reservoir, and the samples you\u2019re normalizing should be in your plate.\n\n",
"protocol-steps": "1. The protocol reads the input .csv file and loads the selected labware and pipettes.\n2. The protocol uses the loaded multi-channel pipette as a single channel pipette to transfer the volume of diluent specified in the csv cell to the given well on the normalization plate. The multi-channel pipette picks up the bottom-most tip in a given tiprack column in order to only pick up one tip at a time. Depending on the `tip usage strategy` the pipette reuses the same tip for every step or it discards the used tip and picks up a new one for every transfer.\n3. The protocol repeats step 2 for each well on the plate until each well on the plate has been normalized.\n\n",
"robot": "* [OT-2](https://opentrons.com/ot-2)\n\n",
"title": "Normalization substituting a multi-channel pipette for a single-channel pipette"
},
"notes": "If you\u2019d like to request a protocol supporting multiple plates or require any other changes to this script, please fill out our Protocol Request Form. You can also modify the Python file directly by following our API Documentation. If you\u2019d like to chat with an applications engineer about changes, please contact us at protocols@opentrons.com.",
- "partner": "AstraZeneca",
+ "partner": "AstraZeneca\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"process": "\nCreate your CSV file according to our instructions.\nUpload your CSV and select all desired settings according to the \u201cSetup\u201d section above to customize your protocol run.\nDownload your customized OT-2 protocol using the blue \u201cDownload\u201d button, located above the deckmap.\nUpload your protocol into the Opentrons App and follow the instructions there to set up your deck, calibrate your labware, and proceed to run.\nMake sure to add reagents to your labware before placing it on the deck! Your diluent should be in your reservoir, and the samples you\u2019re normalizing should be in your plate.\n",
"protocol-steps": "\nThe protocol reads the input .csv file and loads the selected labware and pipettes.\nThe protocol uses the loaded multi-channel pipette as a single channel pipette to transfer the volume of diluent specified in the csv cell to the given well on the normalization plate. The multi-channel pipette picks up the bottom-most tip in a given tiprack column in order to only pick up one tip at a time. Depending on the tip usage strategy the pipette reuses the same tip for every step or it discards the used tip and picks up a new one for every transfer.\nThe protocol repeats step 2 for each well on the plate until each well on the plate has been normalized.\n",
"robot": [
diff --git a/protoBuilds/6e160e/README.json b/protoBuilds/6e160e/README.json
index 802c1cb29b..bb437f75e1 100644
--- a/protoBuilds/6e160e/README.json
+++ b/protoBuilds/6e160e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "6e160e",
"labware": "\nCorning 384 Well Plate 112 \u00b5L Flat\nOpentrons 20\u00b5L Filter Tips\nBio-Rad 96 Well Plate 200 \u00b5L PCR\nNEST 12-Well Reservoirs, 15 mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\nSamples are layed out between plates in the following manner:\n\n\n\n---\n\n",
"description": "This protocol preps a 384 well plate on the temperature module with reaction mix and sample. Reaction mix is held in well one of a 12 well reservoir on the temperature module. Both temperature modules are held at 4C. The protocol uses a multi-channel to fill full plates, and a single channel to fill the last unfilled plate. Samples and reaction mix are filled by row, skipping columns.\n\n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify the number of samples for this run.\n* `P20 Mounts`: Specify which mount (left or right) for each the P20 Single-Channel and P20 Multi-Channel pipettes.\n\n\n---\n\n",
diff --git a/protoBuilds/6e2509-v2/README.json b/protoBuilds/6e2509-v2/README.json
index 11920e09ee..64a61da3e1 100644
--- a/protoBuilds/6e2509-v2/README.json
+++ b/protoBuilds/6e2509-v2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol automates the transfer of PCR mix and samples from up to four 96-well elution plates to a 384-well plate. It follows a specific format detailed in the image below. Columns from the 96 well elution plates correspond to specific wells on the 384 well plate. Each elution plate contains a control that is also transferred into the 384-well plate. For runs that have less than four plates, the controls will need to be added manually for the corresponding elution plate. This protocol is version 2 of the original protocol which can be access here.\n96 Well Elution Plate Format:\n\n384 Well Plate Format:\n\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 4.0.0 or later)\nP20 Multi-Channel Pipette\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nThermofisher 384 well optical PCR plate\nMolgen 96 Well Elution Plate\nOpentrons NEST 12-Well Reservoir\n\n\n\nFor this protocol, be sure that the P20 pipette is attached.\nUsing the customization fields below, set up your protocol.\n* Number of samples: Input the total number of samples in all four plates. Supports up to 384 samples.\nNote About Reagents:\nPCR Mix should be placed into column A1 of the 96 well plate.\nLabware Setup\nPlate 1: Slot 4 (tips: slot 10)\nPlate 2: Slot 5 (tips: slot 11)\nPlate 3: Slot 1 (tips: slot 7)\nPlate 4: Slot 2 (tips: slot 8)\nSlot 3: 384 Well PCR Plate\nSlot 6: 96 Well Plate (PCR Mix in Column A1)\nSlots 7 - 11: Opentrons 96 Filter Tip Rack 20 uL",
"internal": "6e2509-v2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "This protocol automates the transfer of PCR mix and samples from up to four 96-well elution plates to a 384-well plate. It follows a specific format detailed in the image below. Columns from the 96 well elution plates correspond to specific wells on the 384 well plate. Each elution plate contains a control that is also transferred into the 384-well plate. For runs that have less than four plates, the controls will need to be added manually for the corresponding elution plate. This protocol is version 2 of the original protocol which can be access [here](https://protocols.opentrons.com/protocol/6e2509).\n\n**96 Well Elution Plate Format**:\n\n\n\n**384 Well Plate Format**:\n\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 4.0.0 or later)](https://opentrons.com/ot-app/)\n* [P20 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 96 Filter Tip Rack 20 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [Thermofisher 384 well optical PCR plate](https://www.thermofisher.com/order/catalog/product/4309849#/4309849)\n* [Molgen 96 Well Elution Plate](https://molgen.com/)\n* [Opentrons NEST 12-Well Reservoir](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n\n---\n\n\nFor this protocol, be sure that the P20 pipette is attached.\n\nUsing the customization fields below, set up your protocol.\n* Number of samples: Input the total number of samples in all four plates. Supports up to 384 samples.\n\n\n**Note About Reagents:**\nPCR Mix should be placed into column A1 of the 96 well plate.\n\n\n**Labware Setup**\n\nPlate 1: Slot 4 (tips: slot 10)\n\nPlate 2: Slot 5 (tips: slot 11)\n\nPlate 3: Slot 1 (tips: slot 7)\n\nPlate 4: Slot 2 (tips: slot 8)\n\nSlot 3: 384 Well PCR Plate\n\nSlot 6: 96 Well Plate (PCR Mix in Column A1)\n\nSlots 7 - 11: Opentrons 96 Filter Tip Rack 20 uL\n\n",
"internal": "6e2509-v2",
diff --git a/protoBuilds/6e2509/README.json b/protoBuilds/6e2509/README.json
index b22bd5695c..b6ff2c4fa3 100644
--- a/protoBuilds/6e2509/README.json
+++ b/protoBuilds/6e2509/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol automates the transfer of PCR mix and samples from up to four 96-well elution plates to a 384-well plate. It follows a specific format detailed in the image below. Columns from the 96 well elution plates correspond to specific wells on the 384 well plate. Each elution plate contains a control that is also transferred into the 384-well plate. For runs that have less than four plates, the controls will need to be added manually for the corresponding elution plate.\n96 Well Elution Plate Format:\n\n384 Well Plate Format:\n\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 4.0.0 or later)\nP20 Multi-Channel Pipette\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nThermofisher 384 well optical PCR plate\nMolgen 96 Well Elution Plate\nOpentrons NEST 12-Well Reservoir\n\n\n\nFor this protocol, be sure that the P20 pipette is attached.\nUsing the customization fields below, set up your protocol.\n P20 Multichannel Mount: Choose the mount position of your P20 multichannel pipette.\n Number of samples: Input the total number of samples in all four plates. Supports up to 384 samples.\n PCR Mix Transfer Volume: Input the volume of PCR mix to transfer per well. Default is 7.5 uL.\n Sample Transfer Volume: Input the volume of sample to transfer per well. Default is 5 uL.\n Sample Aspiration Height Above Bottom of the Well: Specify the height from the bottom of the well in which the pipette will aspirate.\n Sample Dispensing Height Above Bottom of the Well: Specify the height from the bottom of the well in which the pipette will dispense.\n Master Mix Aspiration Height Above Bottom of the Well: Specify the height from the bottom of the well in which the pipette will aspirate.\n Master Mix Dispensing Height Above Bottom of the Well: Specify the height from the bottom of the well in which the pipette will dispense.\nNote About Reagents:\nPCR Mix should be placed into column A1 of the 96 well plate.\nLabware Setup\nSlot 1: Plate 1\nSlot 2: Plate 2\nSlot 4: Plate 3\nSlot 5: Plate 4\nSlot 3: 384 Well PCR Plate\nSlot 6: 96 Well Plate (PCR Mix in Column A1)\nSlots 7 - 11: Opentrons 96 Filter Tip Rack 20 uL",
"internal": "6e2509",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "This protocol automates the transfer of PCR mix and samples from up to four 96-well elution plates to a 384-well plate. It follows a specific format detailed in the image below. Columns from the 96 well elution plates correspond to specific wells on the 384 well plate. Each elution plate contains a control that is also transferred into the 384-well plate. For runs that have less than four plates, the controls will need to be added manually for the corresponding elution plate.\n\n**96 Well Elution Plate Format**:\n\n\n\n**384 Well Plate Format**:\n\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 4.0.0 or later)](https://opentrons.com/ot-app/)\n* [P20 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 96 Filter Tip Rack 20 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [Thermofisher 384 well optical PCR plate](https://www.thermofisher.com/order/catalog/product/4309849#/4309849)\n* [Molgen 96 Well Elution Plate](https://molgen.com/)\n* [Opentrons NEST 12-Well Reservoir](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n\n---\n\n\nFor this protocol, be sure that the P20 pipette is attached.\n\nUsing the customization fields below, set up your protocol.\n* P20 Multichannel Mount: Choose the mount position of your P20 multichannel pipette.\n* Number of samples: Input the total number of samples in all four plates. Supports up to 384 samples.\n* PCR Mix Transfer Volume: Input the volume of PCR mix to transfer per well. Default is 7.5 uL.\n* Sample Transfer Volume: Input the volume of sample to transfer per well. Default is 5 uL.\n* Sample Aspiration Height Above Bottom of the Well: Specify the height from the bottom of the well in which the pipette will aspirate.\n* Sample Dispensing Height Above Bottom of the Well: Specify the height from the bottom of the well in which the pipette will dispense.\n* Master Mix Aspiration Height Above Bottom of the Well: Specify the height from the bottom of the well in which the pipette will aspirate.\n* Master Mix Dispensing Height Above Bottom of the Well: Specify the height from the bottom of the well in which the pipette will dispense.\n\n\n**Note About Reagents:**\nPCR Mix should be placed into column A1 of the 96 well plate.\n\n\n**Labware Setup**\n\nSlot 1: Plate 1\n\nSlot 2: Plate 2\n\nSlot 4: Plate 3\n\nSlot 5: Plate 4\n\nSlot 3: 384 Well PCR Plate\n\nSlot 6: 96 Well Plate (PCR Mix in Column A1)\n\nSlots 7 - 11: Opentrons 96 Filter Tip Rack 20 uL\n\n",
"internal": "6e2509",
diff --git a/protoBuilds/6ec4dd-2/README.json b/protoBuilds/6ec4dd-2/README.json
index 04b3a8bdec..d44d16ed48 100644
--- a/protoBuilds/6ec4dd-2/README.json
+++ b/protoBuilds/6ec4dd-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Clean Up"
@@ -10,7 +10,7 @@
"internal": "6ec4dd-2",
"labware": "\nKingFisher 96 Well Plate 200 \u00b5L #97002540\nBio-Rad 96 Well Plate 350 \u00b5L #HSS9601\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Clean Up\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a custom NGS library prep cleanup for up to 96 samples.\n\n\n",
diff --git a/protoBuilds/6ec4dd/README.json b/protoBuilds/6ec4dd/README.json
index 00e61deb91..b9e0446b58 100644
--- a/protoBuilds/6ec4dd/README.json
+++ b/protoBuilds/6ec4dd/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "6ec4dd",
"labware": "\nBio-Rad Hard-Shell\u00ae 96-Well PCR Plates, high profile, semi skirted, clear/clear #HSS9601\nThermoFisher KingFisher 96-Well Plate,s 200 \u03bcL #97002540B\nOpentrons 20\u00b5l tipracks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": " \nblue: initial samples \ngreen: mastermix (180\u00b5l per well)\n\n---\n\n",
"description": "This protocol performs part 1 of a custom NGS library prep workflow: PCR Prep 1.\n\nExplanation of complex parameters below:\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n\n---\n\n",
diff --git a/protoBuilds/6f2d9b/README.json b/protoBuilds/6f2d9b/README.json
index 0adb4b6df8..459333b4dc 100644
--- a/protoBuilds/6f2d9b/README.json
+++ b/protoBuilds/6f2d9b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Serial Dilution"
@@ -8,7 +8,7 @@
"description": "This protocol performs preparation of COVID-19 patient samples into a 96 well King Fisher deep well plate for downstream applications. It also has a double extraction mode where each sample will get aliquoted twice for extraction downstream. \n\n\n\nOpentrons 1000uL Filter Tips\nKingFisher Deep Well Plate 2 mL\nP1000 Multichannel GEN2\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nProtocol Steps\nSingle Extraction\n1. Transfer 200 uL (default) of patient sample from patient tube in deck slot 4 position A1 into well A1 of the KingFisher deep well plate.\n2. Transfer 200 uL (default) of patient sample from patient tube in deck slot 4 position B1 into well B1 of the KingFisher deep well plate.\n3. This process continues by going down the column for both tube racks and the deep well plate. Tube racks incremement based on slot number.\nDouble Extraction\n1. Transfer 200 uL (default) of patient sample from patient tube in deck slot 4 position A1 into well A1 of the KingFisher deep well plate.\n2. Transfer 200 uL (default) of patient sample from patient tube in deck slot 4 position A1 into well B1 of the KingFisher deep well plate.\n3. Transfer 200 uL (default) of patient sample from patient tube in deck slot 4 position B1 into well C1 of the KingFisher deep well plate.\n4. Transfer 200 uL (default) of patient sample from patient tube in deck slot 4 position B1 into well D1 of the KingFisher deep well plate.\n5. This process will continue aliquoting the same patient sample twice and will do up to 47 patient samples per run.\nNote: Samples are not aliquoted into wells G12 and H12. Those are reserved for controls.",
"internal": "6f2d9b",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Serial Dilution\n\n",
"description": "This protocol performs preparation of COVID-19 patient samples into a 96 well King Fisher deep well plate for downstream applications. It also has a double extraction mode where each sample will get aliquoted twice for extraction downstream. \n\n---\n\n\n* [Opentrons 1000uL Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [KingFisher Deep Well Plate 2 mL](https://www.thermofisher.com/order/catalog/product/95040450#/95040450)\n* [P1000 Multichannel GEN2](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=5984549142557)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n\n---\n\n\n**Protocol Steps**\n\n**Single Extraction**\n1. Transfer 200 uL (default) of patient sample from patient tube in deck slot 4 position A1 into well A1 of the KingFisher deep well plate.\n2. Transfer 200 uL (default) of patient sample from patient tube in deck slot 4 position B1 into well B1 of the KingFisher deep well plate.\n3. This process continues by going down the column for both tube racks and the deep well plate. Tube racks incremement based on slot number.\n\n**Double Extraction**\n1. Transfer 200 uL (default) of patient sample from patient tube in deck slot 4 position A1 into well A1 of the KingFisher deep well plate.\n2. Transfer 200 uL (default) of patient sample from patient tube in deck slot 4 position A1 into well B1 of the KingFisher deep well plate.\n3. Transfer 200 uL (default) of patient sample from patient tube in deck slot 4 position B1 into well C1 of the KingFisher deep well plate.\n4. Transfer 200 uL (default) of patient sample from patient tube in deck slot 4 position B1 into well D1 of the KingFisher deep well plate.\n5. This process will continue aliquoting the same patient sample twice and will do up to 47 patient samples per run.\n\n**Note**: Samples are not aliquoted into wells G12 and H12. Those are reserved for controls.\n\n",
"internal": "6f2d9b",
diff --git a/protoBuilds/6f4e2c/README.json b/protoBuilds/6f4e2c/README.json
index 5c02a7e3fb..6c2f1e5077 100644
--- a/protoBuilds/6f4e2c/README.json
+++ b/protoBuilds/6f4e2c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Blood Sample"
@@ -8,7 +8,7 @@
"description": "This protocol automates the protocol from Sileks for DNA isolation from whole blood with MP@SiO2 magnetic particles. This protocol utilizes a P300-Multi Channel pipette and gives the user the option to select either 24 or 48 samples and whether to dispense liquid waste in a reservoir in slot 8 or in the fixed trash container. This protocol is also designed for using the Eppendorf 1000\u00b5L deep-well plate.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Magnetic Module\nOpentrons Temperature Module\nOpentrons P300 Multi-Channel Pipette\nOpentrons 50/300\u00b5L Tips or Opentrons 200\u00b5L Filter Tips\nBio-Rad 96-Well Plate, 200\u00b5L\nNEST 12-Well Reservoir, 15mL\nNEST 1-Well Reservoir, 195mL, if using for liquid waste\nEppendorf 1000\u00b5L Deep-Well Plate\nReagents\nSamples\n\n\n\nSlot 1: Opentrons Magnetic Module with Eppendorf Deep-Well Plate and 100\u00b5L of blood sample in corresponding wells (either 24 or 48)\nSlot 2: NEST 12-Well Reservoir, 15mL\n A1: Start Buffer\n A2: Lysis/Binding Buffer\n A3: Lysis/Binding Buffer (48 Samples)\n A4: Wash Buffer 1\n A5: Wash Buffer 1 (48 Samples)\n A6: Wash Buffer 2\n A7: Wash Buffer 2 (48 Samples)\n A8: Wash Buffer 3\n A9: Wash Buffer 3 (48 Samples)\n A10: Elution Buffer\nSlot 3: Opentrons Tips\nSlot 4: Opentrons Temperature Module\nSlot 5: Bio-Rad 96-Well Plate, 200\u00b5L, for final elution\nSlot 6: Opentrons Tips\nSlot 8: NEST 1-Well Reservoir, 195mL, if using for liquid waste\nSlot 9: Opentrons Tips\nSlot 10: Opentrons Tips\nSlot 11: Opentrons Tips\nUsing the customizations fields, below set up your protocol.\n P300 Multi Mount: Select which mount (left or right) the P300 Multi is attached to.\n Tip Type: Select the type of tips for this protocol (non/filter).\n Number of Samples: Specify the number of samples you'd like to run.\n Liquid Waste in Reservoir: Specify if liquid waste should be dispensed in reservoir (or fixed trash container)",
"internal": "6f4e2c",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Blood Sample\n\n\n",
"description": "This protocol automates the protocol from Sileks for [DNA isolation from whole blood with MP@SiO2 magnetic particles](https://www.sileks.com/eu/librarian/librarian_ajax.php?ajaxaction=GetObjectByVName&VName=Manual_DNA_from_Blood_en_20141007.pdf). This protocol utilizes a P300-Multi Channel pipette and gives the user the option to select either 24 or 48 samples and whether to dispense liquid waste in a reservoir in slot 8 or in the fixed trash container. This protocol is also designed for using the Eppendorf 1000\u00b5L deep-well plate.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 50/300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips) or [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Bio-Rad 96-Well Plate, 200\u00b5L](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr)\n* [NEST 12-Well Reservoir, 15mL](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* [NEST 1-Well Reservoir, 195mL](https://labware.opentrons.com/nest_1_reservoir_195ml?category=reservoir), if using for liquid waste\n* Eppendorf 1000\u00b5L Deep-Well Plate\n* Reagents\n* Samples\n\n\n---\n\n\nSlot 1: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with Eppendorf Deep-Well Plate and 100\u00b5L of blood sample in corresponding wells (either 24 or 48)\n\nSlot 2: [NEST 12-Well Reservoir, 15mL](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n* A1: Start Buffer\n* A2: Lysis/Binding Buffer\n* A3: Lysis/Binding Buffer (48 Samples)\n* A4: Wash Buffer 1\n* A5: Wash Buffer 1 (48 Samples)\n* A6: Wash Buffer 2\n* A7: Wash Buffer 2 (48 Samples)\n* A8: Wash Buffer 3\n* A9: Wash Buffer 3 (48 Samples)\n* A10: Elution Buffer\n\nSlot 3: [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 4: [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n\nSlot 5: [Bio-Rad 96-Well Plate, 200\u00b5L](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr), for final elution\n\nSlot 6: [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 8: [NEST 1-Well Reservoir, 195mL](https://labware.opentrons.com/nest_1_reservoir_195ml?category=reservoir), if using for liquid waste\n\nSlot 9: [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 10: [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 11: [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\n**Using the customizations fields, below set up your protocol.**\n* **P300 Multi Mount**: Select which mount (left or right) the P300 Multi is attached to.\n* **Tip Type**: Select the type of tips for this protocol (non/filter).\n* **Number of Samples**: Specify the number of samples you'd like to run.\n* **Liquid Waste in Reservoir**: Specify if liquid waste should be dispensed in reservoir (or fixed trash container)\n\n\n",
"internal": "6f4e2c\n",
diff --git a/protoBuilds/6faa1e/README.json b/protoBuilds/6faa1e/README.json
index 896a361001..aab5157655 100644
--- a/protoBuilds/6faa1e/README.json
+++ b/protoBuilds/6faa1e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -8,7 +8,7 @@
"description": "This protocol automates the steps of transferring mastermix from source plates to destination plates.\n\n\n\nP20 multi-channel GEN2 electronic pipette\nOpentrons 20ul tiprack\nThermo Scientific\u2122 PCR Plate, 96-well, semi-skirted\nEppendorf PCR cooler\n\n\n\nDeck Setup\n Destination Plates (PCR Plate on top of Eppendorf Cooler) (Slots 1-8)\n Source Plate (NEST 12-channel 15ml reservoir on top of ice tray) (Slot 11)\n* Opentrons 20 uL tip rack (Slot 10)",
"internal": "6faa1e",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"description": "This protocol automates the steps of transferring mastermix from source plates to destination plates.\n\n---\n\n\n* [P20 multi-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 20ul tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Thermo Scientific\u2122 PCR Plate, 96-well, semi-skirted](https://www.fishersci.com/shop/products/thermo-scientific-96-well-semi-skirted-plates-flat-deck/ab1400l)\n* [Eppendorf PCR cooler](https://www.daigger.com/eppendorf-pcr-coolers-14616-group?gclid=CjwKCAiAz4b_BRBbEiwA5XlVVkYoJn1xfnsYoEzsrHijqNP-YRCcVBJtWxD9-ENFfB_Pc9RZJUaXYRoCWjQQAvD_BwE)\n\n---\n\n\nDeck Setup\n* Destination Plates (PCR Plate on top of Eppendorf Cooler) (Slots 1-8)\n* Source Plate (NEST 12-channel 15ml reservoir on top of ice tray) (Slot 11)\n* Opentrons 20 uL tip rack (Slot 10)\n\n",
"internal": "6faa1e",
diff --git a/protoBuilds/6fe477-workflow-1/README.json b/protoBuilds/6fe477-workflow-1/README.json
index 33f8c5cea6..5cf3975c67 100644
--- a/protoBuilds/6fe477-workflow-1/README.json
+++ b/protoBuilds/6fe477-workflow-1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Purification"
@@ -8,7 +8,7 @@
"description": "This protocol automates variable pooling of samples and addition of lysis buffer for automated extraction downstream. \nWorkflow 2\n\n\n\nP300 single-channel GEN2 electronic pipette\nP300 multi-channel GEN2 electronic pipette\nOpentrons 200ul filter tiprack\nOpentrons 300ul tiprack\nNunc\u2122 96-Well Polypropylene DeepWell\u2122 Storage Plates\nCaplugs\u2122 30mL Centrifuge Tubes\n\n\n\nDeck Setup\n Opentrons 200ul filter tiprack (Slot 5)\n Opentrons 300ul tiprack (Slot 6)\n Deep Well Plate (Slot 2)\n NEST 195 mL Reservoir (Slot 3)\n* 30 mL Caplug Tubes in Opentrons 6 Tube Rack (Slots 1, 4, 7, 8, 9, 10, 11)",
"internal": "6fe477",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Nucleic Acid Purification\n\n",
"description": "This protocol automates variable pooling of samples and addition of lysis buffer for automated extraction downstream. \n[Workflow 2](https://develop.protocols.opentrons.com/protocol/6fe477-workflow-2)\n\n---\n\n\n* [P300 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [P300 multi-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 200ul filter tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 300ul tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Nunc\u2122 96-Well Polypropylene DeepWell\u2122 Storage Plates](https://www.thermofisher.com/order/catalog/product/260251#/260251)\n* [Caplugs\u2122 30mL Centrifuge Tubes](https://www.fishersci.com/shop/products/evergreen-scientific-30ml-centrifuge-tubes-30ml-freestanding-tubes-with-caps-sterile-500-cs-50-bags-10/22044320)\n\n---\n\n\nDeck Setup\n* Opentrons 200ul filter tiprack (Slot 5)\n* Opentrons 300ul tiprack (Slot 6)\n* Deep Well Plate (Slot 2)\n* NEST 195 mL Reservoir (Slot 3)\n* 30 mL Caplug Tubes in Opentrons 6 Tube Rack (Slots 1, 4, 7, 8, 9, 10, 11)\n\n",
"internal": "6fe477",
diff --git a/protoBuilds/6fe477-workflow-2/README.json b/protoBuilds/6fe477-workflow-2/README.json
index 986174f6f3..395b053b35 100644
--- a/protoBuilds/6fe477-workflow-2/README.json
+++ b/protoBuilds/6fe477-workflow-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Purification"
@@ -8,7 +8,7 @@
"description": "This protocol automates variable pooling of samples and addition of lysis buffer for manual extraction downstream. \nWorkflow 1\n\n\n\nP300 single-channel GEN2 electronic pipette\nP1000 single-channel GEN2 electronic pipette\nOpentrons 200ul filter tiprack\nOpentrons 1000ul tiprack\nVWR 1.5mL Centrifuge\n\n\n\nDeck Setup\n Opentrons 200ul filter tiprack (Slot 5)\n Opentrons 1000ul tiprack (Slot 6)\n VWR 1.5mL Tubes in Opentrons 24 Tube Rack (Slot 2)\n NEST 195 mL Reservoir (Slot 3)\n* 30 mL Caplug Tubes in Opentrons 6 Tube Rack (Slots 1, 4, 7, 8, 9, 10, 11)",
"internal": "6fe477",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Nucleic Acid Purification\n\n",
"description": "This protocol automates variable pooling of samples and addition of lysis buffer for manual extraction downstream. \n[Workflow 1](https://develop.protocols.opentrons.com/protocol/6fe477-workflow-1)\n---\n\n\n* [P300 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [P1000 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 200ul filter tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 1000ul tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* [VWR 1.5mL Centrifuge](https://us.vwr.com/store/product/4674417/vwr-micro-centrifuge-tube-with-flat-screw-cap)\n\n---\n\n\nDeck Setup\n* Opentrons 200ul filter tiprack (Slot 5)\n* Opentrons 1000ul tiprack (Slot 6)\n* VWR 1.5mL Tubes in Opentrons 24 Tube Rack (Slot 2)\n* NEST 195 mL Reservoir (Slot 3)\n* 30 mL Caplug Tubes in Opentrons 6 Tube Rack (Slots 1, 4, 7, 8, 9, 10, 11)\n\n",
"internal": "6fe477",
diff --git a/protoBuilds/6ff9e9/README.json b/protoBuilds/6ff9e9/README.json
index a4d8c4e948..122f33c518 100644
--- a/protoBuilds/6ff9e9/README.json
+++ b/protoBuilds/6ff9e9/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Extraction"
@@ -8,7 +8,7 @@
"description": "This is a flexible protocol accommodating various nucleic acid purifications. The protocol is broken down into 5 main parts:\n binding buffer and magnetic beads addition to samples\n bead wash 2x using magnetic module\n* final elution to chilled PCR plate\nLysed samples should be loaded on the magnetic module in a NEST 96-deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 4) to the elution PCR plate (temperature module, slot 1). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\nExplanation of complex parameters below:\n park tips: If set to yes (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to no, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n track tips across protocol runs: If set to yes, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to no, tips will be picked up from column 1 of the 1st tiprack.\n* flash: If set to yes, the robot rail lights will flash during any automatic pauses in the protocol. If set to no, the lights will not flash.\n\n \nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons magnetic module\nOpentrons temperature module\nCustom 2-channel 170ml reservoirs\nCustom 96 Well Plate 200 \u00b5L PCR\nNEST 96 Deepwell Plate 2mL\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n\n\n\nSlots 11, 8, 5, and 2, from top to bottom:\n",
"internal": "6ffe9",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Nucleic Acid Extraction\n\n",
"description": "This is a flexible protocol accommodating various nucleic acid purifications. The protocol is broken down into 5 main parts:\n* binding buffer and magnetic beads addition to samples\n* bead wash 2x using magnetic module\n* final elution to chilled PCR plate\n\nLysed samples should be loaded on the magnetic module in a NEST 96-deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 4) to the elution PCR plate (temperature module, slot 1). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\nExplanation of complex parameters below:\n* `park tips`: If set to `yes` (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to `no`, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n* `flash`: If set to `yes`, the robot rail lights will flash during any automatic pauses in the protocol. If set to `no`, the lights will not flash.\n\n---\n\n \n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons magnetic module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons temperature module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* Custom 2-channel 170ml reservoirs\n* Custom 96 Well Plate 200 \u00b5L PCR\n* [NEST 96 Deepwell Plate 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep)\n* [Opentrons 96 Filter Tip Rack 200 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n\n---\n\n\nSlots 11, 8, 5, and 2, from top to bottom: \n\n\n",
"internal": "6ffe9\n",
diff --git a/protoBuilds/701319/README.json b/protoBuilds/701319/README.json
index d3975f0ad8..4d387c4fbd 100644
--- a/protoBuilds/701319/README.json
+++ b/protoBuilds/701319/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Mass Spec"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom mass spectrometry sample prep for peptides up to 48 samples.\n\n\n\nOpentrons Magnetic Module\nNEST 96-well PCR plate 100\u00b5l full skirt\nOpentrons 20\u00b5l tiprack\nOpentrons 300\u00b5l tiprack\nOpentrons P20 and P300 GEN2 multi-channel electronic pipette\n\n\n\nDeck Setup\n Nunc 96-well polystyrene V-bottom microwell plate (Slot 1)\n NEST 0.1 mL 96-Well PCR Plate, Full Skirt (Slot 2)\n NEST 12-well reservoir, 15 mL,Opentrons (Slot 3)\n Opentrons Magnetic Module GEN2 (Slot 4)\n Stellar Scientific Cluster Tubes Racked, 1.2 mL (Slot 4, On Magnetic Module)\n Opentrons 300 uL Tip Rack (Slot 5, Slot 8)\n* Opentrons 20 uL Tip Rack (Slot 6, Slot 9)\nNOTE: Switch the Cluster Tubes Plate with the NEST PCR Plate on the Magnetic Module once the Pause step is reached.\nMagnetic Module Engage Height from Well Bottom (mm) refers to the height the magnet will move to from the bottom of the well. A value of 13.5 mm will move the tops of the magnets to level with that height value.",
"internal": "701319",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Mass Spec\n\n",
"description": "This protocol performs a custom mass spectrometry sample prep for peptides up to 48 samples.\n\n---\n\n\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [NEST 96-well PCR plate 100\u00b5l full skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons 20\u00b5l tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* [Opentrons 300\u00b5l tiprack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons P20 and P300 GEN2 multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n\n---\n\n\nDeck Setup\n* Nunc 96-well polystyrene V-bottom microwell plate (Slot 1)\n* NEST 0.1 mL 96-Well PCR Plate, Full Skirt (Slot 2)\n* NEST 12-well reservoir, 15 mL,Opentrons (Slot 3)\n* Opentrons Magnetic Module GEN2 (Slot 4)\n* Stellar Scientific Cluster Tubes Racked, 1.2 mL (Slot 4, On Magnetic Module)\n* Opentrons 300 uL Tip Rack (Slot 5, Slot 8)\n* Opentrons 20 uL Tip Rack (Slot 6, Slot 9)\n\nNOTE: Switch the Cluster Tubes Plate with the NEST PCR Plate on the Magnetic Module once the Pause step is reached.\n\n**Magnetic Module Engage Height from Well Bottom (mm)** refers to the height the magnet will move to from the bottom of the well. A value of 13.5 mm will move the tops of the magnets to level with that height value.\n\n",
"internal": "701319\n",
diff --git a/protoBuilds/70567f/README.json b/protoBuilds/70567f/README.json
index 2afa263f6e..7bbb59b3b1 100644
--- a/protoBuilds/70567f/README.json
+++ b/protoBuilds/70567f/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Ampure XP"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom NGS library prep cleanup using Ampure XP beads in a variable ratio with samples. The samples are mounted on an Opentrons magnetic module, and the final elute is transferred to a fresh PCR plate.\nExplanation of complex parameters below:\n park tips: If set to yes (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to no, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n track tips across protocol runs: If set to yes, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to no, tips will be picked up from column 1 of the 1st tiprack.\n\n\n\nOpentrons Magnetic Module (GEN2)\nNEST 96 Deepwell Plate 2mL\nP300 multi-channel electronic pipette (GEN2)\nOpentrons 200ul filtertipracks\n\n\n\n96-deepwell plate (slot 7)\n column 1: Ampure XP beads\n column 2: 80% ethanol\n column 3: EB buffer\n columns 10-12: liquid waste (loaded empty at start)",
"internal": "70567f",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Ampure XP\n\n",
"description": "This protocol performs a custom NGS library prep cleanup using Ampure XP beads in a variable ratio with samples. The samples are mounted on an Opentrons magnetic module, and the final elute is transferred to a fresh PCR plate.\n\nExplanation of complex parameters below:\n* `park tips`: If set to `yes` (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to `no`, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n\n---\n\n\n* [Opentrons Magnetic Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [NEST 96 Deepwell Plate 2mL](https://shop.opentrons.com/collections/verified-labware/products/nest-0-2-ml-96-well-deep-well-plate-v-bottom)\n* [P300 multi-channel electronic pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 200ul filtertipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n\n---\n\n\n96-deepwell plate (slot 7)\n* column 1: Ampure XP beads\n* column 2: 80% ethanol\n* column 3: EB buffer\n* columns 10-12: liquid waste (loaded empty at start)\n\n",
"internal": "70567f\n",
diff --git a/protoBuilds/7062c9-2/README.json b/protoBuilds/7062c9-2/README.json
index dbaf151845..7491dae38f 100644
--- a/protoBuilds/7062c9-2/README.json
+++ b/protoBuilds/7062c9-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Nucleic Acid Extraction"
@@ -10,7 +10,7 @@
"internal": "7062c9",
"labware": "\nNEST 12 Well Reservoir 15 mL\nNEST 1 Well Reservoir 195 mL\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt\nNEST 96 Deepwell Plate 2mL\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Nucleic Acid Extraction\n\n",
"deck-setup": "This example starting deck state shows the layout for 24 samples: \n\n\n* pink on starting sample plate: 110\u00b5l annealed and digested samples\n* green on reagent reservoir: buffer 2 (wash buffer)\n* blue on reagent reservoir: water\n* purple on reagent reservoir: magnetic beads\n* orange on reagent reservoir: buffer 3 (elution buffer)\n\n---\n\n",
"description": "\nLinks:\n* [Steps 1-2 (annealing preparation and digestion)](./7062c9)\n* [Steps 3-6 (extraction)](7062c9-2)\n\nThis is a flexible protocol accommodating a multi-step nucleic acid extraction. The protocol is broken down into the following main parts:\n* magnetic beads pre-wash\n* beads binding to sample\n* beads washing\n* final elution\n\nBefore running the protocol, 100\u00b5l of beads in buffer should be pre-loaded on the magnetic module in a NEST 96-deepwell plate. For reagent layout, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 4) to the elution PCR plate (temperature module, slot 1). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\nExplanation of complex parameters below:\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n\n---\n\n",
diff --git a/protoBuilds/7062c9-aliquot/README.json b/protoBuilds/7062c9-aliquot/README.json
index e7dd2a44cd..091e477905 100644
--- a/protoBuilds/7062c9-aliquot/README.json
+++ b/protoBuilds/7062c9-aliquot/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Aliquoting"
@@ -10,7 +10,7 @@
"internal": "7062c9-aliquot",
"labware": "\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 15 Tube Rack with Falcon 15 mL Conical\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Aliquoting\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol performs a flexible aliquoting for up to 96 Eppendorf snapcap tubes.\n\n\n",
diff --git a/protoBuilds/7062c9/README.json b/protoBuilds/7062c9/README.json
index 92f306bd06..e313b2369f 100644
--- a/protoBuilds/7062c9/README.json
+++ b/protoBuilds/7062c9/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "7062c9",
"labware": "\nNEST 12 Well Reservoir 15 mL\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt\nOpentrons 24 Tube Rack with NEST 1.5 mL Screwcap Tubes or equivalent\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Normalization\n\n",
"deck-setup": "This example starting deck state shows the layout for 24 samples: \n\n\n* green on sample tuberack: starting samples\n* blue on reagent reservoir: water\n* pink on buffer + probe tuberack: buffer\n* purple on buffer + probe tuberack: protease\n* orange buffer + probe tuberack: available spots for probes\n\n---\n\n",
"description": "\nLinks:\n* [Steps 1-2 (annealing preparation and digestion)](./7062c9)\n* [Steps 3-6 (extraction)](7062c9-2)\n\nThis is a flexible normalization protocol accommodating sample annealing preparation and digestion pre- and post-PCR. Normalization parameters should be input as a .csv file below, and should be formatted as shown in the following template:\n\n```\nsample conc. (mg/ml),sample volume (\u00b5l),water volume (\u00b5l),buffer 1 (\u00b5l),probe volume (\u00b5l),probe tube location (C1-D6),total volume (\u00b5l)\n1,40,50,10,5,A3,105\n2.2,18.2,71.8,10,5,A3,105\n2.03,19.7,70.3,10,5,A4,105\n```\n\nFor sample traceability and consistency, samples are mapped directly from the sample plate (slot 1) to the final normalized plate (slot 2). Sample plate well A1 is transferred to normalized plate A1, Sample plate well B1 to normalized plate B1, ..., D2 to D2, etc.\n\n---\n\n",
diff --git a/protoBuilds/71381b/README.json b/protoBuilds/71381b/README.json
index 589e84ef08..e64dc0cfcc 100644
--- a/protoBuilds/71381b/README.json
+++ b/protoBuilds/71381b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Zymo Kit"
@@ -8,7 +8,7 @@
"description": "This protocol automates the Zymo Quick-DNA HMW MagBead Kit with an additional labelling step that takes place before the purification.\n\nThe parameter section below gives you the ability to select either the Labelling portion of the protocol or the Purification portion of the protocol (you can select both). During the labelling portion, MilliQ Water, CSB, M.Taql, MTC22, and Proteinase K are added to the samples and the samples incubate while on the Temperature Module. During the purification portion, the samples are purified using the Zymo MagBead Kit in conjunction with Opentrons Magnetic Module.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 4.0.0 or later)\nOpentrons Temperatue Module, GEN2\nOpentrons Magnetic Module, GEN2\nOpentrons P20 8-Channel Pipette\nOpentrons P300 8-Channel Pipette\nOpentrons 20\u00b5L Tip Racks\nOpentrons 300\u00b5L Tip Racks\nNEST 12-Well Reservoir, 15mL\nBio-Rad 96-Well PCR Plate, 200\u00b5L\nZymo Quick-DNA HMW MagBead Kit\nReagents for Labelling\nSamples\n\n\n\nDeck Layout\n\nSlot 1: Opentrons Temperatue Module, GEN2 with Bio-Rad 96-Well PCR Plate, 200\u00b5L on 96-Well Aluminum Block, containing samples in columns 1-4\n\nSlot 2: Bio-Rad 96-Well PCR Plate, 200\u00b5L (Labelling Plate)\nColumn 1: MTC22\nColumn 3: CSB\nColumn 5: M.Taql\nColumn 7: Proteinase K\n\nSlot 3: Opentrons 20\u00b5L Tip Racks (for Labelling)\n\nSlot 4: Opentrons Magnetic Module, GEN2 with Bio-Rad 96-Well PCR Plate, 200\u00b5L (empty)\n\nSlot 5: NEST 12-Well Reservoir, 15mL\nColumn 1: MilliQ Water\nColumn 2: MagBeads\nColumn 3: MagBinding Buffer\nColumn 4: Wash Buffer 1\nColumn 5: Wash Buffer 2\nColumn 6: Elution Buffer\n\nSlot 6: Opentrons 20\u00b5L Tip Racks (for Labelling)\n\nSlot 7: Opentrons 300\u00b5L Tip Racks (for Purification)\n\nSlot 8: Opentrons 300\u00b5L Tip Racks (for Purification)\n\nSlot 10: Opentrons 300\u00b5L Tip Racks (for Purification)\n\nSlot 11: Opentrons 20\u00b5L Tip Racks (for Purification)\n\nUsing the customizations field (below), set up your protocol.\n P20-Multi Mount: Specify which mount the Opentrons P20 8-Channel Pipette is attached to.\n P300-Multi Mount: Specify which mount the Opentrons P3000 8-Channel Pipette is attached to.\n Labelling Step: Specify whether or not to run the labelling portion.\n Purification Step: Specify whether or not to run the purification portion.",
"internal": "71381b",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Zymo Kit\n\n\n",
"description": "This protocol automates the [Zymo Quick-DNA HMW MagBead Kit](https://www.zymoresearch.com/products/quick-dna-hmw-magbead-kit) with an additional labelling step that takes place before the purification.\n\nThe parameter section below gives you the ability to select either the **Labelling** portion of the protocol or the **Purification** portion of the protocol (you can select both). During the labelling portion, MilliQ Water, CSB, M.Taql, MTC22, and Proteinase K are added to the samples and the samples incubate while on the Temperature Module. During the purification portion, the samples are purified using the Zymo MagBead Kit in conjunction with Opentrons Magnetic Module.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 4.0.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Temperatue Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Opentrons Magnetic Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P20 8-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons P300 8-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 20\u00b5L Tip Racks](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons 300\u00b5L Tip Racks](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [Bio-Rad 96-Well PCR Plate, 200\u00b5L](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate)\n* [Zymo Quick-DNA HMW MagBead Kit](https://www.zymoresearch.com/products/quick-dna-hmw-magbead-kit)\n* Reagents for Labelling\n* Samples\n\n\n\n---\n\n\n**Deck Layout**\n\n**Slot 1**: [Opentrons Temperatue Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [Bio-Rad 96-Well PCR Plate, 200\u00b5L](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate) on 96-Well Aluminum Block, containing samples in columns 1-4\n\n**Slot 2**: [Bio-Rad 96-Well PCR Plate, 200\u00b5L](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate) (Labelling Plate)\nColumn 1: MTC22\nColumn 3: CSB\nColumn 5: M.Taql\nColumn 7: Proteinase K\n\n**Slot 3**: [Opentrons 20\u00b5L Tip Racks](https://shop.opentrons.com/collections/opentrons-tips) (for Labelling)\n\n**Slot 4**: [Opentrons Magnetic Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [Bio-Rad 96-Well PCR Plate, 200\u00b5L](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate) (empty)\n\n**Slot 5**: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\nColumn 1: MilliQ Water\nColumn 2: MagBeads\nColumn 3: MagBinding Buffer\nColumn 4: Wash Buffer 1\nColumn 5: Wash Buffer 2\nColumn 6: Elution Buffer\n\n**Slot 6**: [Opentrons 20\u00b5L Tip Racks](https://shop.opentrons.com/collections/opentrons-tips) (for Labelling)\n\n**Slot 7**: [Opentrons 300\u00b5L Tip Racks](https://shop.opentrons.com/collections/opentrons-tips) (for Purification)\n\n**Slot 8**: [Opentrons 300\u00b5L Tip Racks](https://shop.opentrons.com/collections/opentrons-tips) (for Purification)\n\n**Slot 10**: [Opentrons 300\u00b5L Tip Racks](https://shop.opentrons.com/collections/opentrons-tips) (for Purification)\n\n**Slot 11**: [Opentrons 20\u00b5L Tip Racks](https://shop.opentrons.com/collections/opentrons-tips) (for Purification)\n\n**Using the customizations field (below), set up your protocol.**\n* **P20-Multi Mount**: Specify which mount the [Opentrons P20 8-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette) is attached to.\n* **P300-Multi Mount**: Specify which mount the [Opentrons P3000 8-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette) is attached to.\n* **Labelling Step**: Specify whether or not to run the labelling portion.\n* **Purification Step**: Specify whether or not to run the purification portion.\n\n\n\n\n\n",
"internal": "71381b\n",
diff --git a/protoBuilds/7363e4/README.json b/protoBuilds/7363e4/README.json
index 7ddef927d1..1ecc0dcd1d 100644
--- a/protoBuilds/7363e4/README.json
+++ b/protoBuilds/7363e4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Aliquoting"
@@ -9,7 +9,7 @@
"description": "This protocol uses a p1000 single channel pipette to divide up to forty eight 3 mL clinical samples each into four aliquots with aliquot volumes of 1 mL, 0.5 mL, 0.5 mL and 1 mL.\nLinks:\n Custom Aliquoting\n Custom Pooling",
"internal": "7363e4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Aliquoting\n\n",
"deck-setup": "\n\n* Opentrons p1000 tips (Deck Slot 11)\n* 15 or 40-well custom rack or empty slot as specified in pulldown lists (Deck Slots 8-9)\n* opentrons_24_tuberack_nest_1.5ml_snapcap or opentrons_24_tuberack_nest_2ml_snapcap as specified with pulldown list (up to 8 racks as displayed in OT app with deck slot fill order 10, 7, 4, 5, 6, 1, 2, 3)\n\n\n",
"description": "\nThis protocol uses a p1000 single channel pipette to divide up to forty eight 3 mL clinical samples each into four aliquots with aliquot volumes of 1 mL, 0.5 mL, 0.5 mL and 1 mL.\n\nLinks:\n* [Custom Aliquoting](https://protocols.opentrons.com/protocol/7363e4)\n* [Custom Pooling](https://protocols.opentrons.com/protocol/7363e4-part-2)\n\n\n",
diff --git a/protoBuilds/74312c/README.json b/protoBuilds/74312c/README.json
index 10d254d8e7..f29a0512ba 100644
--- a/protoBuilds/74312c/README.json
+++ b/protoBuilds/74312c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -9,7 +9,7 @@
"description": "This protocol performs the transfer of buffer to various wells on a custom PCB board. It will delay for 1 minute and repeat the same transfers.\n\n\n\nP20 single-channel GEN2 electronic pipette\n\n\n",
"internal": "74312c",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "* Custom PCB Board with Adapter (Slot 1)\n* Standard Opentrons Trough (Slot 6)\n* Opentrons 20uL Tips (Slot 3)\n\n",
"description": "This protocol performs the transfer of buffer to various wells on a custom PCB board. It will delay for 1 minute and repeat the same transfers.\n\n---\n\n\n\n* [P20 single-channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n\n---\n\n\n",
diff --git a/protoBuilds/743f54/README.json b/protoBuilds/743f54/README.json
index 6d16bdf7b0..a1215f276d 100644
--- a/protoBuilds/743f54/README.json
+++ b/protoBuilds/743f54/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "743f54",
"labware": "\nOpentrons 200\u00b5L Filter Tips\nOpentrons 4-in-1 Tube Rack (with 24 Well Insert)\nReservoirs for Wash Buffers (like the NEST 1-Well Reservoir, 195mL)\nReservoir for Elution Buffer (like the NEST 12-Well Reservoir, 15mL)\n1.5mL Mictrocentrifuge Tube\nKingFisher 96-Deepwell Plate\nKingFisher 96-Well Microplate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n",
"description": "This protocol automates preparation of plates that will be used in the Magnetic Particle Processing protocol on the KingFisher Flex.\n\nThis protocol is still a work in progress and will be updated.\n\n\nExplanation of complex parameters below:\n* **Number of Samples**: Specify the number of samples to be run (1-96).\n* **Pipette Mount**: Select which mount the [P300 Single-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) is attached to.\n\n\n---\n\n\n",
diff --git a/protoBuilds/74750e/README.json b/protoBuilds/74750e/README.json
index 343deaac93..38866064ea 100644
--- a/protoBuilds/74750e/README.json
+++ b/protoBuilds/74750e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Serial Dilution"
@@ -8,7 +8,7 @@
"description": "This protocol performs a simple serial dilution (factor 10) within each well plate for a specified number of well plates. 100ul of solution is mixed 12 times in each column before 20ul is transferred to the following column, with the mix-transfer iteration repeated up to the specified number of columns. New tips are granted between each well plate.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P300 Multi-Channel Pipette\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 300\u00b5L Tips\nCorning 96 Well Plate 360 \u00b5L Flat\n\n\n\nSlot 1: 96 Tip Rack 300 \u00b5L\nSlot 2 - (up to 11): Corning 96 Well Plate 360 \u00b5L Flat\nP300-Multi Mount: Left\n\n\nUsing the customizations field (below), set up your protocol.\n Number of Columns: Specify number of columns with solution in each well plate.\n Number of Plates: Specify number of plates for serial dilution.",
"internal": "74750e",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Serial Dilution\n\n\n",
"description": "This protocol performs a simple serial dilution (factor 10) within each well plate for a specified number of well plates. 100ul of solution is mixed 12 times in each column before 20ul is transferred to the following column, with the mix-transfer iteration repeated up to the specified number of columns. New tips are granted between each well plate.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://labware.opentrons.com/opentrons_96_tiprack_300ul?category=tipRack)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Corning 96 Well Plate 360 \u00b5L Flat](https://labware.opentrons.com/corning_96_wellplate_360ul_flat)\n\n\n\n---\n\n\nSlot 1: 96 Tip Rack 300 \u00b5L\n\nSlot 2 - (up to 11): Corning 96 Well Plate 360 \u00b5L Flat\n\nP300-Multi Mount: Left\n\n\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Columns**: Specify number of columns with solution in each well plate.\n* **Number of Plates**: Specify number of plates for serial dilution.\n\n",
"internal": "74750e\n",
diff --git a/protoBuilds/747d99-assay-1/README.json b/protoBuilds/747d99-assay-1/README.json
index 4589405f33..ede4b687d4 100644
--- a/protoBuilds/747d99-assay-1/README.json
+++ b/protoBuilds/747d99-assay-1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Covid Workstation": [
"RNA Extraction"
@@ -8,7 +8,7 @@
"description": "This protocol is a full COVID-19 diagnostic workflow up to RNA amplification. Samples are loaded in 15ml tubes and transferred to 1.5ml tubes for lysis and temperature incubations on an Opentrons temperature module. The samples are then transferred to a deepwell plate mounted on an Opentrons magnetic module for a 1-wash magnetic bead-based RNA extraction.\nExplanation of protocol parameters below:\n* track tips across protocol runs: If set to yes, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to no, tips will be picked up from column 1 of the 1st tiprack.\n\n \nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons magnetic module GEN2\nOpentrons temperature module GEN2\nNEST 12 Well Reservoir 15 mL\nUSA Scientific PlateOne 96 Deepwell Plate 2mL\nOpentrons 96 Filter Tip Rack 200 and 1000 \u00b5L\nOpentrons P300-Multi GEN2 and P1000-Single GEN2 electronic pipettes\n\n\n \n \nReservoir (slot 2):\n channel 1: lysis and hybridization buffer (HB1)\n channel 2: magnetic beads\n channel 4: wash buffer\n channel 5: master mix\n* channel 12: liquid waste (loaded empty)",
"internal": "747d99",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Covid Workstation\n * RNA Extraction\n\n",
"description": "This protocol is a full COVID-19 diagnostic workflow up to RNA amplification. Samples are loaded in 15ml tubes and transferred to 1.5ml tubes for lysis and temperature incubations on an Opentrons temperature module. The samples are then transferred to a deepwell plate mounted on an Opentrons magnetic module for a 1-wash magnetic bead-based RNA extraction.\n\nExplanation of protocol parameters below:\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n\n---\n\n \n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons magnetic module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons temperature module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [NEST 12 Well Reservoir 15 mL](https://labware.opentrons.com/nest_12_reservoir_15ml)\n* [USA Scientific PlateOne 96 Deepwell Plate 2mL](https://www.usascientific.com/plateone-96-deep-well-2ml/p/PlateOne-96-Deep-Well-2mL)\n* [Opentrons 96 Filter Tip Rack 200 and 1000 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons P300-Multi GEN2 and P1000-Single GEN2 electronic pipettes](https://shop.opentrons.com/collections/ot-2-pipettes)\n\n---\n \n\n \n\nReservoir (slot 2):\n* channel 1: lysis and hybridization buffer (HB1)\n* channel 2: magnetic beads\n* channel 4: wash buffer\n* channel 5: master mix\n* channel 12: liquid waste (loaded empty)\n\n",
"internal": "747d99\n",
diff --git a/protoBuilds/747d99-assay-2/README.json b/protoBuilds/747d99-assay-2/README.json
index 2e50b6a9d8..7fb0226080 100644
--- a/protoBuilds/747d99-assay-2/README.json
+++ b/protoBuilds/747d99-assay-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Covid Workstation": [
"RNA Extraction"
@@ -8,7 +8,7 @@
"description": "This protocol is a full COVID-19 diagnostic workflow up to RNA amplification. Samples are loaded in 15ml tubes and transferred to a 96-wellplate for lysis and temperature incubations on an Opentrons temperature module. The samples are then transferred to a deepwell plate mounted on an Opentrons magnetic module for a 1-wash magnetic bead-based RNA extraction.\nExplanation of protocol parameters below:\n* track tips across protocol runs: If set to yes, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to no, tips will be picked up from column 1 of the 1st tiprack.\n\n \nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons magnetic module GEN2\nOpentrons temperature module GEN2\nNEST 12 Well Reservoir 15 mL\nUSA Scientific PlateOne 96 Deepwell Plate 2mL\nBio-Rad 96 Well Plate 350 \u00b5L PCR\nOpentrons 96 Filter Tip Rack 200 and 1000 \u00b5L\nOpentrons P300-Multi GEN2 and P1000-Single GEN2 electronic pipettes\n\n\n \n \nReservoir (slot 2):\n channel 2: magnetic beads\n channel 4: wash buffer\n channel 5: master mix\n channel 12: liquid waste (loaded empty)",
"internal": "747d99",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Covid Workstation\n * RNA Extraction\n\n",
"description": "This protocol is a full COVID-19 diagnostic workflow up to RNA amplification. Samples are loaded in 15ml tubes and transferred to a 96-wellplate for lysis and temperature incubations on an Opentrons temperature module. The samples are then transferred to a deepwell plate mounted on an Opentrons magnetic module for a 1-wash magnetic bead-based RNA extraction.\n\nExplanation of protocol parameters below:\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n\n---\n\n \n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons magnetic module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons temperature module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [NEST 12 Well Reservoir 15 mL](https://labware.opentrons.com/nest_12_reservoir_15ml)\n* [USA Scientific PlateOne 96 Deepwell Plate 2mL](https://www.usascientific.com/plateone-96-deep-well-2ml/p/PlateOne-96-Deep-Well-2mL)\n* [Bio-Rad 96 Well Plate 350 \u00b5L PCR](https://www.bio-rad.com/en-us/sku/hss9601-hard-shell-96-well-pcr-plates-high-profile-semi-skirted-clear-clear?ID=hss9601)\n* [Opentrons 96 Filter Tip Rack 200 and 1000 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons P300-Multi GEN2 and P1000-Single GEN2 electronic pipettes](https://shop.opentrons.com/collections/ot-2-pipettes)\n\n---\n \n\n \n\nReservoir (slot 2):\n* channel 2: magnetic beads\n* channel 4: wash buffer\n* channel 5: master mix\n* channel 12: liquid waste (loaded empty)\n\n",
"internal": "747d99\n",
diff --git a/protoBuilds/74841a-2/README.json b/protoBuilds/74841a-2/README.json
index 104e47f808..ecc025be8d 100644
--- a/protoBuilds/74841a-2/README.json
+++ b/protoBuilds/74841a-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Swift Normalase Amplicon Panels (SNAP)"
@@ -10,7 +10,7 @@
"internal": "74841a",
"labware": "\nUSA Scientific 12 Well Reservoir 22 mL\nNEST 1 Well Reservoir 195 mL or equivalent for waste\nBio-Rad 96 Well Plate 200 \u00b5L PCR Full Skirt\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Swift Normalase Amplicon Panels (SNAP)\n\n",
"deck-setup": "\n\n",
"description": "\nLinks:\n* [Part 1](./74841a)\n* [Part 2](./74841a-2)\n\nThis is Part 2/2 of the Swift Normalase Amplicon Panels (SNAP) Size Selection and Cleanup protocol.\n\nExplanation of complex parameters below:\n* `park tips`: If set to `yes` (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to `no`, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n* `flash`: If set to `yes`, the robot rail lights will flash during any automatic pauses in the protocol. If set to `no`, the lights will not flash.\n\n---\n\n",
diff --git a/protoBuilds/74841a/README.json b/protoBuilds/74841a/README.json
index 307495148d..181dc8f99c 100644
--- a/protoBuilds/74841a/README.json
+++ b/protoBuilds/74841a/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Swift Normalase Amplicon Panels (SNAP)"
@@ -10,7 +10,7 @@
"internal": "74841a",
"labware": "\nUSA Scientific 12 Well Reservoir 22 mL\nNEST 1 Well Reservoir 195 mL or equivalent for waste\nBio-Rad 96 Well Plate 200 \u00b5L PCR Full Skirt\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Swift Normalase Amplicon Panels (SNAP)\n\n",
"deck-setup": "\n\n",
"description": "\nLinks:\n* [Part 1](./74841a)\n* [Part 2](./74841a-2)\n\nThis is Part 1/2 of the Swift Normalase Amplicon Panels (SNAP) Size Selection and Cleanup protocol.\n\nLysed samples should be loaded on the magnetic module in a Bio-Rad 96-well PCR plate.\n\nExplanation of complex parameters below:\n* `park tips`: If set to `yes` (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to `no`, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n* `flash`: If set to `yes`, the robot rail lights will flash during any automatic pauses in the protocol. If set to `no`, the lights will not flash.\n\n---\n\n",
diff --git a/protoBuilds/74b303/README.json b/protoBuilds/74b303/README.json
index d1b414b32f..ff4bd85caf 100644
--- a/protoBuilds/74b303/README.json
+++ b/protoBuilds/74b303/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Normalization": [
"Normalization and Pooling"
@@ -7,7 +7,7 @@
},
"description": "This protocol automates the normalization of samples provided on four 96 well PCR plates. Normalization of achieved by dispensing custom volumes (specified in the uploaded csv file) of water to each well of the initial pcr plate with the single channel p20, followed by mixing, and then pooling of 5 ul sample aliquots into a separate tube (one for each plate) located in the 24 tube rack.\n\n\n\nOpentrons P20-single channel electronic pipettes (GEN2)\nOpentrons 20ul Filter Tips\ncsv file\nopentrons_24_tuberack_nest_2ml_screwcap\nnest_12_reservoir_15ml\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Normalization\n\t* Normalization and Pooling\n\n",
"description": "This protocol automates the normalization of samples provided on four 96 well PCR plates. Normalization of achieved by dispensing custom volumes (specified in the uploaded csv file) of water to each well of the initial pcr plate with the single channel p20, followed by mixing, and then pooling of 5 ul sample aliquots into a separate tube (one for each plate) located in the 24 tube rack.\n\n---\n\n\n* [Opentrons P20-single channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=31059478970462)\n* [Opentrons 20ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [csv file](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-02-20/9l23lox/4Plate_Norm_ex.csv)\n* [opentrons_24_tuberack_nest_2ml_screwcap](https://labware.opentrons.com/opentrons_24_tuberack_nest_2ml_screwcap?category=tubeRack)\n* [nest_12_reservoir_15ml](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir)\n\n\n",
"process": "1. Transfer appropriate amount (from .csv) water from the NEST 12-well reservoir 15 ml to 96-well PCR Plate 1 (use same tip - single channel 20ul GEN2/right mount).\n2. Repeat for Plate 2, Plate 3 and Plate 4.\n3. Pool using same tip, transfer 5ul from each well in 96-well PCR Plate 1, to A1 of Opentrons 24 Tube Rack with NEST 2 mL Snapcap (include 3x mixing step before adding to 2 ml tube).\n4. Repeat for all plates, with Plate 2 pooled into B1 of tube rack, Plate 3 pooled into C1 and Plate 4 pooled into D1.\n",
diff --git a/protoBuilds/751127/README.json b/protoBuilds/751127/README.json
index 7e89f75857..0b1ead703d 100644
--- a/protoBuilds/751127/README.json
+++ b/protoBuilds/751127/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -8,7 +8,7 @@
"description": "This is a flexible protocol to accommodate .csv file-specified coordinates. You can upload your coordinates for 2 sources and up to 10 locations specified in the .csv template here, and upload your edited template below at the parameter Transfer .csv File. The template has some pre-defined values, but you will likely need to modify these values to suit your specific labware and workflow.\nNote that the coordinates are defined in millimeters from the bottom left-most corner of the Opentrons deck slot 1.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons P20 GEN2 Single-Channel Pipettes and corresponding Tips\nOpentrons 20\u00b5l Tipracks\n\nFor more detailed information on compatible labware, please visit our Labware Library.",
"internal": "751127",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n",
"description": "\nThis is a flexible protocol to accommodate .csv file-specified coordinates. You can upload your coordinates for 2 sources and up to 10 locations specified in the .csv template [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/751127/CSV+Template+-+Sheet1.csv), and upload your edited template below at the parameter `Transfer .csv File`. The template has some pre-defined values, but you will likely need to modify these values to suit your specific labware and workflow.\n\nNote that the coordinates are defined in millimeters from the bottom left-most corner of the Opentrons deck slot 1.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons P20 GEN2 Single-Channel Pipettes](https://shop.opentrons.com/collections/ot-2-pipettes) and corresponding [Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons 20\u00b5l Tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n",
"internal": "751127\n",
diff --git a/protoBuilds/75cfa6/README.json b/protoBuilds/75cfa6/README.json
index 5db226da92..fee7849f7d 100644
--- a/protoBuilds/75cfa6/README.json
+++ b/protoBuilds/75cfa6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "75cfa6",
"labware": "\nBio-Rad 96 Well Plate 200 \u00b5L PCR seated in Opentrons 96-well aluminum block\nOpentrons 4-in-1 tuberack with Eppendorf 1.5ml snapcap microcentrifuge tubes seated in 24-tube adapter\nOpentrons 10/20\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This is a custom PCR prep protocol that transfers up to 24 RNA samples to 4 different mastermixes. The transfer scheme can be shown on the deck layout below.\n\n---\n\n",
diff --git a/protoBuilds/76174b/README.json b/protoBuilds/76174b/README.json
index dfe043b86e..6203543b34 100644
--- a/protoBuilds/76174b/README.json
+++ b/protoBuilds/76174b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "76174b",
"labware": "\nOpentrons Tips for P20 (Standard)\n96-Well Low-Binding Corning Costar Microplate 3363\n384-Well Perkin Elmer OptiPlate (Example)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n\n**Slot 1**: 384-Well Perkin Elmer OptiPlate \n \n**Slot 2**: 384-Well Perkin Elmer OptiPlate \n \n**Slot 3**: 96-Well Low-Binding Microplate \n \n**Slot 5**: 96-Well Low-Binding Microplate \n \n**Slot 6**: 96-Well Low-Binding Microplate \n \n**Slots 4,7,8,9**: Opentrons 20 uL Tips\n\n\n---\n\n",
"description": "This protocol performs an AlphaLISA [Perkin Elmer AlphaLISA](https://resources.perkinelmer.com/lab-solutions/resources/docs/GDE_Quick_AlphaLISA_conversion.pdf) based on the attached experimental protocol with user determined parameters (described below).\n\n\n\n---\n\n\n\n",
diff --git a/protoBuilds/7663a6/README.json b/protoBuilds/7663a6/README.json
index 4e36b54bdd..2ca9c96036 100644
--- a/protoBuilds/7663a6/README.json
+++ b/protoBuilds/7663a6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "7663a6",
"labware": "\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "* Deck Setup running 10 columns. Mastermix is held in A1 of the tube rack on Slot 3.\n\n\n\n---\n\n",
"description": "This protocol preps a 96 well plate by column with mastermix and sample. Mastermix is held in an Opentrons 24-tube aluminum block without the temperature module. The aluminum block is placed in the freezer prior to running to keep mastermix cool.\n\n\nExplanation of complex parameters below:\n* `Number of Columns`: Input number of sample columns for this run.\n* `Source Plate Starting Column`: Specify the source plate starting column for samples.\n* `Destination Plate Starting Column`: Specify the destination plate starting column.\n* `Sample Volume (ul)`: Specify sample volume in ul.\n* `Mastermix Volume (ul)`: Specify mastermix volume in ul.\n* `Source Aspiration Height (Plate)`: Specify aspiration height (in mm) from the bottom of the well in the sample plate.\n* `Source Aspiration Height (Tube)`: Specify aspiration height (in mm) from the bottom of the tube in the mastermix tube.\n* `Source Aspiration Flow Rate Sample (ul/sec)`: Specify the aspiration flow rate for sample.\n* `Delay After Aspiration Mastermix (seconds)`: Specify the delay after aspirating mastermix in the tube. This may be helpful with viscous liquids, allowing the pipette time to achieve the full volume before moving on.\n* `Source Aspiration Flow Rate Mastermix (ul/sec)`: Specify the aspiration flow rate for mastermix.\n* `Dispense Height (from bottom)`: Specify dispense height (in mm) from the bottom of the well in the destination plate.\n* `Dispense Flow Rate Sample`: Specify the dispense flow rate of sample into destination plate.\n* `Dispense Flow Rate Mastermix`: Specify the dispense flow rate of mastermix into destination plate.\n* `Mix Repetitions`: Specify number of mix steps.\n* `Touch Tip?`: Specify whether to include touch tip in this run.\n* `Blowout?`: Specify whether to include blow out in this run.\n* `P20 Single Mount`: Specify left or right mount for the P20 single.\n* `P20 Multi Mount`: Specify left or right mount for the P20 multi.\n\n\n\n\n---\n\n",
diff --git a/protoBuilds/76ab0e/README.json b/protoBuilds/76ab0e/README.json
index 80cfa7cee2..4562fbe9ed 100644
--- a/protoBuilds/76ab0e/README.json
+++ b/protoBuilds/76ab0e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "76ab0e",
"labware": "\nAluminum Block Set\nBio-Rad 96 Well Plate 200 \u00b5L PCR\nNEST 12 well 15 mL reservoir\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"deck-setup": "\n\n",
"description": "This protocol is based on the [ML_normalization protocol](https://protocols.opentrons.com/protocol/ML-normalization) and it performs a custom sample normalization from a source PCR plate to a second PCR plate, diluting with diluent (e.g. buffer or water) from a reservoir. Sample and diluent volumes are specified via .csv file in the following format, including the header line (empty lines ignored):\n\n```\nsource plate well,destination plate well,volume sample (\u00b5l),volume diluent (\u00b5l)\nA1, A1, 2, 28\n```\n\nDiluent is transferred first from the 12 well reservoir to the target plate wells as specified in the csv using the same pipette tip. After finishing, the pipette(s) drop their/its tip(s) and transfer samples to the target wells.\n\nThis protocol loads the sample and destination plate on 2nd generation temperature modules\n\nExplanation of parameters below:\n* `input .csv file`: Here, you should upload a .csv file formatted as described above.\n* `P20 GEN2 mount`: Choose whether to load the p20 in the right or left mount\n* `P300 GEN2 mount`: Choose whether to load the p300 in the right or left mount\n* `Aspiration height from bottom of the plate wells [mm]`: Offset to aspirate from the bottom of the source plate wells (in units of mm)\n* `Dispensing height from bottom of the plate wells [mm]`: Offset for dispenses from the bottom of the destination plate wells (in units of mm)\n* `Aspiration height from bottom of the reservoir wells [mm]`: Offset to aspirate from the bottom of the reservoir wells (in units of mm)\n* `Flow rate multiplier`: By setting this multiplier to a value 0 < x < 1 the rate of sample aspiration and dispensing will be slowed down. The default value is 0.5 resulting in aspiration at half of the regular speed.\n\n---\n\n",
diff --git a/protoBuilds/76ba23-bisulfite_conversion/README.json b/protoBuilds/76ba23-bisulfite_conversion/README.json
index 032c36b0d9..c76039a247 100644
--- a/protoBuilds/76ba23-bisulfite_conversion/README.json
+++ b/protoBuilds/76ba23-bisulfite_conversion/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Zymoresearch: EZ DNA Methylation-Gold Kit"
@@ -9,7 +9,7 @@
"description": "Part 2 of 5: Cell-free DNA (output from step 1) is chemically treated to convert unmethylated cytosine (C to T) in order to reveal patterns of methylated cytosine by DNA sequencing (methylated cytosine is protected and remains unchanged). This bisulfite conversion is followed by bead-clean up, elution, and transfer of the eluate to a fresh PCR plate.\nLinks:\n Part 1: DNA Extraction\n Part 2: Bisulfite Conversion\n Part 3: PCR1\n Part 4: PCR2\n* Part 5: Pooling\nWith this protocol, your robot will use reagents from the Zymoresearch EZ DNA Methylation-Gold Kit Zymoresearch EZ DNA Methylation-Gold Kit to perform the custom steps (DNA extraction, Bisulfite Conversion, PCR1, PCR2, Pooling) detailed in the HKG SOP.HKG SOP.\nThis is part 2 of the protocol: Bisulfite Conversion.\nThis step performs the bisulfite conversion and bead-clean up steps using the output samples from step 1 (DNA extraction).\nAfter the steps carried out in this protocol (part 2), proceed with part 3: PCR1.",
"internal": "76ba23-bisulfite_conversion",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Zymoresearch: EZ DNA Methylation-Gold Kit\n\n",
"deck-setup": "* Opentrons Magnetic Module (Deck Slot 4)\n* Opentrons p300 tips (Deck Slots 6 and 9)\n* 96 cell free DNA samples (\"nest_96_wellplate_100ul_pcr_full_skirt\" Deck slot 1)\n* pcr plate for CT conversion (\"biorad_96_wellplate_200ul_pcr\" Deck slot 2)\n* reservoir for CT conv buffer, beads, water (\"nest_12_reservoir_15ml\" Deck slot 5)\n* reservoir for wash (\"nest_1_reservoir_195ml\" Deck slot 5)\n* reservoir for desulph buffer (\"nest_1_reservoir_195ml\" Deck slot 8)\n* pcr plate for eluate (\"nest_96_wellplate_100ul_pcr_full_skirt\" Deck slot 10)\n* reservoir for waste (\"agilent_1_reservoir_290ml\" Deck slot 11)\n\n\n",
"description": "Part 2 of 5: Cell-free DNA (output from step 1) is chemically treated to convert unmethylated cytosine (C to T) in order to reveal patterns of methylated cytosine by DNA sequencing (methylated cytosine is protected and remains unchanged). This bisulfite conversion is followed by bead-clean up, elution, and transfer of the eluate to a fresh PCR plate.\n\nLinks:\n* [Part 1: DNA Extraction](http://protocols.opentrons.com/protocol/76ba23)\n* [Part 2: Bisulfite Conversion](http://protocols.opentrons.com/protocol/76ba23-bisulfite_conversion)\n* [Part 3: PCR1](http://protocols.opentrons.com/protocol/76ba23-pcr1)\n* [Part 4: PCR2](http://protocols.opentrons.com/protocol/76ba23-pcr2)\n* [Part 5: Pooling](http://protocols.opentrons.com/protocol/76ba23-pooling)\n\nWith this protocol, your robot will use reagents from the Zymoresearch EZ DNA Methylation-Gold Kit [Zymoresearch EZ DNA Methylation-Gold Kit](https://www.zymoresearch.com/collections/ez-dna-methylation-gold-kits) to perform the custom steps (DNA extraction, Bisulfite Conversion, PCR1, PCR2, Pooling) detailed in the HKG SOP.[HKG SOP](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-03-04/vw23kchHKG%20Standard%20Operating%20Procedure%20for%20DNA%20extraction%20Targeted%20next%20generation%20sequencing%20and%20.xlsx).\n\nThis is part 2 of the protocol: Bisulfite Conversion.\n\nThis step performs the bisulfite conversion and bead-clean up steps using the output samples from step 1 (DNA extraction).\n\nAfter the steps carried out in this protocol (part 2), proceed with part 3: PCR1.\n\n\n",
diff --git a/protoBuilds/76ba23-pcr1/README.json b/protoBuilds/76ba23-pcr1/README.json
index 5e1f9ff94a..fe6f69120d 100644
--- a/protoBuilds/76ba23-pcr1/README.json
+++ b/protoBuilds/76ba23-pcr1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Zymoresearch: EZ DNA Methylation-Gold Kit"
@@ -9,7 +9,7 @@
"description": "Part 3 of 5: Bisulfite converted DNA (output from step 2) is set up for a PCR amplification step. For plasma samples (but not saliva) the protocol includes post-PCR clean up steps.\nLinks:\n Part 1: DNA Extraction\n Part 2: Bisulfite Conversion\n Part 3: PCR1\n Part 4: PCR2\n* Part 5: Pooling\nWith this work flow, your robot will use reagents from the Zymoresearch EZ DNA Methylation-Gold Kit Zymoresearch EZ DNA Methylation-Gold Kit to perform the custom steps (DNA extraction, Bisulfite Conversion, PCR1, PCR2, Pooling) detailed in the HKG SOP.HKG SOP.\nThis is part 3 of the work flow: Bisulfite Conversion.\nThis step performs set up for PCR amplification (with additional post-PCR clean up steps for plasma samples only) using the output samples from step 2 (Bisulfite Conversion).\nAfter the steps carried out in this protocol (part 3), proceed with part 4: PCR2.",
"internal": "76ba23-pcr1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Zymoresearch: EZ DNA Methylation-Gold Kit\n\n",
"deck-setup": "* Opentrons Magnetic Module (Deck Slot 4)\n* Opentrons p300 tips (Deck Slots 6, 9, 8, 11)\n* 96 samples from step 2 (\"nest_96_wellplate_100ul_pcr_full_skirt\" Deck slot 1)\n* pcr1 master mix (\"corning_96_wellplate_360ul_flat\" Deck slot 3)\n* trough for beads, water, waste (\"nest_12_reservoir_15ml\" Deck slot 5)\n* pcr plate (\"nest_96_wellplate_100ul_pcr_full_skirt\" Deck slot 2)\n* reservoir for ethanol (\"nest_1_reservoir_195ml\" Deck slot 7)\n\n",
"description": "Part 3 of 5: Bisulfite converted DNA (output from step 2) is set up for a PCR amplification step. For plasma samples (but not saliva) the protocol includes post-PCR clean up steps.\n\nLinks:\n* [Part 1: DNA Extraction](http://protocols.opentrons.com/protocol/76ba23)\n* [Part 2: Bisulfite Conversion](http://protocols.opentrons.com/protocol/76ba23-bisulfite_conversion)\n* [Part 3: PCR1](http://protocols.opentrons.com/protocol/76ba23-pcr1)\n* [Part 4: PCR2](http://protocols.opentrons.com/protocol/76ba23-pcr2)\n* [Part 5: Pooling](http://protocols.opentrons.com/protocol/76ba23-pooling)\n\nWith this work flow, your robot will use reagents from the Zymoresearch EZ DNA Methylation-Gold Kit [Zymoresearch EZ DNA Methylation-Gold Kit](https://www.zymoresearch.com/collections/ez-dna-methylation-gold-kits) to perform the custom steps (DNA extraction, Bisulfite Conversion, PCR1, PCR2, Pooling) detailed in the HKG SOP.[HKG SOP](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-03-04/vw23kchHKG%20Standard%20Operating%20Procedure%20for%20DNA%20extraction%20Targeted%20next%20generation%20sequencing%20and%20.xlsx).\n\nThis is part 3 of the work flow: Bisulfite Conversion.\n\nThis step performs set up for PCR amplification (with additional post-PCR clean up steps for plasma samples only) using the output samples from step 2 (Bisulfite Conversion).\n\nAfter the steps carried out in this protocol (part 3), proceed with part 4: PCR2.\n\n\n",
diff --git a/protoBuilds/76ba23-pcr2/README.json b/protoBuilds/76ba23-pcr2/README.json
index ed0175613c..a6954fd129 100644
--- a/protoBuilds/76ba23-pcr2/README.json
+++ b/protoBuilds/76ba23-pcr2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Zymoresearch: EZ DNA Methylation-Gold Kit"
@@ -9,7 +9,7 @@
"description": "Part 4 of 5: PCR 1 product (output from step 2) is set up for a second PCR step featuring a unique forward primer for each plate (the forward primer is in the pcr2 mix, so different mixes should be used for each plate) and a unique, barcoded reverse primer for each well (there are two plates of unique barcodes for a total of 192).\nLinks:\n Part 1: DNA Extraction\n Part 2: Bisulfite Conversion\n Part 3: PCR1\n Part 4: PCR2\n* Part 5: Pooling\nWith this work flow, your robot will use reagents from the Zymoresearch EZ DNA Methylation-Gold Kit Zymoresearch EZ DNA Methylation-Gold Kit to perform the custom steps (DNA extraction, Bisulfite Conversion, PCR1, PCR2, Pooling) detailed in the HKG SOP.HKG SOP.\nThis is part 4 of the work flow: PCR 2.\nThis step performs set up for a second PCR amplification using the output samples from PCR 1 as the starting template. A unique forward primer is used for each plate (so a different tube of pcr2 mix will be used for each plate). A unique reverse primer will be used for each well. There are 192 unique, barcoded reverse primers between the two barcode plates.\nAfter the steps carried out in this protocol (part 4), proceed with part 5: pooling.",
"internal": "76ba23-pcr2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Zymoresearch: EZ DNA Methylation-Gold Kit\n\n",
"deck-setup": "* Opentrons p10 tips (Deck Slots 8, 11, 7, 10)\n* 1 or 2 plates each with 96 samples (output from PCR 1) (Deck slots 1 and 4)\n* pcr2 mix (\"corning_96_wellplate_360ul_flat\" Deck slot 9)\n* 1 or 2 plates each with 96 unique barcoded primers (Deck slots 2 and 5)\n* 1 or 2 pcr plates for PCR 2 (Deck slots 3 and 6)\n\n",
"description": "Part 4 of 5: PCR 1 product (output from step 2) is set up for a second PCR step featuring a unique forward primer for each plate (the forward primer is in the pcr2 mix, so different mixes should be used for each plate) and a unique, barcoded reverse primer for each well (there are two plates of unique barcodes for a total of 192).\n\nLinks:\n* [Part 1: DNA Extraction](http://protocols.opentrons.com/protocol/76ba23)\n* [Part 2: Bisulfite Conversion](http://protocols.opentrons.com/protocol/76ba23-bisulfite_conversion)\n* [Part 3: PCR1](http://protocols.opentrons.com/protocol/76ba23-pcr1)\n* [Part 4: PCR2](http://protocols.opentrons.com/protocol/76ba23-pcr2)\n* [Part 5: Pooling](http://protocols.opentrons.com/protocol/76ba23-pooling)\n\nWith this work flow, your robot will use reagents from the Zymoresearch EZ DNA Methylation-Gold Kit [Zymoresearch EZ DNA Methylation-Gold Kit](https://www.zymoresearch.com/collections/ez-dna-methylation-gold-kits) to perform the custom steps (DNA extraction, Bisulfite Conversion, PCR1, PCR2, Pooling) detailed in the HKG SOP.[HKG SOP](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-03-04/vw23kchHKG%20Standard%20Operating%20Procedure%20for%20DNA%20extraction%20Targeted%20next%20generation%20sequencing%20and%20.xlsx).\n\nThis is part 4 of the work flow: PCR 2.\n\nThis step performs set up for a second PCR amplification using the output samples from PCR 1 as the starting template. A unique forward primer is used for each plate (so a different tube of pcr2 mix will be used for each plate). A unique reverse primer will be used for each well. There are 192 unique, barcoded reverse primers between the two barcode plates.\n\nAfter the steps carried out in this protocol (part 4), proceed with part 5: pooling.\n\n\n",
diff --git a/protoBuilds/76ba23-pooling/README.json b/protoBuilds/76ba23-pooling/README.json
index 477dbd772f..fefc051e0e 100644
--- a/protoBuilds/76ba23-pooling/README.json
+++ b/protoBuilds/76ba23-pooling/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Zymoresearch: EZ DNA Methylation-Gold Kit"
@@ -9,7 +9,7 @@
"description": "Part 5 of 5: 5 ul aliquots of PCR 2 product (output from step 4) are combined to form a single pool for each plate followed by bead clean up and transfer to a fresh tube.\nLinks:\n Part 1: DNA Extraction\n Part 2: Bisulfite Conversion\n Part 3: PCR1\n Part 4: PCR2\n* Part 5: Pooling\nWith this work flow, your robot will use reagents from the Zymoresearch EZ DNA Methylation-Gold Kit Zymoresearch EZ DNA Methylation-Gold Kit to perform the custom steps (DNA extraction, Bisulfite Conversion, PCR1, PCR2, Pooling) detailed in the HKG SOP.HKG SOP.\nThis is part 5 of the work flow: Pooling.\nThis step combines 5 ul aliquots of 96 post-PCR 2 samples into a single pool for either one or two post-PCR 2 plates. This step is followed by a bead clean up and transfer of the finished pools to clean tubes.",
"internal": "76ba23-pooling",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Zymoresearch: EZ DNA Methylation-Gold Kit\n\n",
"deck-setup": "* Opentrons Magnetic Module (Deck Slot 4)\n* Opentrons p300 tips (Deck Slot 3)\n* Opentrons p10 tips (Deck Slots 8, 7)\n* 1 or 2 pcr plates PCR 2 product from previous step (Deck slots 2 and 5)\n* tube rack for pools (Deck Slot 1)\n* deep well plate on Magnetic Module ('usascientific_96_wellplate_2.4ml_deep') with pool_1_clean_up (pool 1 temporary dispense location and for clean up steps), pool_2_clean_up (pool 2 temporary dispense location and for clean up steps), beads, etoh, water in 'A1', 'A2', 'A3', 'A4', 'A5'\n\n",
"description": "Part 5 of 5: 5 ul aliquots of PCR 2 product (output from step 4) are combined to form a single pool for each plate followed by bead clean up and transfer to a fresh tube.\n\nLinks:\n* [Part 1: DNA Extraction](http://protocols.opentrons.com/protocol/76ba23)\n* [Part 2: Bisulfite Conversion](http://protocols.opentrons.com/protocol/76ba23-bisulfite_conversion)\n* [Part 3: PCR1](http://protocols.opentrons.com/protocol/76ba23-pcr1)\n* [Part 4: PCR2](http://protocols.opentrons.com/protocol/76ba23-pcr2)\n* [Part 5: Pooling](http://protocols.opentrons.com/protocol/76ba23-pooling)\n\nWith this work flow, your robot will use reagents from the Zymoresearch EZ DNA Methylation-Gold Kit [Zymoresearch EZ DNA Methylation-Gold Kit](https://www.zymoresearch.com/collections/ez-dna-methylation-gold-kits) to perform the custom steps (DNA extraction, Bisulfite Conversion, PCR1, PCR2, Pooling) detailed in the HKG SOP.[HKG SOP](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-03-04/vw23kchHKG%20Standard%20Operating%20Procedure%20for%20DNA%20extraction%20Targeted%20next%20generation%20sequencing%20and%20.xlsx).\n\nThis is part 5 of the work flow: Pooling.\n\nThis step combines 5 ul aliquots of 96 post-PCR 2 samples into a single pool for either one or two post-PCR 2 plates. This step is followed by a bead clean up and transfer of the finished pools to clean tubes.\n\n\n",
diff --git a/protoBuilds/76ba23/README.json b/protoBuilds/76ba23/README.json
index 370e975e8e..a71e3a727b 100644
--- a/protoBuilds/76ba23/README.json
+++ b/protoBuilds/76ba23/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Zymoresearch: EZ DNA Methylation-Gold Kit"
@@ -9,7 +9,7 @@
"description": "Part 1 of 5: Extraction and clean-up of cell-free DNA from 96 plasma or saliva patient specimens.\nLinks:\n Part 1: DNA Extraction\n Part 2: Bisulfite Conversion\n Part 3: PCR1\n Part 4: PCR2\n* Part 5: Pooling\nWith this protocol, your robot will use reagents from the Zymoresearch EZ DNA Methylation-Gold Kit Zymoresearch EZ DNA Methylation-Gold Kit to perform the custom steps (DNA extraction, Bisulfite Conversion, PCR1, PCR2, Pooling) detailed in the HKG SOP.HKG SOP.\nThis is part 1 of the protocol: DNA Extraction.\nThis step performs the extraction and bead-clean up of 96 patient samples (plasma or saliva) listed in the sample manifest and follows the HKG SOP described above. A current sample manifest can be uploaded and integrated into this protocol by using the \"Uploaded CSV Copy of Sample Manifest\" parameter below. The OT-2 will read the sample type and sample volume from the uploaded manifest and then set up and perform the experimental steps accordingly.\nAfter the steps carried out in this protocol (part 1), proceed with part 2: Bisulfite Conversion.",
"internal": "76ba23",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Zymoresearch: EZ DNA Methylation-Gold Kit\n\n",
"deck-setup": "* Opentrons Magnetic Module (Deck Slot 4)\n* Opentrons p300 tips (Deck Slots 6 and 9)\n* 96 patient samples (\"usascientific_96_wellplate_2.4ml_deep\" Deck slot 1)\n* DNA extraction plate (\"usascientific_96_wellplate_2.4ml_deep\" Deck slot 2)\n* (if sample volume >= 1 mL) duplicate DNA extraction plate (Deck slot 5)\n* reservoir for bead premix, wash, ethanol (\"nest_1_reservoir_195ml\" Deck slot 3)\n* pcr plate for eluate (\"nest_96_wellplate_100ul_pcr_full_skirt\" Deck slot 10)\n* reservoir for waste (\"nest_1_reservoir_195ml\" Deck slot 11)\n\n\n",
"description": "Part 1 of 5: Extraction and clean-up of cell-free DNA from 96 plasma or saliva patient specimens.\n\nLinks:\n* [Part 1: DNA Extraction](http://protocols.opentrons.com/protocol/76ba23)\n* [Part 2: Bisulfite Conversion](http://protocols.opentrons.com/protocol/76ba23-bisulfite_conversion)\n* [Part 3: PCR1](http://protocols.opentrons.com/protocol/76ba23-pcr1)\n* [Part 4: PCR2](http://protocols.opentrons.com/protocol/76ba23-pcr2)\n* [Part 5: Pooling](http://protocols.opentrons.com/protocol/76ba23-pooling)\n\nWith this protocol, your robot will use reagents from the Zymoresearch EZ DNA Methylation-Gold Kit [Zymoresearch EZ DNA Methylation-Gold Kit](https://www.zymoresearch.com/collections/ez-dna-methylation-gold-kits) to perform the custom steps (DNA extraction, Bisulfite Conversion, PCR1, PCR2, Pooling) detailed in the HKG SOP.[HKG SOP](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-03-04/vw23kchHKG%20Standard%20Operating%20Procedure%20for%20DNA%20extraction%20Targeted%20next%20generation%20sequencing%20and%20.xlsx).\n\nThis is part 1 of the protocol: DNA Extraction.\n\nThis step performs the extraction and bead-clean up of 96 patient samples (plasma or saliva) listed in the sample manifest and follows the HKG SOP described above. A current sample manifest can be uploaded and integrated into this protocol by using the \"Uploaded CSV Copy of Sample Manifest\" parameter below. The OT-2 will read the sample type and sample volume from the uploaded manifest and then set up and perform the experimental steps accordingly.\n\nAfter the steps carried out in this protocol (part 1), proceed with part 2: Bisulfite Conversion.\n\n\n",
diff --git a/protoBuilds/76c562/README.json b/protoBuilds/76c562/README.json
index a8d356e275..13cdf780df 100644
--- a/protoBuilds/76c562/README.json
+++ b/protoBuilds/76c562/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -9,7 +9,7 @@
"description": "This protocol allows for a robust cherrypicking and normalization in one step.\n\nUsing two CSV files (Dilutant CSV and Sample CSV), the OT-2 will make all transfers of dilutant using a single tip (transfers based on Dilutant CSV). Then, using a new tip for each transfer, the OT-2 will make each sample transfers according to the Sample CSV (the dilutant and sample will be mixed after dispensing the sample).\n\nThe two .csv files should be formatted as shown in the example below, including headers:\nSource Labware,Source Slot,Source Well,Source Aspiration Height Above Bottom (in mm),Dest Labware,Dest Slot,Dest Well,Dest Dispense Height Above Bottom (in mm),Volume (in ul),Pipette\nagilent_1_reservoir_290ml,1,A1,1,nest_96_wellplate_100ul_pcr_full_skirt,4,A11,1,5,right\nnest_12_reservoir_15ml,2,A1,1,nest_96_wellplate_2ml_deep,5,A5,5,3,right\nnest_1_reservoir_195ml,3,A1,1,nest_96_wellplate_2ml_deep,5,H12,5,7,right\nAll available empty slots will be filled with the necessary tipracks, and the user will be prompted to refill the tipracks if all are emptied in the middle of the protocol.",
"internal": "76c562",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n\n",
"deck-setup": "* example layout based on example `.csv` above: \n\n\n",
"description": "This protocol allows for a robust cherrypicking and normalization in one step.\n\nUsing two CSV files (Dilutant CSV and Sample CSV), the OT-2 will make all transfers of dilutant using a single tip (transfers based on Dilutant CSV). Then, using a new tip for each transfer, the OT-2 will make each sample transfers according to the Sample CSV (the dilutant and sample will be mixed after dispensing the sample).\n\nThe two .csv files should be formatted as shown in the example below, **including headers**:\n\n```\nSource Labware,Source Slot,Source Well,Source Aspiration Height Above Bottom (in mm),Dest Labware,Dest Slot,Dest Well,Dest Dispense Height Above Bottom (in mm),Volume (in ul),Pipette\nagilent_1_reservoir_290ml,1,A1,1,nest_96_wellplate_100ul_pcr_full_skirt,4,A11,1,5,right\nnest_12_reservoir_15ml,2,A1,1,nest_96_wellplate_2ml_deep,5,A5,5,3,right\nnest_1_reservoir_195ml,3,A1,1,nest_96_wellplate_2ml_deep,5,H12,5,7,right\n```\n\nAll available empty slots will be filled with the necessary tipracks, and the user will be prompted to refill the tipracks if all are emptied in the middle of the protocol.\n\n",
diff --git a/protoBuilds/772041/README.json b/protoBuilds/772041/README.json
index 7d00edf11f..802de4afa0 100644
--- a/protoBuilds/772041/README.json
+++ b/protoBuilds/772041/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "772041",
"labware": "\nNEST 12-Well Reservoirs, 15 mL\nMicroAmp Optical 384-Well Reaction Plate with barcode\n20 \u00b5L filter tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n",
"description": "This protocol distributes PCR mastermix from a source NEST 12 well reservoir to a target Applied Biosystems Microamp optical 384 well 30 \u00b5L plate (or other PCR plate) with an 20 \u00b5L 8-channel pipette loaded in the left mount. The user can choose how many wells to transfer to (rounding up to the nearest number of columns) as well as how much volume to add to each well. The user can also choose whether they want to reuse tips, or pick up new tips for each transfer.\n\nExplanation of parameters below:\n* `Number of wells to distribute mastermix to`: How many wells to transfer mastermix to on the target plate. This will be rounded up to the nearest number of full columns.\n* `Volume of mastermix`: Volume of mastermix to add to each well (\u00b5L)\n* `Reuse tips?`: Yes: Reuse tips when distributing the mastermix. No: Drop the tips after each distribution, and pick up new tips before each aspiration.\n* `Destination plate`: What type of plate to distribute mastermix to\n\n---\n\n",
diff --git a/protoBuilds/776039/README.json b/protoBuilds/776039/README.json
index f57b9544c0..9b12ee5605 100644
--- a/protoBuilds/776039/README.json
+++ b/protoBuilds/776039/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Consolidation"
@@ -8,7 +8,7 @@
"description": "This protocol performs a sample consolidation from .csv file on up to 23 1.5ml snapcap tubes on a 4x6 Opentrons tuberack. All tubes are consolidated into the tube occupying A1 on the rack. The input file should be organized in the following way including header line:\nSample #,Position,Sample ID,Amt Lib (ul),Total Vol. (ul)\n1,A02,GMS,2,131.8\n2,A03,MCS,2.44,\n3,A04,DCS,3.05,\n4,A05,Log Dist,12.45,\n5,A06,NC_NC,15,\n6,B01,NC_Mg,15,\n7,B02,NC_Et,15,\n8,B03,NT_01a,3.83,\n...\nEmpty lines are ignored.\n\n\n\nOpentrons tuberack with 4x6 insert for 1.5ml snapcap tubes\nOpentrons P20 GEN2 single-channel electronic pipette\nOpentrons 20\u00b5l tipracks\nVWR 5ml tubes seated in Opentrons 3x5 tuberack\n",
"internal": "776039",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Consolidation\n\n",
"description": "This protocol performs a sample consolidation from `.csv` file on up to 23 1.5ml snapcap tubes on a 4x6 Opentrons tuberack. All tubes are consolidated into the tube occupying A1 on the rack. The input file should be organized in the following way **including header line**:\n\n```\nSample #,Position,Sample ID,Amt Lib (ul),Total Vol. (ul)\n1,A02,GMS,2,131.8\n2,A03,MCS,2.44,\n3,A04,DCS,3.05,\n4,A05,Log Dist,12.45,\n5,A06,NC_NC,15,\n6,B01,NC_Mg,15,\n7,B02,NC_Et,15,\n8,B03,NT_01a,3.83,\n...\n```\n\nEmpty lines are ignored.\n\n---\n\n\n* [Opentrons tuberack with 4x6 insert for 1.5ml snapcap tubes](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [Opentrons P20 GEN2 single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 20\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-10ul-tips)\n* VWR 5ml tubes seated in [Opentrons 3x5 tuberack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n\n",
"internal": "776039\n",
diff --git a/protoBuilds/7805de-part-2/README.json b/protoBuilds/7805de-part-2/README.json
index 99df5953f6..94e73b582e 100644
--- a/protoBuilds/7805de-part-2/README.json
+++ b/protoBuilds/7805de-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II Directional Library Prep Kit for Illumina"
@@ -9,7 +9,7 @@
"description": "Part 2 of 4: First and second strand cDNA synthesis followed by bead clean up.\nLinks:\n Part 1: RNA Isolation, Fragmentation and Priming\n Part 2: cDNA Synthesis\n Part 3: End Prep and Adapter Ligation\n Part 4: PCR Enrichment\nWith this protocol, your robot can perform the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina protocol described by the NEB Instruction Manual.\nThis is part 2 of the protocol: cDNA Synthesis.\nThis step performs first and second strand cDNA synthesis using the output plate from part 1. This protocol follows Section 1 of the NEB Instruction Manual.",
"internal": "7805de-part-2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * NEBNext Ultra II Directional Library Prep Kit for Illumina\n\n",
"deck-setup": "* Opentrons Temperature Module and opentrons_96_aluminumblock_generic_pcr_strip_200ul (Deck Slot 3)\n* Opentrons Magnetic Module (Deck Slot 4)\n* Opentrons p300 tips (Deck Slots 6 and 9)\n* Opentrons p20 tips (Deck Slots 2 and 5)\n* nest_12_reservoir_15ml (Deck Slot 1)\n* nest_96_wellplate_100ul_pcr_full_skirt (Deck Slot 7)\n\n",
"description": "Part 2 of 4: First and second strand cDNA synthesis followed by bead clean up.\n\nLinks:\n* [Part 1: RNA Isolation, Fragmentation and Priming](http://protocols.opentrons.com/protocol/7805de)\n* [Part 2: cDNA Synthesis](http://protocols.opentrons.com/protocol/7805de-part-2)\n* [Part 3: End Prep and Adapter Ligation](http://protocols.opentrons.com/protocol/7805de-part-3)\n* [Part 4: PCR Enrichment](http://protocols.opentrons.com/protocol/7805de-part-4)\n\nWith this protocol, your robot can perform the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina protocol described by the [NEB Instruction Manual](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-05-20/dv23jqz/NEBNext%20Ultra%20II%20Directional%20RNA%20library%20Prep%20kit%20manualE7760_E7765.pdf).\n\nThis is part 2 of the protocol: cDNA Synthesis.\n\nThis step performs first and second strand cDNA synthesis using the output plate from part 1. This protocol follows Section 1 of the NEB Instruction Manual.\n\n\n",
diff --git a/protoBuilds/7805de-part-3/README.json b/protoBuilds/7805de-part-3/README.json
index bf44e759eb..5dc96e5b44 100644
--- a/protoBuilds/7805de-part-3/README.json
+++ b/protoBuilds/7805de-part-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II Directional Library Prep Kit for Illumina"
@@ -9,7 +9,7 @@
"description": "Part 3 of 4: Perform end-prep and adapter ligation steps with double-stranded cDNA output from part 2 followed by a bead clean up.\nLinks:\n Part 1: RNA Isolation, Fragmentation and Priming\n Part 2: cDNA Synthesis\n Part 3: End Prep and Adapter Ligation\n Part 4: PCR Enrichment\nWith this protocol, your robot can perform the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina protocol described by the NEB Instruction Manual.\nThis is part 3 of the protocol: End Prep and Adapter Ligation.\nThis step prepares ends and ligates indexing primers to the cDNA in preparation for Illumina sequencing. This protocol follows Section 1 of the NEB Instruction Manual.",
"internal": "7805de-part-3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * NEBNext Ultra II Directional Library Prep Kit for Illumina\n\n",
"deck-setup": "* Opentrons Temperature Module and opentrons_96_aluminumblock_generic_pcr_strip_200ul (Deck Slot 3)\n* Opentrons Magnetic Module (Deck Slot 4)\n* Opentrons p300 tips (Deck Slots 6 and 9)\n* Opentrons p20 tips (Deck Slots 2 and 5)\n* nest_12_reservoir_15ml (Deck Slot 1)\n* nest_96_wellplate_100ul_pcr_full_skirt (Deck Slot 7)\n\n",
"description": "Part 3 of 4: Perform end-prep and adapter ligation steps with double-stranded cDNA output from part 2 followed by a bead clean up.\n\nLinks:\n* [Part 1: RNA Isolation, Fragmentation and Priming](http://protocols.opentrons.com/protocol/7805de)\n* [Part 2: cDNA Synthesis](http://protocols.opentrons.com/protocol/7805de-part-2)\n* [Part 3: End Prep and Adapter Ligation](http://protocols.opentrons.com/protocol/7805de-part-3)\n* [Part 4: PCR Enrichment](http://protocols.opentrons.com/protocol/7805de-part-4)\n\nWith this protocol, your robot can perform the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina protocol described by the [NEB Instruction Manual](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-05-20/dv23jqz/NEBNext%20Ultra%20II%20Directional%20RNA%20library%20Prep%20kit%20manualE7760_E7765.pdf).\n\nThis is part 3 of the protocol: End Prep and Adapter Ligation.\n\nThis step prepares ends and ligates indexing primers to the cDNA in preparation for Illumina sequencing. This protocol follows Section 1 of the NEB Instruction Manual.\n\n\n",
diff --git a/protoBuilds/7805de-part-4/README.json b/protoBuilds/7805de-part-4/README.json
index 265f42a18c..9c8a443e94 100644
--- a/protoBuilds/7805de-part-4/README.json
+++ b/protoBuilds/7805de-part-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II Directional Library Prep Kit for Illumina"
@@ -9,7 +9,7 @@
"description": "Part 4 of 4: PCR enrichment and bead clean up of cDNA in preparation for Illumina sequencing.\nLinks:\n Part 1: RNA Isolation, Fragmentation and Priming\n Part 2: cDNA Synthesis\n Part 3: End Prep and Adapter Ligation\n Part 4: PCR Enrichment\nWith this protocol, your robot can perform the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina protocol described by the NEB Instruction Manual.\nThis is part 4 of the protocol: PCR Enrichment.\nThis step amplifies end-prepped and adapter-ligated output from part 3 and then follows up with a bead clean up of the PCR product in preparation for Illumina sequencing. This protocol follows Section 1 of the NEB Instruction Manual.",
"internal": "7805de-part-4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * NEBNext Ultra II Directional Library Prep Kit for Illumina\n\n",
"deck-setup": "* Opentrons Temperature Module and opentrons_96_aluminumblock_generic_pcr_strip_200ul (Deck Slot 3)\n* Opentrons Magnetic Module (Deck Slot 4)\n* Opentrons p300 tips (Deck Slots 6 and 9)\n* Opentrons p20 tips (Deck Slots 2 and 5)\n* nest_12_reservoir_15ml (Deck Slot 1)\n* nest_96_wellplate_100ul_pcr_full_skirt (Deck Slot 7)\n\n",
"description": "Part 4 of 4: PCR enrichment and bead clean up of cDNA in preparation for Illumina sequencing.\n\nLinks:\n* [Part 1: RNA Isolation, Fragmentation and Priming](http://protocols.opentrons.com/protocol/7805de)\n* [Part 2: cDNA Synthesis](http://protocols.opentrons.com/protocol/7805de-part-2)\n* [Part 3: End Prep and Adapter Ligation](http://protocols.opentrons.com/protocol/7805de-part-3)\n* [Part 4: PCR Enrichment](http://protocols.opentrons.com/protocol/7805de-part-4)\n\nWith this protocol, your robot can perform the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina protocol described by the [NEB Instruction Manual](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-05-20/dv23jqz/NEBNext%20Ultra%20II%20Directional%20RNA%20library%20Prep%20kit%20manualE7760_E7765.pdf).\n\nThis is part 4 of the protocol: PCR Enrichment.\n\nThis step amplifies end-prepped and adapter-ligated output from part 3 and then follows up with a bead clean up of the PCR product in preparation for Illumina sequencing. This protocol follows Section 1 of the NEB Instruction Manual.\n\n\n",
diff --git a/protoBuilds/7805de/README.json b/protoBuilds/7805de/README.json
index ae4b3e33ee..f45bd8cbe1 100644
--- a/protoBuilds/7805de/README.json
+++ b/protoBuilds/7805de/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II Directional Library Prep Kit for Illumina"
@@ -9,7 +9,7 @@
"description": "Part 1 of 4: Isolate poly(A) RNA from starting total RNA and perform fragmentation and priming in preparation for cDNA synthesis.\nLinks:\n Part 1: RNA Isolation, Fragmentation and Priming\n Part 2: cDNA Synthesis\n Part 3: End Prep and Adapter Ligation\n Part 4: PCR Enrichment\nWith this protocol, your robot can perform the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina protocol described by the NEB Instruction Manual.\nThis is part 1 of the protocol: RNA Isolation, Fragmentation and Priming.\nThis step isolates poly(A) RNA from up to 48 total RNA samples and then fragments and primes the RNA in preparation for cDNA synthesis (occurs in part 2). This protocol follows Section 1 of the NEB Instruction Manual.\nAfter the steps carried out in this protocol (part 1), it is best to immediately proceed with part 2: cDNA Synthesis.",
"internal": "7805de",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * NEBNext Ultra II Directional Library Prep Kit for Illumina\n\n",
"deck-setup": "* Opentrons Temperature Module and opentrons_96_aluminumblock_generic_pcr_strip_200ul (Deck Slot 3)\n* Opentrons Magnetic Module (Deck Slot 4)\n* Opentrons p300 tips (Deck Slots 6 and 9)\n* Opentrons p20 tips (Deck Slots 2 and 5)\n* nest_12_reservoir_15ml (Deck Slot 1)\n* nest_96_wellplate_100ul_pcr_full_skirt (Deck Slot 7)\n\n",
"description": "Part 1 of 4: Isolate poly(A) RNA from starting total RNA and perform fragmentation and priming in preparation for cDNA synthesis.\n\nLinks:\n* [Part 1: RNA Isolation, Fragmentation and Priming](http://protocols.opentrons.com/protocol/7805de)\n* [Part 2: cDNA Synthesis](http://protocols.opentrons.com/protocol/7805de-part-2)\n* [Part 3: End Prep and Adapter Ligation](http://protocols.opentrons.com/protocol/7805de-part-3)\n* [Part 4: PCR Enrichment](http://protocols.opentrons.com/protocol/7805de-part-4)\n\nWith this protocol, your robot can perform the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina protocol described by the [NEB Instruction Manual](https://s3.amazonaws.com/pf-upload-01/u-4256/0/2021-05-20/dv23jqz/NEBNext%20Ultra%20II%20Directional%20RNA%20library%20Prep%20kit%20manualE7760_E7765.pdf).\n\nThis is part 1 of the protocol: RNA Isolation, Fragmentation and Priming.\n\nThis step isolates poly(A) RNA from up to 48 total RNA samples and then fragments and primes the RNA in preparation for cDNA synthesis (occurs in part 2). This protocol follows Section 1 of the NEB Instruction Manual.\n\nAfter the steps carried out in this protocol (part 1), it is best to immediately proceed with part 2: cDNA Synthesis.\n\n\n",
diff --git a/protoBuilds/7855ef-part2/README.json b/protoBuilds/7855ef-part2/README.json
index e847823a75..eebc2dda4c 100644
--- a/protoBuilds/7855ef-part2/README.json
+++ b/protoBuilds/7855ef-part2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"AgriSeq HTS Library Kit"
@@ -8,7 +8,7 @@
"description": "This protocol is the second of a four part series for performing NGS library prep with the ThermoFisher Scientific AgriSeq kit. 2ul of Pre-ligation mix is distributed to each column of sample that was processed in Part 1 of the protocol.\nNote: This protocol was updated September 28th, 2022\nLinks:\n Part 1: DNA Transfer\n Part 2: Pre-Ligation\n Part 3: Barcoding\n Part 4: Pooling\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP20 Multi-Channel Pipette\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nThermoFisher Scientific 96 Well Plate 200ul (AB-0800)\nThermoFisher Scientific 96 Well Plate 200ul (4483352)\nBioRad Hard-shell 96-well PCR Plate Skirted\nApplied Biosystems 384 Well Plate\nCustom 96 Well Endura Plate\n\nNote About Labware\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\n\n\nUsing the customization fields below, set up your protocol.\n Number of Samples: Specify the number of samples to be processed in this run (max 384).\n P20 single GEN2 mount: Specify which mount to load the P20 single GEN2 pipette.\nNote about 20\u00b5L tip racks\nWhen prompted to replace the 20ul tip racks, be sure to re-load all 4 tip racks as in the original configuration of the deck.\nLabware Setup\nSlot 5: 384 well plate\nSlot 6: MMX Plate with Pre-ligation Mix in Column 3\nSlots 7, 8, 9, 10: Opentrons 20ul Tip Rack",
"internal": "7855ef-part2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* AgriSeq HTS Library Kit\n\n",
"description": "This protocol is the second of a four part series for performing NGS library prep with the [ThermoFisher Scientific AgriSeq kit](https://www.thermofisher.com/order/catalog/product/A34144#/A34144). 2ul of Pre-ligation mix is distributed to each column of sample that was processed in Part 1 of the protocol.\n\n**Note**: This protocol was updated September 28th, 2022\n\nLinks:\n* [Part 1: DNA Transfer](./7855ef)\n* [Part 2: Pre-Ligation](./7855ef-part2)\n* [Part 3: Barcoding](./7855ef-part3)\n* [Part 4: Pooling](./7855ef-part4)\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P20 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 96 Filter Tip Rack 20 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_20ul?category=tipRack)\n* [ThermoFisher Scientific 96 Well Plate 200ul (AB-0800)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fmanuals%2FMAN0014518_96well_pcr_plate_skirted_low_profile_qr.pdf&title=VGVjaG5pY2FsIERyYXdpbmcgLSBQQ1IgUGxhdGUsIDk2LXdlbGwsIExvdyBQcm9maWxlLCBTa2lydGVk)\n* [ThermoFisher Scientific 96 Well Plate 200ul (4483352)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fbrochures%2FEnduraPlate_96Well.pdf&title=RW5naW5lZXJpbmcgRGlhZ3JhbTogTWljcm9BbXAmcmVnOyBFbmR1cmFQbGF0ZSZ0cmFkZTsgT3B0aWNhbCA5Ni13ZWxsIFJlYWN0aW9uIFBsYXRl)\n* [BioRad Hard-shell 96-well PCR Plate Skirted](https://www.bio-rad.com/en-us/sku/hsp9631-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-blue-clear?ID=hsp9631)\n* [Applied Biosystems 384 Well Plate](https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2Fcms_042831.pdf)\n* Custom 96 Well Endura Plate\n\n**Note About Labware**\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\n\n---\n\n\nUsing the customization fields below, set up your protocol.\n* Number of Samples: Specify the number of samples to be processed in this run (max 384).\n* P20 single GEN2 mount: Specify which mount to load the P20 single GEN2 pipette.\n\n\n**Note about 20\u00b5L tip racks**\n\nWhen prompted to replace the 20ul tip racks, be sure to re-load all 4 tip racks as in the original configuration of the deck.\n\n**Labware Setup**\n\n\nSlot 5: 384 well plate\n\nSlot 6: MMX Plate with Pre-ligation Mix in Column 3\n\nSlots 7, 8, 9, 10: Opentrons 20ul Tip Rack\n\n\n\n",
"internal": "7855ef-part2\n",
diff --git a/protoBuilds/7855ef-part3/README.json b/protoBuilds/7855ef-part3/README.json
index fefe3364ae..2f75e04761 100644
--- a/protoBuilds/7855ef-part3/README.json
+++ b/protoBuilds/7855ef-part3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"AgriSeq HTS Library Kit"
@@ -8,7 +8,7 @@
"description": "This protocol is the third of a four part series for performing NGS library prep with the ThermoFisher Scientific AgriSeq kit. 1ul of IonCode Barcode Adapter is 1-1 transferred by well between the sample plate from Parts 1 & 2 and the Ion Code 96 well plate.\nNote: This protocol was updated November 22nd, 2022\nLinks:\n Part 1: DNA Transfer\n Part 2: Pre-Ligation\n Part 3: Barcoding\n Part 4: Pooling\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP20 Multi-Channel Pipette\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nThermoFisher Scientific 96 Well Plate 200ul (AB-0800)\nThermoFisher Scientific 96 Well Plate 200ul (4483352)\nBioRad Hard-shell 96-well PCR Plate Skirted\nApplied Biosystems 384 Well Plate\nCustom 96 Well Endura Plate\n\nNote About Labware\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\n\n\nUsing the customization fields below, set up your protocol.\n Number of Samples: Specify the number of samples to be processed in this run (max 384).\n P20 single GEN2 mount: Specify which mount to load the P20 single GEN2 pipette.\nLabware Setup\nSlots 1, 2, 3: Ion Code Barcode plates (Custom 96 well Endura plate)\nSlot 5: 384 well plate\nSlot 7: MMX Plate with Barcode Reaction Mix in Column 4\nSlot 8, 9, 10, 11: Opentrons 20ul Tip Rack",
"internal": "7855ef-part3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* AgriSeq HTS Library Kit\n\n",
"description": "This protocol is the third of a four part series for performing NGS library prep with the [ThermoFisher Scientific AgriSeq kit](https://www.thermofisher.com/order/catalog/product/A34144#/A34144). 1ul of IonCode Barcode Adapter is 1-1 transferred by well between the sample plate from Parts 1 & 2 and the Ion Code 96 well plate.\n\n**Note**: This protocol was updated November 22nd, 2022\n\nLinks:\n* [Part 1: DNA Transfer](./7855ef)\n* [Part 2: Pre-Ligation](./7855ef-part2)\n* [Part 3: Barcoding](./7855ef-part3)\n* [Part 4: Pooling](./7855ef-part4)\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P20 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 96 Filter Tip Rack 20 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_20ul?category=tipRack)\n* [ThermoFisher Scientific 96 Well Plate 200ul (AB-0800)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fmanuals%2FMAN0014518_96well_pcr_plate_skirted_low_profile_qr.pdf&title=VGVjaG5pY2FsIERyYXdpbmcgLSBQQ1IgUGxhdGUsIDk2LXdlbGwsIExvdyBQcm9maWxlLCBTa2lydGVk)\n* [ThermoFisher Scientific 96 Well Plate 200ul (4483352)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fbrochures%2FEnduraPlate_96Well.pdf&title=RW5naW5lZXJpbmcgRGlhZ3JhbTogTWljcm9BbXAmcmVnOyBFbmR1cmFQbGF0ZSZ0cmFkZTsgT3B0aWNhbCA5Ni13ZWxsIFJlYWN0aW9uIFBsYXRl)\n* [BioRad Hard-shell 96-well PCR Plate Skirted](https://www.bio-rad.com/en-us/sku/hsp9631-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-blue-clear?ID=hsp9631)\n* [Applied Biosystems 384 Well Plate](https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2Fcms_042831.pdf)\n* Custom 96 Well Endura Plate\n\n**Note About Labware**\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\n\n---\n\n\nUsing the customization fields below, set up your protocol.\n* Number of Samples: Specify the number of samples to be processed in this run (max 384).\n* P20 single GEN2 mount: Specify which mount to load the P20 single GEN2 pipette.\n\n\n**Labware Setup**\n\nSlots 1, 2, 3: Ion Code Barcode plates (Custom 96 well Endura plate)\n\nSlot 5: 384 well plate\n\nSlot 7: MMX Plate with Barcode Reaction Mix in Column 4\n\nSlot 8, 9, 10, 11: Opentrons 20ul Tip Rack\n\n\n\n",
"internal": "7855ef-part3\n",
diff --git a/protoBuilds/7855ef-part4/README.json b/protoBuilds/7855ef-part4/README.json
index 7cc6b01a78..5000933006 100644
--- a/protoBuilds/7855ef-part4/README.json
+++ b/protoBuilds/7855ef-part4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"AgriSeq HTS Library Kit"
@@ -8,7 +8,7 @@
"description": "This protocol is the fourth part of a four part series for performing NGS library prep with the ThermoFisher Scientific AgriSeq kit. Samples in plate are pooled into 60ul pools on a well plate. After which, 45ul from each pool is pooled into a single column.\nNote: This protocol was updated on September 28th, 2022\nLinks:\n Part 1: DNA Transfer\n Part 2: Pre-Ligation\n Part 3: Barcoding\n Part 4: Pooling\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 4.7.0 or later)\nP20 8-Channel Pipette\nP300 8-Channel Pipette\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nThermoFisher Scientific 96 Well Plate 200ul (4483352)\nBioRad Hard-shell 96-well PCR Plate Skirted\nCustom 96 Well Endura Plate\nApplied Biosystems 384-Well Plate\n\nNote About Labware\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\n\n\nUsing the customization fields below, set up your protocol.\n Number of Samples: Specify the number of samples to be processed in this run (max 384).\n P20 Mount: Specify which mount to load the P20 GEN2 pipette.\n* Tip Disposal: Set the pipette to return tips to the tip rack immediately after use or in the trash bin first.\nLabware Setup\nSlots 1: Pool Plate (Custom 96 well Endura Plate)\nSlot 5: Reaction Plate (Applied Biosystems 384-Well Plate)\nSlot 7, 8, 9, 10: Opentrons 20ul Filter Tip Rack(s)\nSlot 11: Opentrons 200ul Filter Tip Rack",
"internal": "7855ef-part4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* AgriSeq HTS Library Kit\n\n",
"description": "This protocol is the fourth part of a four part series for performing NGS library prep with the [ThermoFisher Scientific AgriSeq kit](https://www.thermofisher.com/order/catalog/product/A34144#/A34144). Samples in plate are pooled into 60ul pools on a well plate. After which, 45ul from each pool is pooled into a single column.\n\n**Note:** This protocol was updated on September 28th, 2022\n\nLinks:\n* [Part 1: DNA Transfer](./7855ef)\n* [Part 2: Pre-Ligation](./7855ef-part2)\n* [Part 3: Barcoding](./7855ef-part3)\n* [Part 4: Pooling](./7855ef-part4)\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 4.7.0 or later)](https://opentrons.com/ot-app/)\n* [P20 8-Channel Pipette](https://shop.opentrons.com/8-channel-electronic-pipette/)\n* [P300 8-Channel Pipette](https://shop.opentrons.com/8-channel-electronic-pipette/)\n* [Opentrons 96 Filter Tip Rack 20 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_20ul?category=tipRack)\n* [Opentrons 96 Filter Tip Rack 200 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_200ul?category=tipRack)\n* [ThermoFisher Scientific 96 Well Plate 200ul (4483352)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fbrochures%2FEnduraPlate_96Well.pdf&title=RW5naW5lZXJpbmcgRGlhZ3JhbTogTWljcm9BbXAmcmVnOyBFbmR1cmFQbGF0ZSZ0cmFkZTsgT3B0aWNhbCA5Ni13ZWxsIFJlYWN0aW9uIFBsYXRl)\n* [BioRad Hard-shell 96-well PCR Plate Skirted](https://www.bio-rad.com/en-us/sku/hsp9631-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-blue-clear?ID=hsp9631)\n* Custom 96 Well Endura Plate\n* [Applied Biosystems 384-Well Plate](https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2Fcms_042831.pdf)\n\n**Note About Labware**\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\n\n---\n\n\nUsing the customization fields below, set up your protocol.\n* Number of Samples: Specify the number of samples to be processed in this run (max 384).\n* P20 Mount: Specify which mount to load the P20 GEN2 pipette.\n* Tip Disposal: Set the pipette to return tips to the tip rack immediately after use or in the trash bin first.\n\n\n**Labware Setup**\n\nSlots 1: Pool Plate (Custom 96 well Endura Plate)\n\nSlot 5: Reaction Plate (Applied Biosystems 384-Well Plate)\n\nSlot 7, 8, 9, 10: Opentrons 20ul Filter Tip Rack(s)\n\nSlot 11: Opentrons 200ul Filter Tip Rack\n\n",
"internal": "7855ef-part4\n",
diff --git a/protoBuilds/7855ef-plate-part2/README.json b/protoBuilds/7855ef-plate-part2/README.json
index f6668f8b9f..747fef98fd 100644
--- a/protoBuilds/7855ef-plate-part2/README.json
+++ b/protoBuilds/7855ef-plate-part2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"AgriSeq HTS Library Kit"
@@ -8,7 +8,7 @@
"description": "This protocol is the second of a four part series for performing NGS library prep with the ThermoFisher Scientific AgriSeq kit. 2ul of Pre-ligation mix is distributed to each column of sample that was processed in Part 1 of the protocol.\nLinks:\n Part 1: DNA Transfer\n Part 2: Pre-Ligation\n Part 3: Barcoding\n Part 4: Pooling\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP20 Multi-Channel Pipette\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nThermoFisher Scientific 96 Well Plate 200ul (AB-0800)\nThermoFisher Scientific 96 Well Plate 200ul (4483352)\nBioRad Hard-shell 96-well PCR Plate Skirted\nCustom 96 Well Endura Plate\n\nNote About Labware\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\n\n\nUsing the customization fields below, set up your protocol.\n Number of Samples: Specify the number of samples to be processed in this run (max 288).\n P20 single GEN2 mount: Specify which mount to load the P20 single GEN2 pipette.\nNote about 20\u00b5L tip racks\nWhen prompted to replace the 20ul tip racks, be sure to re-load all 3 tip racks as in the original configuration of the deck.\nLabware Setup\nSlots 1, 2, 3: ThermoFisher Scientific (model AB0800) 96 well plate loaded with DNA sample. \nSlot 4, 5, 6: Custom 96 Well Endura Plate with reaction\nSlot 7: MMX Plate with Pre-ligation Mix in Column 2\nSlot 9, 10, 11: Opentrons 20ul Tip Rack",
"internal": "7855ef-96-plate-part2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* AgriSeq HTS Library Kit\n\n",
"description": "This protocol is the second of a four part series for performing NGS library prep with the [ThermoFisher Scientific AgriSeq kit](https://www.thermofisher.com/order/catalog/product/A34144#/A34144). 2ul of Pre-ligation mix is distributed to each column of sample that was processed in Part 1 of the protocol.\n\nLinks:\n* [Part 1: DNA Transfer](http://protocols.opentrons.com/protocol/7855ef-plate)\n* [Part 2: Pre-Ligation](http://protocols.opentrons.com/protocol/7855ef-plate-part2)\n* [Part 3: Barcoding](http://protocols.opentrons.com/protocol/7855ef-plate-part3)\n* [Part 4: Pooling](http://protocols.opentrons.com/protocol/7855ef-plate-part4)\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P20 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 96 Filter Tip Rack 20 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_20ul?category=tipRack)\n* [ThermoFisher Scientific 96 Well Plate 200ul (AB-0800)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fmanuals%2FMAN0014518_96well_pcr_plate_skirted_low_profile_qr.pdf&title=VGVjaG5pY2FsIERyYXdpbmcgLSBQQ1IgUGxhdGUsIDk2LXdlbGwsIExvdyBQcm9maWxlLCBTa2lydGVk)\n* [ThermoFisher Scientific 96 Well Plate 200ul (4483352)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fbrochures%2FEnduraPlate_96Well.pdf&title=RW5naW5lZXJpbmcgRGlhZ3JhbTogTWljcm9BbXAmcmVnOyBFbmR1cmFQbGF0ZSZ0cmFkZTsgT3B0aWNhbCA5Ni13ZWxsIFJlYWN0aW9uIFBsYXRl)\n* [BioRad Hard-shell 96-well PCR Plate Skirted](https://www.bio-rad.com/en-us/sku/hsp9631-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-blue-clear?ID=hsp9631)\n* Custom 96 Well Endura Plate\n\n**Note About Labware**\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\n\n---\n\n\nUsing the customization fields below, set up your protocol.\n* Number of Samples: Specify the number of samples to be processed in this run (max 288).\n* P20 single GEN2 mount: Specify which mount to load the P20 single GEN2 pipette.\n\n\n**Note about 20\u00b5L tip racks**\n\nWhen prompted to replace the 20ul tip racks, be sure to re-load all 3 tip racks as in the original configuration of the deck.\n\n**Labware Setup**\n\nSlots 1, 2, 3: ThermoFisher Scientific (model AB0800) 96 well plate loaded with DNA sample. \n\nSlot 4, 5, 6: Custom 96 Well Endura Plate with reaction\n\nSlot 7: MMX Plate with Pre-ligation Mix in Column 2\n\nSlot 9, 10, 11: Opentrons 20ul Tip Rack\n\n\n\n",
"internal": "7855ef-96-plate-part2\n",
diff --git a/protoBuilds/7855ef-plate-part3/README.json b/protoBuilds/7855ef-plate-part3/README.json
index 6517cf1dea..7420d4caeb 100644
--- a/protoBuilds/7855ef-plate-part3/README.json
+++ b/protoBuilds/7855ef-plate-part3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"AgriSeq HTS Library Kit"
@@ -8,7 +8,7 @@
"description": "This protocol is the third of a four part series for performing NGS library prep with the ThermoFisher Scientific AgriSeq kit. 1ul of IonCode Barcode Adapter is 1-1 transferred by well between the sample plate from Parts 1 & 2 and the Ion Code 96 well plate.\nLinks:\n Part 1: DNA Transfer\n Part 2: Pre-Ligation\n Part 3: Barcoding\n Part 4: Pooling\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP20 Multi-Channel Pipette\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nThermoFisher Scientific 96 Well Plate 200ul (AB-0800)\nThermoFisher Scientific 96 Well Plate 200ul (4483352)\nBioRad Hard-shell 96-well PCR Plate Skirted\nCustom 96 Well Endura Plate\n\nNote About Labware\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\n\n\nUsing the customization fields below, set up your protocol.\n Number of Samples: Specify the number of samples to be processed in this run (max 288).\n P20 Multi GEN2 mount: Specify which mount to load the P20 Multi GEN2 pipette.\n* Overage percent for Barcode Reaction Mix in Slot 7, Column 3\nLabware Setup\nSlots 1, 2, 3: Ion Code Barcode plates (Custom 96 well Endura plate)\nSlot 4, 5, 6: Custom 96 Well Endura Plate with reaction\nSlot 7: MMX Plate with Barcode Reaction Mix in Column 3\nSlot 8, 9, 10: Opentrons 20ul Tip Rack",
"internal": "7855ef-part3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* AgriSeq HTS Library Kit\n\n",
"description": "This protocol is the third of a four part series for performing NGS library prep with the [ThermoFisher Scientific AgriSeq kit](https://www.thermofisher.com/order/catalog/product/A34144#/A34144). 1ul of IonCode Barcode Adapter is 1-1 transferred by well between the sample plate from Parts 1 & 2 and the Ion Code 96 well plate.\n\nLinks:\n* [Part 1: DNA Transfer](http://protocols.opentrons.com/protocol/7855ef-plate)\n* [Part 2: Pre-Ligation](http://protocols.opentrons.com/protocol/7855ef-plate-part2)\n* [Part 3: Barcoding](http://protocols.opentrons.com/protocol/7855ef-plate-part3)\n* [Part 4: Pooling](http://protocols.opentrons.com/protocol/7855ef-plate-part4)\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P20 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 96 Filter Tip Rack 20 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_20ul?category=tipRack)\n* [ThermoFisher Scientific 96 Well Plate 200ul (AB-0800)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fmanuals%2FMAN0014518_96well_pcr_plate_skirted_low_profile_qr.pdf&title=VGVjaG5pY2FsIERyYXdpbmcgLSBQQ1IgUGxhdGUsIDk2LXdlbGwsIExvdyBQcm9maWxlLCBTa2lydGVk)\n* [ThermoFisher Scientific 96 Well Plate 200ul (4483352)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fbrochures%2FEnduraPlate_96Well.pdf&title=RW5naW5lZXJpbmcgRGlhZ3JhbTogTWljcm9BbXAmcmVnOyBFbmR1cmFQbGF0ZSZ0cmFkZTsgT3B0aWNhbCA5Ni13ZWxsIFJlYWN0aW9uIFBsYXRl)\n* [BioRad Hard-shell 96-well PCR Plate Skirted](https://www.bio-rad.com/en-us/sku/hsp9631-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-blue-clear?ID=hsp9631)\n* Custom 96 Well Endura Plate\n\n**Note About Labware**\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\n\n---\n\n\nUsing the customization fields below, set up your protocol.\n* Number of Samples: Specify the number of samples to be processed in this run (max 288).\n* P20 Multi GEN2 mount: Specify which mount to load the P20 Multi GEN2 pipette.\n* Overage percent for Barcode Reaction Mix in Slot 7, Column 3\n\n\n**Labware Setup**\n\nSlots 1, 2, 3: Ion Code Barcode plates (Custom 96 well Endura plate)\n\nSlot 4, 5, 6: Custom 96 Well Endura Plate with reaction\n\nSlot 7: MMX Plate with Barcode Reaction Mix in Column 3\n\nSlot 8, 9, 10: Opentrons 20ul Tip Rack\n\n\n\n",
"internal": "7855ef-part3\n",
diff --git a/protoBuilds/7855ef-plate-part4/README.json b/protoBuilds/7855ef-plate-part4/README.json
index 3e1734fed0..5e006f3002 100644
--- a/protoBuilds/7855ef-plate-part4/README.json
+++ b/protoBuilds/7855ef-plate-part4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"AgriSeq HTS Library Kit"
@@ -8,7 +8,7 @@
"description": "This protocol is the fourth part of a four part series for performing NGS library prep with the ThermoFisher Scientific AgriSeq kit. Samples in plate are pooled into 60ul pools on a well plate. After which, 45ul from each pool is transferred to the final processing plate.\nLinks:\n Part 1: DNA Transfer\n Part 2: Pre-Ligation\n Part 3: Barcoding\n Part 4: Pooling\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP20 Single Channel Pipette\nP300 Single Channel Pipette\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nThermoFisher Scientific 96 Well Plate 200ul (AB-0800)\nThermoFisher Scientific 96 Well Plate 200ul (4483352)\nBioRad Hard-shell 96-well PCR Plate Skirted\nCustom 96 Well Endura Plate\n\nNote About Labware\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\n\n\nUsing the customization fields below, set up your protocol.\n Number of Samples: Specify the number of samples to be processed in this run (max 288).\n P20 single GEN2 mount: Specify which mount to load the P20 single GEN2 pipette.\nLabware Setup\nSlots 1: Pool Plate 1 (Custom 96 well Endura Plate)\nSlot 2: Pool Plate 2 (Custom 96 well Endura Plate)\nSlot 4, 5, 6: Reaction Plates (Custom 96 well Endura Plate)\nSlot 8: Opentrons 200ul Tip Rack\nSlot 9, 10, 11: Opentrons 20ul Tip Rack",
"internal": "7855ef-part4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* AgriSeq HTS Library Kit\n\n",
"description": "This protocol is the fourth part of a four part series for performing NGS library prep with the [ThermoFisher Scientific AgriSeq kit](https://www.thermofisher.com/order/catalog/product/A34144#/A34144). Samples in plate are pooled into 60ul pools on a well plate. After which, 45ul from each pool is transferred to the final processing plate.\n\nLinks:\n* [Part 1: DNA Transfer](http://protocols.opentrons.com/protocol/7855ef-plate)\n* [Part 2: Pre-Ligation](http://protocols.opentrons.com/protocol/7855ef-plate-part2)\n* [Part 3: Barcoding](http://protocols.opentrons.com/protocol/7855ef-plate-part3)\n* [Part 4: Pooling](http://protocols.opentrons.com/protocol/7855ef-plate-part4)\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P20 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [P300 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [Opentrons 96 Filter Tip Rack 20 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_20ul?category=tipRack)\n* [Opentrons 96 Filter Tip Rack 200 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_200ul?category=tipRack)\n* [ThermoFisher Scientific 96 Well Plate 200ul (AB-0800)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fmanuals%2FMAN0014518_96well_pcr_plate_skirted_low_profile_qr.pdf&title=VGVjaG5pY2FsIERyYXdpbmcgLSBQQ1IgUGxhdGUsIDk2LXdlbGwsIExvdyBQcm9maWxlLCBTa2lydGVk)\n* [ThermoFisher Scientific 96 Well Plate 200ul (4483352)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fbrochures%2FEnduraPlate_96Well.pdf&title=RW5naW5lZXJpbmcgRGlhZ3JhbTogTWljcm9BbXAmcmVnOyBFbmR1cmFQbGF0ZSZ0cmFkZTsgT3B0aWNhbCA5Ni13ZWxsIFJlYWN0aW9uIFBsYXRl)\n* [BioRad Hard-shell 96-well PCR Plate Skirted](https://www.bio-rad.com/en-us/sku/hsp9631-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-blue-clear?ID=hsp9631)\n* Custom 96 Well Endura Plate\n\n**Note About Labware**\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\n\n---\n\n\nUsing the customization fields below, set up your protocol.\n* Number of Samples: Specify the number of samples to be processed in this run (max 288).\n* P20 single GEN2 mount: Specify which mount to load the P20 single GEN2 pipette.\n\n\n**Labware Setup**\n\nSlots 1: Pool Plate 1 (Custom 96 well Endura Plate)\n\nSlot 2: Pool Plate 2 (Custom 96 well Endura Plate)\n\nSlot 4, 5, 6: Reaction Plates (Custom 96 well Endura Plate)\n\nSlot 8: Opentrons 200ul Tip Rack\n\nSlot 9, 10, 11: Opentrons 20ul Tip Rack\n\n",
"internal": "7855ef-part4\n",
diff --git a/protoBuilds/7855ef-plate/README.json b/protoBuilds/7855ef-plate/README.json
index 71d00139f3..4dcbe3c2fb 100644
--- a/protoBuilds/7855ef-plate/README.json
+++ b/protoBuilds/7855ef-plate/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"AgriSeq HTS Library Kit"
@@ -8,7 +8,7 @@
"description": "This protocol is the first of a 4 part series for performing NGS library prep with the ThermoFisher Scientific AgriSeq kit. The OT-2 will distribute 7ul of Amplification Mix to each well of 96 well plates up to the number of samples specified by the user. 3ul of DNA is then added to each well containing Ampflication Mix.\nLinks:\n Part 1: DNA Transfer\n Part 2: Pre-Ligation\n Part 3: Barcoding\n Part 4: Pooling\nNote about tips\nThe OT-2 will track tips from Part 1 to Part 4 of the protocol (e.g. tip leaves off in H11 at the end of protocol 1; first tip pick up will be from H12 in Part 2). When tips run out for any particular Part, the user will be prompted to replace all tip racks.\nUpdate (July 18, 2022): The custom touch tip has been adjusted\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nP20 Multi-Channel Pipette\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nThermoFisher Scientific 96 Well Plate 200ul (AB-0800)\nThermoFisher Scientific 96 Well Plate 200ul (4483352)\nBioRad Hard-shell 96-well PCR Plate Skirted\nCustom 96 Well Endura Plate\n\nNote About Labware\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\nNote About Sample Number\nPart 4 of this protocol will pool 5ul from wells until a 60ul pool is achieved (i.e. a full plate would have one pool per row). If there is less than a full plate loaded, the protocol will iterate through wells dependent on the sample number specified until 60ul is reached. Please consider the following examples:\n\n96 sample run (1 plate): 1 column of 60ul pools.\n144 sample run (1.5 plates): 1 column and 4 wells of 60ul pools.\n216 sample run (2.25 plates): 2 columns and 2 wells of 60ul pools.\n\nIt is thus critical to load samples that can be discretely divided by 12.\n\n\nUsing the customization fields below, set up your protocol.\n Number of Samples: Specify the number of samples to be processed in this run (max 288).\n P20 single GEN2 mount: Specify which mount to load the P20 single GEN2 pipette.\nNote about 20\u00b5L tip racks\nWhen prompted to replace the 20ul tip racks, be sure to re-load all 3 tip racks as in the original configuration of the deck.\nLabware Setup\nSlots 1, 2, 3: ThermoFisher Scientific (model AB0800) 96 well plate loaded with DNA sample. \nSlot 4, 5, 6: Custom 96 Well Endura Plate (empty)\nSlot 7: MMX Plate with Amplification Mix in Column 1\nSlot 8, 9, 10, 11: Opentrons 20ul Tip Rack",
"internal": "7855ef",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* AgriSeq HTS Library Kit\n\n",
"description": "This protocol is the first of a 4 part series for performing NGS library prep with the [ThermoFisher Scientific AgriSeq kit](https://www.thermofisher.com/order/catalog/product/A34144#/A34144). The OT-2 will distribute 7ul of Amplification Mix to each well of 96 well plates up to the number of samples specified by the user. 3ul of DNA is then added to each well containing Ampflication Mix.\n\nLinks:\n* [Part 1: DNA Transfer](http://protocols.opentrons.com/protocol/7855ef-plate)\n* [Part 2: Pre-Ligation](http://protocols.opentrons.com/protocol/7855ef-plate-part2)\n* [Part 3: Barcoding](http://protocols.opentrons.com/protocol/7855ef-plate-part3)\n* [Part 4: Pooling](http://protocols.opentrons.com/protocol/7855ef-plate-part4)\n\n**Note about tips**\nThe OT-2 will track tips from Part 1 to Part 4 of the protocol (e.g. tip leaves off in H11 at the end of protocol 1; first tip pick up will be from H12 in Part 2). When tips run out for any particular Part, the user will be prompted to replace all tip racks.\n\n**Update (July 18, 2022):** The custom touch tip has been adjusted\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [P20 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 96 Filter Tip Rack 20 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_20ul?category=tipRack)\n* [ThermoFisher Scientific 96 Well Plate 200ul (AB-0800)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fmanuals%2FMAN0014518_96well_pcr_plate_skirted_low_profile_qr.pdf&title=VGVjaG5pY2FsIERyYXdpbmcgLSBQQ1IgUGxhdGUsIDk2LXdlbGwsIExvdyBQcm9maWxlLCBTa2lydGVk)\n* [ThermoFisher Scientific 96 Well Plate 200ul (4483352)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fbrochures%2FEnduraPlate_96Well.pdf&title=RW5naW5lZXJpbmcgRGlhZ3JhbTogTWljcm9BbXAmcmVnOyBFbmR1cmFQbGF0ZSZ0cmFkZTsgT3B0aWNhbCA5Ni13ZWxsIFJlYWN0aW9uIFBsYXRl)\n* [BioRad Hard-shell 96-well PCR Plate Skirted](https://www.bio-rad.com/en-us/sku/hsp9631-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-blue-clear?ID=hsp9631)\n* Custom 96 Well Endura Plate\n\n**Note About Labware**\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\n\n**Note About Sample Number**\nPart 4 of this protocol will pool 5ul from wells until a 60ul pool is achieved (i.e. a full plate would have one pool per row). If there is less than a full plate loaded, the protocol will iterate through wells dependent on the sample number specified until 60ul is reached. Please consider the following examples:\n\n* 96 sample run (1 plate): 1 column of 60ul pools.\n* 144 sample run (1.5 plates): 1 column and 4 wells of 60ul pools.\n* 216 sample run (2.25 plates): 2 columns and 2 wells of 60ul pools.\n\n**It is thus critical to load samples that can be discretely divided by 12.**\n\n\n---\n\n\nUsing the customization fields below, set up your protocol.\n* Number of Samples: Specify the number of samples to be processed in this run (max 288).\n* P20 single GEN2 mount: Specify which mount to load the P20 single GEN2 pipette.\n\n\n**Note about 20\u00b5L tip racks**\n\nWhen prompted to replace the 20ul tip racks, be sure to re-load all 3 tip racks as in the original configuration of the deck.\n\n**Labware Setup**\n\nSlots 1, 2, 3: ThermoFisher Scientific (model AB0800) 96 well plate loaded with DNA sample. \n\nSlot 4, 5, 6: Custom 96 Well Endura Plate (empty)\n\nSlot 7: MMX Plate with Amplification Mix in Column 1\n\nSlot 8, 9, 10, 11: Opentrons 20ul Tip Rack\n\n\n\n",
"internal": "7855ef\n",
diff --git a/protoBuilds/7855ef/README.json b/protoBuilds/7855ef/README.json
index 4acc61c55d..df3f236d58 100644
--- a/protoBuilds/7855ef/README.json
+++ b/protoBuilds/7855ef/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"AgriSeq HTS Library Kit"
@@ -8,7 +8,7 @@
"description": "This protocol is the first of a 4 part series for performing NGS library prep with the ThermoFisher Scientific AgriSeq kit. The OT-2 will distribute 7ul of Amplification Mix to each well of a 384 well plate up to the number of samples specified by the user. 3ul of DNA is then added to each well containing Ampflication Mix.\nLinks:\n Part 1: DNA Transfer\n Part 2: Pre-Ligation\n Part 3: Barcoding\n Part 4: Pooling\nNote about tips\nThe OT-2 will track tips from Part 1 to Part 4 of the protocol (e.g. tip leaves off in H11 at the end of protocol 1; first tip pick up will be from H12 in Part 2). When tips run out for any particular Part, the user will be prompted to replace all tip racks.\nUpdate: This protocol was updated November 22nd, 2022\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 4.7.0 or later)\nP20 Multi-Channel Pipette\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nThermoFisher Scientific 96 Well Plate 200ul (AB-0800)\nThermoFisher Scientific 96 Well Plate 200ul (4483352)\nBioRad Hard-shell 96-well PCR Plate Skirted\nApplied Biosystems 384 Well Plate\nCustom 96 Well Endura Plate\n\nNote About Labware\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\nNote About Sample Number\nPart 4 of this protocol will pool 5ul from wells until a 60ul pool is achieved (i.e. a full plate would have one pool per row). If there is less than a full plate loaded, the protocol will iterate through wells dependent on the sample number specified until 60ul is reached. Please consider the following examples:\n\n96 sample run (1 plate): 1 column of 60ul pools.\n144 sample run (1.5 plates): 1 column and 4 wells of 60ul pools.\n216 sample run (2.25 plates): 2 columns and 2 wells of 60ul pools.\n\nIt is thus critical to load samples that can be discretely divided by 12.\n\n\nUsing the customization fields below, set up your protocol.\n Number of Samples: Specify the number of samples to be processed in this run (max 384).\n P20 single GEN2 mount: Specify which mount to load the P20 single GEN2 pipette.\nNote about 20\u00b5L tip racks\nWhen prompted to replace the 20ul tip racks, be sure to re-load all 3 tip racks as in the original configuration of the deck.\nLabware Setup\nSlots 1, 2, 3, 4: ThermoFisher Scientific (model AB0800) 96 well plate loaded with DNA sample. \nSlot 5: 384 well plate\nSlot 6: MMX Plate with Amplification Mix in Column 1 and 2\nSlot 7, 8, 9, 10, 11: Opentrons 20ul Tip Rack",
"internal": "7855ef",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* AgriSeq HTS Library Kit\n\n",
"description": "This protocol is the first of a 4 part series for performing NGS library prep with the [ThermoFisher Scientific AgriSeq kit](https://www.thermofisher.com/order/catalog/product/A34144#/A34144). The OT-2 will distribute 7ul of Amplification Mix to each well of a 384 well plate up to the number of samples specified by the user. 3ul of DNA is then added to each well containing Ampflication Mix.\n\nLinks:\n* [Part 1: DNA Transfer](./7855ef)\n* [Part 2: Pre-Ligation](./7855ef-part2)\n* [Part 3: Barcoding](./7855ef-part3)\n* [Part 4: Pooling](./7855ef-part4)\n\n**Note about tips**\nThe OT-2 will track tips from Part 1 to Part 4 of the protocol (e.g. tip leaves off in H11 at the end of protocol 1; first tip pick up will be from H12 in Part 2). When tips run out for any particular Part, the user will be prompted to replace all tip racks.\n\n**Update**: This protocol was updated November 22nd, 2022\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 4.7.0 or later)](https://opentrons.com/ot-app/)\n* [P20 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons 96 Filter Tip Rack 20 \u00b5L](https://labware.opentrons.com/opentrons_96_filtertiprack_20ul?category=tipRack)\n* [ThermoFisher Scientific 96 Well Plate 200ul (AB-0800)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fmanuals%2FMAN0014518_96well_pcr_plate_skirted_low_profile_qr.pdf&title=VGVjaG5pY2FsIERyYXdpbmcgLSBQQ1IgUGxhdGUsIDk2LXdlbGwsIExvdyBQcm9maWxlLCBTa2lydGVk)\n* [ThermoFisher Scientific 96 Well Plate 200ul (4483352)](https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fbrochures%2FEnduraPlate_96Well.pdf&title=RW5naW5lZXJpbmcgRGlhZ3JhbTogTWljcm9BbXAmcmVnOyBFbmR1cmFQbGF0ZSZ0cmFkZTsgT3B0aWNhbCA5Ni13ZWxsIFJlYWN0aW9uIFBsYXRl)\n* [BioRad Hard-shell 96-well PCR Plate Skirted](https://www.bio-rad.com/en-us/sku/hsp9631-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-blue-clear?ID=hsp9631)\n* [Applied Biosystems 384 Well Plate](https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2Fcms_042831.pdf)\n* Custom 96 Well Endura Plate\n\n**Note About Labware**\nThe ThermoFisher 96 well plate (model 4483352) is to be mounted on top of the BioRad Hard-shell plate, making one plate with a moniker of \"Custom 96 Well Endura Plate\".\n\n**Note About Sample Number**\nPart 4 of this protocol will pool 5ul from wells until a 60ul pool is achieved (i.e. a full plate would have one pool per row). If there is less than a full plate loaded, the protocol will iterate through wells dependent on the sample number specified until 60ul is reached. Please consider the following examples:\n\n* 96 sample run (1 plate): 1 column of 60ul pools.\n* 144 sample run (1.5 plates): 1 column and 4 wells of 60ul pools.\n* 216 sample run (2.25 plates): 2 columns and 2 wells of 60ul pools.\n\n**It is thus critical to load samples that can be discretely divided by 12.**\n\n\n---\n\n\nUsing the customization fields below, set up your protocol.\n* Number of Samples: Specify the number of samples to be processed in this run (max 384).\n* P20 single GEN2 mount: Specify which mount to load the P20 single GEN2 pipette.\n\n\n**Note about 20\u00b5L tip racks**\n\nWhen prompted to replace the 20ul tip racks, be sure to re-load all 3 tip racks as in the original configuration of the deck.\n\n**Labware Setup**\n\nSlots 1, 2, 3, 4: ThermoFisher Scientific (model AB0800) 96 well plate loaded with DNA sample. \n\nSlot 5: 384 well plate\n\nSlot 6: MMX Plate with Amplification Mix in Column 1 and 2\n\nSlot 7, 8, 9, 10, 11: Opentrons 20ul Tip Rack\n\n\n\n",
"internal": "7855ef\n",
diff --git a/protoBuilds/78d33c-part-2/README.json b/protoBuilds/78d33c-part-2/README.json
index 211345bcf1..78c1a4e3ad 100644
--- a/protoBuilds/78d33c-part-2/README.json
+++ b/protoBuilds/78d33c-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 6 part workflow, the OT-2 will follow ArcBio Continuous DNA Workflow - Pre-PCR Instrument experimental protocol and Post-PCR Instrument experimental protocol to convert up to 96 input DNA samples into DNA libraries. This is Part 2 of 6: This OT-2 protocol uses the DNA Continuous Workflow, Pre-PCR Instrument section of attached experimental protocol to perform adapter ligation.",
"internal": "78d33c-part-2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul filter tips (deck slots 10, 11)\n* Opentrons 200 ul filter tips (deck slots 7, 8)\n* Opentrons Temperature Module (deck slot 3) with Sample Plate\n* Opentrons Magnetic Module (deck slot 1)\n* Reagents opentrons_96_aluminumblock_generic_pcr_strip_200ul (deck slot 2)\n\n\n\n",
"description": "\nWith this 6 part workflow, the OT-2 will follow [ArcBio Continuous DNA Workflow - Pre-PCR Instrument experimental protocol and Post-PCR Instrument experimental protocol](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/78d33c/ArcBio_DNA_Workflow_020822.xlsx) to convert up to 96 input DNA samples into DNA libraries. This is Part 2 of 6: This OT-2 protocol uses the DNA Continuous Workflow, Pre-PCR Instrument section of attached experimental protocol to perform adapter ligation.\n\n",
diff --git a/protoBuilds/78d33c-part-3/README.json b/protoBuilds/78d33c-part-3/README.json
index 3fc3d016d9..f8d8bfdc9e 100644
--- a/protoBuilds/78d33c-part-3/README.json
+++ b/protoBuilds/78d33c-part-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 6 part workflow, the OT-2 will follow ArcBio Continuous DNA Workflow - Pre-PCR Instrument experimental protocol and Post-PCR Instrument experimental protocol to convert up to 96 input DNA samples into DNA libraries. This is Part 3 of 6: This OT-2 protocol uses the DNA Continuous Workflow, Pre-PCR Instrument section of attached experimental protocol to perform library purification and indexing.",
"internal": "78d33c-part-3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul tips (deck slots 10, 11)\n* Opentrons 300ul tips (deck slots 7, 8, 9)\n* Opentrons Temperature Module (deck slot 3) with Sample Plate opentrons_96_aluminumblock_biorad_wellplate_200ul\n* Opentrons Magnetic Module (deck slot 1) with deep well plate\n* Reservoirs nest_1_reservoir_195ml (deck slots 4, 6)\n* Reagents opentrons_96_aluminumblock_generic_pcr_strip_200ul (deck slot 2)\n* Reservoir nest_12_reservoir_15ml (deck slot 5)\n\n\n\n\n\n",
"description": "\nWith this 6 part workflow, the OT-2 will follow [ArcBio Continuous DNA Workflow - Pre-PCR Instrument experimental protocol and Post-PCR Instrument experimental protocol](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/78d33c/ArcBio_DNA_Workflow_020822.xlsx) to convert up to 96 input DNA samples into DNA libraries. This is Part 3 of 6: This OT-2 protocol uses the DNA Continuous Workflow, Pre-PCR Instrument section of attached experimental protocol to perform library purification and indexing.\n\n",
diff --git a/protoBuilds/78d33c-part-4/README.json b/protoBuilds/78d33c-part-4/README.json
index 2e4d160fd9..f0e44bd956 100644
--- a/protoBuilds/78d33c-part-4/README.json
+++ b/protoBuilds/78d33c-part-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 6 part workflow, the OT-2 will follow ArcBio Continuous DNA Workflow - Pre-PCR Instrument experimental protocol and Post-PCR Instrument experimental protocol to convert up to 96 input DNA samples into DNA libraries. This is Part 4 of 6: This OT-2 protocol uses the DNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform purification post indexing steps.",
"internal": "78d33c-part-4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul tips (deck slots 10, 11)\n* Opentrons 300ul tips (deck slots 7, 8, 9)\n* Opentrons Temperature Module (deck slot 3) with 200 uL PCR plate\n* Opentrons Magnetic Module (deck slot 1) with 200 uL PCR plate\n* Reservoirs nest_1_reservoir_195ml (deck slots 4, 6)\n* Reagents opentrons_96_aluminumblock_generic_pcr_strip_200ul (deck slot 2)\n* Reservoir nest_12_reservoir_15ml (deck slot 5)\n\n\n\n\n\n",
"description": "\nWith this 6 part workflow, the OT-2 will follow [ArcBio Continuous DNA Workflow - Pre-PCR Instrument experimental protocol and Post-PCR Instrument experimental protocol](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/78d33c/ArcBio_DNA_Workflow_020822.xlsx) to convert up to 96 input DNA samples into DNA libraries. This is Part 4 of 6: This OT-2 protocol uses the DNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform purification post indexing steps.\n\n",
diff --git a/protoBuilds/78d33c-part-5/README.json b/protoBuilds/78d33c-part-5/README.json
index 7c801169c8..537e52ca1e 100644
--- a/protoBuilds/78d33c-part-5/README.json
+++ b/protoBuilds/78d33c-part-5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 6 part workflow, the OT-2 will follow ArcBio Continuous DNA Workflow - Pre-PCR Instrument experimental protocol and Post-PCR Instrument experimental protocol to convert up to 96 input DNA samples into DNA libraries. This is Part 5 of 6: This OT-2 protocol uses the DNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform post indexing steps.",
"internal": "78d33c-part-5",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul tips (deck slots 10, 11)\n* Opentrons Temperature Module (deck slot 3) with Sample Plate opentrons_96_aluminumblock_biorad_wellplate_200ul\n* Opentrons Magnetic Module (deck slot 1) with deep well plate\n* Reagents opentrons_96_aluminumblock_generic_pcr_strip_200ul (deck slot 2)\n\n\n\n",
"description": "\nWith this 6 part workflow, the OT-2 will follow [ArcBio Continuous DNA Workflow - Pre-PCR Instrument experimental protocol and Post-PCR Instrument experimental protocol](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/78d33c/ArcBio_DNA_Workflow_020822.xlsx) to convert up to 96 input DNA samples into DNA libraries. This is Part 5 of 6: This OT-2 protocol uses the DNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform post indexing steps.\n\n",
diff --git a/protoBuilds/78d33c-part-6/README.json b/protoBuilds/78d33c-part-6/README.json
index be95aac92f..d1185e5245 100644
--- a/protoBuilds/78d33c-part-6/README.json
+++ b/protoBuilds/78d33c-part-6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 6 part workflow, the OT-2 will follow ArcBio Continuous DNA Workflow - Pre-PCR Instrument experimental protocol and Post-PCR Instrument experimental protocol to convert up to 96 input DNA samples into DNA libraries. This is Part 6 of 6: This OT-2 protocol uses the DNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform library purification steps.",
"internal": "78d33c-part-6",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul tips (deck slots 10, 11)\n* Opentrons 300ul tips (deck slots 7, 8, 9)\n* Opentrons Temperature Module (deck slot 3) with 200 uL PCR plate\n* Opentrons Magnetic Module (deck slot 1) with 200 uL PCR plate\n* Reservoirs nest_1_reservoir_195ml (deck slots 4, 6)\n* Reservoir nest_12_reservoir_15ml (deck slot 5)\n\n\n\n",
"description": "\nWith this 6 part workflow, the OT-2 will follow [ArcBio Continuous DNA Workflow - Pre-PCR Instrument experimental protocol and Post-PCR Instrument experimental protocol](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/78d33c/ArcBio_DNA_Workflow_020822.xlsx) to convert up to 96 input DNA samples into DNA libraries. This is Part 6 of 6: This OT-2 protocol uses the DNA Continuous Workflow, Post-PCR Instrument section of attached experimental protocol to perform library purification steps.\n\n",
diff --git a/protoBuilds/78d33c/README.json b/protoBuilds/78d33c/README.json
index 4cd985b55d..584244bccf 100644
--- a/protoBuilds/78d33c/README.json
+++ b/protoBuilds/78d33c/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -9,7 +9,7 @@
"description": "With this 6 part workflow, the OT-2 will follow ArcBio Continuous DNA Workflow - Pre-PCR Instrument experimental protocol and Post-PCR Instrument experimental protocol to convert up to 96 input DNA samples into DNA libraries. This is Part 1 of 6: This OT-2 protocol uses the DNA Continuous Workflow, Pre-PCR Instrument section of attached experimental protocol to perform concentration and fragmentation.",
"internal": "78d33c",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Custom\n\n",
"deck-setup": "\n\n* Opentrons 20ul tips (deck slots 10, 11)\n* Opentrons 300ul tips (deck slots 7, 8, 9)\n* Opentrons Temperature Module (deck slot 3) with Sample Plate opentrons_96_aluminumblock_biorad_wellplate_200ul\n* Opentrons Magnetic Module (deck slot 1) with deep well plate\n* Reservoirs nest_1_reservoir_195ml (deck slots 4, 6)\n* Reagents opentrons_96_aluminumblock_generic_pcr_strip_200ul (deck slot 2)\n* Reservoir nest_12_reservoir_15ml (deck slot 5)\n\n\n\n\n\n",
"description": "\nWith this 6 part workflow, the OT-2 will follow [ArcBio Continuous DNA Workflow - Pre-PCR Instrument experimental protocol and Post-PCR Instrument experimental protocol](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/78d33c/ArcBio_DNA_Workflow_020822.xlsx) to convert up to 96 input DNA samples into DNA libraries. This is Part 1 of 6: This OT-2 protocol uses the DNA Continuous Workflow, Pre-PCR Instrument section of attached experimental protocol to perform concentration and fragmentation.\n\n",
diff --git a/protoBuilds/796af8/README.json b/protoBuilds/796af8/README.json
index aefd048ab1..e6424534bd 100644
--- a/protoBuilds/796af8/README.json
+++ b/protoBuilds/796af8/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEB Ultra II FS DNA Library Prep"
@@ -8,7 +8,7 @@
"description": "This protocol automates the End Repair, Adapter Ligation and Bead Clean Up for the NEB Ultra II FS DNA Library Prep\n\n\n\nP20-multi channel GEN2 electronic pipette\nP300-multi channel GEN2 electronic pipette\nOpentrons Thermocycler Module\nOpentrons Temperature Module\nOpentrons Magnetic Module\nNEST 100uL PCR Plate\nOpentrons 96 Well Aluminum Block with 200 uL PCR Strips\n\n\n\n\nThermocycler (Slot 7)\nTemperature Module (Slot 4)\nMagnetic Module (Slot 9)\nOpentrons 200 uL Filter Tips (Slot 3)\nOpentrons 20 uL Filter Tips (Slots 1, 2, 5)\n\nInitial Starting State:\n- Samples start in a 96 well PCR plate in the thermocycler.\n- Temperature Block starts at 4C.\n- End Prep Mastermix, Adapter Mastermix, and PCR Mix should be in columns 1-3, respectively, in the 96 Well Aluminum block on the temperature module.\n- Reservoir channel A1 contains ethanol\n- Plate on the magnetic module contains pre-aliquoted magnetic beads and EB",
"internal": "796af8",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEB Ultra II FS DNA Library Prep\n\n\n",
"description": "This protocol automates the End Repair, Adapter Ligation and Bead Clean Up for the [NEB Ultra II FS DNA Library Prep](https://www.neb.com/products/e7805-nebnext-ultra-ii-fs-dna-library-prep-kit-for-illumina#Protocols,%20Manuals%20&%20Usage)\n\n\n---\n\n\n* [P20-multi channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [P300-multi channel GEN2 electronic pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons Thermocycler Module](https://opentrons.com/modules/thermocycler-module/)\n* [Opentrons Temperature Module](https://opentrons.com/modules/temperature-module/)\n* [Opentrons Magnetic Module](https://opentrons.com/modules/temperature-module/)\n* [NEST 100uL PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons 96 Well Aluminum Block with 200 uL PCR Strips](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n\n---\n\n\n* Thermocycler (Slot 7)\n* Temperature Module (Slot 4)\n* Magnetic Module (Slot 9)\n* Opentrons 200 uL Filter Tips (Slot 3)\n* Opentrons 20 uL Filter Tips (Slots 1, 2, 5)\n\nInitial Starting State:\n- Samples start in a 96 well PCR plate in the thermocycler.\n- Temperature Block starts at 4C.\n- End Prep Mastermix, Adapter Mastermix, and PCR Mix should be in columns 1-3, respectively, in the 96 Well Aluminum block on the temperature module.\n- Reservoir channel A1 contains ethanol\n- Plate on the magnetic module contains pre-aliquoted magnetic beads and EB\n\n\n",
"internal": "796af8",
diff --git a/protoBuilds/797dee-normalization/README.json b/protoBuilds/797dee-normalization/README.json
index bbb4df87d8..4ddc8f288a 100644
--- a/protoBuilds/797dee-normalization/README.json
+++ b/protoBuilds/797dee-normalization/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -8,7 +8,7 @@
"description": "\nConcentration normalization is a key component of many genomic and proteomic applications, such as NGS library prep. With this protocol, you can easily normalize the concentrations of samples in a 96 or 384 microwell plate without worrying about missing a well or adding the wrong volume. Just upload your properly formatted CSV file (keep scrolling for an example), customize your parameters, and download your ready-to-run protocol.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Single-Channel Pipette and corresponding Tips\nSamples in a compatible plate (96-well or 384-well)\nOpentrons 4-in-1 Tube Rack Set\nDiluent\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nCSV Format\nYour file must be saved as a comma separated value (.csv) file type. Your CSV must contain values corresponding to volumes in microliters (\u03bcL). It should be formatted in \u201clandscape\u201d orientation, with the value corresponding to well A1 in the upper left-hand corner of the value list.\n\nIn this example, 40\u03bcL will be added to A1, 41\u03bcL will be added to well B1, and so on.\nIf you\u2019d like to follow our template, you can make a copy of this spreadsheet, fill out your values, and export as CSV from there.\nNote about CSV: All values corresponding to wells in the CSV must have a value (zero (0) is a valid value and nothing will be transferred to the corresponding well(s)). Additionally, the CSV can be formatted in \"portrait\" orientation. In portrait orientation, the bottom left corner is treated as A1 and the top right corner would correspond to the furthest well from A1 (H12 in a 96-well plate).\nUsing the customization fields below, set up your protocol.\n Volumes CSV: Upload the CSV (.csv) containing your diluent volumes.\n Pipette Model: Select which pipette you will use for this protocol.\n Pipette Mount: Specify which mount your single-channel pipette is on (left or right)\n Plate Type: Select which (destination) plate you will use for this protocol.\n Volume in 50ml: What is the starting volume of in your 50mL tube of diluent?\n Filter Tips: Specify whether you want to use filter tips.\n* Tip Usage Strategy: Specify whether you'd like to use a new tip for each transfer, or keep the same tip throughout the protocol.",
"internal": "normalization",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"description": "\n\nConcentration normalization is a key component of many genomic and proteomic applications, such as NGS library prep. With this protocol, you can easily normalize the concentrations of samples in a 96 or 384 microwell plate without worrying about missing a well or adding the wrong volume. Just upload your properly formatted CSV file (keep scrolling for an example), customize your parameters, and download your ready-to-run protocol.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes) and corresponding [Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Samples in a compatible plate (96-well or 384-well)](https://labware.opentrons.com/?category=wellPlate)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* Diluent\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n---\n\n\n**CSV Format**\n\nYour file must be saved as a comma separated value (.csv) file type. Your CSV must contain values corresponding to volumes in microliters (\u03bcL). It should be formatted in \u201clandscape\u201d orientation, with the value corresponding to well A1 in the upper left-hand corner of the value list.\n\n\n\nIn this example, 40\u03bcL will be added to A1, 41\u03bcL will be added to well B1, and so on.\n\nIf you\u2019d like to follow our template, you can make a copy of [this spreadsheet](https://opentrons-protocol-library-website.s3.amazonaws.com/Technical+Notes/797dee-normalization/Opentrons+Normalization+Template.xlsx), fill out your values, and export as CSV from there.\n\n*Note about CSV*: All values corresponding to wells in the CSV must have a value (zero (0) is a valid value and nothing will be transferred to the corresponding well(s)). Additionally, the CSV can be formatted in \"portrait\" orientation. In portrait orientation, the bottom left corner is treated as A1 and the top right corner would correspond to the furthest well from A1 (H12 in a 96-well plate).\n\nUsing the customization fields below, set up your protocol.\n* Volumes CSV: Upload the CSV (.csv) containing your diluent volumes.\n* Pipette Model: Select which pipette you will use for this protocol.\n* Pipette Mount: Specify which mount your single-channel pipette is on (left or right)\n* Plate Type: Select which (destination) plate you will use for this protocol.\n* Volume in 50ml: What is the starting volume of in your 50mL tube of diluent?\n* Filter Tips: Specify whether you want to use filter tips.\n* Tip Usage Strategy: Specify whether you'd like to use a new tip for each transfer, or keep the same tip throughout the protocol.\n\n\n\n",
"internal": "normalization\n",
diff --git a/protoBuilds/79e9a1-cleanup/README.json b/protoBuilds/79e9a1-cleanup/README.json
index 02293e29fa..f2c0ae6881 100644
--- a/protoBuilds/79e9a1-cleanup/README.json
+++ b/protoBuilds/79e9a1-cleanup/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Cleanup": [],
"NGS Library Prep": []
@@ -8,7 +8,7 @@
"internal": "0b83b7",
"labware": "\nNEST 0.2 mL 96-Well PCR Plate, Full Skirt\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "\n[Opentrons](https://opentrons.com/)\n\n",
+ "author": "\n[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "\n- NGS Library Prep\n - Cleanup\n\n",
"description": "\nThis protocol is equipped to perform any of 4x cleanups for the Paragon NGS prep protocol.\n\n",
"internal": "\n0b83b7\n",
diff --git a/protoBuilds/7a1ae8/README.json b/protoBuilds/7a1ae8/README.json
index 044a70050d..f50dafb80b 100644
--- a/protoBuilds/7a1ae8/README.json
+++ b/protoBuilds/7a1ae8/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Zymo Kit"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom nucleic acid purification using the Zymobiomics Magbead kit. This protocol was updated from this to APIv2.\n\n\n\nNEST Deep Well Plate or similar\nOpentrons P300 Single-channel electronic pipette\nOpentrons P300 Multi-channel electronic pipette\nOpentrons 300ul tipracks\n\n\n\nSTARTING SETUP for 4x6 2ml screwcap tube aluminum block (slot 2):\n* tubes A1-D6: original samples (up to 24 samples aligned down columns and then across rows)\nMID-PROTOCOL SETUP for 4x6 2ml screwcap tube aluminum block (slot 2):\n tubes A1-C1: molecular grade water tubes (1 tube for each 8 samples); note-- the user is prompted to replace these tubes midway through the protocol*\n12-channel reagent reservoir (slot 4):\n channel 1: mag binding buffer\n channel 2: magnetic beads\n channel 3: magwash 1\n channels 4-5: magwash 2 (1 channel each for 2x washes)\nwaste reservoir (slot 11):\n Note-- the reservoir is programmed as an Agilent 290ml single-channel reservoir, but the user can replace this with any reservoir of choice for containing the liquid waste throughout the protocol by calibrating to the top of the reservoir before the run.*",
"internal": "7a1ae8",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Zymo Kit\n\n",
"description": "This protocol performs a custom nucleic acid purification using the Zymobiomics Magbead kit. This protocol was updated from [this](https://protocol-delivery.protocols.opentrons.com/protocol/0bdecb) to APIv2.\n\n---\n\n\n* [NEST Deep Well Plate](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate) or similar\n* [Opentrons P300 Single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette?variant=5984549109789)\n* [Opentrons P300 Multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 300ul tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n---\n\n\n**STARTING SETUP** for 4x6 2ml screwcap tube aluminum block (slot 2):\n* tubes A1-D6: original samples (up to 24 samples aligned down columns and then across rows)\n\n**MID-PROTOCOL SETUP** for 4x6 2ml screwcap tube aluminum block (slot 2):\n* tubes A1-C1: molecular grade water tubes (1 tube for each 8 samples); *note-- the user is prompted to replace these tubes midway through the protocol*\n\n12-channel reagent reservoir (slot 4):\n* channel 1: mag binding buffer\n* channel 2: magnetic beads\n* channel 3: magwash 1\n* channels 4-5: magwash 2 (1 channel each for 2x washes)\n\nwaste reservoir (slot 11):\n* *Note-- the reservoir is programmed as an Agilent 290ml single-channel reservoir, but the user can replace this with any reservoir of choice for containing the liquid waste throughout the protocol by calibrating to the top of the reservoir before the run.*\n\n",
"internal": "7a1ae8\n",
diff --git a/protoBuilds/7a3e4b/README.json b/protoBuilds/7a3e4b/README.json
index 687b76b631..58dea87506 100644
--- a/protoBuilds/7a3e4b/README.json
+++ b/protoBuilds/7a3e4b/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Featured": [
"Cherrypicking"
@@ -10,7 +10,7 @@
"internal": "7a3e4b",
"labware": "\nStarlab Deepwell Plate\nEppendorf 2mL Tube in Tube Rack\nEppendorf 1.5mL Tube in Tube Rack\nNEST 96-Well, 200\u00b5L Flat\nNEST 96-Well, 100\u00b5L PCR\nBioRad 96-Well, 200\u00b5L PCR\nCorning 96-Well, 360\u00b5L Flat\nCorning 384-Well, 112\u00b5L Flat\nUSA Scientific 96-Deepwell, 2.4mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Featured\n\t* Cherrypicking\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "This protocol utilizes a user-specified CSV to transfer sample using a variety of labware. With this protocol you have the ability to choose between the following:\n\n* Starlab Deepwell Plate\n* Eppendorf 2mL Tube in Tube Rack\n* Eppendorf 1.5mL Tube in Tube Rack\n* NEST 96-Well, 200\u00b5L Flat\n* NEST 96-Well, 100\u00b5L PCR\n* BioRad 96-Well, 200\u00b5L PCR\n* Corning 96-Well, 360\u00b5L Flat\n* Corning 384-Well, 112\u00b5L Flat\n* USA Scientific 96-Deepwell, 2.4mL\n\nWith the destination labware providing the same options as source labware.\n\nExplanation of complex parameters below:\n* `Volumes CSV`: Here, you should upload a .csv file formatted in the following way, being sure to include the header line:\n\n`Source Well` | `Volume` | `Destination Well`\n\n**Note about CSV**: The first column is dedicated to the header of the CSV; thus, the first row of data should occupy cells 'A2', 'B2', and 'C2' in the document.\n\n* `Pipette Model`: Select which pipette you will use for this protocol.\n* `Pipette Mount`: Specify which mount your single-channel pipette is on (left or right).\n* `Source Labware Type`: Select which (destination) labware you will use for this protocol.\n* `Destination Labware Type`: Select which (destination) labware you will use for this protocol.\n* `Tip Use Strategy`: Select whether you would like to use the same tip or a new tip for each transfer.\n\n---\n\n",
diff --git a/protoBuilds/7aa3fd-library-pooling/README.json b/protoBuilds/7aa3fd-library-pooling/README.json
index 790388479d..7f6a332cb2 100644
--- a/protoBuilds/7aa3fd-library-pooling/README.json
+++ b/protoBuilds/7aa3fd-library-pooling/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -8,7 +8,7 @@
"description": "This protocol performs the final library pooling step of NGS library prep. Sample volumes should be specified in a .csv file formatted as followed (including headers line). Transfers will be executed in the order specified in the .csv:\nsource well of elution plate (slot 9),transfer volume(uL)\nA1,4\nA2,3\nA3,5.3\nA4,6.4\n...\n\n\n\nEppendorf twin.tec 96-well PCR plate 150ul #0030128605\nOpentrons temperature module with 4x6-well aluminum block insert\nOpentrons 4-in-1 tuberack with 4x6 insert for 2ml Eppendorf snapcap tubes or equivalent.\nOpentrons 10ul and 200ul filter tipracks\nOpentrons GEN1 P10 and P300 single-channel electronic pipettes\n\n\n\n4x6 tuberack with 2ml Eppendorf snapcap tube (slot 2)\n* tube A1: water\n4x6 aluminum block insert with 2ml Eppendorf snapcap tube (on temperature module, slot 4):\n* tube A1: pool tube (loaded empty)",
"internal": "7aa3fd",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Custom\n\n",
"description": "This protocol performs the final library pooling step of NGS library prep. Sample volumes should be specified in a `.csv` file formatted as followed (**including headers line**). Transfers will be executed in the order specified in the `.csv`:\n\n```\nsource well of elution plate (slot 9),transfer volume(uL)\nA1,4\nA2,3\nA3,5.3\nA4,6.4\n...\n```\n\n---\n\n\n* [Eppendorf twin.tec 96-well PCR plate 150ul #0030128605](https://online-shop.eppendorf.com/OC-en/Laboratory-Consumables-44512/Plates-44516/Eppendorf-twin.tec-PCR-Plates-PF-8180.html)\n* [Opentrons temperature module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [4x6-well aluminum block insert](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set)\n* [Opentrons 4-in-1 tuberack](https://shop.opentrons.com/products/tube-rack-set-1?_ga=2.256255875.900706806.1575911292-1245111371.1550251253) with 4x6 insert for [2ml Eppendorf snapcap tubes](https://online-shop.eppendorf.us/US-en/Laboratory-Consumables-44512/Tubes-44515/Eppendorf-Safe-Lock-Tubes-PF-8863.html) or equivalent.\n* [Opentrons 10ul and 200ul filter tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons GEN1 P10 and P300 single-channel electronic pipettes](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n\n---\n\n\n4x6 tuberack with 2ml Eppendorf snapcap tube (slot 2)\n* tube A1: water\n\n4x6 aluminum block insert with 2ml Eppendorf snapcap tube (on temperature module, slot 4):\n* tube A1: pool tube (loaded empty)\n\n",
"internal": "7aa3fd\n",
diff --git a/protoBuilds/7aa3fd-library-quantification/README.json b/protoBuilds/7aa3fd-library-quantification/README.json
index 46e41fb9a5..4069d61fd6 100644
--- a/protoBuilds/7aa3fd-library-quantification/README.json
+++ b/protoBuilds/7aa3fd-library-quantification/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -8,7 +8,7 @@
"description": "This protocol performs the QPCR plate set-up protocol for library quantification step of NGS library prep.\n\n\n\nEppendorf twin.tec 96-well PCR plate 150ul #0030128605\nOpentrons temperature module with 96-well aluminum block insert\nOpentrons 4-in-1 tuberack with 4x6 insert for 2ml Eppendorf snapcap tubes or equivalent.\nOpentrons 10ul and 200ul filter tipracks\nOpentrons GEN1 P10 and P300 single-channel electronic pipettes\n\n\n\n4x6 tuberack with 2ml Eppendorf snapcap tubes (slot 2)\n tubes A1-A5: water\n tube B1: QPCR mastermix",
"internal": "7aa3fd",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Custom\n\n",
"description": "This protocol performs the QPCR plate set-up protocol for library quantification step of NGS library prep.\n\n---\n\n\n* [Eppendorf twin.tec 96-well PCR plate 150ul #0030128605](https://online-shop.eppendorf.com/OC-en/Laboratory-Consumables-44512/Plates-44516/Eppendorf-twin.tec-PCR-Plates-PF-8180.html)\n* [Opentrons temperature module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [96-well aluminum block insert](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set)\n* [Opentrons 4-in-1 tuberack](https://shop.opentrons.com/products/tube-rack-set-1?_ga=2.256255875.900706806.1575911292-1245111371.1550251253) with 4x6 insert for [2ml Eppendorf snapcap tubes](https://online-shop.eppendorf.us/US-en/Laboratory-Consumables-44512/Tubes-44515/Eppendorf-Safe-Lock-Tubes-PF-8863.html) or equivalent.\n* [Opentrons 10ul and 200ul filter tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons GEN1 P10 and P300 single-channel electronic pipettes](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n\n---\n\n\n4x6 tuberack with 2ml Eppendorf snapcap tubes (slot 2)\n* tubes A1-A5: water\n* tube B1: QPCR mastermix\n\n",
"internal": "7aa3fd\n",
diff --git a/protoBuilds/7aa3fd-size-selection/README.json b/protoBuilds/7aa3fd-size-selection/README.json
index fbf78b1b82..ce3f646672 100644
--- a/protoBuilds/7aa3fd-size-selection/README.json
+++ b/protoBuilds/7aa3fd-size-selection/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Custom"
@@ -8,7 +8,7 @@
"description": "This protocol performs the final size selection step of NGS library prep.\n\n\n\nEppendorf twin.tec 96-well PCR plate 150ul #0030128605\nOpentrons magnetic module\nOpentrons 4-in-1 tuberack with 4x6 insert for 2ml Eppendorf snapcap tubes or equivalent.\nOpentrons 10ul and 200ul filter tipracks\nOpentrons GEN1 P10 and P300 single-channel electronic pipettes\n\n\n\n4x6 tuberack with 2ml Eppendorf snapcap tubes (slot 2)\n tube A1: SPRI beads\n tube B1-B3: 80% ethanol\n* tube C1: Low EDTA TE",
"internal": "7aa3fd",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Custom\n\n",
"description": "This protocol performs the final size selection step of NGS library prep.\n\n---\n\n\n* [Eppendorf twin.tec 96-well PCR plate 150ul #0030128605](https://online-shop.eppendorf.com/OC-en/Laboratory-Consumables-44512/Plates-44516/Eppendorf-twin.tec-PCR-Plates-PF-8180.html)\n* [Opentrons magnetic module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons 4-in-1 tuberack](https://shop.opentrons.com/products/tube-rack-set-1?_ga=2.256255875.900706806.1575911292-1245111371.1550251253) with 4x6 insert for [2ml Eppendorf snapcap tubes](https://online-shop.eppendorf.us/US-en/Laboratory-Consumables-44512/Tubes-44515/Eppendorf-Safe-Lock-Tubes-PF-8863.html) or equivalent.\n* [Opentrons 10ul and 200ul filter tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons GEN1 P10 and P300 single-channel electronic pipettes](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n\n---\n\n\n4x6 tuberack with 2ml Eppendorf snapcap tubes (slot 2)\n* tube A1: SPRI beads\n* tube B1-B3: 80% ethanol\n* tube C1: Low EDTA TE\n\n",
"internal": "7aa3fd\n",
diff --git a/protoBuilds/7aad4e/README.json b/protoBuilds/7aad4e/README.json
index 3dae3c1802..471da62f16 100644
--- a/protoBuilds/7aad4e/README.json
+++ b/protoBuilds/7aad4e/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherry Picking"
@@ -10,7 +10,7 @@
"internal": "7aad4e",
"labware": "\nCorning 96 well plate, flat\nPerkin Elmer 110ul 384 well plate, flat\nOpentrons 200ul Filter tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherry Picking\n\n",
"deck-setup": "Note: the protocol will read the csv source and destination slots to load labware onto the deck. Plates should always be placed in order of smaller deck number to large (i.e. place 384 plates in slots 1, 2, 3, and 4 if running 4 plates. If running one 96 plate, place it in slot 7 as opposed to 8).\n\n\n---\n\n",
"description": "This protocol preps up to two 96 well plates with cell culture media from up to 6 source 384 well plates. The protocol will automatically parse through the csv to determine how many plates are on the deck, giving the user the ability to have flexibility in run size, however plates should still be placed in order of the deck slot numbers. Culture is pre-mixed with an airgap before transfer.\n\n\nExplanation of complex parameters below:\n* `.CSV File`: Here, you should upload a .csv file formatted in the following way, being sure to include the header line (do NOT have \"0\" in between well names i.e. \"A01\", it should be \"A1\")\n\n* `P300 Mount`: Specify which mount (left or right) to host the P300 pipette.\n\n---\n\n\n",
diff --git a/protoBuilds/7ada78-pt2/README.json b/protoBuilds/7ada78-pt2/README.json
index 1d893cae45..3d166b54d3 100644
--- a/protoBuilds/7ada78-pt2/README.json
+++ b/protoBuilds/7ada78-pt2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"Complete PCR Workflow"
@@ -10,7 +10,7 @@
"internal": "7ada78-pt2",
"labware": "\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOpentrons 4-in-1 tube rack with 1.5mL Eppendoft snap cap tubes\nOpentrons 20ul Filter tips\n96-W Abgene Plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* Complete PCR Workflow\n\n",
"deck-setup": "\n\n",
"description": "This protocol preps and runs a PCR on the Opentrons thermocycler using a .csv input. RNA and NF water are added to the PCR plate, and mastermix is then added to the resulting solution and mixed. A pause is included after the RNA is added to the water in the thermocycler plate, and the user is prompted to replace the water plate with the same 96 well plate, but with mastermix in column 1. If the protocol runs out of tips, it will automatically stop and the user is prompted to replace tips. For csv information and formatting notes, see below.\n\n\nExplanation of complex parameters below:\n* `Number of columns`: Specify the number of columns in the thermocycler plate to dispense mastermix into. Mastermix will always come from column 1 of the 96 well plate on slot 3. The mastermix plate will replace with water plate on slot 3 after an automatic pause in the protocol.\n* `.CSV File`: Here, you should upload a .csv file formatted in the following way, being sure to include the header line\n(use slot 7 for the thermocycler slot, and input an \"x\" for values that are not needed in that row):\n\n* `Pipette Mount`: Specify which mount (left or right) to host the P20 Single, and Multi-channel pipettes, respectively.\n\n---\n\n",
diff --git a/protoBuilds/7ada78/README.json b/protoBuilds/7ada78/README.json
index 4cb3030cc6..bff5e4cb66 100644
--- a/protoBuilds/7ada78/README.json
+++ b/protoBuilds/7ada78/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"Complete PCR Workflow"
@@ -10,7 +10,7 @@
"internal": "7ada78-pt2",
"labware": "\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOpentrons 4-in-1 tube rack with 1.5mL Eppendorf snap cap tubes\nOpentrons 20ul Filter tips\n96-W OptiPlate\n96-W Kingfisher plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* Complete PCR Workflow\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This protocol reformats RNA into a 96-well plate per a .csv input. Up to 96 tubes can be reformatted, and fresh tips are granted between each tube. Volume, source slot, source well, destination slot and destination well are all information that is taken from the csv to perform the protocol. Note: the csv format is NOT the same for [Part 2](https://protocols.opentrons.com/protocol/7ada78-pt2). For csv information and formatting notes, see below.\n\n\nExplanation of complex parameters below:\n* `.CSV File`: Here, you should upload a .csv file formatted in the following way, being sure to include the header line (use slot 7 for the thermocycler slot, and input an \"x\" for values that are not needed in that row):\n\n\nIf using a kingfisher plate, the multi-channel pipette will be used. For destination well, include only wells in the first row (A1, A2, A3,.., A12). The protocol will then proceed to perform full column transfers of the volume specified, for all columns specified (feel free to skip columns).\n* `Pipette Mount`: Specify which mount (left or right) to host the P20 single and multi-channel pipettes, respectively.\n\n---\n\n",
diff --git a/protoBuilds/7cfbde/README.json b/protoBuilds/7cfbde/README.json
index 402c4af8fc..522534b0be 100644
--- a/protoBuilds/7cfbde/README.json
+++ b/protoBuilds/7cfbde/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"RNA Extraction"
@@ -8,7 +8,7 @@
"description": "This protocol automates RNA extraction according to the 3D Black Bio RNA Extraction Kit.\n\nUsing the P300 Multi-Channel Pipette, this protocol can accommodate up to 96 samples per run and can be configured to run any multiple of 8 up to 96. Starting with at least 200\u00b5L of sample in a standard 96-well plate, the entire process is automated and ends with 60\u00b5L of elution containing RNA dispensed into a 96-well PCR plate. The elution is then ready for RT/qPCR, Next-Gen Sequencing, hybridization, etc.\n\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 4.0.0 or later)\nOpentrons Magnetic Module, GEN2\nOpentrons P300 Multi-Channel Pipette\nOpentrons 200\u00b5L Filter Tips (recommended) or Opentrons 300\u00b5L Tips\nNEST 96-Deep Well Plate, 2mL\nNEST 96-Well PCR Plate\nNEST 12-Well Reservoir\nNEST 1-Well Reservoir\nOpentrons 96-Well Aluminum Block\nPCR Tubes\n3D Black Bio Extraction Kit\nSamples\n\n\n\nReagent Preparation\nPrepare all reagents according to manual.\n\nProteinase K: At least 10\u00b5L (12\u00b5L is recommended) per column of samples should fill a PCR Strip.\nThe PCR strip should be placed in column 1 of the Opentrons 96-Well Aluminum Block\n\nMagBeads: At least 20\u00b5L (22\u00b5L is recommended) per column of samples should fill 1 (or 2, if processing more than 48 samples) PCR strip(s).\nThe PCR strip(s) should be placed in column(s) 3 (and 4, if processing more than 48 samples) of the Opentrons 96-Well Aluminum Block\n\nLysis-Binding Mix: In slots 1-6 of the NEST 12-Well Reservoir.\nFor every column (8 samples), 7mL of Lysis-Binding Mix should be added to the reservoir. Each slot of the 12 well reservoir can accommodate volume for 2 columns/16 samples and should be loaded sequentially (ex. if running 24 samples, slot 1 would get 14mL lysis-binding mix, slot 2 would get 7mL lysis-binding mix, slot 3 would be empty, etc).\n\nWash 1: In slots 7-12 of the NEST 12-Well Reservoir.\nFor every column (8 samples), 6.5mL of wash 1 should be added to the reservoir. Each slot of the 12 well reservoir can accommodate volume for 2 columns/16 samples and should be loaded sequentially (ex. if running 24 samples, slot 7 would get 13mL wash 1, slot 8 would get 6.5mL wash 1, slot 9 would be empty, etc).\n\nWash 2: In NEST 1-Well Reservoir.\nFor every column (8 samples), 14mL of wash 2 should be added to the reservoir.\n\nElution Buffer: At least 60\u00b5L (65\u00b5L is recommended) per column of samples should fill a PCR Strip. Each PCR strip can accommodate up to two columns worth (16) samples.\nThe PCR strips should be loaded in columns 7-12 of the Opentrons 96-Well Aluminum Block\n\n\nDeck Layout\n\nSlot 1: NEST 96-Well PCR Plate, containing samples\nSlot 2: NEST 1-Well Reservoir containing Wash 2\nSlot 3: Opentrons 200\u00b5L Filter Tips\nSlot 4: Opentrons 96-Well Aluminum Block containing Proteinase K, Magnetic Beads, and Elution Buffer\nSlot 5: NEST 12-Well Reservoir containing Lysis-Binding Mix and Wash 1\nSlot 6: Opentrons 200\u00b5L Filter Tips\nSlot 7: Opentrons Magnetic Module, GEN2 with NEST 96-Deep Well Plate, 2mL\nSlot 8: Opentrons 200\u00b5L Filter Tips\nSlot 9: Opentrons 200\u00b5L Filter Tips\nSlot 10: NEST 1-Well Reservoir, empty for liquid waste\nSlot 11: NEST 1-Well Reservoir, empty for liquid waste\n\nNote about tips: The tips in slot 8 will be used for removing supernatant and used tips will be returned to empty tip racks to prevent overflow of the waste bin.\n\nUsing the customizations field (below), set up your protocol.\n P300-Multi Mount: Select which mount the P300-Multi is attached to\n Number of Samples: Specify the number of samples to run",
"internal": "7cfbde",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* RNA Extraction\n\n\n",
"description": "This protocol automates RNA extraction according to the 3D Black Bio RNA Extraction Kit.\n\nUsing the P300 Multi-Channel Pipette, this protocol can accommodate up to 96 samples per run and can be configured to run any multiple of 8 up to 96. Starting with at least 200\u00b5L of sample in a standard 96-well plate, the entire process is automated and ends with 60\u00b5L of elution containing RNA dispensed into a 96-well PCR plate. The elution is then ready for RT/qPCR, Next-Gen Sequencing, hybridization, etc.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 4.0.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips) (recommended) or [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [NEST 96-Deep Well Plate, 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 1-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* [Opentrons 96-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set)\n* PCR Tubes\n* 3D Black Bio Extraction Kit\n* Samples\n\n\n---\n\n\n**Reagent Preparation**\nPrepare all reagents according to manual.\n\n**Proteinase K**: At least 10\u00b5L (12\u00b5L is recommended) per column of samples should fill a PCR Strip.\nThe PCR strip should be placed in column 1 of the [Opentrons 96-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set)\n\n**MagBeads**: At least 20\u00b5L (22\u00b5L is recommended) per column of samples should fill 1 (or 2, if processing more than 48 samples) PCR strip(s).\nThe PCR strip(s) should be placed in column(s) 3 (and 4, if processing more than 48 samples) of the [Opentrons 96-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set)\n\n**Lysis-Binding Mix**: In slots 1-6 of the [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml).\nFor every column (8 samples), 7mL of Lysis-Binding Mix should be added to the reservoir. Each slot of the 12 well reservoir can accommodate volume for 2 columns/16 samples and should be loaded sequentially (ex. if running 24 samples, slot 1 would get 14mL lysis-binding mix, slot 2 would get 7mL lysis-binding mix, slot 3 would be empty, etc).\n\n**Wash 1**: In slots 7-12 of the [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml).\nFor every column (8 samples), 6.5mL of wash 1 should be added to the reservoir. Each slot of the 12 well reservoir can accommodate volume for 2 columns/16 samples and should be loaded sequentially (ex. if running 24 samples, slot 7 would get 13mL wash 1, slot 8 would get 6.5mL wash 1, slot 9 would be empty, etc).\n\n**Wash 2**: In [NEST 1-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml).\nFor every column (8 samples), 14mL of wash 2 should be added to the reservoir.\n\n**Elution Buffer**: At least 60\u00b5L (65\u00b5L is recommended) per column of samples should fill a PCR Strip. Each PCR strip can accommodate up to two columns worth (16) samples.\nThe PCR strips should be loaded in columns 7-12 of the [Opentrons 96-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set)\n\n\n**Deck Layout**\n\n**Slot 1:** [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt), containing samples\n**Slot 2:** [NEST 1-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml) containing **Wash 2**\n**Slot 3:** [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 4:** [Opentrons 96-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) containing **Proteinase K**, **Magnetic Beads**, and **Elution Buffer**\n**Slot 5:** [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) containing **Lysis-Binding Mix** and **Wash 1**\n**Slot 6:** [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 7:** [Opentrons Magnetic Module, GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [NEST 96-Deep Well Plate, 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate)\n**Slot 8:** [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 9:** [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 10:** [NEST 1-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml), empty for liquid waste\n**Slot 11:** [NEST 1-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml), empty for liquid waste\n\n*Note about tips*: The tips in slot 8 will be used for removing supernatant and used tips will be returned to empty tip racks to prevent overflow of the waste bin.\n\n**Using the customizations field (below), set up your protocol.**\n* **P300-Multi Mount**: Select which mount the P300-Multi is attached to\n* **Number of Samples**: Specify the number of samples to run\n\n\n\n",
"internal": "7cfbde\n",
diff --git a/protoBuilds/7e3b31/README.json b/protoBuilds/7e3b31/README.json
index 1ce01f927d..339de1dfda 100644
--- a/protoBuilds/7e3b31/README.json
+++ b/protoBuilds/7e3b31/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina TruSeq Stranded mRNA Sample Prep"
@@ -9,7 +9,7 @@
"description": "With this 3 part workflow, your OT-2 can use Illumina TruSeq Stranded mRNA Sampe Preparation Guide to convert 8-48 input total RNA samples into libraries having known strand origin to be used for cluster generation and DNA sequencing. This is Part 1 of 3: This OT-2 protocol uses the LT kit and follows the LS protocol found on pages 12-44 of the attached sample prep guide. Oligo-dT magnetic beads are used to isolate poly(A) RNA from input total RNA, followed by fragmentation, priming, and cDNA synthesis. The second-strand synthesis includes dUTP in order to quench the amplification of the second strand.",
"internal": "7e3b31",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Illumina TruSeq Stranded mRNA Sample Prep\n\n",
"deck-setup": "\n\n\n\n\n\n* Opentrons 20ul filter tips (deck slot 3)\n* Opentrons 200ul filter tips (deck slots 6, 9)\n* Opentrons Thermocycler Module (deck slots 7, 8, 10, 11)\n* Opentrons Magnetic Module (deck slot 1)\n* Sample Plate nest_96_wellplate_100ul_pcr_full_skirt (deck slot 4)\n* Reagents opentrons_96_aluminumblock_generic_pcr_strip_200ul (deck slot 5)\n* Reservoir nest_12_reservoir_15ml (deck slot 2)\n\n",
"description": "\nWith this 3 part workflow, your OT-2 can use [Illumina TruSeq Stranded mRNA Sampe Preparation Guide](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/7e3b31/stranded-mrnaTruseq.pdf) to convert 8-48 input total RNA samples into libraries having known strand origin to be used for cluster generation and DNA sequencing. This is Part 1 of 3: This OT-2 protocol uses the LT kit and follows the LS protocol found on pages 12-44 of the attached sample prep guide. Oligo-dT magnetic beads are used to isolate poly(A) RNA from input total RNA, followed by fragmentation, priming, and cDNA synthesis. The second-strand synthesis includes dUTP in order to quench the amplification of the second strand.\n\n",
diff --git a/protoBuilds/7e4c3e-1/README.json b/protoBuilds/7e4c3e-1/README.json
index eb475292f1..24e78d85ff 100644
--- a/protoBuilds/7e4c3e-1/README.json
+++ b/protoBuilds/7e4c3e-1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "Automates first JSON portion of protocol 7e4c3e\n\n\n\nOpentrons P300-multi channel electronic pipettes (GEN2)\nOpentrons 200ul Filter Tips\nMicroAmp Optical 384-Well Reaction Plate with Barcode\nBiorad hard shell 96 well PCR plates low-profile thin-wall skirted\nOpentrons Aluminum Block Set\n",
"internal": "7e4c3e-1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "Automates first JSON portion of protocol [7e4c3e](https://docs.google.com/document/d/1odr2TfSuiJhDRY0qyfBs5fz-swU-nJTulUdvRRDV4ec/edit?ts=5fbbeb03)\n\n---\n\n\n* [Opentrons P300-multi channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [MicroAmp Optical 384-Well Reaction Plate with Barcode](https://www.thermofisher.com/order/catalog/product/4343814#/4343814)\n* [Biorad hard shell 96 well PCR plates low-profile thin-wall skirted](https://www.bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [Opentrons Aluminum Block Set](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n\n",
"internal": "7e4c3e-1\n",
diff --git a/protoBuilds/7e4c3e-2/README.json b/protoBuilds/7e4c3e-2/README.json
index ffeda5996e..03bab1efa6 100644
--- a/protoBuilds/7e4c3e-2/README.json
+++ b/protoBuilds/7e4c3e-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "Automates second JSON portion of protocol 7e4c3e\n\n\n\nOpentrons P300-multi channel electronic pipettes (GEN2)\nOpentrons 200ul Filter Tips\nBiorad hard shell 96 well PCR plates low-profile thin-wall skirted\nOpentrons Aluminum Block Set\n",
"internal": "7e4c3e-2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "Automates second JSON portion of protocol [7e4c3e](https://docs.google.com/document/d/1odr2TfSuiJhDRY0qyfBs5fz-swU-nJTulUdvRRDV4ec/edit?ts=5fbbeb03)\n\n---\n\n\n* [Opentrons P300-multi channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Biorad hard shell 96 well PCR plates low-profile thin-wall skirted](https://www.bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [Opentrons Aluminum Block Set](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n\n",
"internal": "7e4c3e-2\n",
diff --git a/protoBuilds/7e4c3e-3/README.json b/protoBuilds/7e4c3e-3/README.json
index 042e5c6897..f8c00e8989 100644
--- a/protoBuilds/7e4c3e-3/README.json
+++ b/protoBuilds/7e4c3e-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "Automates third JSON portion of protocol 7e4c3e\n\n\n\nOpentrons P300-multi channel electronic pipettes (GEN2)\nOpentrons 200ul Filter Tips\nBiorad hard shell 96 well PCR plates low-profile thin-wall skirted\nOpentrons Aluminum Block Set\n",
"internal": "7e4c3e-3",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "Automates third JSON portion of protocol [7e4c3e](https://docs.google.com/document/d/1odr2TfSuiJhDRY0qyfBs5fz-swU-nJTulUdvRRDV4ec/edit?ts=5fbbeb03)\n\n---\n\n\n* [Opentrons P300-multi channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Biorad hard shell 96 well PCR plates low-profile thin-wall skirted](https://www.bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [Opentrons Aluminum Block Set](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n\n",
"internal": "7e4c3e-3\n",
diff --git a/protoBuilds/7e4c3e-4/README.json b/protoBuilds/7e4c3e-4/README.json
index 0ec58badd3..5551fba4f8 100644
--- a/protoBuilds/7e4c3e-4/README.json
+++ b/protoBuilds/7e4c3e-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "Automates fourth JSON portion of protocol 7e4c3e\n\n\n\nOpentrons P300-multi channel electronic pipettes (GEN2)\nOpentrons 200ul Filter Tips\nNEST 96 Deepwell Plate 2mL\nOpentrons P1000-single channel electronic pipette (GEN2)\nOpentrons 1000ul Filter Tips\nOpentrons 4-in-1 Tube Rack Set\nNEST 15 mL Centrifuge Tube\nNEST 50 mL Centrifuge Tube\n",
"internal": "7e4c3e-4",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "Automates fourth JSON portion of protocol [7e4c3e](https://docs.google.com/document/d/1odr2TfSuiJhDRY0qyfBs5fz-swU-nJTulUdvRRDV4ec/edit?ts=5fbbeb03)\n\n---\n\n\n* [Opentrons P300-multi channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [NEST 96 Deepwell Plate 2mL](http://www.cell-nest.com/page94?product_id=101&_l=en)\n* [Opentrons P1000-single channel electronic pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=5984549142557)\n* [Opentrons 1000ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [NEST 15 mL Centrifuge Tube](https://shop.opentrons.com/collections/tubes/products/nest-15-ml-centrifuge-tube)\n* [NEST 50 mL Centrifuge Tube](https://shop.opentrons.com/collections/tubes/products/nest-50-ml-centrifuge-tube)\n\n",
"internal": "7e4c3e-4\n",
diff --git a/protoBuilds/7e4c3e-5/README.json b/protoBuilds/7e4c3e-5/README.json
index 27e741f230..6381ecfaa0 100644
--- a/protoBuilds/7e4c3e-5/README.json
+++ b/protoBuilds/7e4c3e-5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "Automates fifth JSON portion of protocol 7e4c3e\n\n\n\nOpentrons P300-multi channel electronic pipette (GEN2)\nOpentrons 200ul Filter Tips\nOpentrons P1000-single channel electronic pipette (GEN2)\nOpentrons 1000ul Filter Tips\nOpentrons 4-in-1 Tube Rack Set\nBiorad hard shell 96 well PCR plates low-profile thin-wall skirted\nOpentrons Aluminum Block Set\nGeneric 0.2 ml 12-Tube PCR Strips\n",
"internal": "7e4c3e-5",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "Automates fifth JSON portion of protocol [7e4c3e](https://docs.google.com/document/d/1odr2TfSuiJhDRY0qyfBs5fz-swU-nJTulUdvRRDV4ec/edit?ts=5fbbeb03)\n\n---\n\n\n* [Opentrons P300-multi channel electronic pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons P1000-single channel electronic pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=5984549142557)\n* [Opentrons 1000ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [Biorad hard shell 96 well PCR plates low-profile thin-wall skirted](https://www.bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [Opentrons Aluminum Block Set](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n* Generic 0.2 ml 12-Tube PCR Strips\n\n\n",
"internal": "7e4c3e-5\n",
diff --git a/protoBuilds/7e4c3e-6/README.json b/protoBuilds/7e4c3e-6/README.json
index c8b97c378d..1cb7dc9ab1 100644
--- a/protoBuilds/7e4c3e-6/README.json
+++ b/protoBuilds/7e4c3e-6/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "Automates sixth JSON portion of protocol 7e4c3e\n\n\n\nOpentrons P300-multi channel electronic pipettes (GEN2)\nOpentrons 200ul Filter Tips\nOpentrons P1000-single channel electronic pipette (GEN2)\nOpentrons 1000ul Filter Tips\nOpentrons 4-in-1 Tube Rack Set\nOpentrons Aluminum Block Set\nMicroAmp Optical 384-Well Reaction Plate with Barcode\nGeneric 0.2 ml 12-Tube PCR Strips\n",
"internal": "7e4c3e-6",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"description": "Automates sixth JSON portion of protocol [7e4c3e](https://docs.google.com/document/d/1odr2TfSuiJhDRY0qyfBs5fz-swU-nJTulUdvRRDV4ec/edit?ts=5fbbeb03)\n\n---\n\n\n* [Opentrons P300-multi channel electronic pipettes (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette?variant=5984202489885)\n* [Opentrons 200ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons P1000-single channel electronic pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=5984549142557)\n* [Opentrons 1000ul Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-filter-tips)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [Opentrons Aluminum Block Set](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set)\n* [MicroAmp Optical 384-Well Reaction Plate with Barcode](https://www.thermofisher.com/order/catalog/product/4343814#/4343814)\n* Generic 0.2 ml 12-Tube PCR Strips\n\n\n",
"internal": "7e4c3e-6\n",
diff --git a/protoBuilds/7f0f89/README.json b/protoBuilds/7f0f89/README.json
index 333762a8ca..3ebd8e4143 100644
--- a/protoBuilds/7f0f89/README.json
+++ b/protoBuilds/7f0f89/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -8,7 +8,7 @@
"description": "This protocol transfers a specified volume of developer solution into reservoir cartridges within a custom 216-well plate. The developer solution is aspirated from a Nest 195mL reservoir, then distributed to the 216-well plate down by column. Transfers are distributed in chunks of 3 between the reservoir and plate, with a delay and blow out step for viscous liquid considerations. Extra developer solution is aspirated to ensure that each cartridge receives adequate distribution.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons P300 Single-Channel Pipette\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 300\u00b5L Tips\nNest 1-Well Reservoir 195 mL\nInvoy 216-Well Cartridge Plate\nInvoy Canisters with Fibrous Reservoir\n\n\n\nSlots 1, 2, 4, 5, 7, 8, 10, 11: Invoy 216-Well Cartridge Plate\nSlot 6: Opentrons 96 Tip Rack 300 \u00b5L\nSlot 9: Nest 1-Well Reservoir 195 mL\n\n\nUsing the customizations field (below), set up your protocol.\n Number of Samples: Specify the number of samples in your protocol run.\n Dispense Height Above Cartridge: Specify the height (mm) above the cartridge the pipette will dispense liquid\n Dispense Volume: Specify the volume (\u00b5L) of developer solution dispensed into each cartridge\n Dispense Flow Rate: Specify the rate (\u00b5L/sec) at which the pipette will aspirate solution into each cartridge\n Dispense Flow Rate: Specify the rate (\u00b5L/sec) at which the pipette will dispense solution into each cartridge\n Aspirate Delay Time: Specify the time to delay after each aspiration (in seconds)\n Dispense Delay Time: Specify the time to delay after each dispense (in seconds)\n P300 Single GEN2 Mount: Specify the mount side for the P300 Single GEN2 pipette",
"internal": "7f0f89",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"description": "This protocol transfers a specified volume of developer solution into reservoir cartridges within a custom 216-well plate. The developer solution is aspirated from a Nest 195mL reservoir, then distributed to the 216-well plate down by column. Transfers are distributed in chunks of 3 between the reservoir and plate, with a delay and blow out step for viscous liquid considerations. Extra developer solution is aspirated to ensure that each cartridge receives adequate distribution.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://labware.opentrons.com/opentrons_96_tiprack_300ul?category=tipRack)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Nest 1-Well Reservoir 195 mL](https://shop.opentrons.com/collections/reservoirs/products/nest-1-well-reservoir-195-ml)\n* Invoy 216-Well Cartridge Plate\n* Invoy Canisters with Fibrous Reservoir\n\n\n\n---\n\n\nSlots 1, 2, 4, 5, 7, 8, 10, 11: Invoy 216-Well Cartridge Plate\n\nSlot 6: Opentrons 96 Tip Rack 300 \u00b5L\n\nSlot 9: Nest 1-Well Reservoir 195 mL\n\n\n\n\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Samples**: Specify the number of samples in your protocol run.\n* **Dispense Height Above Cartridge**: Specify the height (mm) above the cartridge the pipette will dispense liquid\n* **Dispense Volume**: Specify the volume (\u00b5L) of developer solution dispensed into each cartridge\n* **Dispense Flow Rate**: Specify the rate (\u00b5L/sec) at which the pipette will aspirate solution into each cartridge\n* **Dispense Flow Rate**: Specify the rate (\u00b5L/sec) at which the pipette will dispense solution into each cartridge\n* **Aspirate Delay Time**: Specify the time to delay after each aspiration (in seconds)\n* **Dispense Delay Time**: Specify the time to delay after each dispense (in seconds)\n* **P300 Single GEN2 Mount**: Specify the mount side for the P300 Single GEN2 pipette\n\n",
"internal": "7f0f89\n",
diff --git a/protoBuilds/7f9595/README.json b/protoBuilds/7f9595/README.json
index a8e7603803..6a267395ff 100644
--- a/protoBuilds/7f9595/README.json
+++ b/protoBuilds/7f9595/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Serial Dilution"
@@ -10,7 +10,7 @@
"internal": "7f9595",
"labware": "\nCorning 96 Well Plate 360 \u00b5L Flat\nNEST 12-Well Reservoirs, 15 mL\nOpentrons 300uL Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Serial Dilution\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This protocol automates the serial dilution of two separate sample columns and also adds PBS buffer to the necessary wells. It will perform additional mixing steps before and after the addition of reagents. Each transfer performs a blow out in the destination well.\n\nSerial Dilution Plate Map:\n\n\nExplanation of parameters below:\n* `P300 Multichannel GEN2 Mount`: Specify which mount (left or right) to load the P300 multi channel pipette.\n* `Reservoir Type`: Specify the type of reservoir being used in Slot 1.\n* `Blowout Height from Well Bottom (mm)`: The height the blowout should occur from the bottom of the well. For example, a volume of 200 uL in this plate will have a height of approximately 5.4 mm from the bottom of the well.\n* `Mix Aspiration Flow Rate (uL/s)`: Flow rate when aspiration occurs in a mixing step.\n* `Mix Dispensing Flow Rate (uL/s)`: Flow rate when dispensing occurs in a mixing step.\n\n---\n\n",
diff --git a/protoBuilds/7fc7b1/README.json b/protoBuilds/7fc7b1/README.json
index 2c5ae47e23..193043839a 100644
--- a/protoBuilds/7fc7b1/README.json
+++ b/protoBuilds/7fc7b1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Featured": [
"Cherrypicking"
@@ -10,7 +10,7 @@
"internal": "7fc7b1",
"labware": "\nGreiner Bio-One 96-Well Plates 200\u00b5l #652290\nSarstedt 384-Well Plates 40\u00b5l #72.1984.202\nNest 12-Well Reservoir 15mL\nOpentrons 20\u00b5l tipracks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Featured\n\t* Cherrypicking\n\n",
"deck-setup": "Example Deck Layout:\n\n\n---\n\n",
"description": "With this protocol, your robot can perform a custom PCR preparation using a P20-multi pipette.\n\nThis protocol allows you to choose which slots and plate (96 or 384 well) will be destination plates, and also gives the option to choose between a reservoir and 96 well plate as source plates for mastermix.\n\nAll available empty slots will be filled with tipracks, and the user will be prompted to refill the tipracks if all are emptied in the middle of the protocol.\n\n\nExplanation of complex parameters below:\n* `.csv file`: Upload a .csv file formatted in the [following way](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/7fc7b1/7fc7b1_csv_template.csv), being sure to include the header line.\n* `Mastermix Labware`: Specify whether you are using a 12-channel NEST 15mL reservoir or Greiner Bio-One 96-Well Plates 200\u00b5l well plate as the mastermix source. If you choose a 12-channel reservoir for mastermix, the mastermix should be filled in the first channel of the reservoir alone. If you choose a 96-well plate for mastermix, the mastermix should be filled in the first columns in as many columns as necessary. The protocol will automatically calculate when the second column should be acccessed for mastermix once the first column has run out of volume, then the third column, etc.\n* `Mastermix Plate Slot`: Specify which slot the mastermix will be on the deck.\n* `Mastermix Volume Loaded in Source`: Specify the volume of mastermix per column in ul.\n* `Mastermix Starting Column`: Specify the column in which populated mastermix columns start.\n* `DNA Volume`: Specify the DNA volume (in ul) to which mastermix will be added.\n* `Transfer Scheme`: Specify whether the transfer scheme will be multi-dispense or single dispense (distribution vs 1-1 transfer).\n* `P20-multi GEN2 mount`: Specify which mount (left or right) the P20 Multi-GEN2 mount will be.\n\n\n\n\n\n\n\n---\n",
diff --git a/protoBuilds/85b0dc/README.json b/protoBuilds/85b0dc/README.json
index 75c64fba35..de4bea4ccc 100644
--- a/protoBuilds/85b0dc/README.json
+++ b/protoBuilds/85b0dc/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"Syber Green Prep"
@@ -10,7 +10,7 @@
"internal": "85b0dc",
"labware": "\nThermoFisher MicroAmp\u2122 Fast Optical 96-Well Reaction Plate with Barcode, 0.1 mL #4346906\nOpentrons 20uL Filter Tips\nOpentrons 4-in-1 tuberack with 4x6 insert for 1.5ml Eppendorf tubes\nOpentrons 96-well aluminum block\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* Syber Green Prep\n\n",
"deck-setup": "\n* green: starting DNA samples\n* blue: possible positions for mastermixes \n\n\n---\n\n",
"description": "\nThis protocol performs a custom Syber Green PCR Prep for up to 96 samples. Mastermix is transferred from 1.5ml Eppendorf tubes, and samples are transferred from source strip tubes mounted in the Opentrons 96-well aluminum block into the MicroAmp PCR plate also mounted in the Opentrons 96-well aluminum block.\n\nThe input .csv file should be formatted in the following way, including the header line:\n\n```\nsource well (A1-H12),mastermix tube (A1-D6),destination well (A1-H12)\nA1,A1,A1\nA2,A2,A2\n...\n```\n\n---\n\n",
diff --git a/protoBuilds/8nhsa0-norm/README.json b/protoBuilds/8nhsa0-norm/README.json
index 373f62378b..c4571294e0 100644
--- a/protoBuilds/8nhsa0-norm/README.json
+++ b/protoBuilds/8nhsa0-norm/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom sample normalization from a source PCR plate to a second PCR plate, diluting with water from a reservoir. Sample and diluent volumes are specified via .csv file in the following format, including the header line (empty lines ignored):\nsource plate well,destination plate well,volume sample (\u00b5l),volume diluent (\u00b5l)\nA1, A1, 2, 28\nYou can download an example .csv to edit directly here.\n\n\n\nBio-Rad 96 Well Plate 200 \u00b5L PCR\nNEST 1-channel reservoir 195ml\nOpentrons 10\u00b5l and 300\u00b5l tipracks\nOpentrons P10 and P300 GEN1 pipettes\n\n\n\nNEST 1-channel reservoir (slot 1)\n* channel 1: water for dilution",
"internal": "8nhsa0-norm",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"description": "This protocol performs a custom sample normalization from a source PCR plate to a second PCR plate, diluting with water from a reservoir. Sample and diluent volumes are specified via .csv file in the following format, including the header line (empty lines ignored):\n\n```\nsource plate well,destination plate well,volume sample (\u00b5l),volume diluent (\u00b5l)\nA1, A1, 2, 28\n```\n\nYou can download an example .csv to edit directly [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/ml-normalization/example_csv.csv).\n\n---\n\n\n* [Bio-Rad 96 Well Plate 200 \u00b5L PCR](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate)\n* [NEST 1-channel reservoir 195ml](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [Opentrons 10\u00b5l and 300\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons P10 and P300 GEN1 pipettes](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n\n---\n\n\nNEST 1-channel reservoir (slot 1)\n* channel 1: water for dilution\n\n",
"internal": "8nhsa0-norm\n",
diff --git a/protoBuilds/8nhsa0/README.json b/protoBuilds/8nhsa0/README.json
index 8d3bede65f..9db86c3c7c 100644
--- a/protoBuilds/8nhsa0/README.json
+++ b/protoBuilds/8nhsa0/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -8,7 +8,7 @@
"description": "This protocol performs cherrypicking from x source plates to y destination plates, both Bio-Rad 96-well low profile PCR plates (200\u00b5l). The user can specify the sources and destinations in any slot, and the protocol will automatically load the plate in that slot. Tipracks will automatically be loaded in the remaining available slots. In the case that all tips are exhausted, the protocol pauses, and the user is prompted to refill the racks before resuming.\nThe CSV should be formatted like so:\nsource slot, source well, destination slot, destination well, volume\n1,A1,3,A1,5\n...\nThe header line should be included (the first line from the .csv file is ignored)\n\n\n\nBio-Rad 96-well low profile PCR plates (200\u00b5l)\nOpentrons P50-single electronic pipette\nOpentrons 300\u00b5l tipracks\n",
"internal": "8nhsa0",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n\n",
"description": "This protocol performs cherrypicking from x source plates to y destination plates, both Bio-Rad 96-well low profile PCR plates (200\u00b5l). The user can specify the sources and destinations in any slot, and the protocol will automatically load the plate in that slot. Tipracks will automatically be loaded in the remaining available slots. In the case that all tips are exhausted, the protocol pauses, and the user is prompted to refill the racks before resuming.\n\nThe CSV should be formatted like so:\n\n```\nsource slot, source well, destination slot, destination well, volume\n1,A1,3,A1,5\n...\n```\n\n**The header line should be included (the first line from the .csv file is ignored)**\n\n---\n\n\n* [Bio-Rad 96-well low profile PCR plates (200\u00b5l)](https://www.bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [Opentrons P50-single electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 300\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n",
"internal": "8nhsa0\n",
diff --git a/protoBuilds/9038af/README.json b/protoBuilds/9038af/README.json
index 0bc28c3f34..3d9018641c 100644
--- a/protoBuilds/9038af/README.json
+++ b/protoBuilds/9038af/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Library Prep": [
"Sequencing"
@@ -10,7 +10,7 @@
"internal": "9038af",
"labware": "\nOpentrons 96 Well Aluminum Block with Bio-Rad Well Plate 200 \u00b5L\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\nOpentrons 24 Well Aluminum Block with NEST 1.5 mL Screwcap\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Library Prep\n\t* Sequencing\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol automates steps 3-10 of the Oxford Nanopore Rapid Barcoding Kit [SQK-RBK110.96](file:///Users/parrishpayne/Downloads/rapid-barcoding-kit-96-sqk-rbk110-96-RBK_9126_v110_revO_24Mar2021-gridion%20(3).pdf). Choose between 16-32 samples per run (> 24 samples requires two 4-in-1 tube racks)\n\n\n",
diff --git a/protoBuilds/925d07-cp/README.json b/protoBuilds/925d07-cp/README.json
index 4793c38434..c361ddc65d 100644
--- a/protoBuilds/925d07-cp/README.json
+++ b/protoBuilds/925d07-cp/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Aliquoting"
@@ -9,7 +9,7 @@
"internal": "925d07",
"labware": "\nEppendorf V-bottom, Lo-Bind 96-well plates #0030603303\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Aliquoting\n\n",
"description": "\nLinks:\n* [Random Aliquoting](./925d07-cp)\n
\n
\n* [Plasmind Luciferase Assay](./925d07-pla)\n
\n
\n* [QIAcuity Plate Transfer](./925d07-q)\n
\n
\n* [PCR Prep](./925d07-v3)\n
\n
\n\nThis protocol performs a custom aliquoting procedure based on a randomly-generated 1-4 plate map. Enough volume to accommodate all 4 destination wells, including overage volume, is aspirated from the source well and distributed to all 4 destination wells. A new tip is obtained for each source-destination set. The layout is shown below: \n\n\n\nThe proper pipette (P20 or P300) is selected automatically based on the input siRNA volume.\n\n---\n\n",
"internal": "925d07\n",
diff --git a/protoBuilds/925d07-pla/README.json b/protoBuilds/925d07-pla/README.json
index 568a71b971..619d71235d 100644
--- a/protoBuilds/925d07-pla/README.json
+++ b/protoBuilds/925d07-pla/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "9250d7",
"labware": "\nCorning 96 Well Plate 360 ul Flat\nCustom 384 Well Plate 100 ul\nNEST 12-Well Reservoirs, 15 mL #999-00076\nOpentrons 20ul tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n* reservoir channel 1: lipofectamine 2000\n* reservoir channel 2: V141 plasmid\n* 96-wellplate: siRNA\n\n---\n\n",
"description": "\nLinks:\n* [Random Aliquoting](./925d07-cp)\n
\n
\n* [Plasmind Luciferase Assay](./925d07-pla)\n
\n
\n* [QIAcuity Plate Transfer](./925d07-q)\n
\n
\n* [PCR Prep](./925d07-v3)\n
\n
\n\n\nThis protocol performs a custom Plasmid Luciferase Assay to a 384-wellplate.\n\n---\n\n",
diff --git a/protoBuilds/925d07-q/README.json b/protoBuilds/925d07-q/README.json
index d8aaab8145..6075d829e9 100644
--- a/protoBuilds/925d07-q/README.json
+++ b/protoBuilds/925d07-q/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "9250d7",
"labware": "\nOpentrons 20ul tips\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nQIAcuity 96-well Nanoplate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Random Aliquoting](./925d07-cp)\n
\n
\n* [Plasmind Luciferase Assay](./925d07-pla)\n
\n
\n* [QIAcuity Plate Transfer](./925d07-q)\n
\n
\n* [PCR Prep](./925d07-v3)\n
\n
\n\nThis protocol performs a custom QIAcuity PCR prep.\n\n---\n\n",
diff --git a/protoBuilds/925d07-v3/README.json b/protoBuilds/925d07-v3/README.json
index b80cdb2624..329a058ed9 100644
--- a/protoBuilds/925d07-v3/README.json
+++ b/protoBuilds/925d07-v3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "9250d7",
"labware": "\nOpentrons 20ul tips\nCorning 96 Well Plate 360 ul Flat\nCustom 384 Well Plate 100 ul\nGeneric PCR Strips in 96-well aluminum block\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n---\n\n",
"description": "\nLinks:\n* [Random Aliquoting](./925d07-cp)\n
\n
\n* [Plasmind Luciferase Assay](./925d07-pla)\n
\n
\n* [QIAcuity Plate Transfer](./925d07-q)\n
\n
\n* [PCR Prep](./925d07-v3)\n
\n
\n\nThis protocol performs a custom PCR prep from 4 source 96-well RNA plate to a single 384-well destination plate. The transfer scheme is shown below.\n\n---\n\n",
diff --git a/protoBuilds/9778eb/README.json b/protoBuilds/9778eb/README.json
index 3dd7d71a92..56a349de51 100644
--- a/protoBuilds/9778eb/README.json
+++ b/protoBuilds/9778eb/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"DNA Extraction"
@@ -10,7 +10,7 @@
"internal": "9778eb",
"labware": "\nNEST 96 Wellplate 2mL\nNEST 12 Reservoir 15mL\nOpentrons 200uL Filter Tips\nThermofisher 96 Well, semi-skirted, black lettering mounted to deck with Opentrons 96 Aluminum block\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* DNA Extraction\n\n",
"deck-setup": "\n\n\n\n\n",
"description": "This is a custom implementation of the Mag-Bind Blood and Tissue kit. Specify the starting sample volume, final elution volume, and number of samples, allowing the OT-2 to automate the procedure with minimal intervention to empty trash and/or refill tip racks as alerted.\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n* `Initial Volume`: Specify starting volume of sample (ul).\n* `Elution Volume`: Specify elution volume (ul) into final plate.\n* `Flash on Robot Pause`: Specify whether the robot will flash on pause. This includes trash or tip rack notifications\n* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n\n---\n\n",
diff --git a/protoBuilds/9778eb_mass_norm/README.json b/protoBuilds/9778eb_mass_norm/README.json
index c710860a33..684d76e152 100644
--- a/protoBuilds/9778eb_mass_norm/README.json
+++ b/protoBuilds/9778eb_mass_norm/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "9778eb_mass_norm",
"labware": "\nOpentrons 6 Tube Rack with Falcon 50 mL Conical\nNEST 96 Deepwell Plate 2mL\nNEST 100uL 96 Well Plate\nThermofisher Semi-Skirted on Adapter 96 Well Plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"deck-setup": "* Reagent Color Code\n\n\n* Starting deck layout with reagents\n\n\n",
"description": "This protocol uses values in a CSV to normalize samples in a 96 well plate. The CSV requires starting and finishing concentrations broken up into mass/moles and volumes. This allows for minimal user calculations before loading samples. Additionally, units can be different from run to run and even sample to sample provided the starting and finishing units are the same. E.g. sample A1 is nanograms/uL and will be diluted to a set concentration in well A1 in the normalized plate while sample B1 is moles/uL and will be diluted to a set molarity in well B1.\n\nThis protocol also checks if normalization is possible with the given concentrations and will list the failed wells at the end of the protocol run. E.g. C1 starting concentration is 15 ng/uL and the stated final concentration target is 20 ng/uL so C1 will be added to the 'failed well' list at the protocol end.\n\nExplanation of complex parameters below:\n* `Volume of Water in Falcon Tube`: Starting volume of nuclease free water in the on-deck tube rack. This is in mL and will be used to track liquid height\n* `Source Plate Type`: 96 well plate samples will start the protocol in\n* `Destination Plate Type`: 96 well plate samples will end the protocol in\n* `P20 Single GEN2 Mount`: Defines which side the P20 single pipette will be mounted on. The P300 single pipette will be mounted on the opposite side\n* `Transfer .csv File`: Here, you should upload a .csv file formatted [like the example CSV located here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/9778eb/csv/example_norm.csv)\n---\n\n",
diff --git a/protoBuilds/9778eb_spri/README.json b/protoBuilds/9778eb_spri/README.json
index f625766e93..49c267580c 100644
--- a/protoBuilds/9778eb_spri/README.json
+++ b/protoBuilds/9778eb_spri/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"SPRI Beads"
@@ -10,7 +10,7 @@
"internal": "9778eb_spri",
"labware": "\nNEST 12 Well Reservoir 15 mL\nNEST 1 Well Reservoir 195 mL\nCustom 3D printed custom adapter for half-skirted plates with below half-skirted plate\nThermofisher PCR Plate, 96-well, semi-skirted, flat deck, black lettering\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* SPRI Beads\n\n",
"deck-setup": "* Reagent Color Code\n\n\n* Starting deck layout with reagents for single bead purification\n\n\n* Slot 1: 96 Filter Tip Rack 20 uL\n* Slot 2: 96 Filter Tip Rack 200 uL\n* Slot 3: Custom 3D printed custom adapter for half-skirted plates with Thermofisher half-skirted plate\n* Slot 4: NEST 1 Well Reservoir 195 mL\n* Slot 5: NEST 12 Well Reservoir 15 mL\n* Slot 6: Empty\n* Slot 7: Magnetic module with custom 3D printed custom adapter for half-skirted plates with Thermofisher half-skirted plate\n* Slot 8: 96 Filter Tip Rack 20 uL\n* Slot 9: 96 Filter Tip Rack 200 uL\n* Slot 10: 96 Filter Tip Rack 20 uL\n* Slot 11: 96 Filter Tip Rack 200 uL\n\n* Starting deck layout with reagents for size selection\n\n\n* Slot 1: 96 Filter Tip Rack 20 uL\n* Slot 2: 96 Filter Tip Rack 200 uL\n* Slot 3: Custom 3D printed custom adapter for half-skirted plates with Thermofisher half-skirted plate\n* Slot 4: NEST 1 Well Reservoir 195 mL\n* Slot 5: NEST 12 Well Reservoir 15 mL\n* Slot 6: Magnetic module with custom 3D printed custom adapter for half-skirted plates with Thermofisher half-skirted plate\n* Slot 7: Magnetic module with custom 3D printed custom adapter for half-skirted plates with Thermofisher half-skirted plate\n* Slot 8: 96 Filter Tip Rack 20 uL\n* Slot 9: 96 Filter Tip Rack 200 uL\n* Slot 10: 96 Filter Tip Rack 20 uL\n* Slot 11: 96 Filter Tip Rack 200 uL\n\n\n---\n\n",
"description": "This protocol purifies samples using SPRI beads with an option to perform a size selection with two separate bead ratios. Size selection requires two magnetic modules.\n\nBeads are added at a specified ratio to the samples, mixed, and left to incubate. If size selection is to be performed, the supernatant is then removed to the second magnetic module for a second bead purification where the resulting supernatant is removed to the liquid waste.\n\nAfter bead purification(s), two 200uL ethanol washes are performed. The beads are left to air dry for a set amount of time before a set volume of elution liquid is added, mixed with the beads, left to incubate for a set time, and separated from the beads. This supernatant is removed to an awaiting plate in slot 3.\n\nExplanation of complex parameters below:\n* `Number of Samples`: How many samples are in the starting 96 well plate, 1-96 allowed. Multiples of 8 for full columns will have the most efficient reagent use\n* `Initial Sample Volume`: Volume in uL for starting samples. This is used to help calculate bead volume addition\n* `Bead Ratio 1`: Bead to sample ratio used to calculate bead volume addition. First addition for size selection and only addition for non-size selection\n* `Bead Ratio 2`: Bead to sample ratio used to calculate bead volume addition for second bead addition during size selection. This is not used during non-size selection protocols with a single bead addition\n* `Transfer Volume`: During size selection, how much volume to transfer from first magnetic module plate to second. This is needed to properly calculate second bead addition volume for proper size selection\n* `Bead Air Dry Time`: How many minutes beads will be left to air dry after ethanol wash steps. Default is 10 minutes\n* `Bead Incubation Time`: How many minutes beads will be left to incubate with samples during first and second addition. Default is 5 minutes\n* `Bead Separation Time`: How many minutes beads will be left to separate on the magnetic module. Default is 5 minutes\n* `Elution Solution Volume`: How much liquid will be added to washed and dried beads for elution. Default volume is 50 uL\n* `Elution Time`: How many minutes the elution solution will incubate with the beads. Default is 10 minutes\n* `Final Plate Volume`: How much liquid will be transferred from the beads mixed with elution liquid to final elution plate in slot 3. Default is 50 uL\n* `Flash Robot on Pause`: Set whether the OT-2 will flash when trash is full, tips need refilling, or general user intervention is needed. Default is flashing.\n* `P300 Multi Channel Pipette Mount`: Select which mount side the p300 multichannel pipette is connected to. Default is left. The p20 multichannel will be set to the opposite side.\n* `Size Selection or Single Purification`: Select whether the OT-2 will perform a single bead purification with one magnetic module or a dual bead ratio-based size selection with two magnetic modules. Default is size selection.\n\n---\n\n",
diff --git a/protoBuilds/979d28-normalization/README.json b/protoBuilds/979d28-normalization/README.json
index 3099cc816c..aa1276f364 100644
--- a/protoBuilds/979d28-normalization/README.json
+++ b/protoBuilds/979d28-normalization/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -10,7 +10,7 @@
"internal": "979d28",
"labware": "\nNEST 12 Well Reservoir 15 mL\nVWR\u00ae PCR Plates, 96-Well 200ul #83007-374 seated in Opentrons 96-well aluminum block\nEppendorf 1.5 mL Safe-Lock Snapcap seated in Opentrons 4-in-1 Tube Rack\nOpentrons 20\u00b5L Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n * Normalization\n\n",
"deck-setup": "\n\n",
"description": "\nLinks:\n
\n[RNA Isolation](./979d28)\n
\n[Normalization](./979d28-normalization)\n
\n[qPCR Prep](./979d28-pcr)\n\nThis protocol performs a custom sample normalization and mastermix addition. You can find a template for file input [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/979d28-normalization/ex.csv).\n\n---\n\n",
diff --git a/protoBuilds/979d28-pcr/README.json b/protoBuilds/979d28-pcr/README.json
index 991f5181f8..c4ed402570 100644
--- a/protoBuilds/979d28-pcr/README.json
+++ b/protoBuilds/979d28-pcr/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "979d28",
"labware": "\nNEST 12 Well Reservoir 15 mL\nAxygen\u00ae 384-well Polypropylene PCR Microplate, Compatible with ABI, Full Skirt, Clear, Nonsterile #PCR-384M2-C\nVWR\u00ae PCR Plates, 96-Well 200ul #83007-374 seated in Opentrons 96-well aluminum block\nOpentrons 200\u00b5L Filter Tips\nOpentrons 20\u00b5L Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * PCR Prep\n\n",
"deck-setup": "\n\n",
"description": "\nLinks:\n
\n[RNA Isolation](./979d28)\n
\n[Normalization](./979d28-normalization)\n
\n[qPCR Prep](./979d28-pcr)\n\nThis protocol performs a custom cDNA dilution and 384-well PCR prep in triplicate.\n\n---\n\n",
diff --git a/protoBuilds/979d28/README.json b/protoBuilds/979d28/README.json
index e3e2bb7739..e938be8506 100644
--- a/protoBuilds/979d28/README.json
+++ b/protoBuilds/979d28/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Omega Mag-Bind\u00ae Total RNA Isolation"
@@ -10,7 +10,7 @@
"internal": "979d28",
"labware": "\nNEST 12 Well Reservoir 15 mL\nNEST 1 Well Reservoir 195 mL\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt\nNEST 96 Deepwell Plate 2mL\nVWR\u00ae PCR Plates, 96-Well 200ul #83007-374 seated in Opentrons 96-well aluminum block\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Omega Mag-Bind\u00ae Total RNA Isolation\n\n",
"deck-setup": "\n\n",
"description": "\nLinks:\n
\n[RNA Isolation](./979d28)\n
\n[Normalization](./979d28-normalization)\n
\n[qPCR Prep](./979d28-pcr)\n\nThis protocol performs the [Omega Mag-Bind\u00ae Total RNA 96 Kit](https://www.omegabiotek.com/product/total-cellular-rna-mag-bind-total-rna-96//?utm_source=google&utm_medium=ppc&utm_campaign=128157293756&utm_term=total%20rna%20extraction%20kit&utm_content=b&gclid=Cj0KCQjw-daUBhCIARIsALbkjSb5Gx-Bz7K1R_Dw_ePhpzhX8NNtwe_9GP1LXR7SfOaeOHWgEnehFE4aAtRVEALw_wcB) protocol.\n\nSamples should be loaded on the magnetic module in a NEST deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\n\nThere are several user-intervention steps to move the deepwell isolation plate to and from the Magnetic Module and QInstruments Bioshake.\n\n---\n\n",
diff --git a/protoBuilds/9ot019/README.json b/protoBuilds/9ot019/README.json
index c008e6a5bf..bfb712a286 100644
--- a/protoBuilds/9ot019/README.json
+++ b/protoBuilds/9ot019/README.json
@@ -1,5 +1,5 @@
{
- "author": "Tim Dobbs",
+ "author": "Tim Dobbs\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"General Liquid Handling": [
"General Liquid Handling"
@@ -10,7 +10,7 @@
"internal": "9ot019",
"labware": "\nCryo 35 Tube Rack with Nunc 1.8 mL #BioArtBot\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons 6 Tube Rack with Falcon 50 mL Conical\nOpentrons 96 Tip Rack 1000 \u00b5L\n",
"markdown": {
- "author": "Tim Dobbs\n\n\n",
+ "author": "Tim Dobbs\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* General Liquid Handling\n\t* General Liquid Handling\n\n\n",
"deck-setup": "[deck](https://drive.google.com/open?id=1KDbr6pEu0CT6NpcVpTAkZRpqyCJ2hkdY)\n\n\n",
"description": "This is a flexible protocol for making glycerol stocks of E. coli cultures for long-term storage in a -80C freezer. The protocol expects that you've grown the strains in liquid culture to an acceptable optical density, and prepared a sterile solution of 50% glycerol, 50% deionized water.\n\nThe protocol loads 500ul of liquid culture and 500ul of glycerol into a cryo tube, which you should then cap, label, and store in your -80C freezer. By default, 3 replicates of each strain are made. The number of samples and the number of replicates can be changed and the protocol will automatically calculate the number of cryo tubes you need. The protocol will throw an error there are too many samples to fit on the deck.\n\nIn our experience, this protocol starts to become worth using if you have more than ~20 total samples. Less than that and it's faster to hand-pipette.\n\nWatchouts:\n- Make sure the de-cap all of your cryo tubes and the 50ml conical glycerol tube before running. If your liquid cultures are in tubes, de-cap those too.\n- The protocol comments will display the final platemap of the cryo tubes. Make sure to label your tubes before taking them out or closing the window.\n\n\n",
diff --git a/protoBuilds/ML-normalization-cdna-library/README.json b/protoBuilds/ML-normalization-cdna-library/README.json
index a7fa4a678f..2259de2fd5 100644
--- a/protoBuilds/ML-normalization-cdna-library/README.json
+++ b/protoBuilds/ML-normalization-cdna-library/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom sample normalization from a source PCR plate to a second PCR plate, diluting with water from a reservoir. Sample and diluent volumes are specified via .csv file in the following format, including the header line (empty lines ignored):\nsource plate well,destination plate well,volume sample (\u00b5l),volume diluent (\u00b5l)\nA1, A1, 2, 28\nYou can download an example .csv to edit directly here.\n\n\n\nEppendorf\u2122 96-Well twin.tec\u2122 PCR Plate 250\u00b5l\nNEST 12-channel reservoir 15ml or USA Scientific 12 Well Reservoir 22 mL\nOpentrons 20\u00b5l and 300\u00b5l tipracks\nOpentrons P20 and P300 GEN2 single-channel pipettes\n\n\n\nNEST 12-channel reservoir (slot 1)\n* channel 1: water for dilution",
"internal": "ML Normalization",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"description": "This protocol performs a custom sample normalization from a source PCR plate to a second PCR plate, diluting with water from a reservoir. Sample and diluent volumes are specified via .csv file in the following format, including the header line (empty lines ignored):\n\n```\nsource plate well,destination plate well,volume sample (\u00b5l),volume diluent (\u00b5l)\nA1, A1, 2, 28\n```\n\nYou can download an example .csv to edit directly [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/ml-normalization/example_csv.csv).\n\n---\n\n\n* [Eppendorf\u2122 96-Well twin.tec\u2122 PCR Plate 250\u00b5l](https://www.fishersci.com/shop/products/eppendorf-96-well-twin-tec-pcr-plates-21/e951020389)\n* [NEST 12-channel reservoir 15ml](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) or [USA Scientific 12 Well Reservoir 22 mL](https://www.usascientific.com/12-channel-automation-reservoir/p/1061-8150)\n* [Opentrons 20\u00b5l and 300\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons P20 and P300 GEN2 single-channel pipettes](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n\n---\n\n\nNEST 12-channel reservoir (slot 1)\n* channel 1: water for dilution\n\n",
"internal": "ML Normalization\n",
diff --git a/protoBuilds/ML-normalization/README.json b/protoBuilds/ML-normalization/README.json
index a7fa4a678f..2259de2fd5 100644
--- a/protoBuilds/ML-normalization/README.json
+++ b/protoBuilds/ML-normalization/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Normalization"
@@ -8,7 +8,7 @@
"description": "This protocol performs a custom sample normalization from a source PCR plate to a second PCR plate, diluting with water from a reservoir. Sample and diluent volumes are specified via .csv file in the following format, including the header line (empty lines ignored):\nsource plate well,destination plate well,volume sample (\u00b5l),volume diluent (\u00b5l)\nA1, A1, 2, 28\nYou can download an example .csv to edit directly here.\n\n\n\nEppendorf\u2122 96-Well twin.tec\u2122 PCR Plate 250\u00b5l\nNEST 12-channel reservoir 15ml or USA Scientific 12 Well Reservoir 22 mL\nOpentrons 20\u00b5l and 300\u00b5l tipracks\nOpentrons P20 and P300 GEN2 single-channel pipettes\n\n\n\nNEST 12-channel reservoir (slot 1)\n* channel 1: water for dilution",
"internal": "ML Normalization",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Normalization\n\n",
"description": "This protocol performs a custom sample normalization from a source PCR plate to a second PCR plate, diluting with water from a reservoir. Sample and diluent volumes are specified via .csv file in the following format, including the header line (empty lines ignored):\n\n```\nsource plate well,destination plate well,volume sample (\u00b5l),volume diluent (\u00b5l)\nA1, A1, 2, 28\n```\n\nYou can download an example .csv to edit directly [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/ml-normalization/example_csv.csv).\n\n---\n\n\n* [Eppendorf\u2122 96-Well twin.tec\u2122 PCR Plate 250\u00b5l](https://www.fishersci.com/shop/products/eppendorf-96-well-twin-tec-pcr-plates-21/e951020389)\n* [NEST 12-channel reservoir 15ml](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) or [USA Scientific 12 Well Reservoir 22 mL](https://www.usascientific.com/12-channel-automation-reservoir/p/1061-8150)\n* [Opentrons 20\u00b5l and 300\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons P20 and P300 GEN2 single-channel pipettes](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n\n---\n\n\nNEST 12-channel reservoir (slot 1)\n* channel 1: water for dilution\n\n",
"internal": "ML Normalization\n",
diff --git a/protoBuilds/Opentrons_Logo/README.json b/protoBuilds/Opentrons_Logo/README.json
index 88d613ad06..287771bfa1 100644
--- a/protoBuilds/Opentrons_Logo/README.json
+++ b/protoBuilds/Opentrons_Logo/README.json
@@ -8,7 +8,7 @@
"description": "This is a demo protocol that will help you to get more familiar with your new OT-2! All you need is some food dye, a 96-well plate, and a 12-row trough or tube rack with 1.5mL or 2mL tubes. Your robot will pipette the Opentrons logo into your plate and you'll be ready to go!\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Single-Channel Pipette and corresponding Tips\n96-Well Microplate\n12-Row Trough or Tube Rack with 1.5mL/2mL Tubes\nWater and Food Dye (Two Colors)\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nFor this demo protocol, you need a clean, empty 96-well plate (where the Opentrons logo will be created) and either a 12-row trough or 1.5mL/2mL tubes in a tube rack to store the water and food dye solution.\nIf using a 12-row trough, the two food dye solutions should be stored in column 1 and column 2. If using the tube rack, dye 1 should be stored in 'A1' & 'B1' and dye 2 should be stored in 'C1' & 'D1'.\nUsing the customization fields below, set up your protocol.\n Pipette Model: Select which pipette you will use for this protocol.\n Pipette Mount: Specify which mount your single-channel pipette is on (left or right)\n Destination Plate Type: Select which (destination) plate you will use for this protocol.\n Dye Labware Type: Select which (source) labware you will use for this protocol.",
"internal": "Demo Protocol 1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/opentrons_logo). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/opentrons_logo). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Getting Started\n\t* Opentrons Logo\n\n",
"description": "This is a demo protocol that will help you to get more familiar with your new OT-2! All you need is some food dye, a 96-well plate, and a 12-row trough or tube rack with 1.5mL or 2mL tubes. Your robot will pipette the Opentrons logo into your plate and you'll be ready to go!\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes) and corresponding [Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [96-Well Microplate](https://labware.opentrons.com/?category=wellPlate)\n* [12-Row Trough](https://labware.opentrons.com/?category=reservoir) or [Tube Rack with 1.5mL/2mL Tubes](https://labware.opentrons.com/?category=tubeRack)\n* Water and Food Dye (Two Colors)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n---\n\n\nFor this demo protocol, you need a clean, empty 96-well plate (where the Opentrons logo will be created) and either a 12-row trough or 1.5mL/2mL tubes in a tube rack to store the water and food dye solution.\n\nIf using a 12-row trough, the two food dye solutions should be stored in column 1 and column 2. If using the tube rack, dye 1 should be stored in 'A1' & 'B1' and dye 2 should be stored in 'C1' & 'D1'.\n\nUsing the customization fields below, set up your protocol.\n* Pipette Model: Select which pipette you will use for this protocol.\n* Pipette Mount: Specify which mount your single-channel pipette is on (left or right)\n* Destination Plate Type: Select which (destination) plate you will use for this protocol.\n* Dye Labware Type: Select which (source) labware you will use for this protocol.\n\n\n",
"internal": "Demo Protocol 1\n",
diff --git a/protoBuilds/arcis-multi-test/README.json b/protoBuilds/arcis-multi-test/README.json
index 12f3f3988f..1442b4d8d0 100644
--- a/protoBuilds/arcis-multi-test/README.json
+++ b/protoBuilds/arcis-multi-test/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Arcis Blood Kit"
@@ -8,7 +8,7 @@
"description": "Note: This is an updated version (apiV1 to apiV2) of this protocol\nThis protocol is designed for running tests with the Arcis Blood Kit with Opentrons Multi-Channel Pipettes and a 12-channel trough. (further instructions about the protocol can be found here). With the Arcis Blood Kit, nucleic acid investigations can be performed easily and efficiently with the two included reagents. This protocol calls for the use of a P50-Multi and P300-Multi pipettes, as well as a 12-channel reservoir (USA Scientific), Corning 96-well, 360\u03bcL flat plate, and two Bioplastics 100\u03bcL plates.\n\n\n\nArcis Blood Kit\nOpentrons P50 Multi-Channel Pipette\nOpentrons P300 Multi-Channel Pipette\nOpentrons 50uL/300uL Tips\nUSA Scientific 12-Channel Reservoir\nCorning 96-Well Plate 360\u00b5L, Flat\nBioplastics 8-Tube Strip Mat, 100\u00b5L\nSamples\n\n\n\nSlot 1: Corning 360\u00b5L Plate, clean and empty\nSlot 2: Bioplastics Plate (in holder), clean and empty\nSlot 3: Bioplastics Plate (in holder), clean and empty\nSlot 4: 12-Channel Reservoir\n Channel 1: Reagent 1 (150\u00b5L per sample, 15mL for full plate)\n Channel 2: Reagent 2 (20\u00b5L per sample, 2mL for full plate)\n Channel 3: MasterMix (20\u00b5L per sample, 2mL for full plate)\n Channel 4: Sample (30\u00b5L per sample, 3mL for full plate)\nSlot 5: Opentrons Tips\nSlot 6: Opentrons Tips",
"internal": "arcis-test-multi",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Arcis Blood Kit\n\n\n",
"description": "**Note: This is an updated version (apiV1 to apiV2) of [this protocol](https://protocol-delivery.protocols.opentrons.com/protocol/arcis-test-multi)**\n\nThis protocol is designed for running tests with the Arcis Blood Kit with Opentrons Multi-Channel Pipettes and a 12-channel trough. (further instructions about the protocol can be found [here](https://arcisbio.com/wp-content/uploads/2019/09/Arcis-Blood-kit-Bulk-kit-UFL005-50rxn-IFU-Rev-6.12.2018.pdf)). With the Arcis Blood Kit, nucleic acid investigations can be performed easily and efficiently with the two included reagents. This protocol calls for the use of a P50-Multi and P300-Multi pipettes, as well as a 12-channel reservoir (USA Scientific), Corning 96-well, 360\u03bcL flat plate, and two Bioplastics 100\u03bcL plates.\n\n---\n\n\n* [Arcis Blood Kit](http://www.arcisbio.com/products/arcis-dna-blood-kit/)\n* [Opentrons P50 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons 50uL/300uL Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [USA Scientific 12-Channel Reservoir](https://labware.opentrons.com/usascientific_12_reservoir_22ml?category=reservoir)\n* [Corning 96-Well Plate 360\u00b5L, Flat](https://labware.opentrons.com/corning_96_wellplate_360ul_flat)\n* [Bioplastics 8-Tube Strip Mat, 100\u00b5L](https://bioplastics.com/productdetails.aspx?code=B59009-1)\n* Samples\n\n---\n\n\nSlot 1: Corning 360\u00b5L Plate, clean and empty\n\nSlot 2: Bioplastics Plate (in holder), clean and empty\n\nSlot 3: Bioplastics Plate (in holder), clean and empty\n\nSlot 4: 12-Channel Reservoir\n* Channel 1: Reagent 1 (150\u00b5L per sample, 15mL for full plate)\n* Channel 2: Reagent 2 (20\u00b5L per sample, 2mL for full plate)\n* Channel 3: MasterMix (20\u00b5L per sample, 2mL for full plate)\n* Channel 4: Sample (30\u00b5L per sample, 3mL for full plate)\n\nSlot 5: Opentrons Tips\n\nSlot 6: Opentrons Tips\n\n\n",
"internal": "arcis-test-multi\n",
diff --git a/protoBuilds/arcis-single-test/README.json b/protoBuilds/arcis-single-test/README.json
index 7aa854d9a9..161a65e8e3 100644
--- a/protoBuilds/arcis-single-test/README.json
+++ b/protoBuilds/arcis-single-test/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Arcis Blood Kit"
@@ -8,7 +8,7 @@
"description": "Note: This is an updated version (apiV1 to apiV2) of this protocol\nThis protocol is designed for running tests with the Arcis Blood Kit (further instructions about the protocol can be found here). With the Arcis Blood Kit, nucleic acid investigations can be performed easily and efficiently with the two included reagents. This protocol calls for the use of a P50-Single and P300-Single pipettes, as well as Opentrons Tube Racks, Corning 96-well, 360\u03bcL flat plate, and two Bioplastics 100\u03bcL plates.\n\n\n\nArcis Blood Kit\nOpentrons P50 Single Channel Pipette\nOpentrons P300 Single Channel Pipette\nOpentrons 50uL/300uL Tips\nOpentrons 4-in-1 Tube Rack Set\nCorning 96-Well Plate 360\u00b5L, Flat\nBioplastics 8-Tube Strip Mat, 100\u00b5L\n1.5mL Eppendorf Safe-Lock Tube\n15mL Conical Tube\n50mL Conical Tube\nSamples\n\n\n\nSlot 1: Opentrons Tiprack\nSlot 2: Corning 360\u00b5L Plate, clean and empty\nSlot 3: Bioplastics Plate (in holder), clean and empty\nSlot 4: Bioplastics Plate (in holder), clean and empty\nSlot 5: Opentrons 24 Tube Rack with 1.5mL Eppendorf Tubes\n A1: Sample (1000\u00b5L)\n A2: PCR Mastermix (1500\u00b5L)\nSlot 6: Opentrons 10 Tube Rack with Conical Tubes\n A1: Reagent 2 (2000\u00b5L)\n A3: Reagent 1 (10000\u00b5L)",
"internal": "arcis-single-test",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Arcis Blood Kit\n\n\n",
"description": "**Note: This is an updated version (apiV1 to apiV2) of [this protocol](https://protocol-delivery.protocols.opentrons.com/protocol/arcis-test)**\n\nThis protocol is designed for running tests with the Arcis Blood Kit (further instructions about the protocol can be found [here](http://www.arcisbio.com/wp-content/uploads/2019/04/Arcis-Blood-kit-Bulk-kit-UFL005-50rxn-IFU-Rev-6.12.2018.pdf)). With the Arcis Blood Kit, nucleic acid investigations can be performed easily and efficiently with the two included reagents. This protocol calls for the use of a P50-Single and P300-Single pipettes, as well as Opentrons Tube Racks, Corning 96-well, 360\u03bcL flat plate, and two Bioplastics 100\u03bcL plates.\n\n---\n\n\n* [Arcis Blood Kit](http://www.arcisbio.com/products/arcis-dna-blood-kit/)\n* [Opentrons P50 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [Opentrons P300 Single Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette)\n* [Opentrons 50uL/300uL Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [Corning 96-Well Plate 360\u00b5L, Flat](https://labware.opentrons.com/corning_96_wellplate_360ul_flat)\n* [Bioplastics 8-Tube Strip Mat, 100\u00b5L](https://bioplastics.com/productdetails.aspx?code=B59009-1)\n* [1.5mL Eppendorf Safe-Lock Tube](https://www.usascientific.com/1.5ml-eppendorf-safe-lock-tube.aspx)\n* 15mL Conical Tube\n* 50mL Conical Tube\n* Samples\n\n---\n\n\nSlot 1: Opentrons Tiprack\n\nSlot 2: Corning 360\u00b5L Plate, clean and empty\n\nSlot 3: Bioplastics Plate (in holder), clean and empty\n\nSlot 4: Bioplastics Plate (in holder), clean and empty\n\nSlot 5: Opentrons 24 Tube Rack with 1.5mL Eppendorf Tubes\n* A1: Sample (1000\u00b5L)\n* A2: PCR Mastermix (1500\u00b5L)\n\nSlot 6: Opentrons 10 Tube Rack with Conical Tubes\n* A1: Reagent 2 (2000\u00b5L)\n* A3: Reagent 1 (10000\u00b5L)\n\n\n",
"internal": "arcis-single-test\n",
diff --git a/protoBuilds/batch-test-plating/README.json b/protoBuilds/batch-test-plating/README.json
index 3faa070d8c..2fc32f3fc2 100644
--- a/protoBuilds/batch-test-plating/README.json
+++ b/protoBuilds/batch-test-plating/README.json
@@ -15,7 +15,7 @@
"internal": "batch-test-plating\n",
"labware": "* [nest_96_wellplate_100ul_pcr_full_skirt](https://shop.opentrons.com/nest-0-1-ml-96-well-pcr-plate-full-skirt/)\n* [opentrons_96_filtertiprack_20ul](https://shop.opentrons.com/opentrons-20ul-filter-tips/)\n* [opentrons_24_tuberack_generic_2ml_screwcap](https://shop.opentrons.com/4-in-1-tube-rack-set/)\n\n\n\n",
"modules": "* [Temperature Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n\n\n",
- "partner": "[Facundo Rodriguez Goren]\n\n",
+ "partner": "[Facundo Rodriguez Goren]\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [p20_single_gen2](https://shop.opentrons.com/single-channel-electronic-pipette-p20/)\n\n\n\n\n---\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n\n",
"protocol-steps": "1. Master Mix dispensing from Temperature Module to pcr plate\n2. Samples dispensing from Tuberack to pcr plate\n3. NTC dispensing from Tuberack to pcr plate\n\n",
@@ -24,7 +24,7 @@
"modules": [
"Temperature Module (GEN2)"
],
- "partner": "[Facundo Rodriguez Goren]",
+ "partner": "[Facundo Rodriguez Goren]\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\np20_single_gen2\n\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nMaster Mix dispensing from Temperature Module to pcr plate\nSamples dispensing from Tuberack to pcr plate\nNTC dispensing from Tuberack to pcr plate\n",
diff --git a/protoBuilds/bc-rnadvance-viral-feat/README.json b/protoBuilds/bc-rnadvance-viral-feat/README.json
index ab52fa500c..4a68560102 100644
--- a/protoBuilds/bc-rnadvance-viral-feat/README.json
+++ b/protoBuilds/bc-rnadvance-viral-feat/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Featured": [
"Beckman Coulter RNAdvance Viral"
@@ -10,7 +10,7 @@
"internal": "bc-rnadvance-viral",
"labware": "\nNEST 12 Well Reservoir 15 mL, 2x\nNEST 1 Well Reservoir 195 mL\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt\nNEST 96 Deepwell Plate 2mL\nOpentrons 96 Filter Tip Rack 200 \u00b5L, up to 10x depending on throughput\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Featured\n * Beckman Coulter RNAdvance Viral\n\n",
"deck-setup": "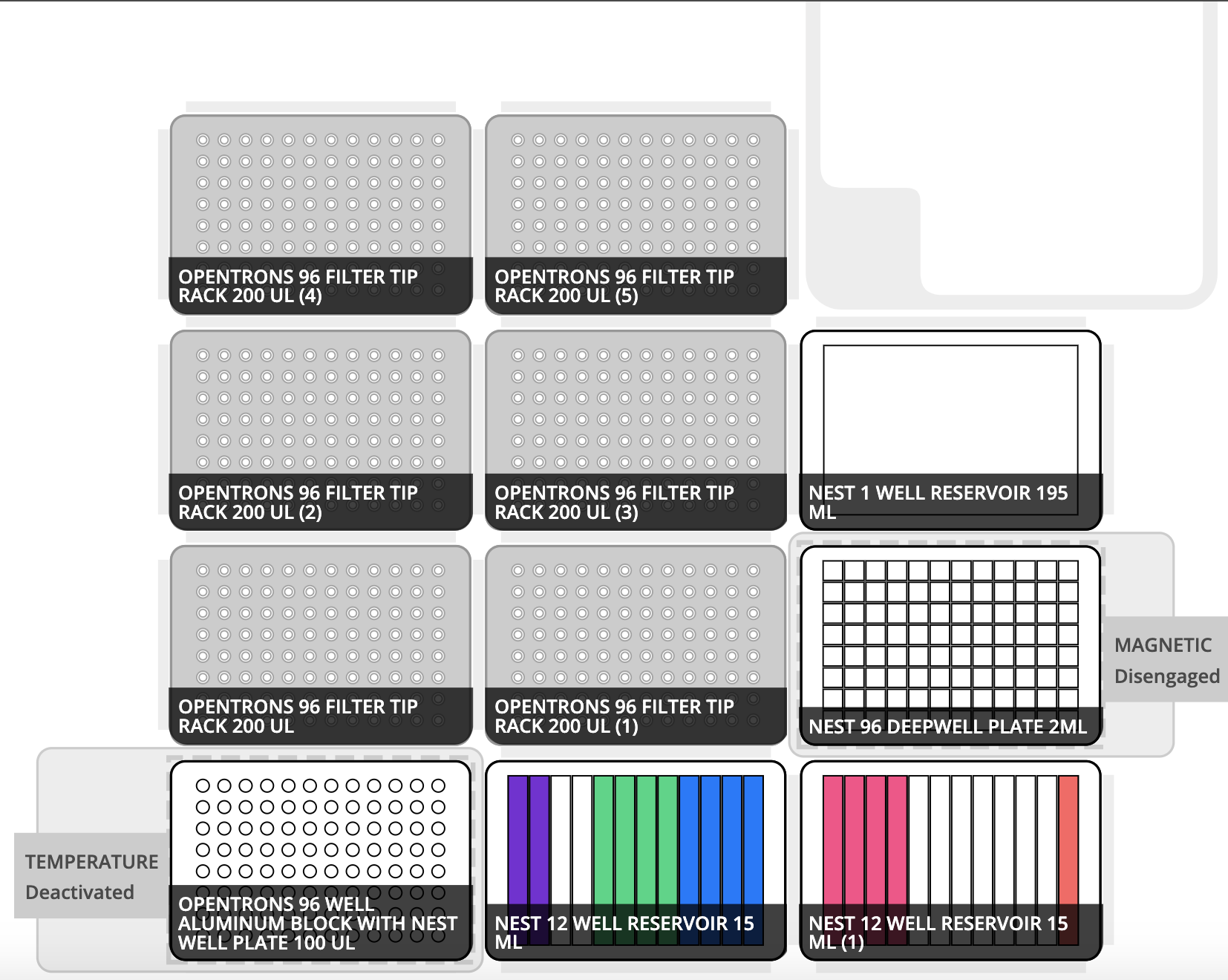\n\n",
"description": "This protocol performs viral RNA isolation on up to 96 samples using the [Beckman Coulter RNAdvance Viral RNA Isolation](https://www.beckman.com/reagents/genomic/rna-isolation/viral/c63510) kit and workflow.\n\nThe protocol begins at the stage of adding binding beads to lysed samples loaded on the magnetic module in a NEST 96-deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 6) to the elution PCR plate (temperature module, slot 1). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\n---\n\n",
diff --git a/protoBuilds/bc-rnadvance-viral/README.json b/protoBuilds/bc-rnadvance-viral/README.json
index e360b538e1..c6ef652aa4 100644
--- a/protoBuilds/bc-rnadvance-viral/README.json
+++ b/protoBuilds/bc-rnadvance-viral/README.json
@@ -18,7 +18,7 @@
"labware": "* [NEST 12 Well Reservoir 15 mL, 2x](https://labware.opentrons.com/nest_12_reservoir_15ml)\n* [NEST 1 Well Reservoir 195 mL](https://labware.opentrons.com/nest_1_reservoir_195ml)\n* [NEST 96 Well Plate 100 \u00b5L PCR Full Skirt](https://labware.opentrons.com/nest_96_wellplate_100ul_pcr_full_skirt)\n* [NEST 96 Deepwell Plate 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep)\n* [Opentrons 96 Filter Tip Rack 200 \u00b5L, up to 10x depending on throughput](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n\n",
"modules": "* [Temperature Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Magnetic Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Beckman Coulter Life Sciences](https://www.beckmancoulter.com/)\n\n",
+ "partner": "[Beckman Coulter Life Sciences](https://www.beckmancoulter.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [Opentrons P300 8-Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"reagent-setup": "* Reservoir 1: slot 2\n* Reservoir 2: slot 3 \n\n\nVolumes per reservoir channel: (for 96-sample run, not including dead volume):\n* 10mL of 100% Isopropanol + 250uL of bead BBD\n* 10mL of Wash WBE\n* 10mL of 70% Ethanol\n* 4mL of nuclease-free water\n\n---\n\n",
@@ -30,7 +30,7 @@
"Magnetic Module (GEN2)"
],
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Beckman Coulter Life Sciences",
+ "partner": "Beckman Coulter Life Sciences\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nOpentrons P300 8-Channel Electronic Pipette (GEN2)\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"reagent-setup": "\nReservoir 1: slot 2\nReservoir 2: slot 3\n\n\nVolumes per reservoir channel: (for 96-sample run, not including dead volume):\n 10mL of 100% Isopropanol + 250uL of bead BBD\n 10mL of Wash WBE\n 10mL of 70% Ethanol\n 4mL of nuclease-free water\n",
diff --git a/protoBuilds/bms-illumina-dna-prep/README.json b/protoBuilds/bms-illumina-dna-prep/README.json
index e19242e6c0..13b7fa5a4c 100644
--- a/protoBuilds/bms-illumina-dna-prep/README.json
+++ b/protoBuilds/bms-illumina-dna-prep/README.json
@@ -18,7 +18,7 @@
"labware": "* [Nest 96 well plate full skirt 100 \u00b5L](https://shop.opentrons.com/nest-0-1-ml-96-well-pcr-plate-full-skirt/)\n* [Opentrons Tough 0.2 mL 96-Well PCR Plate, Full Skirt](https://shop.opentrons.com/tough-0.2-ml-96-well-pcr-plate-full-skirt/)\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/nest-12-well-reservoirs-15-ml/)\n* [Opentrons aluminum block set](https://shop.opentrons.com/aluminum-block-set/)\n\n",
"modules": "* [Temperature Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Magnetic Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Illumina](https://www.illumina.com/)\n\n",
+ "partner": "[Illumina](https://www.illumina.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P300 multi-Channel (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n* [P20 multi-Channel (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n\n**Tips**\n* [Opentrons 20 \u00b5L filter tiprack](https://shop.opentrons.com/opentrons-20ul-filter-tips/)\n* [Opentrons 200 \u00b5L filter tiprack](https://shop.opentrons.com/opentrons-200ul-filter-tips/)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Prepare the thermocycler by setting the block temperature to 4 degrees, and the lid to 100 degrees.\n2. Add tagmentation mix to the samples\n3. User seals the plate and the protocol incubates the samples with the mix in the thermocycler, 55 degrees for 15 minutes.\n4. The thermocycler opens, and the user removes the seal.\n5. Add Tagmentation Stop Buffer to the samples.\n6. Seal and incubate the mix at 37 degrees for 15 minutes.\n7. User removes seal; remove the supernatant and wash the beads three times with Tagmentation Wash Buffer.\n8. Amplification of DNA: Addition of PCR mix and addition of barcodes.\n9. The protocol runs PCR protocol for 5 cycles. This takes 25 minutes to complete. The supernatant is transferred to columns 7, 9 and 11 depending on how many sample columns there are.\n10. Post-PCR cleanup using AMPure beads.\n11. The protocol performs two ethanol washes.\n12. RSB (resuspension buffer) is added to the bead wells and the supernatant is transferred to the output columns (8, 10, and 12).\n\n",
@@ -32,7 +32,7 @@
"Thermocycler Module"
],
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Illumina",
+ "partner": "Illumina\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP300 multi-Channel (GEN2)\nP20 multi-Channel (GEN2)\n\nTips\n Opentrons 20 \u00b5L filter tiprack\n Opentrons 200 \u00b5L filter tiprack",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nPrepare the thermocycler by setting the block temperature to 4 degrees, and the lid to 100 degrees.\nAdd tagmentation mix to the samples\nUser seals the plate and the protocol incubates the samples with the mix in the thermocycler, 55 degrees for 15 minutes.\nThe thermocycler opens, and the user removes the seal.\nAdd Tagmentation Stop Buffer to the samples.\nSeal and incubate the mix at 37 degrees for 15 minutes.\nUser removes seal; remove the supernatant and wash the beads three times with Tagmentation Wash Buffer.\nAmplification of DNA: Addition of PCR mix and addition of barcodes.\nThe protocol runs PCR protocol for 5 cycles. This takes 25 minutes to complete. The supernatant is transferred to columns 7, 9 and 11 depending on how many sample columns there are.\nPost-PCR cleanup using AMPure beads.\nThe protocol performs two ethanol washes.\nRSB (resuspension buffer) is added to the bead wells and the supernatant is transferred to the output columns (8, 10, and 12).\n",
diff --git a/protoBuilds/bpg-rna-extraction/README.json b/protoBuilds/bpg-rna-extraction/README.json
index c13c6c9516..177fc0c68e 100644
--- a/protoBuilds/bpg-rna-extraction/README.json
+++ b/protoBuilds/bpg-rna-extraction/README.json
@@ -13,13 +13,13 @@
"description": "**This protocol is currently being tested and is subject to change**\n\nThis protocol automates the pureBASE RNA protocol by BP Genomics.\n\nAdditionally, it adds the 4ul of internal extraction control RNA as described in the 2019-nCoV Detection Assay protocol.\n\n\n---\n\n\nTo purchase consumables, labware, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons P1000 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml) or [Opentrons 24 Tuberack with 2mL Tubes](https://labware.opentrons.com/opentrons_24_tuberack_eppendorf_2ml_safelock_snapcap?category=tubeRack)\n* NEST Deep Well Plate, 1mL\n* [NEST 1-Well Reservoir, 195mL](https://labware.opentrons.com/nest_1_reservoir_195ml?category=reservoir)\n\n\n\n---\n\n\n*May need to change pending revisions*\n\nSlot 1: [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) (for elutes)\n\nSlot 2: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml) or [Opentrons 24 Tuberack with 2mL Tubes](https://labware.opentrons.com/opentrons_24_tuberack_eppendorf_2ml_safelock_snapcap?category=tubeRack), with Reagents\n**If using NEST 12-Well Reservoir, reagents should be loaded in the following configuration:**\n* A1: Internal Extraction Control RNA\n* A2: Magbeads (in isopropanol; 20ul beads, 560ul isopropanol per sample)\n* A5: Wash Buffer 1\n* A7: Wash Buffer 2\n* A9: Ethanol\n* A10: Ethanol\n* A12: Nuclease-Free Water\n**If using Opentrons 24-Tuberack & 2mL Tubes, reagents should be loaded in the following configuration:**\n* A1: Internal Exctraction Control RNA\n* A3: Magbeads (in isopropanol; 20ul beads, 560ul isopropanol per sample)\n* A5: Wash Buffer 1\n* C1: Wash Buffer 2\n* C3: Ethanol\n* C4: Ethanol\n* C6: Nuclease-Free Water\n\nSlot 3: Opentrons 20ul Filter Tips\n\nSlot 4: MagDeck, with Deep Well Plate containing 400ul of sample\n\nSlot 5: Reservoir for Liquid Waste\n\nSlot 6: Opentrons 1000ul Filter Tips\n\n\n\n\n\n\n__Using the customizations fields, below set up your protocol.__\n* **Number of Samples**: Specify the number of samples you'd like to run.\n* **P20 Single Mount**: Specify which mount (left or right) the P20 Single is attached to.\n* **P1000 Single Mount**: Specify which mount (left or right) the P1000 Single is attached to.\n* **Labware for Reagents**: Specify which labware (NEST 12-Channel Reservoir or Opentrons 24-Tuberack) will house the reagents. This labware (& reagents) should be loaded in slot 2 and reagents should be loaded as described above.\n\n\n\n\n\n",
"internal": "bpg-rna-extraction\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[BP Genomics](https://bpgenomics.com/)\n\n",
+ "partner": "[BP Genomics](https://bpgenomics.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"process": "\n1. Input your protocol parameters.\n2. Download your protocol.\n3. Upload your protocol into the [OT App](https://opentrons.com/ot-app).\n4. Set up your deck according to the deck map.\n5. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n6. Hit \"Run\".\n\n",
"robot": "* [OT-2](https://opentrons.com/ot-2)\n\n",
"title": "Bp Genomics RNA Extraction"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "BP Genomics",
+ "partner": "BP Genomics\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"process": "\nInput your protocol parameters.\nDownload your protocol.\nUpload your protocol into the OT App.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit \"Run\".\n",
"robot": [
"OT-2"
diff --git a/protoBuilds/cepheid/README.json b/protoBuilds/cepheid/README.json
index 7f9bb5eb85..dee22feef8 100644
--- a/protoBuilds/cepheid/README.json
+++ b/protoBuilds/cepheid/README.json
@@ -17,14 +17,14 @@
"internal": "cepheid\n",
"labware": "* [Opentrons 15-Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* 12mL Tubes\n* Cepheid\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[BasisDx](https://www.basisdx.org/)\n\n",
+ "partner": "[BasisDx](https://www.basisdx.org/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P1000 Single-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n\n---\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol bundle containing protocol and custom labware; unzip bundle.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. For each grouping of samples (10), the [P1000 Single-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) will pick up a tip, transfer 62\u00b5L of sample (with 50\u00b5L air gap) to destination well (samples 1-10 --> A1; samples 11-20 --> A5; samples 21-30 --> C1), and drop used tip in the waste bin.\n2. The [P1000 Single-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) will mix the pooled sample and then transfer 300\u00b5L of pooled sample to the corresponding Cepheid device.\n3. This process will repeat for each group of 10 samples, up to 30 samples.\n4. End of the protocol.\n\n",
"title": "Pooling Samples and Distribution to Cepheid"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "BasisDx",
+ "partner": "BasisDx\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP1000 Single-Channel Pipette (GEN2)\n\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol bundle containing protocol and custom labware; unzip bundle.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nFor each grouping of samples (10), the P1000 Single-Channel Pipette (GEN2) will pick up a tip, transfer 62\u00b5L of sample (with 50\u00b5L air gap) to destination well (samples 1-10 --> A1; samples 11-20 --> A5; samples 21-30 --> C1), and drop used tip in the waste bin.\nThe P1000 Single-Channel Pipette (GEN2) will mix the pooled sample and then transfer 300\u00b5L of pooled sample to the corresponding Cepheid device.\nThis process will repeat for each group of 10 samples, up to 30 samples.\nEnd of the protocol.\n",
diff --git a/protoBuilds/cerillo/README.json b/protoBuilds/cerillo/README.json
index 24a0d1d78f..d284b014bc 100644
--- a/protoBuilds/cerillo/README.json
+++ b/protoBuilds/cerillo/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "cerillo",
"labware": "\nCerillo Stratus Armadillo Flat Bottom Plate 96well 200uL\nNEST 96 Deepwell Plate 2mL #503001\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol preps a 96 200ul Armadillo plate to be used directly on the Cerillo Stratus. For detailed protocol steps, please see below.\n\n[For an example csv, please see here.](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/cerillo+csv.xlsx)\n\nNote: keep the x in the top left corner. If you would not like to put a volume in a well (ie skipping a well), then replace any number with an \"x\".\n\n\n",
diff --git a/protoBuilds/checkit/README.json b/protoBuilds/checkit/README.json
index 5dba195670..2ee8a9aec8 100644
--- a/protoBuilds/checkit/README.json
+++ b/protoBuilds/checkit/README.json
@@ -17,7 +17,7 @@
"internal": "checkit\n",
"labware": "* [Next Advance Checkit Go Cartridge](https://www.nextadvance.com/checkit-go/)\n* [Opentrons 20/300ul Tiprack](https://shop.opentrons.com/universal-filter-tips/) (depending on the pipette model used in the test)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "\n\n",
+ "partner": "\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [Opentrons P20 Single-Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/pipettes/)\n* [Opentrons P20 8-Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/pipettes/)\n* [Opentrons P300 Single-Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/pipettes/)\n* [Opentrons P300 8-Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/pipettes/)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "For 8-channel pipettes: \n1. The pipette picks up tips and aspirates the Checkit Go-specific volume from the cartridge reservoir.\n2. The pipette dispenses the entire liquid volume into the wells of the cartridge and drops the tip. If the\n3. The user is prompted to flip the cartridge and record the measurements.\n\nFor single-channel pipettes: \n\n1. The pipette picks up tips and aspirates the Checkit Go-specific volume from the cartridge reservoir.\n2. The pipette dispenses the entire liquid volume into the wells of the cartridge, starting in the first well and drops the tip.\n3. The user is prompted to flip the cartridge and record the measurements.\n4. Steps 1-3 are repeated a total of 8 times for all wells in the plate.\n\n",
@@ -25,7 +25,7 @@
"title": "Next Advance Checkit Go"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "",
+ "partner": "\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nOpentrons P20 Single-Channel Electronic Pipette (GEN2)\nOpentrons P20 8-Channel Electronic Pipette (GEN2)\nOpentrons P300 Single-Channel Electronic Pipette (GEN2)\nOpentrons P300 8-Channel Electronic Pipette (GEN2)\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "For 8-channel pipettes:\n1. The pipette picks up tips and aspirates the Checkit Go-specific volume from the cartridge reservoir.\n2. The pipette dispenses the entire liquid volume into the wells of the cartridge and drops the tip. If the\n3. The user is prompted to flip the cartridge and record the measurements.\nFor single-channel pipettes: \n\nThe pipette picks up tips and aspirates the Checkit Go-specific volume from the cartridge reservoir.\nThe pipette dispenses the entire liquid volume into the wells of the cartridge, starting in the first well and drops the tip.\nThe user is prompted to flip the cartridge and record the measurements.\nSteps 1-3 are repeated a total of 8 times for all wells in the plate.\n",
diff --git a/protoBuilds/cherrypicking/README.json b/protoBuilds/cherrypicking/README.json
index 5ff2182e3e..7c1c639621 100644
--- a/protoBuilds/cherrypicking/README.json
+++ b/protoBuilds/cherrypicking/README.json
@@ -10,7 +10,7 @@
"internal": "cherrypicking",
"labware": "\nAny verified labware found in our Labware Library\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/cherrypicking). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/cherrypicking). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Featured\n\t* Cherrypicking\n\n",
"deck-setup": "* Example deck setup - tip racks loaded onto remining slots.\n\n\n---\n\n",
"description": "\nOur most robust cherrypicking protocol. Specify aspiration height, labware, pipette, as well as source and destination wells with this all inclusive cherrypicking protocol.\n\n\n\nExplanation of complex parameters below:\n\n* `input .csv file`: Here, you should upload a .csv file formatted in the [following way](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/1211/example.csv), making sure to include headers in your csv file. Refer to our [Labware Library](https://labware.opentrons.com/?category=wellPlate) to copy API names for labware to include in the `Source Labware` and `Dest Labware` columns of the .csv.\n* `Pipette Model`: Select which pipette you will use for this protocol.\n* `Pipette Mount`: Specify which mount your single-channel pipette is on (left or right)\n* `Tip Type`: Specify whether you want to use filter tips.\n* `Tip Usage Strategy`: Specify whether you'd like to use a new tip for each transfer, or keep the same tip throughout the protocol.\n\n\n\n---\n\n\n",
diff --git a/protoBuilds/cherrypicking_simple/README.json b/protoBuilds/cherrypicking_simple/README.json
index d75ac9654b..217fedb8b8 100644
--- a/protoBuilds/cherrypicking_simple/README.json
+++ b/protoBuilds/cherrypicking_simple/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Cherrypicking"
@@ -8,7 +8,7 @@
"description": "\nCherrypicking, or hit-picking, is a key component of many workflows from high-throughput screening to microbial transfections. With this protocol, you can easily select specific wells in a 96 or 384 microwell plate without worrying about missing or selecting the wrong well. Just upload your properly formatted CSV file (keep scrolling for an example), customize your parameters, and download your ready-to-run protocol.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Single-Channel Pipette and corresponding Tips\nMicroplates (96-well or 384-well)\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nTipracks should be loaded in Slot 1 (if using more than 96 tips, Slots 4, 7, and 10 can be loaded as well).\nThe Source Plate should be loaded in Slot 2. If using multiple Source Plates, Source Plates 1, 2, 3, and 4 should be loaded into Slot 2, Slot 5, Slot 8, and Slot 11, respectively.\nNote: If using multiple Source Plates, you must specify the Source Plate in the third column of the CSV (see below for examples)\nThe Destination Plate should be loaded in Slot 3.\nNote: The Destination Plate will receive each cherrypicking transfer sequentially, filling up the plate in well A1, then B1, etc.\nCSV Format\nYour cherrypicking transfers must be saved as a comma separated value (.csv) file type. Your CSV must contain values corresponding to volumes in microliters (\u03bcL).\n\nIn the first example, 40\u03bcL will be removed from well A3 in your source plate, and placed in well A1 in your destination plate. 36\u03bcL will be removed from well D4 in your source plate, and placed in well B1 in your destination plate.\n\nIn the second example, 100\u03bcL will be transferred from well E5 in \"Plate 1\" (slot 2) to well A1 in the destination plate (slot 3). After this, 100\u03bcL will be transferred from well G10 in \"Plate 3\" (slot 8) to well B1 in the destination plate (slot 3).\nIf you\u2019d like to follow our template, you can make a copy of this spreadsheet, fill out your values, and export as CSV for use with this protocol.\nUsing the customizations fields, below set up your protocol.\n Volumes CSV: Upload the .csv file containing your well locations, volumes, and source plate (optional).\n Pipette Model: Select which pipette you will use for this protocol.\n Pipette Mount: Specify which mount your single-channel pipette is on (left or right)\n Source Plate Type: Select which (source) plate you will pick samples from.\n Destination Plate Type: Select which (destination) plate you will dispense into.\n Filter Tips: Specify whether you want to use filter tips.\n* Tip Usage Strategy: Specify whether you'd like to use a new tip for each transfer, or keep the same tip throughout the protocol.",
"internal": "cherrypicking_simple",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Cherrypicking\n\n\n",
"description": "\n\nCherrypicking, or hit-picking, is a key component of many workflows from high-throughput screening to microbial transfections. With this protocol, you can easily select specific wells in a 96 or 384 microwell plate without worrying about missing or selecting the wrong well. Just upload your properly formatted CSV file (keep scrolling for an example), customize your parameters, and download your ready-to-run protocol.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes) and corresponding [Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Microplates (96-well or 384-well)](https://labware.opentrons.com/?category=wellPlate)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n\n---\n\n\n[Tipracks](https://shop.opentrons.com/collections/opentrons-tips) should be loaded in **Slot 1** (if using more than 96 tips, Slots 4, 7, and 10 can be loaded as well).\n\nThe Source Plate should be loaded in **Slot 2**. If using multiple Source Plates, Source Plates 1, 2, 3, and 4 should be loaded into **Slot 2**, **Slot 5**, **Slot 8**, and **Slot 11**, respectively.\n*Note*: If using multiple Source Plates, you must specify the Source Plate in the third column of the CSV (see below for examples)\n\nThe Destination Plate should be loaded in **Slot 3**.\n*Note*: The Destination Plate will receive each cherrypicking transfer sequentially, filling up the plate in well A1, then B1, etc.\n\n**CSV Format**\n\nYour cherrypicking transfers must be saved as a comma separated value (.csv) file type. Your CSV must contain values corresponding to volumes in microliters (\u03bcL).\n\n\n\nIn the first example, 40\u03bcL will be removed from well A3 in your source plate, and placed in well A1 in your destination plate. 36\u03bcL will be removed from well D4 in your source plate, and placed in well B1 in your destination plate.\n\n\n\nIn the second example, 100\u03bcL will be transferred from well E5 in \"Plate 1\" (slot 2) to well A1 in the destination plate (slot 3). After this, 100\u03bcL will be transferred from well G10 in \"Plate 3\" (slot 8) to well B1 in the destination plate (slot 3).\n\nIf you\u2019d like to follow our template, you can make a copy of [this spreadsheet](https://opentrons-protocol-library-website.s3.amazonaws.com/Technical+Notes/cherrypicking_simple/Opentrons+Cherrypicking+Simple+Example+Template.xlsx), fill out your values, and export as CSV for use with this protocol.\n\nUsing the customizations fields, below set up your protocol.\n* Volumes CSV: Upload the .csv file containing your well locations, volumes, and source plate (optional).\n* Pipette Model: Select which pipette you will use for this protocol.\n* Pipette Mount: Specify which mount your single-channel pipette is on (left or right)\n* Source Plate Type: Select which (source) plate you will pick samples from.\n* Destination Plate Type: Select which (destination) plate you will dispense into.\n* Filter Tips: Specify whether you want to use filter tips.\n* Tip Usage Strategy: Specify whether you'd like to use a new tip for each transfer, or keep the same tip throughout the protocol.\n\n\n",
"internal": "cherrypicking_simple\n",
diff --git a/protoBuilds/complete-pcr/README.json b/protoBuilds/complete-pcr/README.json
index 6ba85f0a39..abde558e5d 100644
--- a/protoBuilds/complete-pcr/README.json
+++ b/protoBuilds/complete-pcr/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"Complete PCR Workflow"
@@ -8,7 +8,7 @@
"description": "Part 1 of 2: Master Mix Assembly\nLinks:\n Part 1: Master Mix Assembly\n Part 2: Master Mix Distribution and DNA Transfer\n* Part 3: Thermocycler\nThis protocol allows your robot to create a master mix solution using any reagents stored in a 2 mL Eppendorf tube rack or a 2 mL screwcap tube rack. The master mix will be created in well A1 of the trough. The ingredient information will be provided as a CSV file. See Additional Notes for more details.\n\nYou will need:\n 4-in-1 Tube Rack Set\n 12-well Trough",
"internal": "OT-2 PCR Prep v2",
"markdown": {
- "author": "[Opentrons](http://www.opentrons.com/)\n\n",
+ "author": "[Opentrons](http://www.opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * Complete PCR Workflow\n\n",
"description": "Part 1 of 2: Master Mix Assembly\n\nLinks:\n* [Part 1: Master Mix Assembly](./pcr_prep_part_1)\n* [Part 2: Master Mix Distribution and DNA Transfer](./pcr_prep_part_2)\n* [Part 3: Thermocycler](./thermocycler)\n\nThis protocol allows your robot to create a master mix solution using any reagents stored in a 2 mL Eppendorf tube rack or a 2 mL screwcap tube rack. The master mix will be created in well A1 of the trough. The ingredient information will be provided as a CSV file. See Additional Notes for more details.\n\n---\n\nYou will need:\n* [4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* [12-well Trough](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n\n",
"internal": "OT-2 PCR Prep v2\n",
diff --git a/protoBuilds/covid19-rna-extraction/README.json b/protoBuilds/covid19-rna-extraction/README.json
index f0d2ffcfa6..c48e20ff23 100644
--- a/protoBuilds/covid19-rna-extraction/README.json
+++ b/protoBuilds/covid19-rna-extraction/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Covid Workstation": [
"RNA Extraction"
@@ -8,7 +8,7 @@
"description": "This is a flexible protocol accommodating a wide range of commercial RNA extraction workflows for COVID-19 sample processing. The protocol is broken down into 5 main parts:\n binding buffer addition to samples\n bead wash 3x using magnetic module\n* final elution to chilled PCR plate\nLysed samples should be loaded on the magnetic module in a NEST or USA Scientific 96-deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 4) to the elution PCR plate (temperature module, slot 1). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\nExplanation of complex parameters below:\n park tips: If set to yes (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to no, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n track tips across protocol runs: If set to yes, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to no, tips will be picked up from column 1 of the 1st tiprack.\n* flash: If set to yes, the robot rail lights will flash during any automatic pauses in the protocol. If set to no, the lights will not flash.\n\n \nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons magnetic module\nOpentrons temperature module\nNEST 12 Well Reservoir 15 mL or USA Scientific 12 Well Reservoir 22 mL\nNEST 1 Well Reservoir 195 mL\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt\nNEST 96 Deepwell Plate 2mL or USA Scientific 96 Deep Well Plate 2.4 mL\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n\n\n\n\nReservoir 1: slot 5\nReservoir 2: slot 2\n\n",
"internal": "covid-19-rna-extraction",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Covid Workstation\n * RNA Extraction\n\n",
"description": "This is a flexible protocol accommodating a wide range of commercial RNA extraction workflows for COVID-19 sample processing. The protocol is broken down into 5 main parts:\n* binding buffer addition to samples\n* bead wash 3x using magnetic module\n* final elution to chilled PCR plate\n\nLysed samples should be loaded on the magnetic module in a NEST or USA Scientific 96-deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 4) to the elution PCR plate (temperature module, slot 1). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\nExplanation of complex parameters below:\n* `park tips`: If set to `yes` (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to `no`, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n* `flash`: If set to `yes`, the robot rail lights will flash during any automatic pauses in the protocol. If set to `no`, the lights will not flash.\n\n---\n\n \n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons magnetic module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons temperature module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [NEST 12 Well Reservoir 15 mL](https://labware.opentrons.com/nest_12_reservoir_15ml) or [USA Scientific 12 Well Reservoir 22 mL](https://labware.opentrons.com/usascientific_12_reservoir_22ml)\n* [NEST 1 Well Reservoir 195 mL](https://labware.opentrons.com/nest_1_reservoir_195ml)\n* [NEST 96 Well Plate 100 \u00b5L PCR Full Skirt](https://labware.opentrons.com/nest_96_wellplate_100ul_pcr_full_skirt)\n* [NEST 96 Deepwell Plate 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep) or [USA Scientific 96 Deep Well Plate 2.4 mL](https://labware.opentrons.com/usascientific_96_wellplate_2.4ml_deep)\n* [Opentrons 96 Filter Tip Rack 200 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n\n---\n\n\n* Reservoir 1: slot 5\n* Reservoir 2: slot 2 \n\n\n",
"internal": "covid-19-rna-extraction\n",
diff --git a/protoBuilds/csv_transfer/README.json b/protoBuilds/csv_transfer/README.json
index 0e0d860068..b9d3246865 100644
--- a/protoBuilds/csv_transfer/README.json
+++ b/protoBuilds/csv_transfer/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"CSV Transfer"
@@ -10,7 +10,7 @@
"internal": "cherrypicking",
"labware": "\nAny verified labware found in our Labware Library\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* CSV Transfer\n\n",
"deck-setup": "* Example deck setup - tip racks loaded onto remining slots.\n\n\n---\n\n",
"description": "\nAccomplish complex workflows, easily with this Custom CSV Transfer Protocol. This protocol utilizes a CSV to specify aspiration height, labware, pipette, as well as source and destination wells.\n\nTo use, simply upload a CSV (download a template with the proper format [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/1211/example.csv) and select the parameters that match the OT-2 below before downloading the Python protocol. The protocol can then be used on the OT-2 and will make all of the transfers outlined in the CSV.\n\n\n\nExplanation of complex parameters below:\n\n* **input .csv file**: Here, you should upload a .csv file formatted in the [following way](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/1211/example.csv), making sure to include headers in your csv file. Refer to our [Labware Library](https://labware.opentrons.com/?category=wellPlate) to copy API names for labware to include in the *Source Labware* and *Dest Labware* columns of the .csv.\n* **Pipette Model**: Select which pipette you will use for this protocol.\n* **Pipette Mount**: Specify which mount your single-channel pipette is on (left or right)\n* **Tip Type**: Specify whether you want to use filter tips.\n* **Tip Usage Strategy**: Specify whether you'd like to use a new tip for each transfer, or keep the same tip throughout the protocol.\n\n\n\n---\n\n\n",
diff --git a/protoBuilds/customizable_serial_dilution_ot2/README.json b/protoBuilds/customizable_serial_dilution_ot2/README.json
index c270b298e6..097aa89be1 100644
--- a/protoBuilds/customizable_serial_dilution_ot2/README.json
+++ b/protoBuilds/customizable_serial_dilution_ot2/README.json
@@ -8,13 +8,13 @@
"description": "With this protocol, you can do a simple serial dilution across a 96-well plate using either a single-channel or 8-channel pipette. This can be useful for everything from creating a simple standard curve to a concentration-limiting dilution. For more information (including data from the Opentrons Lab and other considerations), please see our Technical Note.\n\n\n\nExample Setup\nThis protocol uses the inputs you define for \"Dilution Factor\" and \"Total Mixing Volume\" to automatically infer the necessary transfer volume for each dilution across your plate. For a 1 in 3 dilution series across an entire plate, as seen above:\n-- Start with your samples/reagents in Column 1 of your plate. In this example, you would pre-add 150 uL of concentrated sample to the first column of your 96-well plate.\n-- Define a Total Mixing Volume of 150uL, a Dilution Factor of 3, and set Number of Dilutions = 11.\n-- Your OT-2 will add 100uL of diluent to each empty well in your plate. Then it will transfer 50uL from Column 1 between each well/column in the plate.\n-- \"Total mixing volume\" = transfer volume + diluent volume.\n\n\n\n-- Opentrons OT-2\n-- Opentrons OT-2 Run App (Version 3.19 or later)\n-- Opentrons Tips for selected Opentrons Pipette\n-- 12-Row, Automation-Friendly Trough\n-- 96-Well Plate (found in our Labware Library)\n-- Diluent (Pre-loaded in row 1 of trough)\n-- Samples/Standards (Pre-loaded in Column 1 of a standard 96-well plate)",
"internal": "Customizable Serial Dilution, v2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/customizable_serial_dilution_ot2). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/customizable_serial_dilution_ot2). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Featured\n * Serial Dilution\n\n",
- "description": "With this protocol, you can do a simple serial dilution across a 96-well plate using either a single-channel or 8-channel pipette. This can be useful for everything from creating a simple standard curve to a concentration-limiting dilution. For more information (including data from the Opentrons Lab and other considerations), please see our [Technical Note](https://s3.amazonaws.com/opentrons-protocol-library-website/Technical+Notes/Serial+Dilution+OT2+Technical+Note.pdf).\n\n---\n\n---\n\n\n\n***Example Setup***\n\nThis protocol uses the inputs you define for \"***Dilution Factor***\" and \"***Total Mixing Volume***\" to automatically infer the necessary transfer volume for each dilution across your plate. For a 1 in 3 dilution series across an entire plate, as seen above:\n\n-- Start with your samples/reagents in Column 1 of your plate. In this example, you would pre-add 150 uL of concentrated sample to the first column of your 96-well plate.\n\n-- Define a ***Total Mixing Volume*** of 150uL, a ***Dilution Factor*** of 3, and set ***Number of Dilutions*** = 11.\n\n-- Your OT-2 will add 100uL of diluent to each empty well in your plate. Then it will transfer 50uL from Column 1 between each well/column in the plate.\n\n-- \"***Total mixing volume***\" = transfer volume + diluent volume.\n\n---\n\n---\n\n\n\n\n-- [Opentrons OT-2](http://opentrons.com/ot-2)\n\n-- [Opentrons OT-2 Run App (Version 3.19 or later)](http://opentrons.com/ot-app)\n\n-- [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips-racks-9-600-tips) for selected Opentrons Pipette\n\n-- [12-Row, Automation-Friendly Trough](https://shop.opentrons.com/nest-12-well-reservoirs-15-ml/)\n\n-- [96-Well Plate](https://shop.opentrons.com/nest-96-well-plate-flat/) (found in our [Labware Library](https://labware.opentrons.com/?category=wellPlate))\n\n-- Diluent (Pre-loaded in row 1 of trough)\n\n-- Samples/Standards (Pre-loaded in Column 1 of a standard 96-well plate)\n\n",
+ "description": "With this protocol, you can do a simple serial dilution across a 96-well plate using either a single-channel or 8-channel pipette. This can be useful for everything from creating a simple standard curve to a concentration-limiting dilution. For more information (including data from the Opentrons Lab and other considerations), please see our [Technical Note](https://s3.amazonaws.com/opentrons-protocol-library-website/Technical+Notes/Serial+Dilution+OT2+Technical+Note.pdf).\n\n---\n\n---\n\n\n\nExample Setup\n\nThis protocol uses the inputs you define for \"Dilution Factor\" and \"Total Mixing Volume\" to automatically infer the necessary transfer volume for each dilution across your plate. For a 1 in 3 dilution series across an entire plate, as seen above:\n\n-- Start with your samples/reagents in Column 1 of your plate. In this example, you would pre-add 150 uL of concentrated sample to the first column of your 96-well plate.\n\n-- Define a Total Mixing Volume of 150uL, a Dilution Factor of 3, and set Number of Dilutions = 11.\n\n-- Your OT-2 will add 100uL of diluent to each empty well in your plate. Then it will transfer 50uL from Column 1 between each well/column in the plate.\n\n-- \"Total mixing volume\" = transfer volume + diluent volume.\n\n---\n\n---\n\n\n\n\n-- [Opentrons OT-2](http://opentrons.com/ot-2)\n\n-- [Opentrons OT-2 Run App (Version 3.19 or later)](http://opentrons.com/ot-app)\n\n-- [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips-racks-9-600-tips) for selected Opentrons Pipette\n\n-- [12-Row, Automation-Friendly Trough](https://shop.opentrons.com/nest-12-well-reservoirs-15-ml/)\n\n-- [96-Well Plate](https://shop.opentrons.com/nest-96-well-plate-flat/) (found in our [Labware Library](https://labware.opentrons.com/?category=wellPlate))\n\n-- Diluent (Pre-loaded in row 1 of trough)\n\n-- Samples/Standards (Pre-loaded in Column 1 of a standard 96-well plate)\n\n",
"internal": "Customizable Serial Dilution, v2\n",
"notes": "Please reference our [Technical Note](https://s3.amazonaws.com/opentrons-protocol-library-website/Technical+Notes/Serial+Dilution+OT2+Technical+Note.pdf) for more information about the expected output of this protocol, in addition to expanded sample data from the Opentrons lab.\n\nWe understand that there are limitations to the use of this protocol, and we plan to make improvements soon. In the meantime, if you'd like to request a more complex dilution workflow, please use our [Protocol Development Request Form](https://opentrons-protocol-dev.paperform.co/). You can also download this Python file and modify it using our [API Documentation](https://docs.opentrons.com/). For additional questions about this protocol, please email .\n\n",
"preview": "Perform a simple serial dilution across a 96-well plate using either a single-channel or multichannel pipette. This can be useful for everything from creating a simple standard curve to a concentration-limiting dilution.\n\n",
- "process": "1. Choose the pipette you want to use from the dropdown menu above and which side it is installed on the OT-2\n2. Set your dilution factor.\n ***Example:*** If you want a 1:2 ratio of sample to total reaction volume, you would set your dilution factor to 2.\n3. Set your number of dilutions (max is 11, 10 if using blank)\n4. Set your total mixing volume. (Total mixing volume = transfer volume + diluent volume). Be careful to make sure this number does not exceed the volume capacity of your plate. To see how this number is used, scroll to the example above.\n5. Set whether a blank will be made or not. The blank will be added to the first available column in the plate.\n ***NOTE:*** 10 dilutions is the max allowed when using a blank\n6. Set your tip reuse strategy.\n ***Note:*** This defaults to no tip changes; adjust only if you want to change tips between each well.\n7. Set your air gap, if desired. This will add a specified amount of air into the tip after aspiration\n8. Download your customized OT-2 Serial Dilution protocol using the blue \"Download\" button.\n9. Upload into the Opentrons Run App and follow the instructions there to set up your deck and proceed to run!\n10. Make sure to add diluent to the first row of your 12-row trough and load your desired samples/standards into column 1 of your plate before running your protocol in the run app!\n\n",
+ "process": "1. Choose the pipette you want to use from the dropdown menu above and which side it is installed on the OT-2\n2. Set your dilution factor.\n Example: If you want a 1:2 ratio of sample to total reaction volume, you would set your dilution factor to 2.\n3. Set your number of dilutions (max is 11, 10 if using blank)\n4. Set your total mixing volume. (Total mixing volume = transfer volume + diluent volume). Be careful to make sure this number does not exceed the volume capacity of your plate. To see how this number is used, scroll to the example above.\n5. Set whether a blank will be made or not. The blank will be added to the first available column in the plate.\n NOTE: 10 dilutions is the max allowed when using a blank\n6. Set your tip reuse strategy.\n Note: This defaults to no tip changes; adjust only if you want to change tips between each well.\n7. Set your air gap, if desired. This will add a specified amount of air into the tip after aspiration\n8. Download your customized OT-2 Serial Dilution protocol using the blue \"Download\" button.\n9. Upload into the Opentrons Run App and follow the instructions there to set up your deck and proceed to run!\n10. Make sure to add diluent to the first row of your 12-row trough and load your desired samples/standards into column 1 of your plate before running your protocol in the run app!\n\n",
"title": "Customizable Serial Dilution for OT-2"
},
"notes": "Please reference our Technical Note for more information about the expected output of this protocol, in addition to expanded sample data from the Opentrons lab.\nWe understand that there are limitations to the use of this protocol, and we plan to make improvements soon. In the meantime, if you'd like to request a more complex dilution workflow, please use our Protocol Development Request Form. You can also download this Python file and modify it using our API Documentation. For additional questions about this protocol, please email support@opentrons.com.",
diff --git a/protoBuilds/demo/README.json b/protoBuilds/demo/README.json
index d2f4227fec..31431ce958 100644
--- a/protoBuilds/demo/README.json
+++ b/protoBuilds/demo/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Getting Started": [
"OT-2 Demo"
@@ -10,7 +10,7 @@
"internal": "demo",
"labware": "\nNEST 12 Well Reservoir 15 mL\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nNEST 96 Well Plate 200 \u00b5L Flat\nOpentrons 4x6 Tube Rack with Eppendorf 2 mL Safe-Lock Snapcap\nAny Opentrons 96 tiprack depending on what pipette is selected.\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Getting Started\n\t* OT-2 Demo\n\n",
"deck-setup": "Here's an example deck setup (note that this will vary based on your selected parameters below): \n\n\n",
"description": "Learn the function OT-2 quickly! This simple plating protocol shows the flexibility of custom protocols hosted on our Protocol Library.\n\nReagents are transferred down columns and then across rows as shown here. \n \n\nIf the source labware is selected to be a 24x tuberack, reagents will be transferred in quadrupliate (ex: reagent tube A1 to destination labware wells A1-D1, reagent tube B1 to destination labware wells E1-H1, reagent tube C1 to destination labware wells A2-D2, etc.)\n\nIf the source labware is selected to be a 12-channel reservoir, reagents will be transferred column to column (ex: reservoir channel 1 to destination labware column 1, reservoir channel 2 to destination labware column 2, etc.)\n\n---\n",
diff --git a/protoBuilds/dinosaur/README.json b/protoBuilds/dinosaur/README.json
index 99139caf7c..3288a5d041 100644
--- a/protoBuilds/dinosaur/README.json
+++ b/protoBuilds/dinosaur/README.json
@@ -8,7 +8,7 @@
"description": "Draw a picture of a dinosaur (Stegosaurus!) on a 96 well plate using food coloring.\n\n\n\n\nOpentrons 300uL Tips\nOpentrons 200uL Filter Tips\nBio-Rad 96 Well Plate 200 uL PCR\nNEST 96 Well Plate 100 uL PCR Full Skirt\nCorning 96 Well Plate 360 \u00b5L Flat\nNEST 96 Well Plate 200 \u00b5L Flat\nP300 Single Channel GEN2\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nDeck Setup\n\nProtocol Steps\n\nDistribute 50 uL of green solution to all the necessary wells.\nDistribute 50 uL of blue solution to all the necessary wells.\nEnjoy the dinosaur!\n",
"internal": "dinosaur",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/dinosaur). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/dinosaur). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Getting Started\n\t* Dinosaur\n\n",
"description": "Draw a picture of a dinosaur (Stegosaurus!) on a 96 well plate using food coloring.\n\n\n\n---\n\n\n* [Opentrons 300uL Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 200uL Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Bio-Rad 96 Well Plate 200 uL PCR](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr/)\n* [NEST 96 Well Plate 100 uL PCR Full Skirt](https://labware.opentrons.com/nest_96_wellplate_100ul_pcr_full_skirt/)\n* [Corning 96 Well Plate 360 \u00b5L Flat](https://labware.opentrons.com/corning_96_wellplate_360ul_flat/)\n* [NEST 96 Well Plate 200 \u00b5L Flat](https://labware.opentrons.com/nest_96_wellplate_200ul_flat/)\n* [P300 Single Channel GEN2](https://shop.opentrons.com/collections/ot-2-robot/products/single-channel-electronic-pipette?variant=5984549109789)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n---\n\n\n**Deck Setup**\n\n\n\n**Protocol Steps**\n\n1. Distribute 50 uL of green solution to all the necessary wells.\n2. Distribute 50 uL of blue solution to all the necessary wells.\n3. Enjoy the dinosaur!\n\n",
"internal": "dinosaur",
diff --git a/protoBuilds/dynabeads_plate_prep_1/README.json b/protoBuilds/dynabeads_plate_prep_1/README.json
index 1b0d4f80c5..d540817eff 100644
--- a/protoBuilds/dynabeads_plate_prep_1/README.json
+++ b/protoBuilds/dynabeads_plate_prep_1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Protein Purification": [
"Thermo Fisher Dynabeads\u2122 Protein A"
@@ -10,7 +10,7 @@
"internal": "dynabeads_plate_prep_1",
"labware": "\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\n4-in-1 Tube Rack Set\nNEST 15 mL Centrifuge Tube\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Protein Purification\n\t* Thermo Fisher Dynabeads\u2122 Protein A\n\n",
"deck-setup": "* Slot 4 \u2013 96 well deepwell plate (reagent plate)\n* Slot 5 - Tube rack/reagent in 15 mL conical tube (reagent stock)\n\n* Green \u2013 beads\n* Blue \u2013 antibody\n\n\n\n",
"description": "This protocol (Plate Prep 1) performs pipetting to transfer reagents (Dynabeads and antibody dilution) from 15 mL conical tubes to a 96 well deepwell plate (the reagent plate) on the OT2. This reagent plate is used for Dynabeads for IP Reagent-In-Plate protocol Part 1. The user can determine the number of samples to be processed.\n\n",
diff --git a/protoBuilds/dynabeads_plate_prep_2/README.json b/protoBuilds/dynabeads_plate_prep_2/README.json
index 2add4d2fb3..2e4e68ba8f 100644
--- a/protoBuilds/dynabeads_plate_prep_2/README.json
+++ b/protoBuilds/dynabeads_plate_prep_2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Protein Purification": [
"Thermo Fisher Dynabeads\u2122 Protein A"
@@ -10,7 +10,7 @@
"internal": "dynabeads_plate_prep_2",
"labware": "\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\n4-in-1 Tube Rack Set\nNEST 15 mL Centrifuge Tube\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Protein Purification\n\t* Thermo Fisher Dynabeads\u2122 Protein A\n\n",
"deck-setup": "* Slot 4 \u2013 96 well deepwell plate (reagent plate)\n* Slot 5 - Tube rack/reagent in 15 mL conical tube (reagent stock)\n\n* Purple \u2013 elution buffer\n\n\n\n",
"description": "This protocol (Plate Prep 2) performs pipetting to transfer elution buffer from 15 mL conical tubes to a 96 well deepwell plate (the reagent plate) on the OT2. This reagent plate is used for Dynabeads for IP Reagent-In-Plate protocol Part 2. The user can determine the number of samples to be processed.\n\n",
diff --git a/protoBuilds/dz1j39/README.json b/protoBuilds/dz1j39/README.json
index 979feddfdb..ce2fe8c82e 100644
--- a/protoBuilds/dz1j39/README.json
+++ b/protoBuilds/dz1j39/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Cell and Tissue Culture": [
"Cell and Tissue Culture"
@@ -10,7 +10,7 @@
"internal": "dz1j39",
"labware": "\nCorning 96 Well Plate 360 \u00b5L Flat #3650\nOpentrons 96 Tip Rack 300 \u00b5L\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 10 Tube Rack with Falcon 4x50 mL, 6x15 mL Conical\n",
"markdown": {
- "author": "Opentrons\n\n\n",
+ "author": "Opentrons\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Cell and Tissue Culture\n\t* Cell and Tissue Culture\n\n\n",
"deck-setup": "[deck]()\n\n\n",
"description": "This protocol can be used to measure the viability and cytotoxicity of two different cell types (suspension and adherent) that have been treated with respective drugs, using Cell Viability assay kit (Cell Titer Glo 2.0) and Cytotoxicity assay kit (CellTox Green) from Promega on the OT-2. This protocol is designed for the 96-well plate format and both assays can be processed in the same plate. In the case of A549 cells, the protocol is broken down into 3 main parts: a) Seeding/ Plating of A549 cells on Day 1 followed by b) treatment with thapsigargin on the second day c) After 72 hours of treatment, completion of both the assays in the same plate i.e., sequential multiplexing of CellTox Green Cytotoxicity Assay and CellTiter Glo 2.0 Assay\n\n\n",
diff --git a/protoBuilds/e54ada/README.json b/protoBuilds/e54ada/README.json
index cfa6118ce5..8a16b71129 100644
--- a/protoBuilds/e54ada/README.json
+++ b/protoBuilds/e54ada/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"DNA/RNA Extraction"
@@ -10,7 +10,7 @@
"internal": "e54ada",
"labware": "\nNEST 12 Reservoir 15mL\nOpentrons 200uL Filter Tips\nOpentrons 4-in-1 Tube Rack\nBioRad 96 Well Plate\nGreiner Microplate, 96 Well, PP, V-Bottom\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* DNA/RNA Extraction\n\n",
"deck-setup": "\n* Tube rack in slot 6 is only used when under 7 samples are specified. Beads are loaded in A1 of tube rack for 1-6 samples. If samples are 7-96 beads are loaded in slot 3, well 12\n\n\n\n",
"description": "This is a bead-based purification of nucleic acids using either BioRad 96 well plates or Grenier 96 well plates. Using P300 single and multi channel pipettes we have flexibility using 4-in-1 tube racks or 12 well reservoirs to hold beads. The beads can be diluted with 40% PEG 8000 mixtures or left neat from the reagent bottle.\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96)\n* `Reaction Volume`: Specify the starting sample volume in uL for the first plate\n* `Bead Ratio`: Specify the ratio of beads to sample volume with a float. Default is 1.8\n* `Elution Volume`: Specify the volume of elution liquid to use and subsequently transfer to the final plate in uL\n* `P300 Multi Channel Pipette Mount`: Specify which side the multi-channel P300 is mounted to. The single-channel P300 will default to the opposite side\n* `Well Plate Type`: Change between the BioRad plate in the OT-2 default labware library and the custom definition for Greiner 96 well plates with a circular well\n* `Flash`: Specify whether the robot will flash on pauses for tip rack refills and trash alerts\n\n---\n\n",
diff --git a/protoBuilds/e54ada_rt/README.json b/protoBuilds/e54ada_rt/README.json
index a8afd3abb0..f4a0f859ef 100644
--- a/protoBuilds/e54ada_rt/README.json
+++ b/protoBuilds/e54ada_rt/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR Prep": [
"Reverse Transcriptase"
@@ -10,7 +10,7 @@
"internal": "e54ada",
"labware": "\nOpentrons 20uL Filter Tips\nOpentrons 4-in-1 Tube Rack\nBioRad 96 Well Plate\nGreiner Microplate, 96 Well, PP, V-Bottom\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR Prep\n\t* Reverse Transcriptase\n\n",
"deck-setup": "\n* Tube racks in slots 1, 2, 4, and 5 map directly to the 96 well plate. I.e. if 95 samples are specified all four tube racks will load in, RT mix will be in the bottom right well, C4, for slot 2. Well G12 will have primers added from slot 2, tube C3. If 13 samples are specified, the tube racks in slot 4 and 1 will load in, RT mix will be in well B2 in slot 1 (the 14th tube). Slot 1 A1's primer will add to well E1 in slot 3's well plate. Click [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/e54ada_rt/Tube+to+Plate+Mapping+for+RT.xlsx) for an explanatory spreadsheet\n\n\n\n",
"description": "This is a plate filling protocol for a reverse transcriptase procedure. Up to 95 samples can be prepped using 4-in-1 tube racks filled with barcode primer mixes and a prepared enzyme mix in the final tube.\n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify the number of samples this run (1-95)\n* `Reagent Volume to Add`: Specify the volume of RT mix to add. Default is 4.7 uL\n* `Well Plate Type`: Change between the BioRad plate in the OT-2 default labware library and the custom definition for Greiner 96 well plates with a circular well\n* `P20 Mount`: Specify which side the single-channel P20 is mounted to\n\n---\n\n",
diff --git a/protoBuilds/e9ff8d/README.json b/protoBuilds/e9ff8d/README.json
index b5f010f923..fb61476a3c 100644
--- a/protoBuilds/e9ff8d/README.json
+++ b/protoBuilds/e9ff8d/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"Pre-Amplification"
@@ -10,7 +10,7 @@
"internal": "e9ff8d",
"labware": "\nEppendorf 96 Well Plate 200 \u00b5L on Opentrons Semi-Skirted Adapter\nEppendorf 96 Well Plate 200 \u00b5L on Aluminum Block 200 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 24 Well Aluminum Block with NEST 2 mL Snapcap\nOpentrons 96 Tip Rack 300 \u00b5L or Opentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* Pre-Amplification\n\n\n",
"deck-setup": "\n\n\n",
"description": "This is the first of four parts for a custom PCR prep followed by two stage cleanup.\n\n\n",
diff --git a/protoBuilds/e9ff8d_part2/README.json b/protoBuilds/e9ff8d_part2/README.json
index 0c672c52cc..3aaa553e37 100644
--- a/protoBuilds/e9ff8d_part2/README.json
+++ b/protoBuilds/e9ff8d_part2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Purification": [
"DNA Cleanup"
@@ -10,7 +10,7 @@
"internal": "e9ff8d_part2",
"labware": "\nBiorad 96 Well Plate 200 \u00b5L to hold Generic PCR Strips\nEppendorf 96 Well Plate 200 \u00b5L on Opentrons Semi-Skirted Adapter\nEppendorf 96 Well Plate 200 \u00b5L on Aluminum Block 200 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 24 Well Aluminum Block with NEST 2 mL Snapcap\nOpentrons 96 Tip Rack 300 \u00b5L or Opentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Purification\n\t* DNA Cleanup\n\n\n",
"deck-setup": "\n\n\n",
"description": "This is the second of four parts for a custom PCR prep followed by two stage cleanup. This is the first of two cleanups.\n\n\n",
diff --git a/protoBuilds/e9ff8d_part3/README.json b/protoBuilds/e9ff8d_part3/README.json
index 559f8d9180..3c35655258 100644
--- a/protoBuilds/e9ff8d_part3/README.json
+++ b/protoBuilds/e9ff8d_part3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "e9ff8d_part3",
"labware": "\nFoil-Covered Index Adapter 96 Well Plate, 200 \u00b5L\nEppendorf 96 Well Plate 200 \u00b5L on Opentrons Semi-Skirted Adapter\nEppendorf 96 Well Plate 200 \u00b5L on Aluminum Block 200 \u00b5L\nOpentrons 24 Well Aluminum Block with NEST 2 mL Snapcap\nOpentrons 96 Tip Rack 300 \u00b5L or Opentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n\n",
"deck-setup": "\n\n\n",
"description": "This is the third of four parts for a custom PCR prep followed by two stage cleanup.\n\n\n",
diff --git a/protoBuilds/e9ff8d_part4/README.json b/protoBuilds/e9ff8d_part4/README.json
index ce5415c0d7..bc9a2b87d2 100644
--- a/protoBuilds/e9ff8d_part4/README.json
+++ b/protoBuilds/e9ff8d_part4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Purification": [
"DNA Cleanup"
@@ -10,7 +10,7 @@
"internal": "e9ff8d_part4",
"labware": "\nBiorad 96 Well Plate 200 \u00b5L to hold Generic PCR Strips\nEppendorf 96 Well Plate 200 \u00b5L on Opentrons Semi-Skirted Adapter\nEppendorf 96 Well Plate 200 \u00b5L on Aluminum Block 200 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons 24 Well Aluminum Block with NEST 2 mL Snapcap\nOpentrons 96 Tip Rack 300 \u00b5L or Opentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Purification\n\t* DNA Cleanup\n\n\n",
"deck-setup": "\n\n\n",
"description": "This is the fourth of four parts for a custom PCR prep followed by two stage cleanup. This is the second of two cleanups.\n\n\n",
diff --git a/protoBuilds/ff5763/README.json b/protoBuilds/ff5763/README.json
index 72725b6ce6..f3e83d8564 100644
--- a/protoBuilds/ff5763/README.json
+++ b/protoBuilds/ff5763/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina DNA Prep"
@@ -10,7 +10,7 @@
"internal": "ff5763",
"labware": "\nCustom 96 Well Plate with AB Gene and NEST 96 well plate\nPCR Strip Tubes for Master Mix\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Illumina DNA Prep\n\n",
"deck-setup": "* Deck Layout for 96 samples\n\n---\n\n",
"description": "This is part one of a five part protocol for the [Illumina DNA Prep kit](https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/nextera-dna-flex.html)\n\n[Part 2](https://develop.protocols.opentrons.com/protocol/ff5763_part2)\n\n[Part 3](https://develop.protocols.opentrons.com/protocol/ff5763_part3)\n\n[Part 4](https://develop.protocols.opentrons.com/protocol/ff5763_part4)\n\n[Part 5](https://develop.protocols.opentrons.com/protocol/ff5763_part5)\n\nPart 1: Tagmentation\nMaster mix is prepared offdeck and added to DNA samples. DNA samples are 30 uL in total volume. After master mix addition, the sample plate is put in a thermocycler pre-programmed as outlined in the reference manual.\n\nOnce the thermocycler is complete, part 2 can be run.\n\nVariables:\n* `Number of Samples`: Total number of samples from 1 to 48\n\n\n---\n\n",
diff --git a/protoBuilds/ff5763_part2/README.json b/protoBuilds/ff5763_part2/README.json
index f8072c587c..58f9f965bf 100644
--- a/protoBuilds/ff5763_part2/README.json
+++ b/protoBuilds/ff5763_part2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina DNA Prep"
@@ -10,7 +10,7 @@
"internal": "ff5763",
"labware": "\nPCR Strip Tubes for Tagmentation Stop Buffer (TSB) in slot 1, row 5\nCustom 96 Well Plate with AB Gene and NEST 96 well plate with samples, slot 2\nNEST 2ml Deep Well Plate on Magnetic Module in slot 4\nNEST 12-Well 15ml Reservoir in slot 5\nNEST 195ml Reservoir in slot 6\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Illumina DNA Prep\n\n",
"deck-setup": "* Deck Layout for 96 samples\n\n---\n\n",
"description": "This is part two of a five part protocol for the [Illumina DNA Prep kit](https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/nextera-dna-flex.html)\n\n[Part 1](https://develop.protocols.opentrons.com/protocol/ff5763)\n\n[Part 3](https://develop.protocols.opentrons.com/protocol/ff5763_part3)\n\n[Part 4](https://develop.protocols.opentrons.com/protocol/ff5763_part4)\n\n[Part 5](https://develop.protocols.opentrons.com/protocol/ff5763_part5)\n\nPart 2: Post-Tagmentation Clean-Up\nTagmentation Stop Buffer (TSB) is added to each sample with a multi-channel 20ul pipette, using a fresh tip each time. The multi-channel 300ul pipette is then used to mix the samples before the plate is tranferred to an off-deck thermocycler. Once the post-tagmentation thermocycler program has completed, the plate is returned to slot 2 and 'Resume' is clicked on the OT-2 application\n\nSamples are subsequently moved to a deep well plate locked onto the magnetic module in slot 4. The magnetic module is engaged and left for 3 minutes. After the 3 minutes the supernatant is discarded into the liquid trash in slot 6.\n\nTagmentation Wash Buffer (TWB) is utilized to wash the samples in the magnetic module twice as follows:\n100ul TWB is added to the samples, the magnetic module is engaged for 3 minutes, then supernatant is removed to liquid trash in slot 6.\n\nAfter the two TWB washes a third TWB wash is partially done. 100ul TWB is added to the samples and the magnetic module is engaged. Part 3 should be initiated as soon as possible after this step.\n\nExplanation of complex parameters below:\n* `Number of Samples`: Total number of samples from 1 to 48\n\n---\n\n",
diff --git a/protoBuilds/ff5763_part3/README.json b/protoBuilds/ff5763_part3/README.json
index 88da4345ce..f043f7cdbc 100644
--- a/protoBuilds/ff5763_part3/README.json
+++ b/protoBuilds/ff5763_part3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina DNA Prep"
@@ -10,7 +10,7 @@
"internal": "ff5763",
"labware": "\nCustom 96 Well Plate with AB Gene and NEST 96 well plate with samples, slot 2\nNEST 2ml Deep Well Plate on Magnetic Module in slot 4\nNEST 12-Well 15ml Reservoir in slot 5\nNEST 195ml Reservoir in slot 6\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Illumina DNA Prep\n\n",
"deck-setup": "* Deck Layout for 96 samples, barcoding primers\n\n\n\n* Deck Layout for 96 samples, non-barcoding primers\n\n---\n\n",
"description": "This is part three of a five part protocol for the [Illumina DNA Prep kit](https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/nextera-dna-flex.html)\n\n[Part 1](https://develop.protocols.opentrons.com/protocol/ff5763)\n\n[Part 2](https://develop.protocols.opentrons.com/protocol/ff5763_part2)\n\n[Part 4](https://develop.protocols.opentrons.com/protocol/ff5763_part4)\n\n[Part 5](https://develop.protocols.opentrons.com/protocol/ff5763_part5)\n\nPart 3: Amplify Tagmented DNA\nSupernatant is discarded from the sample plate on the magnetic module into the liquid trash in slot 6\n\n40 uL of PCR Master Mix is added to the samples\n\nSamples are transferred from the magnetic module deep well plate to the thermocycler plate in slot 2 before being centrifuged. Index adapters are added to the samples and mixed. The sample plate in slot 2 is moved to an off-deck thermocycler for a pre-programed PCR cycle as outlined in the Illumina DNA Prep reference material.\n\nExplanation of complex parameters below:\n* `Number of Samples`: Total number of samples from 1 to 48\n\n---\n\n",
diff --git a/protoBuilds/ff5763_part4/README.json b/protoBuilds/ff5763_part4/README.json
index 89b21d246a..a4adb06f0b 100644
--- a/protoBuilds/ff5763_part4/README.json
+++ b/protoBuilds/ff5763_part4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina DNA Prep"
@@ -10,7 +10,7 @@
"internal": "ff5763",
"labware": "\nCustom 96 Well Plate with AB Gene and NEST 96 well plate with samples, slot 2\nNew NEST 2ml Deep Well Plate in slot 3\nNEST 2ml Deep Well Plate containing samples from previous steps on Magnetic Module in slot 4\nNEST 12-Well 15ml Reservoir in slot 5 (reagent reservoirs)\nNEST 195ml Reservoir in slot 6 (liquid trash)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Illumina DNA Prep\n\n",
"deck-setup": "* Deck Layout for 96 samples\n\n---\n\n",
"description": "This is part four of a five part protocol for the [Illumina DNA Prep kit](https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/nextera-dna-flex.html)\n\n[Part 1](https://develop.protocols.opentrons.com/protocol/ff5763)\n\n[Part 2](https://develop.protocols.opentrons.com/protocol/ff5763_part2)\n\n[Part 3](https://develop.protocols.opentrons.com/protocol/ff5763_part3)\n\n[Part 5](https://develop.protocols.opentrons.com/protocol/ff5763_part5)\n\nPart 4: Cleanup Libraries First Half\n\nSamples are centrifuged down before adding the sample tray to slot 2 on the OT-2's deck. Tray contents are transferred to the available wells in the deep well plate on the magnetic module and left until clear (~5 minutes). 45ul of the resulting supernatant is moved to the second deep well plate in slot 3. 40ul of NFW is added to this supernatant for each sample. 45ul of IPB mix is then added to the now diluted samples and mixed 10x at 100ul each time. The deep well plate is left to incubate for 5 minutes. This ends part 4.\nExplanation of complex parameters below:\n* `Number of Samples`: Total number of samples from 1 to 48\n\n---\n\n",
diff --git a/protoBuilds/ff5763_part5/README.json b/protoBuilds/ff5763_part5/README.json
index f7bf62e00f..413db4184b 100644
--- a/protoBuilds/ff5763_part5/README.json
+++ b/protoBuilds/ff5763_part5/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina DNA Prep"
@@ -10,7 +10,7 @@
"internal": "ff5763",
"labware": "\nNew Custom 96 Well Plate with AB Gene and NEST 96 well plate with samples, slot 2\nNEST 2ml Deep Well Plate containing samples from previous step on Magnetic Module in slot 4\nNEST 12-Well 15ml Reservoir in slot 5 (reagent reservoirs)\nNEST 195ml Reservoir in slot 6 (liquid trash)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n * Illumina DNA Prep\n\n",
"deck-setup": "* Deck Layout for 96 samples\n\n---\n",
"description": "This is part five of a five part protocol for the [Illumina DNA Prep kit](https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/nextera-dna-flex.html)\n\n[Part 1](https://develop.protocols.opentrons.com/protocol/ff5763)\n\n[Part 2](https://develop.protocols.opentrons.com/protocol/ff5763_part2)\n\n[Part 3](https://develop.protocols.opentrons.com/protocol/ff5763_part3)\n\n[Part 4](https://develop.protocols.opentrons.com/protocol/ff5763_part4)\n\nPart 5: Cleanup Libraries 2\nMagnetic module is engaged for 5 minutes. IPB is added to the right side of the magnetic module. 125ul of supernatant is transferred from the left side of the magnetic module to the right side containing the bead mixture and mixed 10x. 5 minute incubation occurs followed by discarding of supernatant. Beads are then washed with 80% ethyl alcohol twice and allowed to airdry for 5 minutes. RSB is added to the samples and a 2 minute delay occurs. The magnetic module is now engaged for 2 minutes followed by a final transfer of resulting supernatant to an awaiting 96 well plate in slot 2.\n\nExplanation of complex parameters below:\n* `Number of Samples`: Total number of samples from 1 to 48\n\n---\n\n",
diff --git a/protoBuilds/generic_ngs_prep/README.json b/protoBuilds/generic_ngs_prep/README.json
index ea0991d7ca..02032a0ff3 100644
--- a/protoBuilds/generic_ngs_prep/README.json
+++ b/protoBuilds/generic_ngs_prep/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Generic"
@@ -10,7 +10,7 @@
"internal": "generic_ngs_prep",
"labware": "Plates\n Bio-Rad 96 Well Plate 200 \u00b5L PCR\n Opentrons 96 Well Aluminum Block with Generic PCR Strip 200 \u00b5L\n Opentrons 96 Well Aluminum Block with Bio-Rad Well Plate 200 \u00b5L\n Opentrons 96 Well Aluminum Block with NEST Well Plate 100 \u00b5L\n* NEST 96 Well Plate 100 \u00b5L PCR Full Skirt\nTips\n Opentrons 96 Filter Tip Rack 200 \u00b5L\n Opentrons 96 Filter Tip Rack 20 \u00b5L\n12 well reservoir options\n NEST 12 Well Reservoir 15 mL\n USA Scientific 12 Well Reservoir 22 mL\nreservoir options\n Agilent 1 Well Reservoir 290 mL\n Axygen 1 Well Reservoir 90 mL\n* NEST 1 Well Reservoir 195 mL\nPipettes\n P20 Multichannel GEN2 Pipette\n P300 Multichannel GEN2 Pipette\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Generic\n\n",
"deck-setup": "**Note: The deck setup changes throughout the course of the protocol. The initial calibration layout may differ from the final setup. Please follow the initial calibration setup and any instructions during the pauses.**\n\nInitial deck state:\n\n\n\n**Step 1**\n* Slot 1: Temperature Module slot 1 + Sample Plate\n* Slot 2: Reagent reservoir (SPRI, Buffer I, Mastermix)\n* Slot 3: Magnetic Module (Empty)\n* Slot 4: Temperature Module slot 4 + Reagent 1 Plate\n* Slot 6: 1 Well Reservoir (Ethanol)\n* Slot 7: Temperature Module slot 7 + Reagent 2 plate\n* Slot 8: Opentrons 200 uL Filter Tips\n* Slot 9: Opentrons 200 uL Filter Tips\n* Slot 10: Opentrons 20 uL Filter Tips\n* Slot 11: Opentrons 20 uL Filter Tips\n\n**Step 2**\n* Slot 1: Temperature Module slot 1\n* Slot 2: Reagent reservoir (SPRI, Buffer I, Mastermix)\n* Slot 3: Magnetic Module + Sample plate\n* Slot 4: Temperature Module slot 4 + Primers\n* Slot 6: 1 Well Reservoir (Ethanol Reservoir)\n* Slot 7: Temperature Module slot 7 + Indexing plate\n* Slot 8: Opentrons 200 uL Filter Tips\n* Slot 9: Opentrons 200 uL Filter Tips\n* Slot 10: Opentrons 20 uL Filter Tips\n* Slot 11: Opentrons 20 uL Filter Tips\n\n---\n\n",
"description": "This protocol performs a generic NGS Library Prep. It utilizes up to three temperature modules in Slot 1, 4 and 7 for the cooling of reagents. It also utilizes the magnetic module for bead based purification of samples.\n\nExplanation of parameters below:\n* `P20 Multichannel GEN2 Mount Position`: Choose the mount position of the P20 Multichannel GEN2 pipette. (left or right)\n* `P300 Multichannel GEN2 Mount Position`: Choose the mount position of the P300 Multichannel GEN2 pipette. (left or right\n* `Number of Samples`: Enter total number of samples in the protocol run. **Note: Because both pipettes are 8-channels, the number of samples should be a multiple of 8 or close if possible, otherwise it will use up additional tips for an entire column.**\n* `Temperature module in slot 1` (Optional parameter) Specify which temperature module to use in slot 1 (or None). This temperature module would allow the user to control the temperature of the sample plate in part 1 of the protocol.\n* `Temperature module in slot 1` (Optional parameter) Specify which temperature module to use in slot 4 (or None). This temperature module would allow the user to control the temperature of reagent plate 1 in part 1 of the protocol, and the primer plate in part 2.\n* `Temperature module in slot 7` (Optional parameter) Specify which temperature module to use in slot 7 (or None). This temperature module would allow the user to control the temperature of reagent plate 2 in part 1 of the protocol, and the Indexing plate in part 2.\n* `Sample plate` The sample plate holds DNA samples and is the site of mixing with buffers and reagents in part 1.\n* `Reagent 1 plate` The reagent 1 plate holds the first mixing reagent in column 1 (e.g. a buffer or an enzyme) to be mixed with the samples\n* `Reagent 2 plate` The reagent 2 plate holds the second mixing reagent in column 1 (e.g. a buffer or an enzyme) to be mixed with the samples.\n* `Primer plate` Contains primers to be mixed with DNA samples in step 2\n* `Reagent reservoir` The (12 well) reservoir contains paramagnetic bead solution, PCR buffer, and PCR mastermix.\n* `Ethanol reservoir` Ethanol solution for washing beads, e.g. 80:20 ethanol:water. Uses 180 \u00b5L per sample\n* `Number of Samples` The number of DNA samples (up to 96)\n* `Temperature (deg C) for temperature module on slot x in part 1` These parameters for each of the temperature modules allow the user to set the temperatures for all/any temperature module in part 1.\n* `Temperature (deg C) for temperature module on slot x in part 2` These parameters for each of the temperature modules allow the user to set the temperatures for all/any temperature module in part 2.\n* `Volume of reagent 1 to add` Volume of reagent 1 to add to samples, e.g. an enzyme or a reaction buffer\n* `Volume of reagent 2 to add` Volume of reagent 2 to add to samples, e.g. an enzyme or a reaction buffer\n* `Ethanol wash volume` The volume of ethanol solution to use for bead washing\n* `Mastermix volume` Volume of PCR mastermix for the PCR reaction in step 2, this is added to the indexing plate\n* `Primer mix volume` Volume of primer mix for the PCR reaction in step 2, this is added to the indexing plate\n* `Bead volume` Volume of paramagnetic bead solution to use for cleaning up the DNA from the enzymatic reaction in part 1 of the protocol\n* `Elution buffer volume` The volume of elution buffer to use for eluting the DNA after the ethanol cleanup\n* `Sample volume` The initial volume of DNA on the sample plate\n* `DNA supernatant volume` The volume of clean DNA to be transferred from the magnetic deck plate to the indexing plate to be used in the PCR reaction\n\n---\n\n",
diff --git a/protoBuilds/generic_pcr_prep_1/README.json b/protoBuilds/generic_pcr_prep_1/README.json
index 0fd383e427..f4ad659e6e 100644
--- a/protoBuilds/generic_pcr_prep_1/README.json
+++ b/protoBuilds/generic_pcr_prep_1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"Generic Mastermix Assembly"
@@ -10,7 +10,7 @@
"internal": "generic_pcr_prep_1",
"labware": "\nTube racks\nAluminum blocks\n12 well reservoirs\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * Generic Mastermix Assembly\n\n",
"deck-setup": "* Example setup\n\n\n\n\n\n\n* Slot 1: Tube rack or well plate 1\n* Slot 2: Tube rack or well plate 2\n* Slot 3: 12-Channel reservoir: Well 1 - Mastermix target\n* Slot 4: Tiprack for the left pipette\n* Slot 5: Tiprack for the right pipette\n* Slot 6: Empty\n* Slot 7: Empty\n* Slot 8: Empty\n* Slot 9: Empty\n* Slot 10: Empty\n* Slot 11: Empty\n\n---\n\n",
"description": "Part 1 of 2: Master Mix Assembly\n\nThis protocol allows your robot to create a master mix solution using any reagents stored in one to three different pieces of labware such as a tube racks, well plates, and 12 well reservoirs. The master mix will be created in well A1 of the trough of the chosen reservoir. The ingredient information will be provided as a CSV file. See Additional Notes for more details.\n\nLinks:\n* [Part 1: Master Mix Assembly](./generic_pcr_prep_1)\n* [Part 2: Master Mix Distribution and DNA Transfer](./generic_pcr_prep_2)\n\n\nExplanation of parameters below:\n* `right pipette type`: Pipette for the right mount\n* `Left pipette type`: Pipette for the left mount\nFor the pipette choices it is important that the pipettes selected will cover the range of volumes used in the protocol\n* `master mix .csv file`: Here, you should upload a .csv file formatted in the following way, being sure to include the header line:\n\n\n* `Left pipette: Filter or regular tips?` Select whether you want unfiltered or filtered tips. The protocol will load the appropriate tips, e.g. 20 uL tips for a 20 uL pipette, 1000 uL filtered/nonfiltered for a 1000 uL pipette etc.\n* `Right pipette: Filter or regular tips?` Select whether you want unfiltered or filtered tips. The protocol will load the appropriate tips, e.g. 20 uL tips for a 20 uL pipette, 1000 uL filtered/nonfiltered for a 1000 uL pipette etc.\n* `Reagent labware 1` Choose the appropriate labware (such as tube racks or well plates) for your reagents, you may choose aluminum blocks if you intend to use a temperature module. This is referred to as \"slot 1\" in the .csv\n* `Reagent labware 2` (Optional) Choose a secondary unit of labware for your reagents, you may choose aluminum blocks if you intend to use a temperature module. This is referred to as \"slot 2\" in the .csv\n* `Twelve well Reservoir and mastermix destination` Select the type of 12 well reservoir to use. The mastermix will be created in well A1. You may use the other wells as sources for reagents (slot 3 in the .csv)\n\n\n---\n\n",
diff --git a/protoBuilds/generic_pcr_prep_2/README.json b/protoBuilds/generic_pcr_prep_2/README.json
index 10053de1c9..3db00bbfc0 100644
--- a/protoBuilds/generic_pcr_prep_2/README.json
+++ b/protoBuilds/generic_pcr_prep_2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"Generic PCR Prep"
@@ -10,7 +10,7 @@
"internal": "generic_pcr_prep_2",
"labware": "\nReservoirs\nDeep Well plates which may also be used as mastermix reservoirs\nWell plates\nAlternatively: Well plate/PCR strips on aluminum blocks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n * Generic PCR Prep\n\n",
"deck-setup": "* Example setup with a 384 well destination plate and four 96 well plates\n\n\n\n\n\n* Slot 1: Template DNA plate 2 (optional: for transferring to a 384 well plate)\n* Slot 2: Template DNA plate 3 (optional: for transferring to a 384 well plate)\n* Slot 3: Reservoir: or Deep well plate: Well 1 (to 8)- Mastermix source\n* Slot 4: Tiprack 1 for the left pipette\n* Slot 5: Tiprack 1 for the right pipette\n* Slot 6: Destination plate (where mastermix and template DNA is combined, optionally on a temperature module)\n* Slot 7: Tiprack 2 for the right pipette\n* Slot 8: Tiprack 2 for the left pipette\n* Slot 9: Template DNA plate 1 (optionally on a temperature module)\n* Slot 10: Tiprack 3 for the left pipette\n* Slot 11: Template DNA plate 4 (optional: for transferring to a 384 well plate)\n\n\n",
"description": "Part 2 of 2: Master Mix Distribution and DNA Transfer\n\nLinks:\n* [Part 1: Master Mix Assembly](./generic_pcr_prep_1)\n* [Part 2: Master Mix Distribution and DNA Transfer](./generic_pcr_prep_2)\n\n\nThis protocol allows your robot to distribute a master mix solution from well A1 of a reservoir or deep well plate to a target (a plate or a set of PCR strips on an aluminum block). The robot will then transfer DNA samples to the destination. At this point the samples can be mixed if so desired. The protocol works with both single- and multi-channel pipettes, just be sure that the minimum end of their combined volume range covers the smallest volume. There is also an option to place the DNA template plate, and the target plate on temperature modules. The protocol allows a user to 1) transfer the DNA samples of a 96 well plate to the target 96 well plate or 2) transfer up to four 96 well plates containing template DNA to a 384 well plate.\n\nIf only one pipette is selected it can use all 5 tipracks located on slots 4, 5, 7, 8, and 10. If two pipettes are loaded 4,7, and 10 are used by the left pipette and 5, and 8 by the right pipette.\n\nWhen 96 well plates are transferred with a multi-channel pipette to 384 well plates their columns are intercalated on the 384 well plate, e.g. column 1 and 2 of the 96 well plate go into odd the and even rows of the 1st column of the 384 well plate respectively. Then column 3, and 4 of the 96 well plate go into the next column of the 384 well plate etc.\n\nIf a single channel pipette is used the DNA will be transferred sequentially from 96 well plates to the 384 well plate\n\nExplanation of parameters below:\n* `Number of samples` : The number of DNA template samples to mix with PCR mastermix on the target well plate. This parameter controls how many plate columns the mastermix is transferred to as well as how many columns of samples are transferred to the destination plate.\n* `Number of mixes`: How many times to mix the DNA template samples with the mastermix after adding them together\n* `Aspiration rate multiplier`: A multiplier for controlling the flow rate of aspiration: Less than 1 to slow down, or greater than one to speed up aspiration.\n* `Dispensation rate multiplier`: A multiplier for controlling the flow rate of dispensation, less than 1 to slow down, or greater than one to speed up dispensation.\n* `Right pipette type`: Pipette in the right mount, it can be either a single channel or a multi-channel pipette\n* `Left pipette type`: Pipette in the left mount, it can be either a single channel or a multi-channel pipette.\n* `Filtered or unfiltered tips for the left pipette?`: Whether the left pipette is using filter or regular tips. for P20s and P10s the options are Opentrons 20 \u00b5L filtered tips, or 20 \u00b5L unfiltered.\nfor P50 and P300s the filtered tips are 200 \u00b5L Opentrons filtered tips, and the non-filtered are 300 \u00b5L Opentrons regular tips. For the P1000 the options is either Opentrons 1000 \u00b5L filtered or non-filtered tips.\n* `Filtered or unfiltered tips for the right pipette?`: Whether the right pipette is using filter or regular Opentrons tips\n* `Mastermix volume (in \u00b5l)`: The volume of mastermix to transfer to each well on the destination plate (in \u00b5L)\n* `DNA volume (in \u00b5l)`: The amount of DNA template to transfer to each destination plate from the DNA template plate (in \u00b5L)\n* `Mastermix reservoir`: A 1 well, or 12 well reservoir, or a deep well plate containing your PCR mastermix in well `A1` (or the column spanning `A1` to `H1`)\n* `PCR well plate (or PCR strips) containing template DNA`: Your source of template DNA, such as a 96 well plate (up to 4 when the destination plate is a 384 well plate)\n* `Destination PCR well plate or PCR strips`: This is the well plate where the PCR mastermix and the DNA template is transferred to and mixed. This can be a 96 well plate, PCR strips on an aluminum block, or a 384 well plate.\n* `Temperature module for the 1st DNA template well plate`: (Optional) You can load a temperature module for your (1st) DNA template well plate if you want to control its temperature.\n* `Temperature module for the destination well plate`: (Optional) You can load a temperature module for your destination well plate if you want to control its temperature.\n\n\n---\n\n\n",
diff --git a/protoBuilds/generic_protein_purification/README.json b/protoBuilds/generic_protein_purification/README.json
index dc6ae137a9..f1d9600c99 100644
--- a/protoBuilds/generic_protein_purification/README.json
+++ b/protoBuilds/generic_protein_purification/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Protein Purification": [
"Generic Protein Purification"
@@ -10,7 +10,7 @@
"internal": "generic_protein_purification",
"labware": "\n12-well Reservoirs\nNEST 12 well reservoir, 15 mL\n\n\nWell plates\nNEST 2 mL 96-Well Deep Well Plate\n\n\nTarget plate\nNEST 96 Well Plate Flat\n\n\nTube racks\n4-in-1 Tube Rack Set\n\n\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Protein Purification\n * Generic Protein Purification\n\n",
"deck-setup": "\n\n",
"description": "\nThe generic protein purification is a flexible protocol that starts from bacterial\nmedia ready for lysis on a well plate. From there the protocol adds lysis buffer\nand then proceeds to bind the target his-tagged proteins with nickel linked paramagnetic beads. After a variable amount of washes the user may choose to\nelute the proteins using elution buffer, or with SDS buffer if the purpose is to\nrun the samples on a gel.\n\nThe user is given extensive control of the protocol from the parameters. Pipettes, labware, modules (i.e. temperature and magnetic modules), protocol steps, and volumes are examples of things that can be customized to the users specifications.\n\nThe protocol is based on Promega's MagneHis\u2122 system, but should work for any other similar system.\n\n**Parameters**\n\n* `Magnet engagment time for bead binding` How long to engage magnets in order to attract the beads before doing any pipetting steps\n* `Number of samples` How many bacterial media samples there are on the sample plate\n* `Small pipette` The smaller pipette, can be a multi-channel or single\n* `Large pipette`The larger pipette, can be a multi-channel or single\n* `Small pipette tip-racks` Tips to use with the small pipette\n* `Large pipette tip-racks` Tips to use with the large pipette\n* `Reagent reservoir` 12 well reservoir for reagents: Lysis buffer, NaCl (optional), Wash buffer, elution buffer, SDS buffer (optional), paramegnetic beads.\n* `Destination plate` Plate where eluted proteins are transferred after adding elution or SDS buffer\n* `Sample plate` Plate containing wells with bacterial media\n* `Magnetic module` Which magnetic module to use for the bead binding steps\n* `Tube rack (optional)` Tube rack for DNAse I (well 1) if used.\n* `Destination plate temperature module (optional)` A temperature module for controlling the temperature of the destination plate\n* `Pause robot operation to allow user to vortex magnetic beads?` If this parameter is on, the robot will pause before the paramagnetic bead addition step to allow the user to resuspend the beads by vortexing the solution before adding it to the reservoir.\n* `Elute proteins in SDS buffer instead of elution buffer` Allows the user to elute proteins with SDS buffer instead of elution buffer if the proteins are intended to be run on a gel\n* `Add DNAse I to lysis sample from a tube?` If yes, then DNAse will be added from well A1 of the (optional) tube rack. If no, then the DNAse should be added to the lysis buffer in the reservoir prior to running the protocol\n* `Add NaCl to samples to a conc. of 500 mM during wash steps to enhance protein binding?` Optional; If you want to do this step, Add a 4M NaCl solution to the reservoir (see picture in the deck setup section)\n* `Volume of paramagnetic beads to use with each sample (\u00b5L)` Volume of paramagnetic beads to add to each sample well after the lysis step\n* `Initial volume (of bacteria containing media in each sample well) (\u00b5L)` Initial volume of sample in each sample plate well containing bacterial culture to be lysed.\n* `Wash buffer volume for the washing steps (\u00b5L)` Volume of wash buffer to use in each washing step\n* `Elution buffer volume to elute proteins with (\u00b5L)` How much elution buffer to elute the purified protein with.\n* `Number of washing steps` How many repetitions of washing the beads to remove unwanted proteins and other contaminants.\n* `Number of times to mix the wash buffer with the beads for each wash step` Number of times to mix (pipette solution up and down) for each washing step in order to mix the solution.\n* `Number of times to mix the elution buffer with the beads` Number of times to mix the elution buffer with the samples before transferring the protein containing supernatant.\n* `Bead incubation time for binding and eluting protein (before magnets are engaged)` How many minutes to incubate beads with the lysed bacterial solution before engaging magnets, also the amount of time to incubate the beads with elution buffer before engaging magnets.\n* `Number of times to mix the bead solution before transferring beads to the samples` Optional number of times to use the large pipette to mix the bead solution before transferring the bead solution to the samples\n* `SDS buffer volume to elute proteins with (optional: this only matters if you decide to elute with SDS buffer)` In the final step you can choose whether to elute with elution buffer or SDS buffer if you want to run a gel on the samples, this parameter specifies the volume in microliter of SDS buffer.\n\n---\n\n",
diff --git a/protoBuilds/generic_station_A/README.json b/protoBuilds/generic_station_A/README.json
index 492f4f1c28..aad5c66482 100644
--- a/protoBuilds/generic_station_A/README.json
+++ b/protoBuilds/generic_station_A/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Covid Workstation": [
"Sample Plating"
@@ -8,7 +8,7 @@
"description": "This protocol automates sample plating from collection tubes to a 96-well plate. After sample plating (Station A), the plate containing samples can be used on Station B for RNA extraction as outlined in our article on automating Covid-19 testing.\n\nUsing a Single-Channel Pipette, this protocol will transfer the user-specified volume from sample tubes placed in the Opentrons Tube Rack into a 96-well plate. Users can specify which pipette, the type of source tube, the type of destination plate, the number of samples to transfer, and the volume of each sample being transferred.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons Single-Channel Pipette\nOpentrons Filter Tips\nOpentrons 4-in-1 Tube Rack Set\n96-Well Plate\nSample Tubes\n\n\n\nThe Opentrons tiprack should be placed in slot 3 and the 96-well plate should be placed in slot 2.\nWith this protocol, the Opentrons tube rack with samples will be accessed in the following order: slot 1, 4, 7, 10, 5, 8, 11 -- see below for examples.\n\nUsing the customizations field (below), set up your protocol.\n Number of Samples: Specify the number of samples to run (1-96)\n Volume of Samples (in \u00b5L): Specify the volume to be transferred from sample tubes to destination plate (this volume should fall in the range of the selected pipette)\n Destination Plate Labware: Specify whether to use the NEST Deepwell Plate, 2mL or the NEST PCR Plate as the destination plate\n Source Tube Labware: Specify whether to use the Opentrons Tubebrack (24) with 2mL, 1.5mL, or 0.5mL tubes or the Opentrons Tuberack (15) with 15mL conical tubes.\n* Pipette Type: Specify which pipette (p300/p1000 GEN1/GEN2) to use (should be mounted on the right gantry)\n\n\nExample layout: 15 samples with 2mL tubes\n\n\n\nExample layout: 15 samples with 15mL tubes\n\n\n\nExample layout: 96 samples with 2mL tubes\n\n\n\nExample layout: 96 samples with 15mL tubes\n\n\n",
"internal": "generic_station_A",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Covid Workstation\n\t* Sample Plating\n\n\n",
"description": "This protocol automates sample plating from collection tubes to a 96-well plate. After sample plating (Station A), the plate containing samples can be used on Station B for RNA extraction as outlined in our [article on automating Covid-19 testing](https://blog.opentrons.com/how-to-use-opentrons-to-test-for-covid-19/).\n\nUsing a [Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette), this protocol will transfer the user-specified volume from sample tubes placed in the [Opentrons Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) into a 96-well plate. Users can specify which pipette, the type of source tube, the type of destination plate, the number of samples to transfer, and the volume of each sample being transferred.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons Filter Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons 4-in-1 Tube Rack Set](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* 96-Well Plate\n* Sample Tubes\n\n\n\n---\n\n\nThe [Opentrons tiprack](https://shop.opentrons.com/collections/opentrons-tips) should be placed in slot 3 and the 96-well plate should be placed in slot 2.\n\nWith this protocol, the [Opentrons tube rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with samples will be accessed in the following order: slot 1, 4, 7, 10, 5, 8, 11 -- see below for examples.\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Samples**: Specify the number of samples to run (1-96)\n* **Volume of Samples (in \u00b5L)**: Specify the volume to be transferred from sample tubes to destination plate (this volume should fall in the range of the selected pipette)\n* **Destination Plate Labware**: Specify whether to use the [NEST Deepwell Plate, 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate) or the [NEST PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) as the destination plate\n* **Source Tube Labware**: Specify whether to use the Opentrons Tubebrack (24) with [2mL](https://labware.opentrons.com/opentrons_24_tuberack_nest_2ml_screwcap?category=tubeRack), [1.5mL](https://labware.opentrons.com/opentrons_24_tuberack_nest_1.5ml_screwcap?category=tubeRack), or [0.5mL](https://labware.opentrons.com/opentrons_24_tuberack_nest_0.5ml_screwcap?category=tubeRack) tubes or the Opentrons Tuberack (15) with [15mL](https://labware.opentrons.com/opentrons_15_tuberack_falcon_15ml_conical?category=tubeRack) conical tubes.\n* **Pipette Type**: Specify which pipette (p300/p1000 GEN1/GEN2) to use (should be mounted on the right gantry)\n\n\n**Example layout: 15 samples with 2mL tubes**\n\n\n\n**Example layout: 15 samples with 15mL tubes**\n\n\n\n**Example layout: 96 samples with 2mL tubes**\n\n\n\n**Example layout: 96 samples with 15mL tubes**\n\n\n\n\n\n",
"internal": "generic_station_A\n",
diff --git a/protoBuilds/generic_station_C/README.json b/protoBuilds/generic_station_C/README.json
index 63b7a26dbb..4ba2b69f2d 100644
--- a/protoBuilds/generic_station_C/README.json
+++ b/protoBuilds/generic_station_C/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Covid Workstation": [
"qPCR Setup"
@@ -8,7 +8,7 @@
"description": "This protocol automates setting up a plate for (reverse transcriptase) qPCR. Using the purified nucleic acid samples from Station B (RNA Extraction), the samples are then aliquoted and mixed with the reaction mix of the assay as outlined in our article on automating Covid-19 testing.\n\nUsing a Single-Channel Pipette, this protocol will begin by transferring reaction mix (combination of master mix and primers/probes) from a 1.5mL tube in the 24-well aluminum block to the specified wells of a plate for qPCR. Then, using the Single-Channel Pipette or optional Multi-Channel Pipette, samples will be transferred from their plate to the qPCR plate and mixed with the reaction mix.\n\nThis protocol uses a custom labware definition. For more information on using labware with the OT-2, please see this support article.\n\nIf you have any questions about this protocol, please email our Applications Engineering team at protocols@opentrons.com.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons Temperature Module with Aluminum Block Set)\nOpentrons Single-Channel Pipette\nOpentrons Multi-Channel Pipette, optional\nOpentrons Filter Tips\nNEST PCR Plate containing purified nucleic acid samples\nNEST 1.5mL Microcentrifuge Tube containing reaction mix(es)\nOptical 96-Well qPCR Plate\n\n\n\nThe Opentrons Temperature Module should be placed in slot 4 with the 96-Well Aluminum Block and qPCR plate on top. The temperature module can be pre-cooled through the Opentrons OT-2 Run App\n\nThe reaction mix(es) (master mix/primers/probes/etc) should kept in the microcentrifuge tube and placed in the 24-Well Aluminum Block (which can be put in the freezer beforehand to keep reagents cold) in slot 5. Up to three different reaction mixes can be used per plate and should be loaded in A1, B1, and C1.\n\nThe NEST PCR Plate containing purified nucleic acid samples should be placed in slot 1. The samples should be filled in column order (ie, 3 samples would go in wells A1, B1, and C1).\n\nThe Opentrons Tiprack for the Opentrons Single-Channel Pipette should be placed in slot 6.\n\nIf also using an Opentrons Multi-Channel Pipette that is the same volumetrically as the single-channel pipette and the total number of tips that will be used is less than 96, then the same tiprack in slot 6 can be utilized. Otherwise, another tiprack should be placed in slot 3.\n\n\nUsing the customizations field (below), set up your protocol.\n Number of Samples: Specify the number of samples to run. If using more than one reaction mix, the number of samples would be the same for each reaction mix. If using 1 reaction mix, the maximum number of samples would be 96; if using 2 reaction mixes, the maximum would be 48; and if using 3 reaction mixes, the maxium is 32.\n Number of Reaction Mixes: Select the number of reaction mixes (1, 2, or 3) that will be used.\n Volume of Reaction Mix (\u00b5L): Specify the volume of the reaction mix(es) that should be added to each well of the qPCR plate.\n Volume of Sample (\u00b5L): Specify the volume of the sample that should be transferred to each well of the qPCR plate.\n Single-Channel Pipette Type (right mount): Select which single-channel pipette (p10, p50, or p20) that will be used. This pipette should be mounted to the right mount.\n Multi-Channel Pipette Type (left mount): Select which multi-channel pipette (p10, p50, or p20) that will be used, if using (can also select none).\n\n\nExample layout: 48 samples with 2 reaction mixes\nnote: this configuration requires 98 tips, thus two tipracks are needed\n\n\n\nExample layout: 48 samples with 1 reaction mix and different pipettes\n\n\n",
"internal": "generic_station_A",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Covid Workstation\n\t* qPCR Setup\n\n\n",
"description": "This protocol automates setting up a plate for (reverse transcriptase) qPCR. Using the purified nucleic acid samples from Station B (RNA Extraction), the samples are then aliquoted and mixed with the reaction mix of the assay as outlined in our [article on automating Covid-19 testing](https://blog.opentrons.com/how-to-use-opentrons-to-test-for-covid-19/).\n\nUsing a [Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette), this protocol will begin by transferring reaction mix (combination of master mix and primers/probes) from a 1.5mL tube in the 24-well aluminum block to the specified wells of a plate for qPCR. Then, using the [Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) or optional [Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette), samples will be transferred from their plate to the qPCR plate and mixed with the reaction mix.\n\nThis protocol uses a custom labware definition. For more information on using labware with the OT-2, please see this [support article](https://support.opentrons.com/en/articles/3136506-using-labware-in-your-protocols).\n\nIf you have any questions about this protocol, please email our Applications Engineering team at [protocols@opentrons.com](mailto:protocols@opentrons.com).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [Aluminum Block Set](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set))\n* [Opentrons Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette), *optional*\n* [Opentrons Filter Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) containing purified nucleic acid samples\n* [NEST 1.5mL Microcentrifuge Tube](https://shop.opentrons.com/collections/tubes/products/nest-microcentrifuge-tubes) containing reaction mix(es)\n* [Optical 96-Well qPCR Plate](https://www.thermofisher.com/order/catalog/product/N8010560#/N8010560)\n\n\n\n---\n\n\nThe [Opentrons Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) should be placed in **slot 4** with the [96-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) and [qPCR plate](https://www.thermofisher.com/order/catalog/product/N8010560#/N8010560) on top. The temperature module can be pre-cooled through the [Opentrons OT-2 Run App](https://opentrons.com/ot-app/)\n\nThe reaction mix(es) (master mix/primers/probes/etc) should kept in the [microcentrifuge tube](https://shop.opentrons.com/collections/tubes/products/nest-microcentrifuge-tubes) and placed in the [24-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) (which can be put in the freezer beforehand to keep reagents cold) in **slot 5**. Up to three different reaction mixes can be used per plate and should be loaded in A1, B1, and C1.\n\nThe [NEST PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) containing purified nucleic acid samples should be placed in **slot 1**. The samples should be filled in column order (ie, 3 samples would go in wells A1, B1, and C1).\n\nThe [Opentrons Tiprack](https://shop.opentrons.com/collections/opentrons-tips) for the [Opentrons Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) should be placed in **slot 6**.\n\nIf also using an [Opentrons Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette) that is the same volumetrically as the single-channel pipette **and** the total number of tips that will be used is less than 96, then the same tiprack in slot 6 can be utilized. Otherwise, another tiprack should be placed in **slot 3**.\n\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Samples**: Specify the number of samples to run. If using more than one reaction mix, the number of samples would be the same for each reaction mix. If using 1 reaction mix, the maximum number of samples would be 96; if using 2 reaction mixes, the maximum would be 48; and if using 3 reaction mixes, the maxium is 32.\n* **Number of Reaction Mixes**: Select the number of reaction mixes (1, 2, or 3) that will be used.\n* **Volume of Reaction Mix (\u00b5L)**: Specify the volume of the reaction mix(es) that should be added to each well of the qPCR plate.\n* **Volume of Sample (\u00b5L)**: Specify the volume of the sample that should be transferred to each well of the qPCR plate.\n* **Single-Channel Pipette Type (right mount)**: Select which single-channel pipette (p10, p50, or p20) that will be used. This pipette should be mounted to the right mount.\n* **Multi-Channel Pipette Type (left mount)**: Select which multi-channel pipette (p10, p50, or p20) that will be used, if using (can also select *none*).\n\n\n**Example layout: 48 samples with 2 reaction mixes**\n*note*: this configuration requires 98 tips, thus two tipracks are needed\n\n\n\n**Example layout: 48 samples with 1 reaction mix and different pipettes**\n\n\n\n\n",
"internal": "generic_station_A\n",
diff --git a/protoBuilds/gna_octea/README.json b/protoBuilds/gna_octea/README.json
index a16fd60dd3..de238720db 100644
--- a/protoBuilds/gna_octea/README.json
+++ b/protoBuilds/gna_octea/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Covid"
@@ -10,7 +10,7 @@
"internal": "gna_octea",
"labware": "\nOpentrons 200\u00b5L Filter Tips\nNEST 96-Deep Well Plate, 2mL\nBio-Rad 96-Well Plate\nNEST 12-Well Reservoir, 15mL\nCustom Plate for GNA Octea Chip\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Covid\n\n",
"deck-setup": "\n\n",
"description": "This protocol automates GNA Octea prep. Using the [P300 Multi-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette) and the [P300 Single-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette), all the reagents necessary for the sample prep are transferred to the samples and the samples are incubated on the [Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) and [Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck). Once all sample prep has been completed, the samples are transferred to a custom plate containing the labware needed to test the samples on the [GNA analyzer](https://www.gna-bio.com/products/).\n\nThis protocol is still a work in progress and will be updated.\n\n**Update (06/07/2021)**: This protocol has been updated per conversation and has several modifications (slight), such as, changing the distribution of reagent to multi-dispense and allowing the temperature module to heat up while pipetting occurs.\n\n**Update (06/10/2021)**: This protocol has been updated and now includes new mixing functions and incorporated air gaps.\n\nExplanation of complex parameters below:\n* **P300-Multi Mount**: Select which mount the [P300 Multi-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette) is attached to.\n* **P300-Single Mount**: Select which mount the [P300 Single-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) is attached to.\n* **Number of Samples**: Specify the number of samples to be run. Currently, 8 is the only number allowed.\n\n\n---\n\n",
diff --git a/protoBuilds/gna_octea_v2/README.json b/protoBuilds/gna_octea_v2/README.json
index b77c056112..e1b68041b3 100644
--- a/protoBuilds/gna_octea_v2/README.json
+++ b/protoBuilds/gna_octea_v2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Covid"
@@ -10,7 +10,7 @@
"internal": "gna_octea_v2",
"labware": "\nOpentrons 200\u00b5L Filter Tips\nNEST 96-Deep Well Plate, 2mL\nNEST 12-Well Reservoir, 15mL\nOpentrons Tube Rack\n1.5mL (and/or 2mL) Microcentrigue Tubes\nCustom Plate for GNA Octea Chip\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Covid\n\n",
"deck-setup": "\n\n",
"description": "This protocol automates GNA Octea prep. Using the [P300 Multi-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette) and the [P300 Single-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette), all the reagents necessary for the sample prep are transferred to the samples and the samples are incubated on the [Temperature Module](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) and [Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck). Once all sample prep has been completed, the samples are transferred to a custom plate containing the labware needed to test the samples on the [GNA analyzer](https://www.gna-bio.com/products/).\n\nThis protocol is still a work in progress and will be updated.\n\n\nExplanation of complex parameters below:\n* **P300-Multi Mount**: Select which mount the [P300 Multi-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette) is attached to.\n* **P300-Single Mount**: Select which mount the [P300 Single-Channel Pipette (GEN2)](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) is attached to.\n* **Labware on Temp Deck + Al Block**: Select which tube type will contain the samples.\n* **Reconstitue MagBeads/PCA?**: Specify whether the OT-2 will reconstitute the MagBeads and PCA.\n\n\n---\n\n",
diff --git a/protoBuilds/gsdx/README.json b/protoBuilds/gsdx/README.json
index 05037d5f0f..5e7c6f7901 100644
--- a/protoBuilds/gsdx/README.json
+++ b/protoBuilds/gsdx/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Screening"
@@ -9,7 +9,7 @@
"internal": "gsdx",
"labware": "\nBIOPlastics BV 96 Aluminum Block 200 \u00b5L #K58001-B58009\nOpentrons x Gold Standard 17 Tube Rack\nGoldStandardDx 96 Tip Rack 200 \u00b5L\nBIOPlastics BV 96 Aluminum Block 100 \u00b5L #B59001-1\nKingFisher 96 Tube Rack with GoldStandard 1.1 mL #10536617\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Screening\n\n\n",
"description": "This protocol runs the BACGene protocol for up to 96 target samples for listeria and/or salmonella pathogens. Upon upload to the Opentrons App, the user will be prompted with the volumes and positions of all labware and liquids on the OT-2 deck. Lysis buffer will automatically be prepped with enzyme addition when needed before distribution to the lysis buffer plate.\n\n\n",
"internal": "gsdx\n",
diff --git a/protoBuilds/heater-shaker-beta/README.json b/protoBuilds/heater-shaker-beta/README.json
index 40778b5d8b..4e57af4a36 100644
--- a/protoBuilds/heater-shaker-beta/README.json
+++ b/protoBuilds/heater-shaker-beta/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Heater Shaker": [
"Heater Shaker Beta Test"
@@ -10,7 +10,7 @@
"internal": "heater-shaker-test",
"labware": "\nOpentrons Tips for the Selected Pipette (https://shop.opentrons.com)\nOpentrons Heater Shaker Module-compatible labware (see five options in the protocol parameters section of the python script)\nSelected Source and Destination Labware ([see lists in the protocol parameters section of the python script] https://labware.opentrons.com/)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Heater Shaker\n\t* Heater Shaker Beta Test\n\n",
"deck-setup": "\n\n\n**Slot 1**: Opentrons Heater Shaker Module with selected labware \n**Slot 3**: Source Labware (well plate or reservoir) \n**Slot 6**: Destination Labware (well plate or reservoir) \n**Slot 11**: Opentrons Tips\n\n\n---\n\n",
"description": "This protocol performs beta-testing steps of the Opentrons Heater Shaker Module with user-determined parameters for selection of the target temperature of the heater shaker (37-95 degrees Celsius), the target shaking speed (recommended 200-2000 rpm although it is possible to go up to 3000 rpm), duration of time for shaking (minutes), n (dispense to every nth column of the heater shaker labware - to save run-time), the labware on the heater shaker (five options listed in the protocol parameters section of the python script), the source labware (a well plate or reservoir from the Opentrons Labware Library - see list in protocol parameters section of the python script), the destination labware (a well plate or reservoir from the Opentrons Labware Library - see list in protocol parameters section of the python script), the transfer volume, and the pipette to be used (any single- or multi-channel pipette). The beta-test protocol will open and close the heater shaker latch and report latch status to the log, set target temperature and proceed without waiting, report the current temperature to the log, report the selected target wells to the log, perform liquid transfers from the source labware to the heater shaker labware, wait to ensure the previously set temperature has been reached, start shaking and capture the start time, perform a pipette mix in the destination labware to demo a pipetting step NOT targeting the heater shaker but occurring during the timed shake, wait until the shake time has elapsed, stop shaking and report current rpm to the log, perform liquid transfer of 80 percent of the previously-dispensed volume from the heater shaker to the destination plate, deactivate the heater, open the latch, process complete.\n\n\n\n---\n\n\n\n",
diff --git a/protoBuilds/i7g1ym/README.json b/protoBuilds/i7g1ym/README.json
index 7d4493d6e8..a939c9933f 100644
--- a/protoBuilds/i7g1ym/README.json
+++ b/protoBuilds/i7g1ym/README.json
@@ -1,5 +1,5 @@
{
- "author": "Tim Fallon",
+ "author": "Tim Fallon\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"General Liquid Handling": [
"General Liquid Handling"
@@ -10,7 +10,7 @@
"internal": "i7g1ym",
"labware": "\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "Tim Fallon\n\n\n",
+ "author": "Tim Fallon\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* General Liquid Handling\n\t* General Liquid Handling\n\n\n",
"deck-setup": "[deck](https://drive.google.com/open?id=1549-GH5Qyzv7JzDqJG7ZLGkNzckp_PLq)\n\n\n",
"description": "Rearranges pipette tips from their standard spacing in a full box, to two empty tip box putting the pipette tips in every other row. This allows the P300/P20-multipipette to be used for pipetting from 24 well plates.\n\n\n",
diff --git a/protoBuilds/illumina-nextera-XT-library-prep-part1/README.json b/protoBuilds/illumina-nextera-XT-library-prep-part1/README.json
index a7509e7f1e..6cf58e15e2 100644
--- a/protoBuilds/illumina-nextera-XT-library-prep-part1/README.json
+++ b/protoBuilds/illumina-nextera-XT-library-prep-part1/README.json
@@ -8,7 +8,7 @@
"description": "Part 1 of 4: Tagment Genomic DNA and Amplify Libraries\nLinks:\n Part 1: Tagment and Amplify\n Part 2: Clean Up Libraries\n Part 3: Normalize Libraries\n Part 4: Pool Libraries\nWith this protocol, your robot can perform the Nextera XT DNA Library Prep Kit protocol described by the Illumina Reference Guide.\nThis is part 1 of the protocol, which includes the steps (1) Tagment Genomic DNA and (2) Amplify Libraries.\nThe tagmentation step uses Nextera transposase to fragment DNA into sizes suitable for sequencing, and then tags the DNA with adapter sequences. The library amplification step increases the yield of the tagmented DNA using PCR. PCR adds the Index 1 (i7), Index 2 (i5), and full adapter sequences to the tagmented DNA from the previous step. This protocol assumes you are taking your plate off the OT-2 and thermocycling on a stand-alone PCR machine according to the Illumina Reference Guide.\nAfter the two steps carried out in this protocol, you can safely stop work and return to it at a later point. If you are stopping, seal the plate and store at 2\u00b0C to 8\u00b0C for up to 2 days.",
"internal": "bU7eUGEh\n872",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/illumina-nextera-xt-library-prep-part1). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/illumina-nextera-xt-library-prep-part1). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Featured\n * NGS Library Prep: Illumina Nextera XT\n\n",
"description": "Part 1 of 4: Tagment Genomic DNA and Amplify Libraries\n\nLinks:\n* [Part 1: Tagment and Amplify](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part1)\n* [Part 2: Clean Up Libraries](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part2)\n* [Part 3: Normalize Libraries](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part3)\n* [Part 4: Pool Libraries](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part4)\n\nWith this protocol, your robot can perform the Nextera XT DNA Library Prep Kit protocol described by the [Illumina Reference Guide](https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_nextera/nextera-xt/nextera-xt-library-prep-reference-guide-15031942-06.pdf).\n\nThis is part 1 of the protocol, which includes the steps (1) Tagment Genomic DNA and (2) Amplify Libraries.\n\nThe tagmentation step uses Nextera transposase to fragment DNA into sizes suitable for sequencing, and then tags the DNA with adapter sequences. The library amplification step increases the yield of the tagmented DNA using PCR. PCR adds the Index 1 (i7), Index 2 (i5), and full adapter sequences to the tagmented DNA from the previous step. This protocol assumes you are taking your plate off the OT-2 and thermocycling on a stand-alone PCR machine according to the [Illumina Reference Guide](https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_nextera/nextera-xt/nextera-xt-library-prep-reference-guide-15031942-05.pdf).\n\nAfter the two steps carried out in this protocol, you can safely stop work and return to it at a later point. If you are stopping, seal the plate and store at 2\u00b0C to 8\u00b0C for up to 2 days.\n\n",
"internal": "bU7eUGEh\n872\n",
diff --git a/protoBuilds/illumina-nextera-XT-library-prep-part2/README.json b/protoBuilds/illumina-nextera-XT-library-prep-part2/README.json
index 3633857451..39a8116c07 100644
--- a/protoBuilds/illumina-nextera-XT-library-prep-part2/README.json
+++ b/protoBuilds/illumina-nextera-XT-library-prep-part2/README.json
@@ -8,7 +8,7 @@
"description": "Part 2 of 4: Clean Up Libraries\nLinks:\n Part 1: Tagment and Amplify\n Part 2: Clean Up Libraries\n Part 3: Normalize Libraries\n Part 4: Pool Libraries\nWith this protocol, your robot can perform the Nextera XT DNA Library Prep Kit protocol describe by the Illumina Reference Guide.\nThis is Part 2 of the protocol, which consists of just step (3) of the overall process: clean up libraries. This step uses AMPure XP beads to purify the library DNA and remove short library fragments after the previous step, library amplification.\nAfter this step, it is safe to stop the workflow and return to it at a later point. If you are stopping, seal the plate and store at -15\u00b0C to -25\u00b0C for up to seven days.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons P300 or P50 Pipette (Single or 8-Channel) and corresponding Tips\nOpentrons Magnetic Module\nBio-Rad 96-Well Plate, 200\u03bcl containing samples from Part 1\nBio-Rad 96-Well Plate, 200\u03bcl, clean and empty (x2)\nUSA Scientific 12-Channel Reservoir\nNextera XT DNA Library Prep Kit\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nThis protocol requires specific labware in a specific set-up.\nSlot 1: Bio-Rad Plate (clean and empty); final elution will be transferred to this plate.\nSlot 2: USA Scientific 12-Channel Reservoir\n A1: Resuspension Buffer\n A2: AMPure XP Beads\n A3: 80% Ethanol\n A4: 80% Ethanol (if needed)\n A5: 80% Ethanol (if needed)\n A9: Liquid Waste\n A10: Liquid Waste\n A11: Liquid Waste\n* A12: Liquid Waste\nSlot 4: Magnetic Module with Bio-Rad Plate (clean and empty)\nSlot 5: Bio-Rad Plate containing samples from Part 1\nSlot 6: Opentrons Tip Rack\nSlot 7: Opentrons Tip Rack\nSlot 8: Opentrons Tip Rack\n* Note: If your protocol requires more tips due to the parameters you set, you will need to fill subsequent slots with tip racks.\nUsing the customization fields below, set up your protocol.\n Pipette Model: Select which pipette (P50/P300; Single/8-Channel) you will use for this protocol.\n Pipette Mount: Specify which mount your single-channel pipette is on (left or right).\n Magnetic Module Gen: Specify which Magnetic Module is in use\n Number of Samples: Select the number of samples (1-96) to be run in the protocol.\n Initial Product Volume (\u00b5l): Select the starting volume of PCR product to be used in the protocol.\n Bead Ratio: Select the bead ratio. 1.8 is recommended for small (300-500 bp) inputs, and 0.6 is recommended for larger (>500 bp) samples. Please see documentation for more information.\n Resuspension Buffer Volume (\u00b5L): Specify how much Resuspension Buffer is added to the wells.\n Final PCR Product Volume (\u00b5L): Specify the final elution volume to be transferred.\n* Dry Time (minutes): Specify how long (in minutes) you would like to let the beads dry after the wash steps and before adding the resuspension buffer.",
"internal": "bU7eUGEh\n872",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/illumina-nextera-xt-library-prep-part2). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/illumina-nextera-xt-library-prep-part2). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Featured\n * NGS Library Prep: Illumina Nextera XT\n\n",
"description": "Part 2 of 4: Clean Up Libraries\n\nLinks:\n* [Part 1: Tagment and Amplify](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part1)\n* [Part 2: Clean Up Libraries](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part2)\n* [Part 3: Normalize Libraries](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part3)\n* [Part 4: Pool Libraries](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part4)\n\nWith this protocol, your robot can perform the Nextera XT DNA Library Prep Kit protocol describe by the [Illumina Reference Guide](https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_nextera/nextera-xt/nextera-xt-library-prep-reference-guide-15031942-06.pdf).\n\nThis is Part 2 of the protocol, which consists of just step (3) of the overall process: clean up libraries. This step uses AMPure XP beads to purify the library DNA and remove short library fragments after the previous step, library amplification.\n\nAfter this step, it is safe to stop the workflow and return to it at a later point. If you are stopping, seal the plate and store at -15\u00b0C to -25\u00b0C for up to seven days.\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 or P50 Pipette (Single or 8-Channel)](https://shop.opentrons.com/collections/ot-2-pipettes) and corresponding [Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Bio-Rad 96-Well Plate, 200\u03bcl](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr) containing samples from [Part 1](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part1)\n* [Bio-Rad 96-Well Plate, 200\u03bcl](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr), clean and empty (x2)\n* [USA Scientific 12-Channel Reservoir](https://labware.opentrons.com/usascientific_12_reservoir_22ml?category=reservoir)\n* [Nextera XT DNA Library Prep Kit](https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/nextera-xt-dna.html)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n---\n\n\nThis protocol requires specific labware in a specific set-up.\n\nSlot 1: [Bio-Rad Plate](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate) (clean and empty); final elution will be transferred to this plate.\n\nSlot 2: [USA Scientific 12-Channel Reservoir](https://labware.opentrons.com/usascientific_12_reservoir_22ml?category=reservoir)\n* A1: Resuspension Buffer\n* A2: AMPure XP Beads\n* A3: 80% Ethanol\n* A4: 80% Ethanol (if needed)\n* A5: 80% Ethanol (if needed)\n* A9: Liquid Waste\n* A10: Liquid Waste\n* A11: Liquid Waste\n* A12: Liquid Waste\n\nSlot 4: [Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [Bio-Rad Plate](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate) (clean and empty)\n\nSlot 5: [Bio-Rad Plate](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr) containing samples from [Part 1](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part1)\n\nSlot 6: [Opentrons Tip Rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 7: [Opentrons Tip Rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 8: [Opentrons Tip Rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* Note: If your protocol requires more tips due to the parameters you set, you will need to fill subsequent slots with tip racks.\n\n\nUsing the customization fields below, set up your protocol.\n* Pipette Model: Select which pipette (P50/P300; Single/8-Channel) you will use for this protocol.\n* Pipette Mount: Specify which mount your single-channel pipette is on (left or right).\n* Magnetic Module Gen: Specify which Magnetic Module is in use\n* Number of Samples: Select the number of samples (1-96) to be run in the protocol.\n* Initial Product Volume (\u00b5l): Select the starting volume of PCR product to be used in the protocol.\n* Bead Ratio: Select the bead ratio. 1.8 is recommended for small (300-500 bp) inputs, and 0.6 is recommended for larger (>500 bp) samples. Please see [documentation](https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_nextera/nextera-xt/nextera-xt-library-prep-reference-guide-15031942-05.pdf) for more information.\n* Resuspension Buffer Volume (\u00b5L): Specify how much Resuspension Buffer is added to the wells.\n* Final PCR Product Volume (\u00b5L): Specify the final elution volume to be transferred.\n* Dry Time (minutes): Specify how long (in minutes) you would like to let the beads dry after the wash steps and before adding the resuspension buffer.\n\n\n\n\n\n",
"internal": "bU7eUGEh\n872\n",
diff --git a/protoBuilds/illumina-nextera-XT-library-prep-part3/README.json b/protoBuilds/illumina-nextera-XT-library-prep-part3/README.json
index 7cfa293d1f..15ea8a4be5 100644
--- a/protoBuilds/illumina-nextera-XT-library-prep-part3/README.json
+++ b/protoBuilds/illumina-nextera-XT-library-prep-part3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Featured": [
"NGS Library Prep: Illumina Nextera XT"
@@ -8,7 +8,7 @@
"description": "Part 3 of 4: Normalize Libraries\nLinks:\n Part 1: Tagment and Amplify\n Part 2: Clean Up Libraries\n Part 3: Normalize Libraries\n Part 4: Pool Libraries\nWith this protocol, your robot can perform the Nextera XT DNA Library Prep Kit protocol describe by the Illumina Reference Guide.\nThis is part 3 of the protocol, which is step (4) of the overall workflow: normalize libraries. This step normalizes the concentration of each library to ensure correct library representation in the donwstream pooled libraries.\nIt is safe to stop after this step and re-start work at a later point. If you are stopping, seal the plate and store at -25\u00b0C to -15\u00b0C for up to seven days.\n--\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons P300 or P50 Pipette (Single or 8-Channel) and corresponding Tips\nOpentrons Magnetic Module\nBio-Rad 96-Well Plate, 200\u03bcl containing samples from Part 1\nBio-Rad 96-Well Plate, 200\u03bcl, clean and empty (x2)\nUSA Scientific 12-Channel Reservoir\nNextera XT DNA Library Prep Kit\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nThis protocol requires specific labware in a specific set-up.\nSlot 1: Bio-Rad Plate (clean and empty); final elution will be transferred to this plate.\nSlot 2: USA Scientific 12-Channel Reservoir\n A1: Library Normalization Additive 1 (LNA1)\n A2: Library Normalization Beads 1 (LNB1)\n A3: Library Normalization Wash 1 (LNW1)\n A4: Library Normalization Storage Buffer 1 (LNS1)\n A5: 0.1 N NaOH\n A6: Empty - LNA1 & LNB1 will be combined here, if not pre-mixed\n* A12: Liquid Waste\nSlot 4: Magnetic Module with Bio-Rad Plate (clean and empty)\nSlot 5: Bio-Rad Plate containing samples from Part 2\nSlot 7: Opentrons Tip Rack\nSlot 8: Opentrons Tip Rack\nSlot 9: Opentrons Tip Rack\nUsing the customization fields below, set up your protocol.\n Pipette Model: Select which pipette (P50/P300) you will use for this protocol.\n Pipette Mount: Specify which mount your single-channel pipette is on (left or right).\n Number of Samples: Select the number of samples (1-96) to be run in the protocol. NOTE ~ selecting 24 or less samples will result in only the top-left corner of the plates being used (A1 - D6).\n Mix LNA1 & LNB1 with OT-2: Select whether the OT-2 should mix the LNA1 or LNB1 together. If yes, the OT-2 will transfer the reagents from A1 and A2 of the reservoir, respectively, and mix in A6. Otherwise, the pre-mixed reagent should be loaded directly in A6 of the 12-well reservoir.",
"internal": "bU7eUGEh\n872",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Featured\n * NGS Library Prep: Illumina Nextera XT\n\n",
"description": "Part 3 of 4: Normalize Libraries\n\nLinks:\n* [Part 1: Tagment and Amplify](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part1)\n* [Part 2: Clean Up Libraries](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part2)\n* [Part 3: Normalize Libraries](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part3)\n* [Part 4: Pool Libraries](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part4)\n\nWith this protocol, your robot can perform the Nextera XT DNA Library Prep Kit protocol describe by the [Illumina Reference Guide](https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_nextera/nextera-xt/nextera-xt-library-prep-reference-guide-15031942-06.pdf).\n\nThis is part 3 of the protocol, which is step (4) of the overall workflow: normalize libraries. This step normalizes the concentration of each library to ensure correct library representation in the donwstream pooled libraries.\n\nIt is safe to stop after this step and re-start work at a later point. If you are stopping, seal the plate and store at -25\u00b0C to -15\u00b0C for up to seven days.\n\n--\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons P300 or P50 Pipette (Single or 8-Channel)](https://shop.opentrons.com/collections/ot-2-pipettes) and corresponding [Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Bio-Rad 96-Well Plate, 200\u03bcl](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr) containing samples from [Part 1](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part1)\n* [Bio-Rad 96-Well Plate, 200\u03bcl](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr), clean and empty (x2)\n* [USA Scientific 12-Channel Reservoir](https://labware.opentrons.com/usascientific_12_reservoir_22ml?category=reservoir)\n* [Nextera XT DNA Library Prep Kit](https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/nextera-xt-dna.html)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n---\n\n\nThis protocol requires specific labware in a specific set-up.\n\nSlot 1: [Bio-Rad Plate](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate) (clean and empty); final elution will be transferred to this plate.\n\nSlot 2: [USA Scientific 12-Channel Reservoir](https://labware.opentrons.com/usascientific_12_reservoir_22ml?category=reservoir)\n* A1: Library Normalization Additive 1 (LNA1)\n* A2: Library Normalization Beads 1 (LNB1)\n* A3: Library Normalization Wash 1 (LNW1)\n* A4: Library Normalization Storage Buffer 1 (LNS1)\n* A5: 0.1 N NaOH\n* A6: Empty - LNA1 & LNB1 will be combined here, if not pre-mixed\n* A12: Liquid Waste\n\nSlot 4: [Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [Bio-Rad Plate](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr?category=wellPlate) (clean and empty)\n\nSlot 5: [Bio-Rad Plate](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr) containing samples from [Part 2](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part2)\n\nSlot 7: [Opentrons Tip Rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 8: [Opentrons Tip Rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\nSlot 9: [Opentrons Tip Rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n\nUsing the customization fields below, set up your protocol.\n* Pipette Model: Select which pipette (P50/P300) you will use for this protocol.\n* Pipette Mount: Specify which mount your single-channel pipette is on (left or right).\n* Number of Samples: Select the number of samples (1-96) to be run in the protocol. NOTE ~ selecting 24 or less samples will result in only the top-left corner of the plates being used (A1 - D6).\n* Mix LNA1 & LNB1 with OT-2: Select whether the OT-2 should mix the LNA1 or LNB1 together. If **yes**, the OT-2 will transfer the reagents from A1 and A2 of the reservoir, respectively, and mix in A6. Otherwise, the pre-mixed reagent should be loaded directly in A6 of the 12-well reservoir.\n\n\n\n",
"internal": "bU7eUGEh\n872\n",
diff --git a/protoBuilds/illumina-nextera-XT-library-prep-part4/README.json b/protoBuilds/illumina-nextera-XT-library-prep-part4/README.json
index 415e83edc1..6d0fd19e4e 100644
--- a/protoBuilds/illumina-nextera-XT-library-prep-part4/README.json
+++ b/protoBuilds/illumina-nextera-XT-library-prep-part4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Featured": [
"NGS Library Prep: Illumina Nextera XT"
@@ -8,7 +8,7 @@
"description": "Part 4 of 4: Pool Libraries\nLinks:\n Part 1: Tagment and Amplify\n Part 2: Clean Up Libraries\n Part 3: Normalize Libraries\n Part 4: Pool Libraries\nWith this protocol, your robot can perform the Nextera XT DNA Library Prep Kit protocol described by the Illumina Reference Guide.\nThis is part 4 of the protocol, which is step (4) pool libraries. This step combines equal volumes of normalized libraries in a single tube. After pooling, dilute and heat-denature the library pool before loading libraries for the sequencing run.\nStore unused pooled libraries in the PAL tube and SGP plate at -25\u00b0C to -15\u00b0C for up to 7 days.\n--\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Single-Channel Pipette and corresponding Tips\nBio-Rad 96-Well Plate, 200\u03bcl containing samples from Part 3\nOpentrons Tube Rack with 2mL tubes\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nThis protocol requires specific labware in a specific set-up.\nSlot 1: Bio-Rad Plate containing samples from Part 3\nSlot 2: Tube Rack with 2mL tubes (for collecting pools)\nSlot 3: Opentrons Tip Rack\n* Note: If your protocol requires more tips due to the parameters you set, you will need to fill subsequent slots with tip racks.\nUsing the customization fields below, set up your protocol.\n Pipette Model: Select which pipette you will use for this protocol.\n Pipette Mount: Specify which mount your single-channel pipette is on (left or right).\n Number of Samples: Select the number of samples (1-96) to be run in the protocol. NOTE ~ selecting 24 or less samples will result in only the top-left corner of the plates being used (A1 - D6).\n Number of Pools: Select the number of pools (2mL tubes) that should be created from the sample plate.\n* Pool Volume (\u03bcl): Select the amount (in microliters) of sample material to transfer to each pooling tube.",
"internal": "bU7eUGEh\n872",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Featured\n * NGS Library Prep: Illumina Nextera XT\n\n",
"description": "Part 4 of 4: Pool Libraries\n\nLinks:\n* [Part 1: Tagment and Amplify](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part1)\n* [Part 2: Clean Up Libraries](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part2)\n* [Part 3: Normalize Libraries](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part3)\n* [Part 4: Pool Libraries](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part4)\n\nWith this protocol, your robot can perform the Nextera XT DNA Library Prep Kit protocol described by the [Illumina Reference Guide](https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_nextera/nextera-xt/nextera-xt-library-prep-reference-guide-15031942-06.pdf).\n\nThis is part 4 of the protocol, which is step (4) pool libraries. This step combines equal volumes of normalized libraries in a single tube. After pooling, dilute and heat-denature the library pool before loading libraries for the sequencing run.\n\nStore unused pooled libraries in the PAL tube and SGP plate at -25\u00b0C to -15\u00b0C for up to 7 days.\n\n--\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes) and corresponding [Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Bio-Rad 96-Well Plate, 200\u03bcl](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr) containing samples from [Part 3](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part3)\n* [Opentrons Tube Rack](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1) with 2mL tubes\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n---\n\n\nThis protocol requires specific labware in a specific set-up.\n\nSlot 1: [Bio-Rad Plate](https://labware.opentrons.com/biorad_96_wellplate_200ul_pcr) containing samples from [Part 3](http://protocols.opentrons.com/protocol/illumina-nextera-XT-library-prep-part3)\n\nSlot 2: [Tube Rack](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1) with 2mL tubes (for collecting pools)\n\nSlot 3: [Opentrons Tip Rack](https://shop.opentrons.com/collections/opentrons-tips)\n* Note: If your protocol requires more tips due to the parameters you set, you will need to fill subsequent slots with tip racks.\n\n\nUsing the customization fields below, set up your protocol.\n* Pipette Model: Select which pipette you will use for this protocol.\n* Pipette Mount: Specify which mount your single-channel pipette is on (left or right).\n* Number of Samples: Select the number of samples (1-96) to be run in the protocol. NOTE ~ selecting 24 or less samples will result in only the top-left corner of the plates being used (A1 - D6).\n* Number of Pools: Select the number of pools (2mL tubes) that should be created from the sample plate.\n* Pool Volume (\u03bcl): Select the amount (in microliters) of sample material to transfer to each pooling tube.\n\n\n\n",
"internal": "bU7eUGEh\n872\n",
diff --git a/protoBuilds/macherey-nagel-nucleomag-DNA-microbiome/README.json b/protoBuilds/macherey-nagel-nucleomag-DNA-microbiome/README.json
index f34788f18c..fcd22f2e03 100644
--- a/protoBuilds/macherey-nagel-nucleomag-DNA-microbiome/README.json
+++ b/protoBuilds/macherey-nagel-nucleomag-DNA-microbiome/README.json
@@ -17,7 +17,7 @@
"internal": "macherey-nagal-nucleomag-dna-microbiome\n",
"labware": "* Macherey Nagel 96 Well Square Well Block\n* Macherey Nagel 96 Well Elution Plate U-bottom\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 24 Tube Rack with Generic 2 mL Screwcap](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* [USA Scientific 12 Well Reservoir 22 mL #1061-8150](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n* [Opentrons 96 Tip Rack 1000 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* [Agilent 1 Well Reservoir 290 mL #201252-100](https://www.agilent.com/store/en_US/Prod-201252-100/201252-100)\n\n\n",
"modules": "* [Opentrons Magnetic Module (GEN2)](https://shop.opentrons.com/magnetic-module-gen2/)\n\n\n",
- "partner": "[Opentrons](https://opentrons.com/)\n\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-dna-microbiome). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "partner": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-dna-microbiome). This page won\u2019t be available after January 31st, 2024.\n\n",
"pipettes": "* [Opentrons P300 8 Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n* [Opentrons P1000 Single Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/single-channel-electronic-pipette-p20/)\n\n\n",
"protocol-steps": "1. Binding Step\n2. Wash 1\n3. Wash 2\n4. Wash 3\n5. Wash 4\n6. Delay for drying\n7. Elution\n\n\n* Note\nThe default values for all volumes, incubation times and mix repetitions were pretested and validated.\nWe do not recommend to change them. If you still decide to change them please scale all volumes proportionally.\nFor change recommendations please contact automation-bio@mn-net.de\n\n\n\n**Process**\n1. Input your protocol parameters below.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit \"Run\".\n\n\n**Additional Notes**\n\nIf you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n**Disclaimer**: MACHEREY-NAGEL GmbH & Co. KG makes every effort to include accurate and up-to-date information within this publication; however, it is possible that omissions or errors might have occurred. MACHEREY-NAGEL GmbH & Co. KG cannot, therefore, make any representations or warranties, expressed or implied, as to the accuracy or completeness of the information provided in this publication. Changes in this publication can be made at any time without notice. For technical details and detailed procedures of the specifications provided in this document please contact your MACHEREY-NAGEL representative. This publication may contain reference to applications and products which are not available in all markets. Please check with your local sales representative.\nAll mentioned trademarks are protected by law. All used names and denotations can be brands, trademarks, or registered labels of their respective owner \u2013 also if they are not special denotation. To mention products and brands is only a kind of information (i.e., it does not offend against trademarks and brands and can not be seen as a kind of recommendation or assessment). Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or support by MACHEREY-NAGEL GmbH & Co. KG or Opentrons. Any views or opinions expressed herein by the authors' do not necessarily state or reflect those of MACHEREY-NAGEL or the Eppendorf AG. NucleoMag\u00ae is a registered trademark of MACHEREY-NAGEL GmbH & Co. KG, D\u00fcren, Germany. Opentrons OT-2 is a device from Opentrons, New York, USA.\n\n* The Download links includes all Protocols in their respective subfolders as well as the .json files for the two labware files.\n\n\n",
"reagent-setup": "\n\n\n",
diff --git a/protoBuilds/macherey-nagel-nucleomag-clean-up/README.json b/protoBuilds/macherey-nagel-nucleomag-clean-up/README.json
index b6b96269ef..5f2da7d8d3 100644
--- a/protoBuilds/macherey-nagel-nucleomag-clean-up/README.json
+++ b/protoBuilds/macherey-nagel-nucleomag-clean-up/README.json
@@ -17,7 +17,7 @@
"internal": "macherey-nagel-nucleomag-clean-up\n",
"labware": "* 96 Well PCR Plate\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 24 Tube Rack with Generic 2 mL Screwcap](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* [USA Scientific 12 Well Reservoir 22 mL #1061-8150](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n* [Opentrons 96 Tip Rack 1000 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* 96 Deepwell Plate 2mL\n* [Agilent 1 Well Reservoir 290 mL #201252-100](https://www.agilent.com/store/en_US/Prod-201252-100/201252-100)\n\n\n",
"modules": "* [Opentrons Magnetic Module (GEN2)](https://shop.opentrons.com/magnetic-module-gen2/)\n\n\n",
- "partner": "[Opentrons](https://opentrons.com/)\n\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-clean-up). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "partner": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-clean-up). This page won\u2019t be available after January 31st, 2024.\n\n",
"pipettes": "* [Opentrons P300 8 Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n\n\n",
"protocol-steps": "1. Binding\n2. Wash 1\n3. Wash 2\n4. Elution\n\n\n* Note\nThe default values for all volumes, incubation times and mix repetitions were pretested and validated.\nWe do not recommend to change them. If you still decide to change them please scale all volumes proportionally.\nFor change recommendations please contact automation-bio@mn-net.de\n\n\n\n**Process**\n1. Input your protocol parameters below.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit \"Run\".\n\n\n**Additional Notes**\n\nIf you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n**Disclaimer**: MACHEREY-NAGEL GmbH & Co. KG makes every effort to include accurate and up-to-date information within this publication; however, it is possible that omissions or errors might have occurred. MACHEREY-NAGEL GmbH & Co. KG cannot, therefore, make any representations or warranties, expressed or implied, as to the accuracy or completeness of the information provided in this publication. Changes in this publication can be made at any time without notice. For technical details and detailed procedures of the specifications provided in this document please contact your MACHEREY-NAGEL representative. This publication may contain reference to applications and products which are not available in all markets. Please check with your local sales representative.\nAll mentioned trademarks are protected by law. All used names and denotations can be brands, trademarks, or registered labels of their respective owner \u2013 also if they are not special denotation. To mention products and brands is only a kind of information (i.e., it does not offend against trademarks and brands and can not be seen as a kind of recommendation or assessment). Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or support by MACHEREY-NAGEL GmbH & Co. KG or Opentrons. Any views or opinions expressed herein by the authors' do not necessarily state or reflect those of MACHEREY-NAGEL or the Eppendorf AG. NucleoMag\u00ae is a registered trademark of MACHEREY-NAGEL GmbH & Co. KG, D\u00fcren, Germany. Opentrons OT-2 is a device from Opentrons, New York, USA.\n\n* The Download links includes all Protocols in their respective subfolders as well as the .json files for the two labware files.\n\n\n",
"reagent-setup": "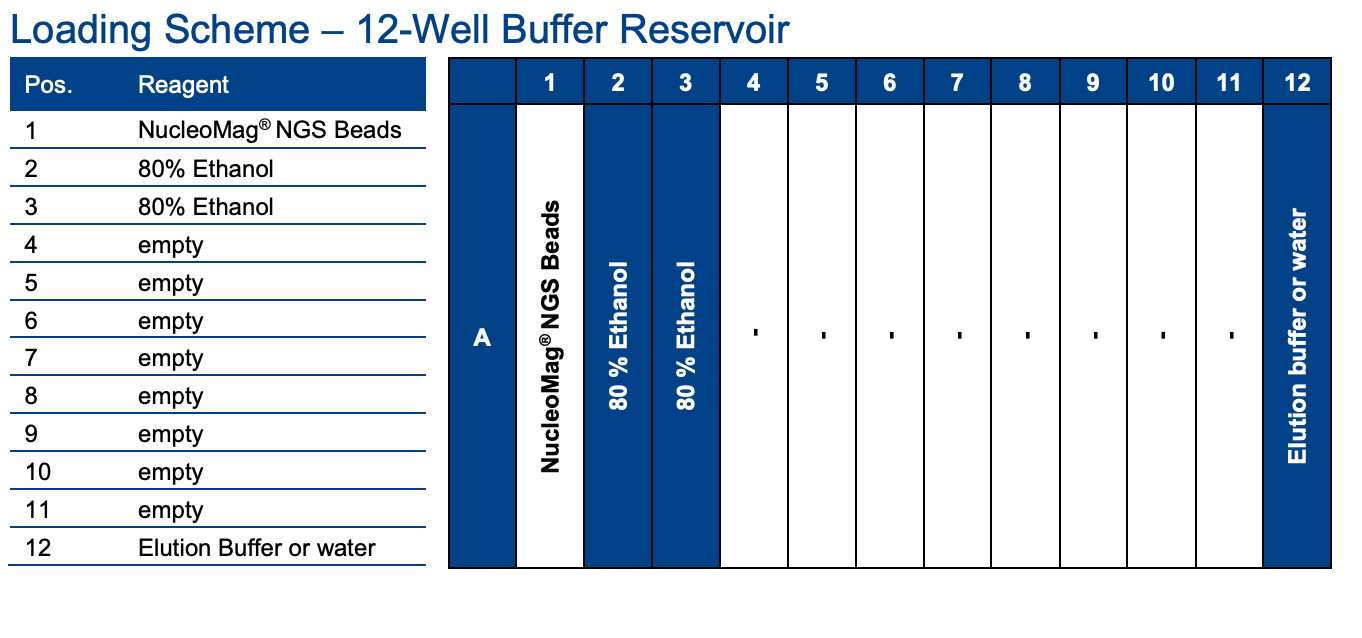\n\n\n",
diff --git a/protoBuilds/macherey-nagel-nucleomag-dna-food/README.json b/protoBuilds/macherey-nagel-nucleomag-dna-food/README.json
index 46b4beb0e8..08f4849c4c 100644
--- a/protoBuilds/macherey-nagel-nucleomag-dna-food/README.json
+++ b/protoBuilds/macherey-nagel-nucleomag-dna-food/README.json
@@ -17,7 +17,7 @@
"internal": "macherey-nagal-nucleomag-dna-food\n",
"labware": "* Macherey Nagel 96 Well Square Well Block\n* Macherey Nagel 96 Well Elution Plate U-bottom\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 24 Tube Rack with Generic 2 mL Screwcap](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* [USA Scientific 12 Well Reservoir 22 mL #1061-8150](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n* [Opentrons 96 Tip Rack 1000 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* [Agilent 1 Well Reservoir 290 mL #201252-100](https://www.agilent.com/store/en_US/Prod-201252-100/201252-100)\n\n\n",
"modules": "* [Opentrons Magnetic Module (GEN2)](https://shop.opentrons.com/magnetic-module-gen2/)\n\n\n",
- "partner": "[Opentrons](https://opentrons.com/)\n\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-dna-food). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "partner": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-dna-food). This page won\u2019t be available after January 31st, 2024.\n\n",
"pipettes": "* [Opentrons P300 8 Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n* [Opentrons P1000 Single Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/single-channel-electronic-pipette-p20/)\n\n\n",
"protocol-steps": "1. Binding Step\n2. Wash 1\n3. Wash 2\n4. Wash 3\n5. Delay for drying\n6. Elution\n\n\n* Note\nThe default values for all volumes, incubation times and mix repetitions were pretested and validated.\nWe do not recommend to change them. If you still decide to change them please scale all volumes proportionally.\nFor change recommendations please contact automation-bio@mn-net.de\n\n\n\n**Process**\n1. Input your protocol parameters below.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit \"Run\".\n\n\n**Additional Notes**\n\nIf you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n**Disclaimer**: MACHEREY-NAGEL GmbH & Co. KG makes every effort to include accurate and up-to-date information within this publication; however, it is possible that omissions or errors might have occurred. MACHEREY-NAGEL GmbH & Co. KG cannot, therefore, make any representations or warranties, expressed or implied, as to the accuracy or completeness of the information provided in this publication. Changes in this publication can be made at any time without notice. For technical details and detailed procedures of the specifications provided in this document please contact your MACHEREY-NAGEL representative. This publication may contain reference to applications and products which are not available in all markets. Please check with your local sales representative.\nAll mentioned trademarks are protected by law. All used names and denotations can be brands, trademarks, or registered labels of their respective owner \u2013 also if they are not special denotation. To mention products and brands is only a kind of information (i.e., it does not offend against trademarks and brands and can not be seen as a kind of recommendation or assessment). Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or support by MACHEREY-NAGEL GmbH & Co. KG or Opentrons. Any views or opinions expressed herein by the authors' do not necessarily state or reflect those of MACHEREY-NAGEL or the Eppendorf AG. NucleoMag\u00ae is a registered trademark of MACHEREY-NAGEL GmbH & Co. KG, D\u00fcren, Germany. Opentrons OT-2 is a device from Opentrons, New York, USA.\n\n* The Download links includes all Protocols in their respective subfolders as well as the .json files for the two labware files.\n\n\n",
"reagent-setup": "\n\n\n",
diff --git a/protoBuilds/macherey-nagel-nucleomag-pathogen/README.json b/protoBuilds/macherey-nagel-nucleomag-pathogen/README.json
index 3bd1520808..16877bd453 100644
--- a/protoBuilds/macherey-nagel-nucleomag-pathogen/README.json
+++ b/protoBuilds/macherey-nagel-nucleomag-pathogen/README.json
@@ -17,7 +17,7 @@
"internal": "macherey-nagal-nucleomag-pathogen\n",
"labware": "* Macherey Nagel 96 Well Square Well Block\n* Macherey Nagel 96 Well Elution Plate U-bottom\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 24 Tube Rack with Generic 2 mL Screwcap](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* [USA Scientific 12 Well Reservoir 22 mL #1061-8150](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n* [Opentrons 96 Tip Rack 1000 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* [Agilent 1 Well Reservoir 290 mL #201252-100](https://www.agilent.com/store/en_US/Prod-201252-100/201252-100)\n\n\n",
"modules": "* [Opentrons Magnetic Module (GEN2)](https://shop.opentrons.com/magnetic-module-gen2/)\n\n\n",
- "partner": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "partner": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [Opentrons P300 8 Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n* [Opentrons P1000 Single Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/single-channel-electronic-pipette-p20/)\n\n\n",
"protocol-steps": "1. Binding Step\n2. Wash 1\n3. Wash 2\n4. Wash 3\n5. Delay for drying\n6. Elution\n\n\n* Note\nThe default values for all volumes, incubation times and mix repetitions were pretested and validated.\nWe do not recommend to change them. If you still decide to change them please scale all volumes proportionally.\nFor change recommendations please contact automation-bio@mn-net.de\n\n\n\n**Process**\n1. Input your protocol parameters below.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit \"Run\".\n\n\n**Additional Notes**\n\nIf you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n**Disclaimer**: MACHEREY-NAGEL GmbH & Co. KG makes every effort to include accurate and up-to-date information within this publication; however, it is possible that omissions or errors might have occurred. MACHEREY-NAGEL GmbH & Co. KG cannot, therefore, make any representations or warranties, expressed or implied, as to the accuracy or completeness of the information provided in this publication. Changes in this publication can be made at any time without notice. For technical details and detailed procedures of the specifications provided in this document please contact your MACHEREY-NAGEL representative. This publication may contain reference to applications and products which are not available in all markets. Please check with your local sales representative.\nAll mentioned trademarks are protected by law. All used names and denotations can be brands, trademarks, or registered labels of their respective owner \u2013 also if they are not special denotation. To mention products and brands is only a kind of information (i.e., it does not offend against trademarks and brands and can not be seen as a kind of recommendation or assessment). Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or support by MACHEREY-NAGEL GmbH & Co. KG or Opentrons. Any views or opinions expressed herein by the authors' do not necessarily state or reflect those of MACHEREY-NAGEL or the Eppendorf AG. NucleoMag\u00ae is a registered trademark of MACHEREY-NAGEL GmbH & Co. KG, D\u00fcren, Germany. Opentrons OT-2 is a device from Opentrons, New York, USA.\n\n* The Download links includes all Protocols in their respective subfolders as well as the .json files for the two labware files.\n\n\n",
"reagent-setup": "\n\n\n",
@@ -26,7 +26,7 @@
"modules": [
"Opentrons Magnetic Module (GEN2)"
],
- "partner": "Opentrons",
+ "partner": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nOpentrons P300 8 Channel Electronic Pipette (GEN2)\nOpentrons P1000 Single Channel Electronic Pipette (GEN2)\n",
"protocol-steps": "\nBinding Step\nWash 1\nWash 2\nWash 3\nDelay for drying\nElution\n\n\nNote\nThe default values for all volumes, incubation times and mix repetitions were pretested and validated.\nWe do not recommend to change them. If you still decide to change them please scale all volumes proportionally.\nFor change recommendations please contact automation-bio@mn-net.de\n\n\n\nProcess\n1. Input your protocol parameters below.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the OT App in the Protocol tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\n7. Hit \"Run\".\n\n\nAdditional Notes\n\nIf you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.\n\nDisclaimer: MACHEREY-NAGEL GmbH & Co. KG makes every effort to include accurate and up-to-date information within this publication; however, it is possible that omissions or errors might have occurred. MACHEREY-NAGEL GmbH & Co. KG cannot, therefore, make any representations or warranties, expressed or implied, as to the accuracy or completeness of the information provided in this publication. Changes in this publication can be made at any time without notice. For technical details and detailed procedures of the specifications provided in this document please contact your MACHEREY-NAGEL representative. This publication may contain reference to applications and products which are not available in all markets. Please check with your local sales representative.\nAll mentioned trademarks are protected by law. All used names and denotations can be brands, trademarks, or registered labels of their respective owner \u2013 also if they are not special denotation. To mention products and brands is only a kind of information (i.e., it does not offend against trademarks and brands and can not be seen as a kind of recommendation or assessment). Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or support by MACHEREY-NAGEL GmbH & Co. KG or Opentrons. Any views or opinions expressed herein by the authors' do not necessarily state or reflect those of MACHEREY-NAGEL or the Eppendorf AG. NucleoMag\u00ae is a registered trademark of MACHEREY-NAGEL GmbH & Co. KG, D\u00fcren, Germany. Opentrons OT-2 is a device from Opentrons, New York, USA.\n\n* The Download links includes all Protocols in their respective subfolders as well as the .json files for the two labware files.",
"reagent-setup": "",
diff --git a/protoBuilds/macherey-nagel-nucleomag-rna/README.json b/protoBuilds/macherey-nagel-nucleomag-rna/README.json
index 55c2f50d79..28949dc010 100644
--- a/protoBuilds/macherey-nagel-nucleomag-rna/README.json
+++ b/protoBuilds/macherey-nagel-nucleomag-rna/README.json
@@ -17,7 +17,7 @@
"internal": "macherey-nagal-nucleomag-rna\n",
"labware": "* 96 Well PCR Plate\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 24 Tube Rack with Generic 2 mL Screwcap](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* [USA Scientific 12 Well Reservoir 22 mL #1061-8150](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n* [Opentrons 96 Tip Rack 1000 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* 96 Deepwell Plate 2mL\n* [Agilent 1 Well Reservoir 290 mL #201252-100](https://www.agilent.com/store/en_US/Prod-201252-100/201252-100)\n\n\n",
"modules": "* [Opentrons Magnetic Module (GEN2)](https://shop.opentrons.com/magnetic-module-gen2/)\n\n\n",
- "partner": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "partner": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [Opentrons P300 8 Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n* [Opentrons P1000 Single Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/single-channel-electronic-pipette-p20/)\n\n\n",
"protocol-steps": "1. Binding Step\n2. rDNase Digestion\n3. Wash 1\n4. Wash 2\n5. Wash 3\n6. Elution\n\n\n* Note\nThe default values for all volumes, incubation times and mix repetitions were pretested and validated.\nWe do not recommend to change them. If you still decide to change them please scale all volumes proportionally.\nFor change recommendations please contact automation-bio@mn-net.de\n\n\n\n**Process**\n1. Input your protocol parameters below.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit \"Run\".\n\n\n**Additional Notes**\n\nIf you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n**Disclaimer**: MACHEREY-NAGEL GmbH & Co. KG makes every effort to include accurate and up-to-date information within this publication; however, it is possible that omissions or errors might have occurred. MACHEREY-NAGEL GmbH & Co. KG cannot, therefore, make any representations or warranties, expressed or implied, as to the accuracy or completeness of the information provided in this publication. Changes in this publication can be made at any time without notice. For technical details and detailed procedures of the specifications provided in this document please contact your MACHEREY-NAGEL representative. This publication may contain reference to applications and products which are not available in all markets. Please check with your local sales representative.\nAll mentioned trademarks are protected by law. All used names and denotations can be brands, trademarks, or registered labels of their respective owner \u2013 also if they are not special denotation. To mention products and brands is only a kind of information (i.e., it does not offend against trademarks and brands and can not be seen as a kind of recommendation or assessment). Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or support by MACHEREY-NAGEL GmbH & Co. KG or Opentrons. Any views or opinions expressed herein by the authors' do not necessarily state or reflect those of MACHEREY-NAGEL or the Eppendorf AG. NucleoMag\u00ae is a registered trademark of MACHEREY-NAGEL GmbH & Co. KG, D\u00fcren, Germany. Opentrons OT-2 is a device from Opentrons, New York, USA.\n\n* The Download links includes all Protocols in their respective subfolders as well as the .json files for the two labware files.\n\n\n",
"reagent-setup": "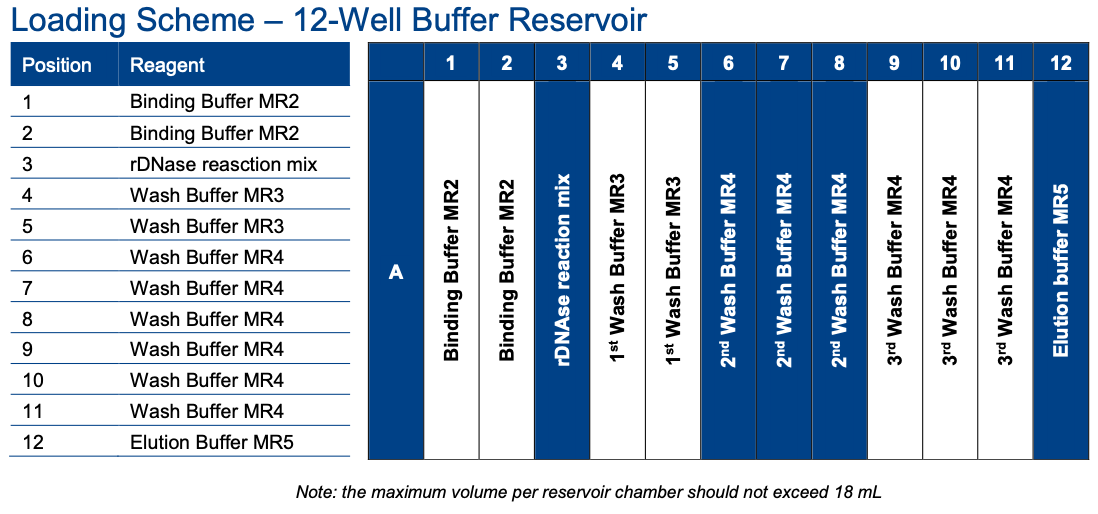\n\n\n",
@@ -26,7 +26,7 @@
"modules": [
"Opentrons Magnetic Module (GEN2)"
],
- "partner": "Opentrons",
+ "partner": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nOpentrons P300 8 Channel Electronic Pipette (GEN2)\nOpentrons P1000 Single Channel Electronic Pipette (GEN2)\n",
"protocol-steps": "\nBinding Step\nrDNase Digestion\nWash 1\nWash 2\nWash 3\nElution\n\n\nNote\nThe default values for all volumes, incubation times and mix repetitions were pretested and validated.\nWe do not recommend to change them. If you still decide to change them please scale all volumes proportionally.\nFor change recommendations please contact automation-bio@mn-net.de\n\n\n\nProcess\n1. Input your protocol parameters below.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the OT App in the Protocol tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\n7. Hit \"Run\".\n\n\nAdditional Notes\n\nIf you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.\n\nDisclaimer: MACHEREY-NAGEL GmbH & Co. KG makes every effort to include accurate and up-to-date information within this publication; however, it is possible that omissions or errors might have occurred. MACHEREY-NAGEL GmbH & Co. KG cannot, therefore, make any representations or warranties, expressed or implied, as to the accuracy or completeness of the information provided in this publication. Changes in this publication can be made at any time without notice. For technical details and detailed procedures of the specifications provided in this document please contact your MACHEREY-NAGEL representative. This publication may contain reference to applications and products which are not available in all markets. Please check with your local sales representative.\nAll mentioned trademarks are protected by law. All used names and denotations can be brands, trademarks, or registered labels of their respective owner \u2013 also if they are not special denotation. To mention products and brands is only a kind of information (i.e., it does not offend against trademarks and brands and can not be seen as a kind of recommendation or assessment). Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or support by MACHEREY-NAGEL GmbH & Co. KG or Opentrons. Any views or opinions expressed herein by the authors' do not necessarily state or reflect those of MACHEREY-NAGEL or the Eppendorf AG. NucleoMag\u00ae is a registered trademark of MACHEREY-NAGEL GmbH & Co. KG, D\u00fcren, Germany. Opentrons OT-2 is a device from Opentrons, New York, USA.\n\n* The Download links includes all Protocols in their respective subfolders as well as the .json files for the two labware files.",
"reagent-setup": "",
diff --git a/protoBuilds/macherey-nagel-nucleomag-size-select/README.json b/protoBuilds/macherey-nagel-nucleomag-size-select/README.json
index bbd493c9d5..e98e082a9a 100644
--- a/protoBuilds/macherey-nagel-nucleomag-size-select/README.json
+++ b/protoBuilds/macherey-nagel-nucleomag-size-select/README.json
@@ -17,7 +17,7 @@
"internal": "macherey-nagel-nucleomag-size-select\n",
"labware": "* 96 Well PCR Plate\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 24 Tube Rack with Generic 2 mL Screwcap](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* [USA Scientific 12 Well Reservoir 22 mL #1061-8150](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n* [Opentrons 96 Tip Rack 1000 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* 96 Deepwell Plate 2mL\n* [Agilent 1 Well Reservoir 290 mL #201252-100](https://www.agilent.com/store/en_US/Prod-201252-100/201252-100)\n\n\n",
"modules": "* [Opentrons Magnetic Module (GEN2)](https://shop.opentrons.com/magnetic-module-gen2/)\n\n\n",
- "partner": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "partner": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [Opentrons P300 8 Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n\n\n",
"protocol-steps": "1. Removal of large fragments\n2. Removal of small fragments\n3. Wash 1\n4. Wash 2\n5. Elution\n\n\n* Note\nThe default values for all volumes, incubation times and mix repetitions were pretested and validated.\nWe do not recommend to change them. If you still decide to change them please scale all volumes proportionally.\nFor change recommendations please contact automation-bio@mn-net.de\n\n\n\n**Process**\n1. Input your protocol parameters below.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit \"Run\".\n\n\n**Additional Notes**\n\nIf you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n**Disclaimer**: MACHEREY-NAGEL GmbH & Co. KG makes every effort to include accurate and up-to-date information within this publication; however, it is possible that omissions or errors might have occurred. MACHEREY-NAGEL GmbH & Co. KG cannot, therefore, make any representations or warranties, expressed or implied, as to the accuracy or completeness of the information provided in this publication. Changes in this publication can be made at any time without notice. For technical details and detailed procedures of the specifications provided in this document please contact your MACHEREY-NAGEL representative. This publication may contain reference to applications and products which are not available in all markets. Please check with your local sales representative.\nAll mentioned trademarks are protected by law. All used names and denotations can be brands, trademarks, or registered labels of their respective owner \u2013 also if they are not special denotation. To mention products and brands is only a kind of information (i.e., it does not offend against trademarks and brands and can not be seen as a kind of recommendation or assessment). Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or support by MACHEREY-NAGEL GmbH & Co. KG or Opentrons. Any views or opinions expressed herein by the authors' do not necessarily state or reflect those of MACHEREY-NAGEL or the Eppendorf AG. NucleoMag\u00ae is a registered trademark of MACHEREY-NAGEL GmbH & Co. KG, D\u00fcren, Germany. Opentrons OT-2 is a device from Opentrons, New York, USA.\n\n* The Download links includes all Protocols in their respective subfolders as well as the .json files for the two labware files.\n\n\n",
"reagent-setup": "\n\n\n",
@@ -26,7 +26,7 @@
"modules": [
"Opentrons Magnetic Module (GEN2)"
],
- "partner": "Opentrons",
+ "partner": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nOpentrons P300 8 Channel Electronic Pipette (GEN2)\n",
"protocol-steps": "\nRemoval of large fragments\nRemoval of small fragments\nWash 1\nWash 2\nElution\n\n\nNote\nThe default values for all volumes, incubation times and mix repetitions were pretested and validated.\nWe do not recommend to change them. If you still decide to change them please scale all volumes proportionally.\nFor change recommendations please contact automation-bio@mn-net.de\n\n\n\nProcess\n1. Input your protocol parameters below.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the OT App in the Protocol tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\n7. Hit \"Run\".\n\n\nAdditional Notes\n\nIf you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.\n\nDisclaimer: MACHEREY-NAGEL GmbH & Co. KG makes every effort to include accurate and up-to-date information within this publication; however, it is possible that omissions or errors might have occurred. MACHEREY-NAGEL GmbH & Co. KG cannot, therefore, make any representations or warranties, expressed or implied, as to the accuracy or completeness of the information provided in this publication. Changes in this publication can be made at any time without notice. For technical details and detailed procedures of the specifications provided in this document please contact your MACHEREY-NAGEL representative. This publication may contain reference to applications and products which are not available in all markets. Please check with your local sales representative.\nAll mentioned trademarks are protected by law. All used names and denotations can be brands, trademarks, or registered labels of their respective owner \u2013 also if they are not special denotation. To mention products and brands is only a kind of information (i.e., it does not offend against trademarks and brands and can not be seen as a kind of recommendation or assessment). Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or support by MACHEREY-NAGEL GmbH & Co. KG or Opentrons. Any views or opinions expressed herein by the authors' do not necessarily state or reflect those of MACHEREY-NAGEL or the Eppendorf AG. NucleoMag\u00ae is a registered trademark of MACHEREY-NAGEL GmbH & Co. KG, D\u00fcren, Germany. Opentrons OT-2 is a device from Opentrons, New York, USA.\n\n* The Download links includes all Protocols in their respective subfolders as well as the .json files for the two labware files.",
"reagent-setup": "",
diff --git a/protoBuilds/macherey-nagel-nucleomag-tissue/README.json b/protoBuilds/macherey-nagel-nucleomag-tissue/README.json
index 4dcab4f77a..60dc3c03e9 100644
--- a/protoBuilds/macherey-nagel-nucleomag-tissue/README.json
+++ b/protoBuilds/macherey-nagel-nucleomag-tissue/README.json
@@ -17,7 +17,7 @@
"internal": "macherey-nagel-nucleomag-tissue\n",
"labware": "* Macherey Nagel 96 Well Square Well Block\n* Macherey Nagel 96 Well Elution Plate U-bottom\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 24 Tube Rack with Generic 2 mL Screwcap](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* [USA Scientific 12 Well Reservoir 22 mL #1061-8150](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n* [Opentrons 96 Tip Rack 1000 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* [Agilent 1 Well Reservoir 290 mL #201252-100](https://www.agilent.com/store/en_US/Prod-201252-100/201252-100)\n\n\n",
"modules": "* [Opentrons Magnetic Module (GEN2)](https://shop.opentrons.com/magnetic-module-gen2/)\n\n\n",
- "partner": "[Opentrons](https://opentrons.com/)\n\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-tissue). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "partner": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-tissue). This page won\u2019t be available after January 31st, 2024.\n\n",
"pipettes": "* [Opentrons P300 8 Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n* [Opentrons P1000 Single Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/single-channel-electronic-pipette-p20/)\n\n\n",
"protocol-steps": "1. Binding Step\n2. Wash 1\n3. Wash 2\n4. Wash 3\n5. Delay for drying\n6. Elution\n\n\n* Note\nThe default values for all volumes, incubation times and mix repetitions were pretested and validated.\nWe do not recommend to change them. If you still decide to change them please scale all volumes proportionally.\nFor change recommendations please contact automation-bio@mn-net.de\n\n\n\n**Process**\n1. Input your protocol parameters below.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit \"Run\".\n\n\n**Additional Notes**\n\nIf you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n**Disclaimer**: MACHEREY-NAGEL GmbH & Co. KG makes every effort to include accurate and up-to-date information within this publication; however, it is possible that omissions or errors might have occurred. MACHEREY-NAGEL GmbH & Co. KG cannot, therefore, make any representations or warranties, expressed or implied, as to the accuracy or completeness of the information provided in this publication. Changes in this publication can be made at any time without notice. For technical details and detailed procedures of the specifications provided in this document please contact your MACHEREY-NAGEL representative. This publication may contain reference to applications and products which are not available in all markets. Please check with your local sales representative.\nAll mentioned trademarks are protected by law. All used names and denotations can be brands, trademarks, or registered labels of their respective owner \u2013 also if they are not special denotation. To mention products and brands is only a kind of information (i.e., it does not offend against trademarks and brands and can not be seen as a kind of recommendation or assessment). Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or support by MACHEREY-NAGEL GmbH & Co. KG or Opentrons. Any views or opinions expressed herein by the authors' do not necessarily state or reflect those of MACHEREY-NAGEL or the Eppendorf AG. NucleoMag\u00ae is a registered trademark of MACHEREY-NAGEL GmbH & Co. KG, D\u00fcren, Germany. Opentrons OT-2 is a device from Opentrons, New York, USA.\n\n* The Download links includes all Protocols in their respective subfolders as well as the .json files for the two labware files.\n\n\n",
"reagent-setup": "\n\n\n",
diff --git a/protoBuilds/macherey-nagel-nucleomag-virus/README.json b/protoBuilds/macherey-nagel-nucleomag-virus/README.json
index d5fef902fb..7848f86eca 100644
--- a/protoBuilds/macherey-nagel-nucleomag-virus/README.json
+++ b/protoBuilds/macherey-nagel-nucleomag-virus/README.json
@@ -17,7 +17,7 @@
"internal": "macherey-nagel-nucleomag-virus\n",
"labware": "* Macherey Nagel 96 Well Square Well Block\n* Macherey Nagel 96 Well Elution Plate U-bottom\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 24 Tube Rack with Generic 2 mL Screwcap](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n* [USA Scientific 12 Well Reservoir 22 mL #1061-8150](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n* [Opentrons 96 Tip Rack 1000 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-1000ul-tips)\n* [Agilent 1 Well Reservoir 290 mL #201252-100](https://www.agilent.com/store/en_US/Prod-201252-100/201252-100)\n\n\n",
"modules": "* [Opentrons Magnetic Module (GEN2)](https://shop.opentrons.com/magnetic-module-gen2/)\n\n\n",
- "partner": "[Opentrons](https://opentrons.com/)\n\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-virus). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "partner": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-virus). This page won\u2019t be available after January 31st, 2024.\n\n",
"pipettes": "* [Opentrons P300 8 Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n* [Opentrons P1000 Single Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/single-channel-electronic-pipette-p20/)\n\n\n",
"protocol-steps": "1. Binding Step\n2. Wash 1\n3. Wash 2\n4. Delay for drying\n5. Elution\n\n\n* Note\nThe default values for all volumes, incubation times and mix repetitions were pretested and validated.\nWe do not recommend to change them. If you still decide to change them please scale all volumes proportionally.\nFor change recommendations please contact automation-bio@mn-net.de\n\n\n\n**Process**\n1. Input your protocol parameters below.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit \"Run\".\n\n\n**Additional Notes**\n\nIf you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n**Disclaimer**: MACHEREY-NAGEL GmbH & Co. KG makes every effort to include accurate and up-to-date information within this publication; however, it is possible that omissions or errors might have occurred. MACHEREY-NAGEL GmbH & Co. KG cannot, therefore, make any representations or warranties, expressed or implied, as to the accuracy or completeness of the information provided in this publication. Changes in this publication can be made at any time without notice. For technical details and detailed procedures of the specifications provided in this document please contact your MACHEREY-NAGEL representative. This publication may contain reference to applications and products which are not available in all markets. Please check with your local sales representative.\nAll mentioned trademarks are protected by law. All used names and denotations can be brands, trademarks, or registered labels of their respective owner \u2013 also if they are not special denotation. To mention products and brands is only a kind of information (i.e., it does not offend against trademarks and brands and can not be seen as a kind of recommendation or assessment). Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or support by MACHEREY-NAGEL GmbH & Co. KG or Opentrons. Any views or opinions expressed herein by the authors' do not necessarily state or reflect those of MACHEREY-NAGEL or the Eppendorf AG. NucleoMag\u00ae is a registered trademark of MACHEREY-NAGEL GmbH & Co. KG, D\u00fcren, Germany. Opentrons OT-2 is a device from Opentrons, New York, USA.\n\n* The Download links includes all Protocols in their respective subfolders as well as the .json files for the two labware files.\n\n\n",
"reagent-setup": "\n\n\n",
diff --git a/protoBuilds/magnesil/README.json b/protoBuilds/magnesil/README.json
index 22c0387dd1..7e118df726 100644
--- a/protoBuilds/magnesil/README.json
+++ b/protoBuilds/magnesil/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"MagneSil"
@@ -10,7 +10,7 @@
"internal": "magnesil",
"labware": "\nOpentrons Tips for P300 (Filter or Standard)\n96-Deepwell Plate (containing samples) (Example)\n12-Well Reservoir (Example)\n96-Well Plate (Example)\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* MagneSil\n\n",
"deck-setup": "\n\n\n**Slot 2**: 12-Well Reservoir (See Reagent Setup below for more information)\n\n**Slot 4**: [Opentrons Magnetic Module](https://shop.opentrons.com/magnetic-module-gen2/) with 96-Deepwell Plate containing samples in odd columns (during the protocol, samples will be transferred to the even columns)\n\n\n**Slot 6**: 96-Well Plate for elutes\n\n**Slots 7-11**: Opentrons Tips (Each column of samples requires 15 columns of tips)\n\n",
"description": "This protocol performs a nucleic acid purification protocol based on the MagneSil Plasmid Purification System from Promega. Using flexible parameters (described below), one can adapt this protocol to the equipment in their lab and modify to their workflow.\n\n\nExplanation of complex parameters below:\n* **Number of Samples**: Specify the number of samples (1-32)\n* **Pipette Model**: Select between the GEN1 and GEN2 [P300 8-Channel Pipette](https://shop.opentrons.com/8-channel-electronic-pipette/)\n* **Pipette Mount**: Select which mount (Right, Left) the pipette is attached to\n* **Use Filter Tips?**: Select whether to use [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/opentrons-200ul-filter-tips/) or [Opentrons 300\u00b5L Standard Tips](https://shop.opentrons.com/opentrons-300ul-tips-1000-refills/)\n* **Magnetic Module Model**: Select between the GEN1 and GEN2 [Opentrons Magnetic Module](https://shop.opentrons.com/magnetic-module-gen2/)\n* **Sample Plate Type**: Select the type of plate containing samples (will be placed on Magnetic Module)\n* **Reservoir Type**: Select which 12-Well Reservoir will be used to hold reagents\n* **Elution Plate Type**: Select the type of plate that will be used for the elutions\n\n\n---\n\n",
diff --git a/protoBuilds/nextera-flex-library-prep-amplify-tagmented-dna/README.json b/protoBuilds/nextera-flex-library-prep-amplify-tagmented-dna/README.json
index 5b1d9e74f8..8e228a013e 100644
--- a/protoBuilds/nextera-flex-library-prep-amplify-tagmented-dna/README.json
+++ b/protoBuilds/nextera-flex-library-prep-amplify-tagmented-dna/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Nextera DNA Flex Library Prep"
@@ -7,7 +7,7 @@
},
"description": "This protocol performs the 'Amplify Tagmented DNA' section of the Nextera DNA Flex Library Prep protocol.\nLinks:\n Cherrypick Samples\n Tagment DNA\n Post Tagmentation Cleanup\n Amplify Tagmented DNA\n* Cleanup Libraries\n\n\n\nOpentrons magnetic module\nBio-Rad Hard Shell 96-well low profile PCR plate 200ul #hsp9601\nAgilent single-channel reservoir 290ml #201252-100\nOpentrons 4-in-1 tuberack with 2ml Eppendorf tubes\nP50 Single-channel electronic pipette\nP300 Single/multi-channel electronic pipette\n300ul Opentrons tipracks\n\n\n\nPreprogram the thermocycler according to the BLT PCR program parameters described in the kit manual.\n4x6 aluminum block tuberack (slot 4, mounted on temperature module)\n A1-C1: mastermix 1.5ml snapcap tubes (loaded empty, 1 tube needed per 32 samples)\n A2-D2: EPM in 0.5ml false-bottom tubes (1 tube needed per 24 samples)\n* A3-B3: nuclease-free water in 1.5ml snapcap tubes (1 tube needed per 48 samples)",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Nextera DNA Flex Library Prep\n\n\n",
"description": "This protocol performs the 'Amplify Tagmented DNA' section of the [Nextera DNA Flex Library Prep protocol](https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/nextera-dna-flex.html).\n\nLinks:\n* [Cherrypick Samples](./nextera-flex-library-prep-cherrypick-samples)\n* [Tagment DNA](./nextera-flex-library-prep-tagment-dna)\n* [Post Tagmentation Cleanup](./nextera-flex-library-prep-post-tag-cleanup)\n* [Amplify Tagmented DNA](./nextera-flex-library-prep-amplify-tagmented-dna)\n* [Cleanup Libraries](./nextera-flex-library-prep-cleanup-libraries)\n\n---\n\n\n* [Opentrons magnetic module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Bio-Rad Hard Shell 96-well low profile PCR plate 200ul #hsp9601](bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [Agilent single-channel reservoir 290ml #201252-100](https://www.agilent.com/store/en_US/Prod-201252-100/201252-100)\n* [Opentrons 4-in-1 tuberack with 2ml Eppendorf tubes](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [P50 Single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [P300 Single/multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [300ul Opentrons tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n---\n\n\nPreprogram the thermocycler according to the BLT PCR program parameters described in the kit manual.\n\n4x6 aluminum block tuberack (slot 4, mounted on temperature module)\n* A1-C1: mastermix 1.5ml snapcap tubes (loaded empty, 1 tube needed per 32 samples)\n* A2-D2: EPM in 0.5ml false-bottom tubes (1 tube needed per 24 samples)\n* A3-B3: nuclease-free water in 1.5ml snapcap tubes (1 tube needed per 48 samples)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n",
diff --git a/protoBuilds/nextera-flex-library-prep-cherrypick-samples/README.json b/protoBuilds/nextera-flex-library-prep-cherrypick-samples/README.json
index 5b3f67b397..ed21a81cac 100644
--- a/protoBuilds/nextera-flex-library-prep-cherrypick-samples/README.json
+++ b/protoBuilds/nextera-flex-library-prep-cherrypick-samples/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Nextera DNA Flex Library Prep"
@@ -7,7 +7,7 @@
},
"description": "This protocol cherrypicks samples from up to 9 Olympus PCR source plates to 1 Bio-Rad Hard Shell PCR destination plate as preparation for the the Nextera DNA Flex Library Prep protocol. The CSV should be formatted in the following format, including the header line:\nSource plate (1-9), Source well (A1-H12), Destination well (A1-H12)\n1, A1, A1\n1, C5, A2\n3, H9, A3\n9, E3, B1\nLinks:\n Cherrypick Samples\n Tagment DNA\n Post Tagmentation Cleanup\n Amplify Tagmented DNA\n* Cleanup Libraries\n\n\n\nOlympus 96-well PCR plates 200ul\nBio-Rad Hard Shell 96-well low profile PCR plate 200ul #hsp9601\nP50/P300 Single-channel electronic pipette\n300ul Opentrons tipracks\n\n\n\nPreprogram the thermocycler according to the BLT PCR program parameters described in the kit manual.\n4x6 aluminum block tuberack (slot 4, mounted on temperature module)\n A1-C1: mastermix 1.5ml snapcap tubes (loaded empty, 1 tube needed per 32 samples)\n A2-D2: EPM in 0.5ml false-bottom tubes (1 tube needed per 24 samples)\n* A3-B3: nuclease-free water in 1.5ml snapcap tubes (1 tube needed per 48 samples)",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Nextera DNA Flex Library Prep\n\n\n",
"description": "This protocol cherrypicks samples from up to 9 Olympus PCR source plates to 1 Bio-Rad Hard Shell PCR destination plate as preparation for the the [Nextera DNA Flex Library Prep protocol](https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/nextera-dna-flex.html). The CSV should be formatted in the following format, **including the header line:**\n\n```\nSource plate (1-9), Source well (A1-H12), Destination well (A1-H12)\n1, A1, A1\n1, C5, A2\n3, H9, A3\n9, E3, B1\n```\n\nLinks:\n* [Cherrypick Samples](./nextera-flex-library-prep-cherrypick-samples)\n* [Tagment DNA](./nextera-flex-library-prep-tagment-dna)\n* [Post Tagmentation Cleanup](./nextera-flex-library-prep-post-tag-cleanup)\n* [Amplify Tagmented DNA](./nextera-flex-library-prep-amplify-tagmented-dna)\n* [Cleanup Libraries](./nextera-flex-library-prep-cleanup-libraries)\n\n---\n\n\n* Olympus 96-well PCR plates 200ul\n* [Bio-Rad Hard Shell 96-well low profile PCR plate 200ul #hsp9601](bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [P50/P300 Single-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [300ul Opentrons tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n---\n\n\nPreprogram the thermocycler according to the BLT PCR program parameters described in the kit manual.\n\n4x6 aluminum block tuberack (slot 4, mounted on temperature module)\n* A1-C1: mastermix 1.5ml snapcap tubes (loaded empty, 1 tube needed per 32 samples)\n* A2-D2: EPM in 0.5ml false-bottom tubes (1 tube needed per 24 samples)\n* A3-B3: nuclease-free water in 1.5ml snapcap tubes (1 tube needed per 48 samples)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n",
diff --git a/protoBuilds/nextera-flex-library-prep-cleanup-libraries/README.json b/protoBuilds/nextera-flex-library-prep-cleanup-libraries/README.json
index ed12a1191f..a794923771 100644
--- a/protoBuilds/nextera-flex-library-prep-cleanup-libraries/README.json
+++ b/protoBuilds/nextera-flex-library-prep-cleanup-libraries/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Nextera DNA Flex Library Prep"
@@ -7,7 +7,7 @@
},
"description": "This protocol performs the 'Cleanup Libraries' section of the Nextera DNA Flex Library Prep protocol.\nLinks:\n Cherrypick Samples\n Tagment DNA\n Post Tagmentation Cleanup\n Amplify Tagmented DNA\n* Cleanup Libraries\n\n\n\nOpentrons magnetic module\nBio-Rad Hard Shell 96-well low profile PCR plate 200ul #hsp9601\nUSA Scientific 12-channel reservoir 22ml #1061-8150\nOpentrons 4-in-1 tuberack with 2ml Eppendorf tubes\nP50 Single/multi-channel electronic pipette\nP300 Single/multi-channel electronic pipette\n300ul Opentrons tipracks\n\n\n\nCentrifuge the PCR plate from the Amplify Tagmented DNA step. Place the plate on the magnetic module.\n12-channel reservoir (slot 3)\n channel 1: SPB (user is prompted to vortex before adding mid-protocol)\n channel 2: nuclease-free water\n channel 3: RSB\n channel 4-5: EtOH\n* channels 10-12: liquid waste (loaded empty)",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Nextera DNA Flex Library Prep\n\n",
"description": "This protocol performs the 'Cleanup Libraries' section of the [Nextera DNA Flex Library Prep protocol](https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/nextera-dna-flex.html).\n\nLinks:\n* [Cherrypick Samples](./nextera-flex-library-prep-cherrypick-samples)\n* [Tagment DNA](./nextera-flex-library-prep-tagment-dna)\n* [Post Tagmentation Cleanup](./nextera-flex-library-prep-post-tag-cleanup)\n* [Amplify Tagmented DNA](./nextera-flex-library-prep-amplify-tagmented-dna)\n* [Cleanup Libraries](./nextera-flex-library-prep-cleanup-libraries)\n\n---\n\n\n* [Opentrons magnetic module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Bio-Rad Hard Shell 96-well low profile PCR plate 200ul #hsp9601](bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [USA Scientific 12-channel reservoir 22ml #1061-8150](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n* [Opentrons 4-in-1 tuberack with 2ml Eppendorf tubes](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [P50 Single/multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [P300 Single/multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [300ul Opentrons tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n---\n\n\nCentrifuge the PCR plate from the Amplify Tagmented DNA step. Place the plate on the magnetic module.\n\n12-channel reservoir (slot 3)\n* channel 1: SPB (user is prompted to vortex before adding mid-protocol)\n* channel 2: nuclease-free water\n* channel 3: RSB\n* channel 4-5: EtOH\n* channels 10-12: liquid waste (loaded empty)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n",
diff --git a/protoBuilds/nextera-flex-library-prep-post-tag-cleanup/README.json b/protoBuilds/nextera-flex-library-prep-post-tag-cleanup/README.json
index db69ab2d74..4b7df34231 100644
--- a/protoBuilds/nextera-flex-library-prep-post-tag-cleanup/README.json
+++ b/protoBuilds/nextera-flex-library-prep-post-tag-cleanup/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Nextera DNA Flex Library Prep"
@@ -7,7 +7,7 @@
},
"description": "This protocol performs the 'Post Tagmentation Cleanup' section of the Nextera DNA Flex Library Prep protocol.\nLinks:\n Cherrypick Samples\n Tagment DNA\n Post Tagmentation Cleanup\n Amplify Tagmented DNA\n* Cleanup Libraries\n\n\n\nOpentrons magnetic module\nBio-Rad Hard Shell 96-well low profile PCR plate 200ul #hsp9601\nUSA Scientific 12-channel reservoir 22ml #1061-8150\nP300 Single/multi-channel electronic pipette\n300ul Opentrons tipracks\n\n\n\nAdd 10ul of TSB to each sample, seal the plate with Microseal B, place on the preprogrammed thermal cycler, and run the PTC program.\n12-channel reservoir (slot 3)\n channel 1: TWB\n channel 2: TWB (only needed if number of samples > 48)\n* channel 12: liquid waste (loaded empty)",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Nextera DNA Flex Library Prep\n\n\n",
"description": "This protocol performs the 'Post Tagmentation Cleanup' section of the [Nextera DNA Flex Library Prep protocol](https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/nextera-dna-flex.html).\n\nLinks:\n* [Cherrypick Samples](./nextera-flex-library-prep-cherrypick-samples)\n* [Tagment DNA](./nextera-flex-library-prep-tagment-dna)\n* [Post Tagmentation Cleanup](./nextera-flex-library-prep-post-tag-cleanup)\n* [Amplify Tagmented DNA](./nextera-flex-library-prep-amplify-tagmented-dna)\n* [Cleanup Libraries](./nextera-flex-library-prep-cleanup-libraries)\n\n---\n\n\n* [Opentrons magnetic module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Bio-Rad Hard Shell 96-well low profile PCR plate 200ul #hsp9601](bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [USA Scientific 12-channel reservoir 22ml #1061-8150](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n* [P300 Single/multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [300ul Opentrons tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n---\n\n\nAdd 10ul of TSB to each sample, seal the plate with Microseal B, place on the preprogrammed thermal cycler, and run the PTC program.\n\n12-channel reservoir (slot 3)\n* channel 1: TWB\n* channel 2: TWB **(only needed if number of samples > 48)**\n* channel 12: liquid waste (loaded empty)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n",
diff --git a/protoBuilds/nextera-flex-library-prep-tagment-dna/README.json b/protoBuilds/nextera-flex-library-prep-tagment-dna/README.json
index 627c44aace..861064ded2 100644
--- a/protoBuilds/nextera-flex-library-prep-tagment-dna/README.json
+++ b/protoBuilds/nextera-flex-library-prep-tagment-dna/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Nextera DNA Flex Library Prep"
@@ -7,7 +7,7 @@
},
"description": "This protocol performs the 'Tagment DNA' section of the Nextera DNA Flex Library Prep protocol.\nLinks:\n Cherrypick Samples\n Tagment DNA\n Post Tagmentation Cleanup\n Amplify Tagmented DNA\n* Cleanup Libraries\n\n\n\nBio-Rad Hard Shell 96-well low profile PCR plate 200ul #hsp9601\nOpentrons 4-in-1 tuberack with 1.5ml Eppendorf tubes\n200ul PCR strips\nP50 Single/multi-channel electronic pipette\n300ul Opentrons tipracks\n\n\n\nVortex BLT and TB1 vigorously for 10 seconds before placing in tube rack wells B1 and C1, respectively, and resuming. Preprogram the thermocycler according to the TAG program parameters described in the kit manual.\n1.5ml Eppendorf Tuberack (slot 2)\n A1-B1: mastermix 1.5ml snapcap tubes (loaded empty, 1 tube needed per 48 samples)\n A2-B2: BLT in 0.5ml false-bottom tubes (1 tube needed per 48 samples)\n* A3-B3: TB1 in 0.5ml false-bottom tubes (1 tube needed per 48 samples)\n200ul PCR strips (slot 3 only needed if using P50 multi-channel pipette\n* strip column 1: mastermix strip (loaded empty)",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Nextera DNA Flex Library Prep\n\n\n",
"description": "This protocol performs the 'Tagment DNA' section of the [Nextera DNA Flex Library Prep protocol](https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/nextera-dna-flex.html).\n\nLinks:\n* [Cherrypick Samples](./nextera-flex-library-prep-cherrypick-samples)\n* [Tagment DNA](./nextera-flex-library-prep-tagment-dna)\n* [Post Tagmentation Cleanup](./nextera-flex-library-prep-post-tag-cleanup)\n* [Amplify Tagmented DNA](./nextera-flex-library-prep-amplify-tagmented-dna)\n* [Cleanup Libraries](./nextera-flex-library-prep-cleanup-libraries)\n\n---\n\n\n* [Bio-Rad Hard Shell 96-well low profile PCR plate 200ul #hsp9601](bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n* [Opentrons 4-in-1 tuberack with 1.5ml Eppendorf tubes](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* 200ul PCR strips\n* [P50 Single/multi-channel electronic pipette](https://shop.opentrons.com/collections/ot-2-pipettes)\n* [300ul Opentrons tipracks](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n\n---\n\n\nVortex BLT and TB1 vigorously for 10 seconds before placing in tube rack wells B1 and C1, respectively, and resuming. Preprogram the thermocycler according to the TAG program parameters described in the kit manual.\n\n1.5ml Eppendorf Tuberack (slot 2)\n* A1-B1: mastermix 1.5ml snapcap tubes (loaded empty, 1 tube needed per 48 samples)\n* A2-B2: BLT in 0.5ml false-bottom tubes (1 tube needed per 48 samples)\n* A3-B3: TB1 in 0.5ml false-bottom tubes (1 tube needed per 48 samples)\n\n200ul PCR strips (slot 3 **only needed if using P50 multi-channel pipette**\n* strip column 1: mastermix strip (loaded empty)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n",
diff --git a/protoBuilds/normalization/README.json b/protoBuilds/normalization/README.json
index bd61e44cb8..67d790df1d 100644
--- a/protoBuilds/normalization/README.json
+++ b/protoBuilds/normalization/README.json
@@ -8,7 +8,7 @@
"description": "\nConcentration normalization is a key component of many genomic and proteomic applications, such as NGS library prep. With this protocol, you can easily normalize the concentrations of samples in a 96 or 384 microwell plate without worrying about missing a well or adding the wrong volume. Just upload your properly formatted CSV file (keep scrolling for an example), customize your parameters, and download your ready-to-run protocol.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Single-Channel Pipette and corresponding Tips\nSamples in a compatible plate (96-well or 384-well)\nAutomation-friendly reservoir\nDiluent\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nCSV Format\nYour file must be saved as a comma separated value (.csv) file type. Your CSV must contain values corresponding to volumes in microliters (\u03bcL). It should be formatted in \u201clandscape\u201d orientation, with the value corresponding to well A1 in the upper left-hand corner of the value list.\n\nIn this example, 40\u03bcL will be added to A1, 41\u03bcL will be added to well B1, and so on.\nIf you\u2019d like to follow our template, you can make a copy of this spreadsheet, fill out your values, and export as CSV from there.\nNote about CSV: All values corresponding to wells in the CSV must have a value (zero (0) is a valid value and nothing will be transferred to the corresponding well(s)). Additionally, the CSV can be formatted in \"portrait\" orientation. In portrait orientation, the bottom left corner is treated as A1 and the top right corner would correspond to the furthest well from A1 (H12 in a 96-well plate).\nUsing the customization fields below, set up your protocol.\n Volumes CSV: Upload the CSV (.csv) containing your diluent volumes.\n Pipette Model: Select which pipette you will use for this protocol.\n Pipette Mount: Specify which mount your single-channel pipette is on (left or right)\n Plate Type: Select which (destination) plate you will use for this protocol.\n Reservoir Type: Select which (source) reservoir you will use for this protocol.\n Filter Tips: Specify whether you want to use filter tips.\n* Tip Usage Strategy: Specify whether you'd like to use a new tip for each transfer, or keep the same tip throughout the protocol.",
"internal": "normalization",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/normalization). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/normalization). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Featured\n\t* Normalization\n\n",
"description": "\n\nConcentration normalization is a key component of many genomic and proteomic applications, such as NGS library prep. With this protocol, you can easily normalize the concentrations of samples in a 96 or 384 microwell plate without worrying about missing a well or adding the wrong volume. Just upload your properly formatted CSV file (keep scrolling for an example), customize your parameters, and download your ready-to-run protocol.\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes) and corresponding [Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Samples in a compatible plate (96-well or 384-well)](https://labware.opentrons.com/?category=wellPlate)\n* [Automation-friendly reservoir](https://labware.opentrons.com/?category=reservoir)\n* Diluent\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n---\n\n\n**CSV Format**\n\nYour file must be saved as a comma separated value (.csv) file type. Your CSV must contain values corresponding to volumes in microliters (\u03bcL). It should be formatted in \u201clandscape\u201d orientation, with the value corresponding to well A1 in the upper left-hand corner of the value list.\n\n\n\nIn this example, 40\u03bcL will be added to A1, 41\u03bcL will be added to well B1, and so on.\n\nIf you\u2019d like to follow our template, you can make a copy of [this spreadsheet](https://opentrons-protocol-library-website.s3.amazonaws.com/Technical+Notes/normalization/Opentrons+Normalization+Template.xlsx), fill out your values, and export as CSV from there.\n\n*Note about CSV*: All values corresponding to wells in the CSV must have a value (zero (0) is a valid value and nothing will be transferred to the corresponding well(s)). Additionally, the CSV can be formatted in \"portrait\" orientation. In portrait orientation, the bottom left corner is treated as A1 and the top right corner would correspond to the furthest well from A1 (H12 in a 96-well plate).\n\nUsing the customization fields below, set up your protocol.\n* Volumes CSV: Upload the CSV (.csv) containing your diluent volumes.\n* Pipette Model: Select which pipette you will use for this protocol.\n* Pipette Mount: Specify which mount your single-channel pipette is on (left or right)\n* Plate Type: Select which (destination) plate you will use for this protocol.\n* Reservoir Type: Select which (source) reservoir you will use for this protocol.\n* Filter Tips: Specify whether you want to use filter tips.\n* Tip Usage Strategy: Specify whether you'd like to use a new tip for each transfer, or keep the same tip throughout the protocol.\n\n\n\n",
"internal": "normalization\n",
diff --git a/protoBuilds/nucleic_acid_purification_with_magnetic_beads/README.json b/protoBuilds/nucleic_acid_purification_with_magnetic_beads/README.json
index a542c7a5eb..c57db9346b 100644
--- a/protoBuilds/nucleic_acid_purification_with_magnetic_beads/README.json
+++ b/protoBuilds/nucleic_acid_purification_with_magnetic_beads/README.json
@@ -8,7 +8,7 @@
"description": "With this protocol, you can perform high-quality nucleic acid purifications using magnetic beads and the Opentrons Magnetic Module. This protocol contains flexible parameters that you can customize for many different magnetic bead and nucleic acid types. Use this setup to rapidly iterate and optimize your magbead-based workflows!\nYou can use any magnetic beads you prefer with this protocol, but we have included some reagent recommendations in the Materials Needed section below to help you get started. For more detailed information on how to use this protocol, please see our Technical Note.\n\n\n\n-- Opentrons OT-2\n-- Opentrons Magnetic Module\n-- Opentrons OT-2 Run App (Version 3.1.2 or later)\n-- 200uL or 300 uL Tiprack (Opentrons tips suggested)\n-- 12-row automation-friendly trough\n-- BioRad HardShell 96-Well PCR Plate\n-- Magnetic Beads (Looking for a kit? We recommend trying Omega Bio-tek Mag-Bind\u00ae TotalPure NGS)\n-- Ethanol\n-- Elution Buffer (Typically 10 mM Tris pH 8.0, TE Buffer, or nuclease-free water)\n\n\n\nUsing the customization fields below, set up your protocol as follows:\n\nPipette: Specify your pipette. We recommend using a p50 or p300 multi- or single-channel.\nPipette Mount: Specify which mount (left or right) your pipette is on.\nSample number: Customize the number of samples to run per protocol. A multiple of 8 is recommended when you are using a multichannel pipette.\nSample volume: Specify the starting volume (in uL) of the input sample.\nBead Ratio: Customize the ratio of beads for left or right side size-selection of fragments. The default bead ratio is 1.8x the input sample volume.\nElution Volume: Specify the final volume (in uL) to elute the purified nucleic acid. The Opentrons MagDeck supports elution volumes above 10 \u00b5L.\nIncubation Time: Specify the amount of time (in minutes) that the bead solution and input sample interact.\nSettling Time: Specify the amount of time (in minutes) needed to pellet the beads. Higher volumes may require a longer settling time.\nDrying Time: Specify the drying time (in minutes) needed after wash steps.\n\n\n",
"internal": "Nucleic Acid Purification, v1",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/nucleic_acid_purification_with_magnetic_beads). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/nucleic_acid_purification_with_magnetic_beads). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Featured\n * Nucleic Acid Purification with Magnetic Beads (Universal)\n\n",
"description": "With this protocol, you can perform high-quality nucleic acid purifications using magnetic beads and the [Opentrons Magnetic Module](https://shop.opentrons.com/products/magdeck]). This protocol contains flexible parameters that you can customize for many different magnetic bead and nucleic acid types. Use this setup to rapidly iterate and optimize your magbead-based workflows!\n\nYou can use any magnetic beads you prefer with this protocol, but we have included some reagent recommendations in the **Materials Needed** section below to help you get started. For more detailed information on how to use this protocol, please see our [Technical Note](https://s3.amazonaws.com/opentrons-protocol-library-website/Technical+Notes/Nucleic+Acid+Purification+with+Magnetic+Module+OT2+Technical+Note.pdf).\n\n---\n\n---\n\n\n\n-- [Opentrons OT-2](http://opentrons.com/ot-2)\n\n-- [Opentrons Magnetic Module](https://shop.opentrons.com/products/magdeck?_ga=2.171718441.823190023.1542396855-403439593.1535387376)\n\n-- [Opentrons OT-2 Run App (Version 3.1.2 or later)](http://opentrons.com/ot-app)\n\n-- 200uL or 300 uL Tiprack ([Opentrons tips suggested](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips-racks-9-600-tips))\n\n-- [12-row automation-friendly trough](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n\n-- [BioRad HardShell 96-Well PCR Plate](http://www.bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n\n-- Magnetic Beads (Looking for a kit? We recommend trying [Omega Bio-tek Mag-Bind\u00ae TotalPure NGS](https://shop.opentrons.com/products/mag-bind-total-pure-ngs))\n\n-- Ethanol\n\n-- Elution Buffer (Typically 10 mM Tris pH 8.0, TE Buffer, or nuclease-free water)\n\n---\n\n---\n\n\n\nUsing the customization fields below, set up your protocol as follows:\n\n * **Pipette:** Specify your pipette. We recommend using a p50 or p300 multi- or single-channel.\n * **Pipette Mount:** Specify which mount (left or right) your pipette is on.\n * **Sample number:** Customize the number of samples to run per protocol. A multiple of 8 is recommended when you are using a multichannel pipette.\n * **Sample volume:** Specify the starting volume (in uL) of the input sample.\n * **Bead Ratio:** Customize the ratio of beads for left or right side size-selection of fragments. *The default bead ratio is 1.8x the input sample volume.*\n * **Elution Volume:** Specify the final volume (in uL) to elute the purified nucleic acid. *The Opentrons MagDeck supports elution volumes above 10 \u00b5L.*\n * **Incubation Time:** Specify the amount of time (in minutes) that the bead solution and input sample interact.\n * **Settling Time:** Specify the amount of time (in minutes) needed to pellet the beads. *Higher volumes may require a longer settling time.*\n * **Drying Time:** Specify the drying time (in minutes) needed after wash steps.\n\n---\n\n---\n\n",
"internal": "Nucleic Acid Purification, v1\n\n",
diff --git a/protoBuilds/omega-biotek-xpress/README.json b/protoBuilds/omega-biotek-xpress/README.json
index 8235730f4a..bac1b6807e 100644
--- a/protoBuilds/omega-biotek-xpress/README.json
+++ b/protoBuilds/omega-biotek-xpress/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Covid Workstation": [
"RNA Extraction"
@@ -10,7 +10,7 @@
"internal": "omega-biotek-xpress",
"labware": "NEST 2mL deep well plate (input with samples)\n200\u03bcl filter tip racks (10 x 200\u03bcl if you select false for tiprack parking)\nNEST 1 well reservoir\nNEST 12 well reservoir\nNEST 96 well aluminum block 100ul",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Covid Workstation\n\t* RNA Extraction\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol fully automates the [Omega Biotek Mag-Bind Viral RNA XPress Kit](https://www.omegabiotek.com/product/viral-rna-extraction-kit-mag-bind-viral-rna-xpress/?gclid=Cj0KCQjwlOmLBhCHARIsAGiJg7l7b_wVehYVQaXLe_wBJzEiE91FvrAfySaQaLjZ6VpLZzkCRcJLl6oaAoSjEALw_wcB). This specific protocol allows the user to manipulate the number of samples, elution volume, as well as tip parking for ultimate reuse (saving up to 4 tip boxes per run).\n\n\n\nBefore you begin:\n1. Pre-cool the Temperature Module in the Opentrons App to 4\u00b0C\n2. Create the Binding MasterMix\n\n3. Add the Binding Mastermix, RMP Buffer, Nuclease Free H20, and Lysis Buffer to the 12 well\nreservoir\n4. Create the freshly diluted 80% ethanol and add it to the 1 well reservoir in slot 2\n5. Place the deep well plate filled with samples on top of the magnetic module in slot 4.\n6. Add a 96 well aluminum block and the 96 well PCR plate or PCR strip tubes on top of\nthe Temperature Module in slot 1\nThe final plate of eluates/extractions will be found on top of the temperature module in slot 1.\n\nExplanation of complex parameters below:\n* `Number of Samples`: Select the number of samples for this run.\n* `Park tips?`: if True for \u201ctiprack parking,\u201d tips used for the same buffers with the same samples will be\nreused where 1 tiprack turns into a tiprack where used tips are \u201cparked\u201d. This method has low risk of\ncontamination and is highly recommended to avoid pauses to reuse tips. If selected, the parked tiprack slot is on slot 7.\n* `Elution Volume`: Specify the elution volume for this run.\n* `P300 Multi-Channel Mount`: Specify which mount (left or right) to host the P300 multi-channel pipette.\n\n\n\n---\n\n",
diff --git a/protoBuilds/omega_biotek_magbind_totalpure_NGS/README.json b/protoBuilds/omega_biotek_magbind_totalpure_NGS/README.json
index 80a878f805..075e4650e1 100644
--- a/protoBuilds/omega_biotek_magbind_totalpure_NGS/README.json
+++ b/protoBuilds/omega_biotek_magbind_totalpure_NGS/README.json
@@ -13,7 +13,7 @@
"description": "\n\nWith this protocol, you can perform high-quality nucleic acid purifications using [Omega Bio-tek Mag-Bind\u00ae TotalPure NGS](https://shop.opentrons.com/products/mag-bind-total-pure-ngs) magnetic beads and the [Opentrons Magnetic Module](https://shop.opentrons.com/products/magdeck). This setup yields high quality PCR product and other nucleic acids without the use of centrifugation or vacuum separation.\n\nThis kit is widely used in NGS cleanup for its affordability and simplicity. It is also well-adapted for nucleic acid size selection by varying bead ratios for the isolation of a wide array of fragment sizes. For more detailed information on how to use this protocol, please see our [Application Note](https://s3.amazonaws.com/opentrons-protocol-library-website/Technical+Notes/Omega_Application_Note.pdf).\n\nPlease note this protocol is currently being updated.\n\n---\n\n---\n\n\n\nTo purchase tips, reagents, or our Magnetic Module, please [visit our online store](https://shop.opentrons.com/) or contact our Sales team at .\n\n * [Omega Bio-tek Mag-Bind\u00ae TotalPure NGS Kit](https://shop.opentrons.com/products/mag-bind-total-pure-ngs)\n * [Opentrons OT-2](http://opentrons.com/ot-2)\n * [Opentrons Magnetic Module](https://shop.opentrons.com/products/magdeck)\n * [Opentrons OT-2 Run App (Version 3.1.2 or later)](http://opentrons.com/ot-app)\n * 200uL or 300 uL Tiprack ([Opentrons tips suggested](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips-racks-9-600-tips))\n * [12-row automation-friendly trough](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n * [BioRad HardShell 96-Well PCR Plates](http://www.bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n * Ethanol\n * Elution Buffer (Typically 10 mM Tris pH 8.0, TE Buffer, or nuclease-free water)\n\n---\n\n---\n\n\n\n\nUsing the customization fields below, set up your protocol as follows:\n\n * **Pipette:** Specify your pipette. We recommend using a p50 or p300 multi- or single-channel.\n * **Pipette Mount:** Specify which mount (left or right) your pipette is on.\n * **Sample number:** Customize the number of samples to run per protocol. A multiple of 8 is recommended when you are using a multichannel pipette.\n * **Sample volume:** Specify the starting volume (in uL) of the input sample.\n * **Bead Ratio:** Customize the ratio of beads for left or right side size-selection of fragments. *The default bead ratio is 1.8x the input sample volume.*\n * **Elution Volume:** Specify the final volume (in uL) to elute the purified nucleic acid. *The Opentrons MagDeck supports elution volumes above 10 \u00b5L.*\n\nMake sure to add reagents to your labware before placing it on the deck! You can see where to place your reagents below.\n\n\n\n---\n\n---\n\n",
"internal": "Omega Nucleic Acid Purification, v2\n\n",
"notes": "Please reference our [Application Note](https://s3.amazonaws.com/opentrons-protocol-library-website/Technical+Notes/Omega_Application_Note.pdf) for more information about the expected output of this protocol, in addition to expanded sample data from the Opentrons and Omega Bio-tek labs.\n\nIf you'd like to request a more complex purification workflow, please use our [Protocol Development Request Form](https://opentrons-protocol-dev.paperform.co/). You can also download the Python file from this page and modify it using our [API Documentation](https://docs.opentrons.com/). For additional questions about this protocol, please email .\n\nIf you are interested in purchasing the Opentrons Magnetic Module or trying out the Omega Bio-tek Mag-Bind\u00ae beads, please contact our Sales Team at to learn more!\n\n",
- "partner": "[Omega Bio-tek](http://omegabiotek.com/store/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/omega_biotek_magbind_totalpure_ngs). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "partner": "[Omega Bio-tek](http://omegabiotek.com/store/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/omega_biotek_magbind_totalpure_ngs). This page won\u2019t be available after January 31st, 2024.\n\n",
"preview": "With this protocol, you can perform high-quality nucleic acid purifications using the [Opentrons Magnetic Module](https://shop.opentrons.com/products/magdeck) and [Omega Bio-tek Mag-Bind\u00ae TotalPure NGS](https://shop.opentrons.com/products/mag-bind-total-pure-ngs) magnetic beads. This kit is widely used in NGS cleanup for its affordability and simplicity. You can select specific sizes of nucleic acids by varying the bead-to-DNA ratio across a wide array of fragment sizes. For reagent and module purchasing details contact .\n",
"process": "1. Select all desired settings according to the \"Setup\" section above to create your customized protocol.\n2. Download your customized OT-2 protocol using the blue \"Download\" button.\n3. Upload your protocol file into the Opentrons Run App and follow the instructions there to set up your deck and proceed to run!\n4. Make sure to add reagents to your labware before placing it on the deck! You can see where to place your reagents in the \"Setup\" section above.\n\n",
"title": "NGS Cleanup and Size Selection with Omega Bio-tek Mag-Bind\u00ae TotalPure NGS"
diff --git a/protoBuilds/onsite-bms-48plate/README.json b/protoBuilds/onsite-bms-48plate/README.json
index 6fe386cad8..4b2d820ea6 100644
--- a/protoBuilds/onsite-bms-48plate/README.json
+++ b/protoBuilds/onsite-bms-48plate/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "onsite-bms",
"labware": "\nOpentrons 6 Tube Rack with NEST 50 mL Conical\nNEST 96 Well Plate Flat\nOpentrons 20ul Tips\nOpentrons 1000ul Tips\nCustom 40 Tube Rack\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This protocol transfers sample and diluent to a custom 40 tube rack on the temperature module. Liquid height is tracked in the diluent tube rack, and the temperature module is set to 4C. A fresh tip is granted for each diluent and sample transfer, and mixed after sample is dispensed.\n\nExplanation of complex parameters below:\n* `input .csv file`: Here, you should upload a .csv file formatted in the following way, being sure to include the header line:\n```\nDiluent Transfer Volume (ul), Dispense Tube (rack), Source Well, Sample Transfer Volume\n1000, A1, A1, 20\n```\n* `Track tips?`: Specify whether to start at A1 of both tip racks, or to start picking up from where the last protocol left off.\n* `Initial Volume Diluent (mL)`: Specify the initial volume of diluent in A1 of the 6 tube rack.\n* `P20/P1000 Mounts`: Specify which mount (left or right) to host the P20 and P1000 pipettes.\n\n---\n\n",
diff --git a/protoBuilds/onsite-bms/README.json b/protoBuilds/onsite-bms/README.json
index 3da7682bd5..2acac314fa 100644
--- a/protoBuilds/onsite-bms/README.json
+++ b/protoBuilds/onsite-bms/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "onsite-bms",
"labware": "\nOpentrons 6 Tube Rack with NEST 50 mL Conical\nNEST 96 Well Plate Flat\nOpentrons 20ul Tips\nOpentrons 1000ul Tips\nCustom 40 Tube Rack\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\n---\n\n",
"description": "This protocol transfers sample and diluent to a custom 40 tube rack on the temperature module. Liquid height is tracked in the diluent tube rack, and the temperature module is set to 4C. A fresh tip is granted for each diluent and sample transfer, and mixed after sample is dispensed.\n\nExplanation of complex parameters below:\n* `input .csv file`: Here, you should upload a .csv file formatted in the following way, being sure to include the header line:\n```\nDiluent Transfer Volume (ul), Dispense Tube (rack), Source Well, Sample Transfer Volume\n1000, A1, A1, 20\n```\n* `Track tips?`: Specify whether to start at A1 of both tip racks, or to start picking up from where the last protocol left off. \n* `Initial Volume Diluent (mL)`: Specify the initial volume of diluent in A1 of the 6 tube rack.\n* `P20/P1000 Mounts`: Specify which mount (left or right) to host the P20 and P1000 pipettes.\n\n---\n\n",
diff --git a/protoBuilds/onsite-circlelabs/README.json b/protoBuilds/onsite-circlelabs/README.json
index 0710fcaa06..52ef7ebc7e 100644
--- a/protoBuilds/onsite-circlelabs/README.json
+++ b/protoBuilds/onsite-circlelabs/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS LIBRARY PREP": [
"Custom"
@@ -10,7 +10,7 @@
"internal": "onsite-circlelabs",
"labware": "\nBulldog 96 Aluminum Block 200 \u00b5L\nFisher 96 Aluminum Block 200 \u00b5L\nBulldog 96 Well Plate 200 \u00b5L\nFisher 96 Well Plate 200 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons 24 Well Aluminum Block with NEST 2 mL Snapcap\nOpentrons 96 Filter Tip Rack 200 \u00b5L\nOpentrons 96 Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS LIBRARY PREP\n\t* Custom\n\n\n",
"deck-setup": "\n\n\n\n",
"description": "This protocol performs an extraction for up to 96 samples, and for 3 plate types. Plates should be consistent for slots 1, 3, and 6. For protocol details and parameters, please see below.\n\n",
diff --git a/protoBuilds/onsite-dmso/README.json b/protoBuilds/onsite-dmso/README.json
index 19382661c0..6e9a841127 100644
--- a/protoBuilds/onsite-dmso/README.json
+++ b/protoBuilds/onsite-dmso/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -9,7 +9,7 @@
"internal": "onsite-dmso",
"labware": "\nCustom Opentrons 4-in-1 tube rack\nAltemis lab 48 plate\nMicronic 96 well plate\nNest 1-well Reservoir\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\nExplanation of complex parameters below:\n* `csv`: Here, you should upload a .csv file formatted in the following way, being sure to include the header line, with volume in the 6th column:\n\n* `Tubes on Slot 4`: Specify which tube rack to use on slot 4. \n* `P1000 Mount`: Specify which mount (left or right) to host the P1000 single channel pipette\n\n---\n\n\n",
"deck-setup": "\n\n---\n\n",
"internal": "onsite-dmso\n",
diff --git a/protoBuilds/onsite-ganda-1/README.json b/protoBuilds/onsite-ganda-1/README.json
index 661b8e3c69..634ecd68b0 100644
--- a/protoBuilds/onsite-ganda-1/README.json
+++ b/protoBuilds/onsite-ganda-1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"rhAmpSeq Library Prep"
@@ -10,7 +10,7 @@
"internal": "onsite-ganda-lab",
"labware": "\nNEST 96 Well Plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* rhAmpSeq Library Prep\n\n",
"deck-setup": "* If the deck layout of a particular protocol is more or less static, it is often helpful to attach a preview of the deck layout, most descriptively generated with Labware Creator. Example:\n\n\n",
"description": "This is part 1 of a 4 part protocol for the rhAmpSeq kit.\n* [Part 2](https://develop.protocols.opentrons.com/protocol/onsite-ganda-2)\n\n* [Part 3](https://develop.protocols.opentrons.com/protocol/onsite-ganda-3)\n* [Part 4](https://develop.protocols.opentrons.com/protocol/onsite-ganda-4)\n\nThis portion adds three components to 30 uL samples as outlined in the rhAmpSeq manual.\n* 10X rhAmp PCR Panel - Forward Pool\n* 10X rhAmp PCR Panel - Reverse Pool\n* 4X rhAmpSeq Library Mix 1\n\nExplanation of complex parameters below:\n* `Number of Samples`: How many samples are to be run, from 1 to 96\n* `P20 Multi GEN2 mount`: Which side the P20 Multi pipette is attached to\n* `Flash on Protocol Completion?`: Will the OT-2 lights flash on and off when the protocol is complete?\n\n---\n\n",
diff --git a/protoBuilds/onsite-ganda-2/README.json b/protoBuilds/onsite-ganda-2/README.json
index 1193231f8e..21415d91a7 100644
--- a/protoBuilds/onsite-ganda-2/README.json
+++ b/protoBuilds/onsite-ganda-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"rhAmpSeq Library Prep"
@@ -10,7 +10,7 @@
"internal": "onsite-ganda-lab",
"labware": "\nNEST 96 Well Plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* rhAmpSeq Library Prep\n\n",
"deck-setup": "* For 96 Samples\n\n\n",
"description": "This is part 2 of a 4 part protocol for the rhAmpSeq kit.\n* [Part 1](https://develop.protocols.opentrons.com/protocol/onsite-ganda-1)\n\n* [Part 3](https://develop.protocols.opentrons.com/protocol/onsite-ganda-3)\n* [Part 4](https://develop.protocols.opentrons.com/protocol/onsite-ganda-4)\n\nThis portion performs a cleanup on the PCR amplification product. A reagent list is below:\n* 80% Freshly Prepared Ethanol\n* IDTE Buffer\n* Agencourt AMPure XP Beads\n\nExplanation of complex parameters below:\n* `Number of Samples`: How many samples are to be run, from 1 to 96\n* `P20 Multi GEN2 mount`: Which side the P20 Multi pipette is attached to\n* `Flash on Protocol Completion?`: Will the OT-2 lights flash on and off when the protocol is complete?\n\n---\n\n",
diff --git a/protoBuilds/onsite-ganda-3/README.json b/protoBuilds/onsite-ganda-3/README.json
index 52a176ddca..a4bf7247f1 100644
--- a/protoBuilds/onsite-ganda-3/README.json
+++ b/protoBuilds/onsite-ganda-3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"rhAmpSeq Library Prep"
@@ -10,7 +10,7 @@
"internal": "onsite-ganda-lab",
"labware": "\nNEST 96 Well Plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* rhAmpSeq Library Prep\n\n",
"deck-setup": "\n\n\n",
"description": "This is part 3 of a 4 part protocol for the rhAmpSeq kit.\n* [Part 1](https://develop.protocols.opentrons.com/protocol/onsite-ganda-1)\n\n* [Part 2](https://develop.protocols.opentrons.com/protocol/onsite-ganda-2)\n* [Part 4](https://develop.protocols.opentrons.com/protocol/onsite-ganda-4)\n\nThis portion adds three components to 30 uL samples as outlined in the rhAmpSeq manual.\n* Indexing PCR Primer i5, prepared in 96 well plate with i7 primer\n* Indexing PCR Primer i7 prepared in 96 well plate with i5 primer\n* 4X rhAmpSeq Library Mix 2\n\nExplanation of complex parameters below:\n* `Number of Samples`: How many samples are to be run, from 1 to 96\n* `P20 Multi GEN2 mount`: Which side the P20 Multi pipette is attached to\n* `Flash on Protocol Completion?`: Will the OT-2 lights flash on and off when the protocol is complete?\n\n---\n\n",
diff --git a/protoBuilds/onsite-ganda-4/README.json b/protoBuilds/onsite-ganda-4/README.json
index 8e34c4ea11..2eed8a097a 100644
--- a/protoBuilds/onsite-ganda-4/README.json
+++ b/protoBuilds/onsite-ganda-4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"rhAmpSeq Library Prep"
@@ -10,7 +10,7 @@
"internal": "onsite-ganda-lab",
"labware": "\nNEST 96 Well Plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* rhAmpSeq Library Prep\n\n",
"deck-setup": "* For 96 Samples\n\n\n",
"description": "This is part 4 of a 4 part protocol for the rhAmpSeq kit.\n* [Part 1](https://develop.protocols.opentrons.com/protocol/onsite-ganda-1)\n\n* [Part 2](https://develop.protocols.opentrons.com/protocol/onsite-ganda-2)\n* [Part 3](https://develop.protocols.opentrons.com/protocol/onsite-ganda-3)\n\nThis portion performs a cleanup on the PCR amplification product. A reagent list is below:\n* 80% Freshly Prepared Ethanol\n* IDTE Buffer\n* Agencourt AMPure XP Beads\n\nExplanation of complex parameters below:\n* `Number of Samples`: How many samples are to be run, from 1 to 96\n* `P20 Multi GEN2 mount`: Which side the P20 Multi pipette is attached to\n* `Flash on Protocol Completion?`: Will the OT-2 lights flash on and off when the protocol is complete?\n\n---\n\n",
diff --git a/protoBuilds/onsite_atila_nasal/README.json b/protoBuilds/onsite_atila_nasal/README.json
index daeb183441..4d69e5300c 100644
--- a/protoBuilds/onsite_atila_nasal/README.json
+++ b/protoBuilds/onsite_atila_nasal/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "atila-nasal",
"labware": "\nOpentrons 4-in-1 Tube Racks\nOpentrons 20ul Filter Tips\nCustom 0.12 and 0.2mL 96 well plates\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n",
"description": "* This protocol preps a 96 well plate with buffer, sample, and mastermix for PCR. See below for details on reagent setup, protocol steps, etc. Tube racks should be placed in order of the slot numbers on the deck, (1, 2, 3,...,7). The pipette will access each tube rack in this order beginning from tube rack 1 on slot 1. The pipettes will access each tuberack by column (A1, B1, C1, A2, B2, C2...), meaning a 94 sample run would have all tube racks in slots 1-6 filled, with 4 tubes in A1, B1, C1, A2 of slot 7.\n\n* If running a full plate (94 samples), the protocol will use the multi-channel pipette to dispense mastermix to all 96 wells. For SBM, it will use the multi-channel to dispense into the first 11 columns, and then the single-channel to dispense into the first 6 wells of the 12th column. If running less than 94 samples, say 12 for example, the protocol will use the multi-channel to dispense mastermix and SBM into the first column, then the single channel to dispense to the first four wells of column 2, then finally mastermix and SBM into G12, H12. Note, that extra SBM and mastermix should be placed in wells 1-7 depending on the number of samples.\n\nExplanation of complex parameters below:\n* `Number of Samples (1-94)`: Specify the number of samples for this run (1-94).\n* `Reaction plate`: Specify whether using the non-skirted or half-skirted reaction plate on slot 9. Slot 8 should always use a non-skirted plate.\n* `P20 Single/Multi Pipette Mounts`: Specify which mount (left or right) to host each pipette.\n\n---\n\n",
diff --git a/protoBuilds/onsite_atila_saliva/README.json b/protoBuilds/onsite_atila_saliva/README.json
index a81bfc81d7..2b0bfa203d 100644
--- a/protoBuilds/onsite_atila_saliva/README.json
+++ b/protoBuilds/onsite_atila_saliva/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "atila-saliva",
"labware": "\nOpentrons 4-in-1 Tube Racks\nOpentrons 200ul Filter Tips\nOpentrons 20ul Filter Tips\nCustom 0.12 and 0.2mL 96 well plates\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n",
"description": "* This protocol preps a 96 well plate with sample and mastermix for PCR. See below for details on reagent setup, protocol steps, etc. Tube racks should be placed in order of the slot numbers on the deck, (1, 2, 3,...,7). The pipette will access each tube rack in this order beginning from tube rack 1 on slot 1. The pipettes will access each tuberack by column (A1, B1, C1, A2, B2, C2...), meaning a 94 sample run would have all tube racks in slots 1-6 filled, with 4 tubes in A1, B1, C1, A2 of slot 7.\n\n* If running a full plate (94 samples), the protocol will use the multi-channel pipette to dispense mastermix to all 96 wells. If running less than 94 samples, say 12 for example, the protocol will use the multi-channel to dispense mastermix into the first column, then the single channel to dispense the first four wells of column 2, then finally mastermix into G12, H12. Note, in this example, that the pipette will be taking mastermix from whichever column it is taking from (1 or 2, dependent on the number of samples running), in which case extra mastermix should be provided for the first four wells of column 1.\n\nExplanation of complex parameters below:\n* `Number of Samples (1-94)`: Specify the number of samples for this run (1-94).\n* `Reaction plate`: Specify whether using the non-skirted or half-skirted reaction plate on slot 9. Slot 8 should always use a non-skirted plate. \n* `P20/P300 Pipette Mounts`: Specify which mount (left or right) to host each pipette.\n\n---\n\n",
diff --git a/protoBuilds/onsite_atila_saliva384_pt1/README.json b/protoBuilds/onsite_atila_saliva384_pt1/README.json
index a208e28c82..a2e7b73edb 100644
--- a/protoBuilds/onsite_atila_saliva384_pt1/README.json
+++ b/protoBuilds/onsite_atila_saliva384_pt1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "atila-saliva-pt1",
"labware": "\nOpentrons 4-in-1 Tube Racks\nOpentrons 20ul Filter Tips\nCustom 0.12 and 0.2mL 96 well plates\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "\n\n",
"description": "* This protocol reformats 10ul of saliva samples from tubes to a 96 well plate. Tube racks should be placed in order of the slot numbers on the deck, (1, 2, 3,...,7). The pipette will access each tube rack in this order beginning from tube rack 1 on slot 1. The pipettes will access each tuberack by column (A1, B1, C1, A2, B2, C2...), meaning a 94 sample run would have all tube racks in slots 1-6 filled, with 4 tubes in A1, B1, C1, A2 of slot 7.\n\nExplanation of complex parameters below:\n* `Number of Samples (1-96)`: Specify the number of samples for this run (1-96).\n* `Reaction plate`: Specify whether using the non-skirted or half-skirted reaction plate on slot 9. Slot 8 should always use a non-skirted plate.\n* `P20 Pipette Mount`: Specify which mount (left or right) to host the P20 Single-Channel Pipette.\n\n---\n\n",
diff --git a/protoBuilds/onsite_atila_saliva384_pt2/README.json b/protoBuilds/onsite_atila_saliva384_pt2/README.json
index cd63c72755..00d445fe58 100644
--- a/protoBuilds/onsite_atila_saliva384_pt2/README.json
+++ b/protoBuilds/onsite_atila_saliva384_pt2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR": [
"PCR Prep"
@@ -10,7 +10,7 @@
"internal": "atila-saliva-pt2",
"labware": "\nOpentrons 20ul Filter Tips\nNEST 2mL 96, Deepwell Plate\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR\n\t* PCR Prep\n\n",
"deck-setup": "Plate 4 (slot 2) should only have 94 samples to allow room for controls. Please refer to the following diagram for how samples are loaded from each of the four quadrants onto the 284 plate with the multi-channel. Any unfilled columns will switch to the single channel. SRM is added to control wells O24 and P24.\n\n\n",
"description": "* This protocol transfer SRM and saliva sample to a 384 well plate with one mix repetition. 4 plates are loaded into one 384 well plate. If running less than 4 full plates, SRM will still be transferred to the last two wells in the 384 well plate (controls). NOTE: positive and negative controls are to be added manually after the protocol into the last two wells. See below for plate setup.\n\nExplanation of complex parameters below:\n* `Number of Samples`: Specify the number of samples for each plate.\n* `Reaction plate`: Specify whether using the non-skirted or half-skirted reaction plate on slot 9. Slot 8 should always use a non-skirted plate.\n* `P20 Pipette Mount`: Specify which mount (left or right) to host the P20 Single and Multi-Channel Pipettes.\n\n---\n\n",
diff --git a/protoBuilds/onsite_egenesis/README.json b/protoBuilds/onsite_egenesis/README.json
index c899200887..52c31e6721 100644
--- a/protoBuilds/onsite_egenesis/README.json
+++ b/protoBuilds/onsite_egenesis/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "onsite_egenesis",
"labware": "\nEppendorf 96 Well Plate 200 \u00b5L\nUSA Scientific 12 Well Reservoir 22 mL #1061-8150\nOpentrons 24 Well Aluminum Block with NEST 1.5 mL Screwcap\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "For a detailed description of this protocol, please refer to the [Nanoporo Direct RNA sequencing (SQK-RNA002) SOP](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/onsite_egenesis/direct-rna-sequencing-sqk-rna002-DRS_9080_v2_revO_14Aug2019-minion+(2).pdf).\n\nNote: wash buffer should be split evenly between wells A5 and A6 on slot 5.\n\n\n",
diff --git a/protoBuilds/onsite_egenesis_part2/README.json b/protoBuilds/onsite_egenesis_part2/README.json
index 1929f104bd..3236b146b6 100644
--- a/protoBuilds/onsite_egenesis_part2/README.json
+++ b/protoBuilds/onsite_egenesis_part2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "onsite_egenesis_part2",
"labware": "\nBarcode 96 Well Plate 200 \u00b5L\nAxygen 96 Deep Well Plate 2000 \u00b5L\nEppendorf 96 Well Plate 200 \u00b5L\nUSA Scientific 12 Well Reservoir 22 mL #1061-8150\nOpentrons 24 Well Aluminum Block with NEST 1.5 mL Screwcap\nOpentrons 96 Filter Tip Rack 20 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "For a detailed description of this protocol, please refer to the [Ligation Sequencing Amplicons Native Barcoding](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/onsite_egenesis_part2/manual.pdf)\n\nNote: select the barcode numbered well to start from counting by column. \"1\" would be A1, \"8\" would be H1, \"10\" would be B2, so on and so forth.\n\n\n",
diff --git a/protoBuilds/onsite_galatea_mag_bind/README.json b/protoBuilds/onsite_galatea_mag_bind/README.json
index 61f6f3c995..73ca8c663c 100644
--- a/protoBuilds/onsite_galatea_mag_bind/README.json
+++ b/protoBuilds/onsite_galatea_mag_bind/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons (verified)",
+ "author": "Opentrons (verified)\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"DNA Extraction"
@@ -10,7 +10,7 @@
"internal": "onsite_galatea_mag_bind",
"labware": "\nNEST 96 Wellplate 2mL\nUSA Scientific 96 Wellplate 2.4mL\nNEST 12 Reservoir 15mL\nUSA Scientific 12 Reservoir 22mL\nOpentrons 200uL Filter Tips\nOpentrons 96 Aluminum block Nest Wellplate 100ul\n",
"markdown": {
- "author": "[Opentrons (verified)](https://opentrons.com/)\n\n",
+ "author": "[Opentrons (verified)](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* DNA Extraction\n\n",
"deck-setup": "\n* Tip rack on Slot 1 is used for tip parking if selected.\n\n\n\n\n---\n\n",
"description": "Your OT-2 can fully automate the entire Mag-Bind\u00ae Blood & Tissue DNA HDQ 96 Kit.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below:\n\n\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n* `Deepwell type`: Specify which well plate will be mounted on the magnetic module.\n* `Reservoir Type`: Specify which reservoir will be employed.\n* `Starting Volume`: Specify starting volume of sample (ul).\n* `Binding Buffer Volume`: Specify the volume of binding buffer to use (ul).\n* `Elution Volume`: Specify elution volume (ul).\n* `Park Tips`: Specify whether to park tips or drop tips.\n* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n\n---\n\n",
diff --git a/protoBuilds/onsite_galatea_normalization/README.json b/protoBuilds/onsite_galatea_normalization/README.json
index 300c1076db..e2e32a9620 100644
--- a/protoBuilds/onsite_galatea_normalization/README.json
+++ b/protoBuilds/onsite_galatea_normalization/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "onsite_galatea_normalization",
"labware": "\nNEST 12 Well Reservoir 15 mL #360102\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nBio-Rad 96 Well Plate 200 \u00b5L PCR #hsp9601\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n\n\n",
"description": "* This protocol normalized a 96 well plate with samples and water, then moves them to a barcode plate along with two buffers for further processing. A csv is uploaded to the protocol in which the destination well in the final plate should be column 1 (source well and destination well are the same, so should be 1:1), the water transfer volume should be in column 5, and the dna transfer volume should be in column 4. Note: if \"top half of plate\" or \"bottom half of plate\" is selcted, then the multi-channel pipette will pick up four tips and will always aspirate from the top 4 rows of the final plate on slot 3 to dispense into the barcode plate, either in the top or bottom half (always from top half to either top or bottom half). The csv will run for as many rows as there are, and there should only be one row at the top for the header.\n\n* For water addition to the plate, if the tranfser volume is greater or equal to 5ul, it will transfer 5ul.\n\n* For DNA addition to plate, the pipette will transfer 6ul if the transfer volume is greater than 6ul. It will also transfer 1 if the transfer volume is less than 1ul.\n\n* A slow aspiration rate and delay is employed for Buffer B, since it is a viscous liquid. \n\n\n",
diff --git a/protoBuilds/onsite_galatea_pre_normalization/README.json b/protoBuilds/onsite_galatea_pre_normalization/README.json
index 6036141837..c84a8c556c 100644
--- a/protoBuilds/onsite_galatea_pre_normalization/README.json
+++ b/protoBuilds/onsite_galatea_pre_normalization/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "onsite_galatea_pre_normalization",
"labware": "\nNEST 12 Well Reservoir 15 mL #360102\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt #402501\nOpentrons 96 Filter Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n\n",
"description": "The csv should include a header in row 1. Source/destination well should be in column 1. Qubit reading should be in column 3. The transfer volume is determined by the Qubit reading. \n\n\n",
diff --git a/protoBuilds/pcr_prep_part_1/README.json b/protoBuilds/pcr_prep_part_1/README.json
index 5cf15992a3..56f14fc17c 100644
--- a/protoBuilds/pcr_prep_part_1/README.json
+++ b/protoBuilds/pcr_prep_part_1/README.json
@@ -10,7 +10,7 @@
"internal": "OT-2 PCR Prep v2",
"labware": "\n4-in-1 Tube Rack Set\n12-well Trough\n",
"markdown": {
- "author": "[Opentrons (verified)](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/pcr_prep_part_1). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons (verified)](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/pcr_prep_part_1). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* PCR\n * Mastermix Assembly\n\n",
"deck-setup": "* Slot 1: Option of Opentrons tuberack/tube combo 1, or none\n* Slot 2: Option of Opentrons tuberack/tube combo 2, or none\n* Slot 3: Choice of Opentrons labware library 12-well reservoir\n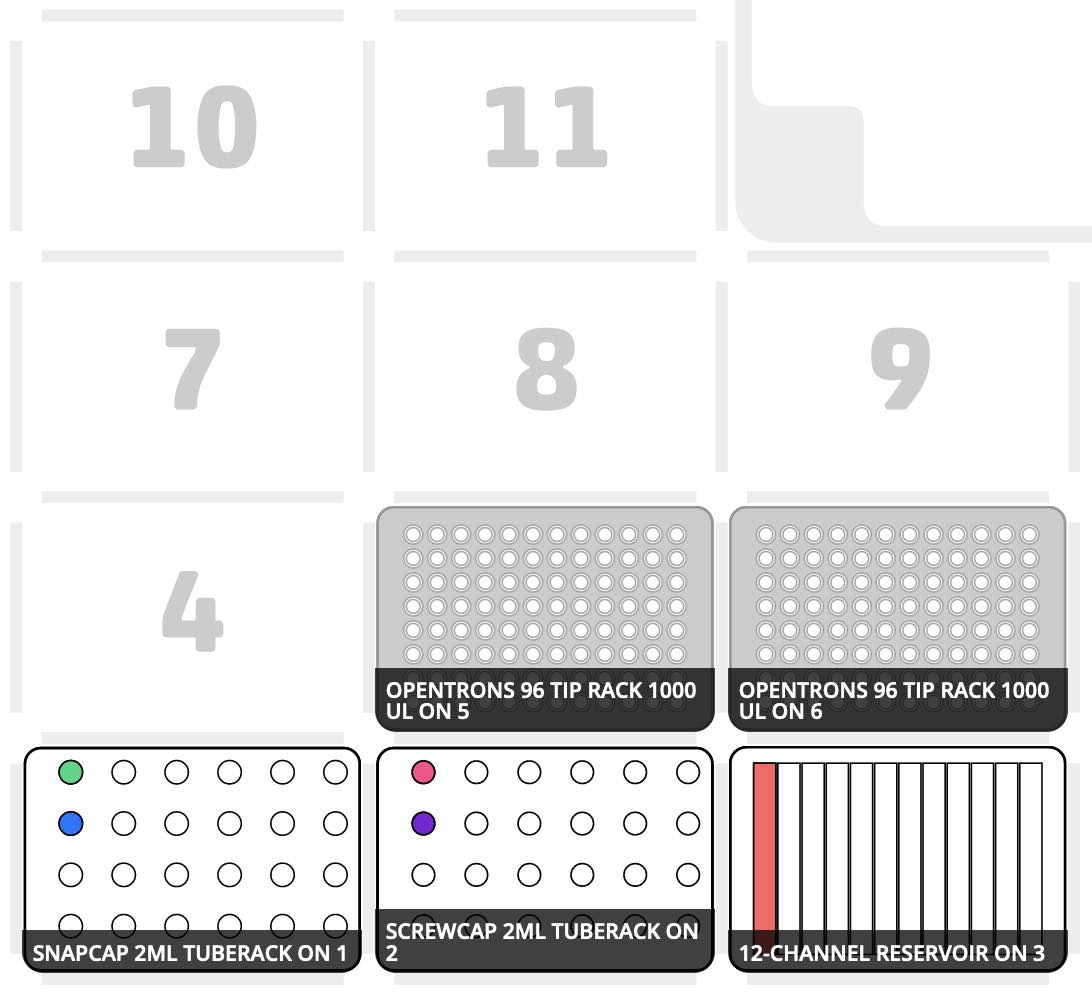\n\n",
"description": "Part 1 of 2: Master Mix Assembly\n\nLinks:\n* [Part 1: Master Mix Assembly](./pcr_prep_part_1)\n* [Part 2: Master Mix Distribution and DNA Transfer](./pcr_prep_part_2)\n\nThis protocol allows your robot to create a master mix solution using any reagents stored in one or two different types of tube racks, or reservoir well A2 to A12. The master mix will be created in well A1 of the trough. The ingredient information will be provided as a CSV file. See Additional Notes for more details.\n\nParameters:\n* `right pipette type`: Which single channel pipette to use in the right mount\n* `left pipette type`: Which single channel pipette to use in the left mount\n* `Filter or regular tips`: Use filter tips or non-filtered.\n* `Tuberack 1`: Tuberack 1 for reagents (optional)\n* `Tuberack 2`: Tuberack 2 for reagents (optional)\n* `12-well reservoir`: 12 well reservoir for mastermix target and optionally reagents in well A2-A12\n* `master mix .csv file`: Input csv file (see format below)\n\n---\n\n",
diff --git a/protoBuilds/pcr_prep_part_2/README.json b/protoBuilds/pcr_prep_part_2/README.json
index 9e48a7dfaa..aab56fd4b2 100644
--- a/protoBuilds/pcr_prep_part_2/README.json
+++ b/protoBuilds/pcr_prep_part_2/README.json
@@ -8,7 +8,7 @@
"description": "Part 2 of 2: Master Mix Distribution and DNA Transfer\nLinks:\n Part 1: Master Mix Assembly\n Part 2: Master Mix Distribution and DNA Transfer\nThis protocol allows your robot to distribute a master mix solution from well A1 of a trough to PCR strips. Robot will then transfer DNA samples to the master mix solution.\n\nYou will need:\n 12-channel reservoir\n 96-well PCR plate",
"internal": "OT-2 PCR Prep v2",
"markdown": {
- "author": "[Opentrons (verified)](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/pcr_prep_part_2). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons (verified)](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/pcr_prep_part_2). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* PCR\n * PCR Prep\n\n",
"description": "Part 2 of 2: Master Mix Distribution and DNA Transfer\n\nLinks:\n* [Part 1: Master Mix Assembly](./pcr_prep_part_1)\n* [Part 2: Master Mix Distribution and DNA Transfer](./pcr_prep_part_2)\n\n\nThis protocol allows your robot to distribute a master mix solution from well A1 of a trough to PCR strips. Robot will then transfer DNA samples to the master mix solution.\n\n---\n\nYou will need:\n* [12-channel reservoir](https://www.usascientific.com/12-channel-automation-reservoir.aspx)\n* [96-well PCR plate](https://www.bio-rad.com/en-us/sku/hsp9601-hard-shell-96-well-pcr-plates-low-profile-thin-wall-skirted-white-clear?ID=hsp9601)\n\n",
"internal": "OT-2 PCR Prep v2\n",
diff --git a/protoBuilds/pcr_test_plan/README.json b/protoBuilds/pcr_test_plan/README.json
index 93b1dcea89..16b5d6b7d3 100644
--- a/protoBuilds/pcr_test_plan/README.json
+++ b/protoBuilds/pcr_test_plan/README.json
@@ -14,7 +14,7 @@
"internal": "[Link To Google Doc](https://docs.google.com/presentation/d/1T90uIaz3ci-UPK3x-OkrLX0x0_hDDQClusc5U2qAYWs/edit#slide=id.p7)",
"modules": "* [MagDeck](https://shop.opentrons.com/collections/labware/products/magdeck)\n* [CoolDeck](https://shop.opentrons.com/collections/labware/products/cold-deck)\n* [HeatDeck](https://shop.opentrons.com/collections/labware/products/heat-deck)\n\n",
"notes": "Lorem ipsum dolor sit amet, consectetur adipiscing elit. Nam sed purus eu urna vehicula ultrices at vel quam. Aliquam erat volutpat. Quisque imperdiet ut urna ut dapibus. Sed placerat, libero at ullamcorper pharetra, massa tellus sagittis eros, in feugiat est libero et turpis. Sed ac efficitur mi, at porttitor elit.\n\n\n\n",
- "partner": "[Stanford](https://www.stanford.edu/)\n\n",
+ "partner": "[Stanford](https://www.stanford.edu/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"process": "1. Suspendisse ultrices elit ex, sit amet porttitor justo convallis eget. Integer quis felis eleifend, venenatis.\n2. Sed ultricies lacus eu justo laoreet, sed maximus libero faucibus. Nulla sit amet imperdiet erat. Pellentesque ultrices, felis at auctor.\n\n\n",
"reagents": "* [Reagent Name](url)\n* [Reagent Name](url)\n\n",
"robot": "* [OT PRO](https://opentrons.com/ot-one-pro)\n* [OT Standard](https://opentrons.com/ot-one-standard) \n* [OT Hood](https://opentrons.com/ot-one-hood) \n\n",
@@ -27,7 +27,7 @@
"HeatDeck"
],
"notes": "Lorem ipsum dolor sit amet, consectetur adipiscing elit. Nam sed purus eu urna vehicula ultrices at vel quam. Aliquam erat volutpat. Quisque imperdiet ut urna ut dapibus. Sed placerat, libero at ullamcorper pharetra, massa tellus sagittis eros, in feugiat est libero et turpis. Sed ac efficitur mi, at porttitor elit.",
- "partner": "Stanford",
+ "partner": "Stanford\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"process": "\nSuspendisse ultrices elit ex, sit amet porttitor justo convallis eget. Integer quis felis eleifend, venenatis.\nSed ultricies lacus eu justo laoreet, sed maximus libero faucibus. Nulla sit amet imperdiet erat. Pellentesque ultrices, felis at auctor.\n",
"reagents": [
"Reagent Name",
diff --git a/protoBuilds/pooling/README.json b/protoBuilds/pooling/README.json
index 3dc3977275..8ad0cbe8a9 100644
--- a/protoBuilds/pooling/README.json
+++ b/protoBuilds/pooling/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Pooling"
@@ -10,7 +10,7 @@
"internal": "pooling",
"labware": "\nEppendorf\u2122 96-Well twin.tec\u2122 PCR Plate 250\u00b5l\nOpentrons 20\u00b5l and 300\u00b5l tipracks\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Pooling\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "This protocol performs a custom sample pooling from a source PCR plate to a 1.5, 2, or 5ml Eppendorf tube. Sample locations and volumes are specified via .csv file in the following format, including the header line (empty lines ignored):\n\n```\nsource plate well,volume sample (\u00b5l)\nA1, 28\nC4, 12\n```\n\nThe proper pipette for a given volume is automatically calculated. Tips are changed for each transfer.\n\nYou can download an example .csv to edit directly [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/pooling/ex.csv).\n\n---\n\n",
diff --git a/protoBuilds/primer_hydration/README.json b/protoBuilds/primer_hydration/README.json
index 87f8471c39..da527449eb 100644
--- a/protoBuilds/primer_hydration/README.json
+++ b/protoBuilds/primer_hydration/README.json
@@ -1,5 +1,5 @@
{
- "author": "REM Analytics",
+ "author": "REM Analytics\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR Prep": [
"Primer hydration"
@@ -10,7 +10,7 @@
"internal": "protocol-hex-code",
"labware": "\nOpentrons 10 Tube Rack with Falcon 4x50 mL, 6x15 mL Conical\nOpentrons 24 Well Aluminum Block with Generic 2 mL Screwcap\n",
"markdown": {
- "author": "[REM Analytics](https://www.remanalytics.ch/)\n\n",
+ "author": "[REM Analytics](https://www.remanalytics.ch/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR Prep\n\t* Primer hydration\n\n",
"deck-setup": "* To start the protocol, it is necessary to use the middle row to avoid any damage to the pipette. The water 15mL falcon tubes will be placed in the middle slot, and the primer pellet 2mL screw cap tubes will be placed in the right slot.\n \n\n",
"description": "The goal of this protocol is to hydrate dry pellets of primers using water or tris-edta buffer. The user should provide the desired transfer volumes and well coordinates in a csv file. \n\nThis protocol uses a multi-channel 300uL pipette as a single-channel pipette, which makes it necessary to place all the equipment in the middle row of the deck. \n\nExplanation of complex parameters below:\n* `input .csv file`: Here, you should upload a .csv file formatted in the following way, being sure to include the header line:\n```\nsource,dest,vol\nA1,B1,400\n```\nThe above input will make the robot pipette 400 uL from well A01 of 15mL falcon tube rack on slot 5 to well B01 of the 2mL generic tube rack on slot 6.The volume of each row can be above 300uL, but be careful not to surpass 5000uL for a single falcon tube. If you need to transfer more than 5000uL, ensure you add more falcon tubes and update the 'source' value in the csv\n\n---\n\n\n",
diff --git a/protoBuilds/promega-maxwell/README.json b/protoBuilds/promega-maxwell/README.json
index 031824655d..e90edcd32c 100644
--- a/protoBuilds/promega-maxwell/README.json
+++ b/protoBuilds/promega-maxwell/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Viral RNA"
@@ -10,7 +10,7 @@
"internal": "promega-maxwell",
"labware": "\nNEST 12 Well Reservoir 15 mL, 2x\nNEST 1 Well Reservoir 195 mL\nNEST 96 Well Plate 100 \u00b5L PCR Full Skirt\nNEST 96 Deepwell Plate 2mL\nOpentrons 96 Filter Tip Rack 200 \u00b5L, up to 10x depending on throughput\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Viral RNA\n\n",
"deck-setup": "\n\n",
"description": "This protocol performs viral RNA isolation on up to 96 samples using the [Promega Maxwell\u00ae HT Viral TNA Kit](https://www.promega.com/products/nucleic-acid-extraction/viral-rna-extraction-viral-dna-extraction/maxwell-ht-viral-tna-kit/?catNum=AX2340) and [workflow](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/promega-maxwell-extraction/viral-rna-universal-transport-medium-maxwell-ht-viral-tna-kit-96-well-manual-format.pdf).\n\nThe protocol begins at the stage of adding binding beads to lysed samples loaded on the magnetic module in a NEST 96-deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 6) to the elution PCR plate (temperature module, slot 1). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\n---\n\n",
diff --git a/protoBuilds/quintara_onsite_nanopore/README.json b/protoBuilds/quintara_onsite_nanopore/README.json
index 66d6a1a1fd..c763ba09f3 100644
--- a/protoBuilds/quintara_onsite_nanopore/README.json
+++ b/protoBuilds/quintara_onsite_nanopore/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Nanopore"
@@ -10,7 +10,7 @@
"internal": "quintara_onsite_nanopore",
"labware": "\nBarcode 96 Well Plate 200 \u00b5L\nCombo 96 Well Plate Black Label on Nest Plate 300\u00b5L\nOpentrons 96 Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Nanopore\n\n\n",
"deck-setup": "\n\n\n\n",
"description": "This protocol preps 1-7 destination plates from one source barcode plate. Destination plates should loaded onto the deck in order of lowest to highest, depending on the number of plates selected (up to 7). For detailed protocol steps, see below.\n\n\n",
diff --git a/protoBuilds/quintara_onsite_nanopore_pooling/README.json b/protoBuilds/quintara_onsite_nanopore_pooling/README.json
index 6bbd42e3c8..173d5d98ff 100644
--- a/protoBuilds/quintara_onsite_nanopore_pooling/README.json
+++ b/protoBuilds/quintara_onsite_nanopore_pooling/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Nanopore"
@@ -10,7 +10,7 @@
"internal": "quintara_onsite_nanopore",
"labware": "\nBarcode 96 Well Plate 200 \u00b5L\nCombo 96 Well Plate Black Label on Nest Plate 300\u00b5L\nOpentrons 96 Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Nanopore\n\n\n",
"deck-setup": "\n\n\n\n",
"description": "This protocol pools 1-6 destination plates from all columns to the first 4 wells in column 1 for each respective plate. Destination plates should loaded onto the deck in order of lowest to highest, depending on the number of plates selected (up to 6). That is, for two plates, load the plates in slots 4 and 5. For detailed protocol steps, see below.\n\n\n",
diff --git a/protoBuilds/quintara_onsite_part1/README.json b/protoBuilds/quintara_onsite_part1/README.json
index 4d39a62d69..970f885c13 100644
--- a/protoBuilds/quintara_onsite_part1/README.json
+++ b/protoBuilds/quintara_onsite_part1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "quintara_onsite_part1",
"labware": "\nQuintara Vertical Plate 192 Wells\nQuintara 12 Reservoir 15000 \u00b5L\nDouble Pcr 96 Well Plate 300 \u00b5L\nAppliedbiosystem 384 Well Plate 40 \u00b5L\nQuintara 96 Well Plate 300 \u00b5L\nQuintara Vertical Plate 192 Wells\nDeepwell 96 Well Plate 2000 \u00b5L\nOpentrons 96 Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "Four 96 wells are consolidated in one 96 PCR plate. Number of sets is equal to a group of 4 columns, taking 20ul from the 1st and fourth column of that set, and 2ul from columns 2 and 3 from that set.\n\n\n",
diff --git a/protoBuilds/quintara_onsite_part1_384/README.json b/protoBuilds/quintara_onsite_part1_384/README.json
index cac5ececa9..cd49e24ba8 100644
--- a/protoBuilds/quintara_onsite_part1_384/README.json
+++ b/protoBuilds/quintara_onsite_part1_384/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "quintara_onsite_part1_384",
"labware": "\nQuintara Vertical Plate 192 Wells\nQuintara 12 Reservoir 15000 \u00b5L\nDouble Pcr 96 Well Plate 300 \u00b5L\nAppliedbiosystem 384 Well Plate 40 \u00b5L\nQuintara 96 Well Plate 300 \u00b5L\nQuintara Vertical Plate 192 Wells\nDeepwell 96 Well Plate 2000 \u00b5L\nOpentrons 96 Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "This protocol preps a 96 plate from a 384 source plate.\n\n\n",
diff --git a/protoBuilds/quintara_onsite_part2/README.json b/protoBuilds/quintara_onsite_part2/README.json
index a081f0037d..e732ff3ea3 100644
--- a/protoBuilds/quintara_onsite_part2/README.json
+++ b/protoBuilds/quintara_onsite_part2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "quintara_onsite_part2",
"labware": "\nQuintara Vertical Plate 192 Wells\nQuintara 12 Reservoir 15000 \u00b5L\nDouble Pcr 96 Well Plate 300 \u00b5L\nAppliedbiosystem 384 Well Plate 40 \u00b5L\nQuintara 96 Well Plate 300 \u00b5L\nQuintara Vertical Plate 192 Wells\nDeepwell 96 Well Plate 2000 \u00b5L\nOpentrons 96 Tip Rack 20 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "Two 96 wells are consolidated in one 96 PCR plate. Number of sets is equal to a group of 4 columns, taking 20ul from the 1st and fourth column of that set, and 2ul from columns 2 and 3 from that set. Then, 4 tips are picked up from tip rack on slot 6, and the exact same process is repeated for the remaining four wells under the 8 filled wells in both the source and destination plates.\n\n\n",
diff --git a/protoBuilds/quintara_onsite_part3/README.json b/protoBuilds/quintara_onsite_part3/README.json
index 93e15a963c..4e95d3e820 100644
--- a/protoBuilds/quintara_onsite_part3/README.json
+++ b/protoBuilds/quintara_onsite_part3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "quintara_onsite_part3",
"labware": "\nQuintara Vertical Plate 192 Wells\nQuintara 12 Reservoir 15000 \u00b5L\nDouble Pcr 96 Well Plate 300 \u00b5L\nAppliedbiosystem 384 Well Plate 40 \u00b5L\nQuintara 96 Well Plate 300 \u00b5L\nQuintara Vertical Plate 192 Wells\nDeepwell 96 Well Plate 2000 \u00b5L\nOpentrons 96 Tip Rack 20 \u00b5L\nNEST 96 Deepwell Plate 2mL #503001\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "1 of 6 mastermixes is filled into all 108 columns in all 9 plates from two wells of a reservoir. Pipette will automatically be assigned depending on the mastermix selected, and multi-dispensing is installed when possible to the pipette maximum volume. Mastermix is taken back and forth between the two mastermix wells (aspirate A1, aspirate A2, aspirate A1,..., etc.), so fill mastermix source columns in the reservoir evenly. Mastermix details are included in the dropdown menu.\n\n\n\n",
diff --git a/protoBuilds/quintara_onsite_part4/README.json b/protoBuilds/quintara_onsite_part4/README.json
index 120f8b93b9..bfddf064ca 100644
--- a/protoBuilds/quintara_onsite_part4/README.json
+++ b/protoBuilds/quintara_onsite_part4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "quintara_onsite_part4",
"labware": "\nQuintara Vertical Plate 192 Wells\nQuintara 12 Reservoir 15000 \u00b5L\nDouble Pcr 96 Well Plate 300 \u00b5L\nAppliedbiosystem 384 Well Plate 40 \u00b5L\nQuintara 96 Well Plate 300 \u00b5L\nQuintara Vertical Plate 192 Wells\nDeepwell 96 Well Plate 2000 \u00b5L\nOpentrons 96 Filter Tip Rack 200 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "Water is filled into all 108 columns in all 9 plates from a reservoir. Multi-dispensing is installed to pipette's maximum volume.\n\n",
diff --git a/protoBuilds/rem-pcr-plate/README.json b/protoBuilds/rem-pcr-plate/README.json
index 0ffa032e94..decebf1550 100644
--- a/protoBuilds/rem-pcr-plate/README.json
+++ b/protoBuilds/rem-pcr-plate/README.json
@@ -1,5 +1,5 @@
{
- "author": "REM Analytics",
+ "author": "REM Analytics\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"PCR Prep": []
},
@@ -7,7 +7,7 @@
"description": "Protocol to dispense master mix (with or without DNA) in PCR plates. The current protocol dispenses only one mix per \nplate. You input the number of plates that you'd like to\nprepare and the protocol pauses after preparing each plate. This gives you the opportunity to wait for the previous \nplate to finish, prepare the following master mix etc. The protocol will tell you which trough you need to pour the mix\ninto for the following plate.\nExplanation of parameters below:\n Master Mix Source: Do you have one master mix for the whole plate (Reservoir) or one for each row (Plate). NB the\nDeck Setup diagram shows both the Plate and Reservoir (troughs), you only need to put the one\nyou require in place.\n Number of Plates: The total number of plates you'd like to prepare (one master mix needs to be prepared for each \nplate)\n Starting Trough: The first trough you'd like to pour the master mix into (e.g. if some troughs have already been\nused). NB the subsequent troughs will be used for the following plates so they also need to be free. Make sure\nthat Number of Plates is not greater than the number of available troughs, if so use a new reservoir plate.\n Volume to dispense (uL): Typically 9 (without DNA in mix) or 10 (with DNA in mix).\n* Touch Tip on the...: Which side should the tip touch the well on (the opposite side to which you touched the tip\nwhen adding the DNA (if applicable)) \n",
"labware": "\nNEST 12 Well Reservoir 15 mL\nShallow Well Plate 200 uL (x2)\n",
"markdown": {
- "author": "[REM Analytics](https://www.remanalytics.ch/)\n\n",
+ "author": "[REM Analytics](https://www.remanalytics.ch/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* PCR Prep\n\t\n\n",
"deck-setup": "* After each plate is prepared, the protocol pauses, the plate must be removed and the following plate must be placed\n in the same slot.\n \n\n---\n\n",
"description": "Protocol to dispense master mix (with or without DNA) in PCR plates. The current protocol dispenses only one mix per \nplate. You input the number of plates that you'd like to\nprepare and the protocol pauses after preparing each plate. This gives you the opportunity to wait for the previous \nplate to finish, prepare the following master mix etc. The protocol will tell you which trough you need to pour the mix\ninto for the following plate.\n\nExplanation of parameters below:\n* `Master Mix Source`: Do you have one master mix for the whole plate (Reservoir) or one for each row (Plate). NB the\nDeck Setup diagram shows both the Plate and Reservoir (troughs), you only need to put the one\nyou require in place.\n* `Number of Plates`: The total number of plates you'd like to prepare (one master mix needs to be prepared for each \nplate)\n* `Starting Trough`: The first trough you'd like to pour the master mix into (e.g. if some troughs have already been\nused). NB the subsequent troughs will be used for the following plates so they also need to be free. Make sure\nthat Number of Plates is *not* greater than the number of available troughs, if so use a new reservoir plate.\n* `Volume to dispense (uL)`: Typically 9 (without DNA in mix) or 10 (with DNA in mix).\n* `Touch Tip on the...`: Which side should the tip touch the well on (the opposite side to which you touched the tip\nwhen adding the DNA (if applicable)) \n\n\n---\n\n\n",
diff --git a/protoBuilds/sci-amplex-red/README.json b/protoBuilds/sci-amplex-red/README.json
index 411000a64d..c30b305705 100644
--- a/protoBuilds/sci-amplex-red/README.json
+++ b/protoBuilds/sci-amplex-red/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Standard curve for Hydrogen Peroxide": [
"Measurement of peroxides/hydrogen peroxide from THP-1 cells"
@@ -10,7 +10,7 @@
"internal": "Amplex Red Hydrogen Peroxide Assay",
"labware": "\n\nGreiner-Bio One 96-Well White TC plate with clear bottom\n\n\nOpentrons 96 Filter Tip rack 200\u03bcL\n\n\nOpentrons 96 Filter Tip rack 20\u03bcL\n\n\nOpentrons 10 Tube Rack with Falcon 4X50 mL, 6X15mL Conical\n\n\n(2) Opentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\n\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Standard curve for Hydrogen Peroxide\n\t* Measurement of peroxides/hydrogen peroxide from THP-1 cells\n\n",
"deck-setup": "* Slot 3 Temperature Module- Plate initially at 23\u00b0C(deactivated) and after reagent addition, target temperature is 37\u00b0 C.\n* Slot 4 Opentrons 96 Filter Tip rack 200\u03bcl\n* Slot 5 Opentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap- PMA Dilutions from 10mM to 10\u03bcM.\n* Slot 7 Opentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap- Concentrations of H2O2 for standard curve (0.1, 0.2, 0.3, 0.4, 0.5, 1 and 2 \u03bcM) and concentrations of PMA from 0-150 ng/mL\n* Slot 10 Opentrons 96 Filter Tip rack 20\u03bcl\n* Slot 11 Opentrons 10 Tube Rack with Falcon 4X50 mL, 6X15mL Conical-Rack for holding 5X Reaction buffer, 1X Reaction buffer and Amplex Red Working Reagent\n\n\n\n* Figure 1 Deck layout after the reagent additions is carried out.\nSlot 9 A1 tube contains (5X reaction buffer)\nA3 falcon tube contains Distilled water for the 5X reaction buffer dilution. After addition of 5X to the tube, results in 1X Reaction Buffer being present in the same\nB1 contains empty tube in which 1X Reaction buffer, Amplex red dye and HRP are dispensed during the protocol run.\nSlot 7 Opentrons 24 tube rack Eppendorfs A1 to A6 has 20mM,0.0\u03bcM, 0.1\u03bcM,0.2\u03bcM, 0.3\u03bcM, 0.4\u03bcM concentrations of H2O2\nEppendorfs B1 to B6 has 0.5\u03bcM,1\u03bcM 2\u03bcM, 1mM, 100\u03bcM and 20\u03bcM of H2O2\nEppendorfs C1 to D1 have PMA concentrations of 0,10,25,50,75,100 and 150 ng/mL\nrespectively.\nEppendorfs D3, D4 and D5 have reagents 3% H2O2, HRP (Horse Radish Peroxidase) and Amplex red dye respectively\nSlot 5 Opentrons 24 tube rack Eppendorfs from A1 to A3 have PMA concentrations of 1mM,100\u03bcM and 10\u03bcM respectively.\nSlot3 The 96 well plate on the temperature module has the layout where Rows 0 and 2 have different concentrations of H2O2 standards ranging from 0 to 2\u03bcM added to wells (50\u03bcl). The rows 4 to 6 (columns 1 to 7; E1, F1 and G1 have cells which are untreated followed by E2, F2, G2 having cells treated with 10ng/mL of PMA till E7, F7 and G7 being cells treated with 150ng/mL of PMA (50\u03bcl)\n\n\n\n* Figure 2 Deck Layout after addition of Amplex red reagent to the 96-well plate that sits on the temperature module (Slot 3).\n\n\n\n* Figure 3 This representation shows the addition of 50\u03bcl of Amplex red working solution to the Blank wells A1 and A2 (Figure shows first well A1 containing 1X Reaction Buffer, to which the Amplex red working solution is added). Same steps are followed for the H2O2 standards (Figure shows representative well A9 displaying the addition of Amplex red). Likewise, the steps are same for wells in which THP-1 cells are present and contain different concentrations of PMA (one representative well, E6 is shown above)\n\n---\n\n",
"description": "This protocol can be used to measure the levels of hydrogen peroxide from THP-1 cells using Amplex Red reagent kit on the OT-2. 96-well plate format is used for this protocol.\n\nExplanation of complex parameters below:\n* `Number of Samples (5-12)`: Specify the number of samples for this run (from 5 to 12 samples).\n* `P300/P20 Mount`: Specify which mount (left or right) for the P300 and P20 single channel pipettes. \n\n---\n\n",
diff --git a/protoBuilds/sci-bead-based-elisa-for-sars-cov-2-surrogate-virus-neutralization-test/README.json b/protoBuilds/sci-bead-based-elisa-for-sars-cov-2-surrogate-virus-neutralization-test/README.json
index 94f695c358..645e459ac2 100644
--- a/protoBuilds/sci-bead-based-elisa-for-sars-cov-2-surrogate-virus-neutralization-test/README.json
+++ b/protoBuilds/sci-bead-based-elisa-for-sars-cov-2-surrogate-virus-neutralization-test/README.json
@@ -1,5 +1,5 @@
{
- "author": "Boren Lin",
+ "author": "Boren Lin\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins": [
"Proteins"
@@ -10,7 +10,7 @@
"internal": "sci-bead-based-elisa-for-sars-cov-2-surrogate-virus-neutralization-test",
"labware": "\nOpentrons 96 Tip Rack 300 \u00b5L\nNEST 96 Deepwell Plate 2mL #503001\nNEST 1 Well Reservoir 195 mL #360103\nCorning 96 Well Plate 360 \u00b5L Flat #3650\n",
"markdown": {
- "author": "Boren Lin\n\n\n",
+ "author": "Boren Lin\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins\n\t* Proteins\n\n\n",
"deck-setup": "[deck](https://drive.google.com/open?id=1lEBa2fcXszBBB_ndEMUCY1tMzY6r2aJq)\n\n\n",
"description": "ACROBiosystems SARS-CoV-2 Spike RBD-coupled Magnetic Beads (Cat. #: MBS-K002, Newark, DE, USA) are streptavidin conjugated magnetic beads attached with biotinylated SARS-CoV-2 Spike RBD protein. Using RBD protein as the target antigen for neutralizing antibodies to bind, a bead-based ELISA is developed to detect anti-SARS-CoV-2 antibodies in serum samples. \n\nThe protocol performs mixing and incubation of the RBD-conjugated magnetic beads with serum samples to enable specific binding of the RBD and neutralizing antibodies. The Magnetic Module renders magnetic separation of the beads from unbound molecules in the samples, which are removed by washing. Then, an HRP-conjugated anti-human IgG antibody is added. This antibody binds to neutralizing antibodies which are immobilized on the beads through the interaction with RBD. The complex formed on the beads is a typical format of a sandwich ELISA (antibody-antigen-antibody) which after another round of washes, can be exposed to a substrate of HRP to generate a photometric signal that can be measured on a microplate reader at 450 nm.\n\nReagents:\n1.\tSARS-CoV-2 Spike RBD-coupled Magnetic Beads (ACROBiosystems, Cat. #: MBS-K002, Newark, DE, USA)\n2.\tMouse Monoclonal Anti-Human IgG Fc (HRP) (Abcam, Cat. #: ab99759, Cambridge, UK)\n3.\tWash Buffer (20 mM Tris HCl, 130 mM NaCl, 0.05% Tween-20, pH 7.3)\n4.\tDilution Buffer for sample or antibody preparation (20 mM Tris HCl, 130 mM NaCl, 0.05% Tween-20, 0.25% BSA pH 7.3)\n5.\tTMB Substrate Solution\n6.\tStop Solution\n\nLabware:\n1.\tSlit seal (BioChromato, Fujisawa, Japan)\n2.\tNEST 1 Well Reservoir 195 mL \n3.\tNEST 96 Deep Well Plate 2mL\n4.\tOpentrons Tip Rack, 300 \u00b5L\n5.\tCorning 96 Well Plate 360 \u00b5L Flat or compatible \n6.\tMicroplate Reader\n\nPipettes and Modules\n1.\tOpentrons P300 8-Channel Pipette (GEN2)\n2.\tHeater Shaker Module (GEN1) with Aluminum Adaptor for 96-well Deep Well Plate\n3.\tMagnetic Moddule (GEN2)\n\nDeck setup:\nPlease see the deck layout as the general guideline for labware setup. \n1.\tThe Heater Shaker Module is loaded at Slot 3. \n2.\tSamples or sample dilutions are pre-loaded in a NEST 96 Deep Well Plate 2mL (the working plate) placed on the Magnetic Module (Slot 9)\n3.\tThe REAGENTS plate (Slot 11) is filled with bead suspension in Column 1, Anti-Human IgG Antibody in Column 2, TMB Substrate Solution in Column 3, and Stop Solution in Column 4\nNote: See Reagent Setup for reagent plate preparation\n4.\tFor WASH (Slot 5), sufficient wash buffer is filled in each well for 6 washes of 250 \u03bcL.\n5.\tThree boxes of Opentrons 96 tips (Slot 3, 4, and 10) are sufficient to run a full plate of 96 samples. \n6.\tThe WASTE plate is loaded at Slot 6.\n7.\tA Corning 96 Well Plate 360 \u00b5L Flat (the FINAL plate) is loaded at Slot 7.\n\n\n\n",
diff --git a/protoBuilds/sci-dynabeads-ip-1/README.json b/protoBuilds/sci-dynabeads-ip-1/README.json
index 8fb1a71199..a477e62a1c 100644
--- a/protoBuilds/sci-dynabeads-ip-1/README.json
+++ b/protoBuilds/sci-dynabeads-ip-1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Protein Purification": [
"ThermoFisher Dynabeads Protein A/G"
@@ -10,7 +10,7 @@
"internal": "protocol-hex-code",
"labware": "\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nNEST 1-Well Reservoirs, 195 mL\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Protein Purification\n\t* ThermoFisher Dynabeads Protein A/G\n\n",
"deck-setup": "Slot 1 - Magnetic module/96 deepwell plate (working plate)\nSlot 3 - Temperature module\nSlot 4 \u2013 Reagent in 96 well deepwell plate\n a.\tGreen \u2013 beads\n b.\tBlue \u2013 antibody\nSlot 5 - Samples in 96 well deepwell plate (red)\nSlot 7 - Tiprack1\nSlot 8 - Tiprack2 \nSlot 9 \u2013 Liquid waste\n\n\n\n",
"description": "This protocol (Part 1) performs pipetting and mixing of reagents and samples on the OT2 as the first part of automated immunoprecipitation using Thermo Fisher Dynabeads Protein A or Protein G.\n\nThe user can determine the number of samples to be processed.\n\nThe samples (e.g. cell lysates), Dynabeads and the antibody specific for the protein of interest will be transferred to and mixed in a 96-well deepwell working plate on the OT2. The first part of the protocol completes at this point allowing the user to move the working plate to an agitation device, an antibody/target protein incubation period determined by the user. After incubation, the process proceeds with the second part of the protocol on the OT2. Dynabeads/antibody/target protein complex will be washed, eluted, and heated to denature. The final product is ready for SDS-PAGE. \n\n---\n\n",
diff --git a/protoBuilds/sci-dynabeads-ip-2/README.json b/protoBuilds/sci-dynabeads-ip-2/README.json
index 77fdbcedb8..b6ff87e249 100644
--- a/protoBuilds/sci-dynabeads-ip-2/README.json
+++ b/protoBuilds/sci-dynabeads-ip-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Protein Purification": [
"ThermoFisher Dynabeads Protein A/G"
@@ -10,7 +10,7 @@
"internal": "sci-dynabeads-ip-2",
"labware": "\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nNEST 1-Well Reservoirs, 195 mL\nNEST 12-Well Reservoirs, 15 mL\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Protein Purification\n\t* ThermoFisher Dynabeads Protein A/G\n\n",
"deck-setup": "Slot 1 - Magnetic module/sample + beads + antibody (red) in 96 deepwell plate (working plate)\nSlot 2 \u2013 Wash buffer in 12 well reservoir (orange)\nSlot 3 - Temperature module/96 well PCR plate (final plate)\nSlot 4 \u2013 Reagent in 96 well deepwell plate/elution buffer (purple)\nSlot 5 \u2013 Tiprack3 (Note: this tiprack is assigned for use in 3 wash steps. The tips are returned to the tiprack and reused and will not be discarded.)\nSlot 6 \u2013 Tiprack4\nSlot 7 - Tiprack1\nSlot 8 - Tiprack2\nSlot 9 \u2013 Liquid waste\n\n\n\n",
"description": "This protocol (Part 2) performs washing and elution on the OT2 as the second part of automated immunoprecipitation using Thermo Fisher Dynabeads Protein A or Protein G.\n\nThe user can determine the number of samples to be processed.\n\nThe samples (e.g. cell lysates), Dynabeads and the antibody specific for the protein of interest will be transferred to and mixed in a 96-well deepwell working plate on the OT2. The first part of the protocol completes at this point allowing the user to move the working plate to an agitation device, an antibody/target protein incubation period determined by the user. After incubation, the process proceeds with the second part of the protocol on the OT2. Dynabeads/antibody/target protein complex will be washed, eluted and heated to denature. The final product is ready for SDS-PAGE. \n\n\n---\n\n",
diff --git a/protoBuilds/sci-dynabeads-tube-1/README.json b/protoBuilds/sci-dynabeads-tube-1/README.json
index 00d8fda0c9..239b448ba0 100644
--- a/protoBuilds/sci-dynabeads-tube-1/README.json
+++ b/protoBuilds/sci-dynabeads-tube-1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Protein Purification": [
"Thermo Fisher Dynabeads\u2122 Protein A/G"
@@ -10,7 +10,7 @@
"internal": "sci-dynabeads-tube-1",
"labware": "\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nNEST 1-Well Reservoirs, 195 mL\n4-in-1 Tube Rack Set\nNEST 15 mL Centrifuge Tube\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Protein Purification\n\t* Thermo Fisher Dynabeads\u2122 Protein A/G\n\n",
"deck-setup": " \n* Beads: 50 uL per sample\n* Antibody: diluted in phosphate-buffered saline with 0.1% Tween 20 (PBS-T), 50 uL per sample \n\n\n",
"description": "This protocol (Part 1) performs pipetting and mixing of reagents and samples on the OT2 as the first part of automated immunoprecipitation using Thermo Fisher Dynabeads Protein A or Protein G.\n\nThe user can determine the number of samples to be processed.\n\nThe samples (e.g. cell lysates), Dynabeads and the antibody specific for the protein of interest will be transferred to and mixed in a 96-well deepwell working plate on the OT2. The first part of the protocol completes at this point allowing the user to move the working plate to an agitation device, an antibody/target protein incubation period determined by the user. After incubation, the process proceeds with the second part of the protocol on the OT2. Dynabeads/antibody/target protein complex will be washed, eluted, and heated to denature. The final product is ready for SDS-PAGE. \n\n",
diff --git a/protoBuilds/sci-dynabeads-tube-2/README.json b/protoBuilds/sci-dynabeads-tube-2/README.json
index 8b76ff123e..42cef204ac 100644
--- a/protoBuilds/sci-dynabeads-tube-2/README.json
+++ b/protoBuilds/sci-dynabeads-tube-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons(https://opentrons.com/)",
+ "author": "Opentrons(https://opentrons.com/)\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Protein Purification": [
"Thermo Fisher Dynabeads\u2122 Protein A/G"
@@ -10,7 +10,7 @@
"internal": "sci-dynabeads-tube-2",
"labware": "\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nNEST 1-Well Reservoirs, 195 mL\n4-in-1 Tube Rack Set\nNEST 15 mL Centrifuge Tube\nNEST 12-Well Reservoirs, 15 mL\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOpentrons 300\u00b5L Tips\n",
"markdown": {
- "author": "Opentrons(https://opentrons.com/)\n\n",
+ "author": "Opentrons(https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Protein Purification\n\t* Thermo Fisher Dynabeads\u2122 Protein A/G\n\n",
"deck-setup": "\n* Slot 1 - Magnetic module/sample + beads + antibody (red) in 96 deepwell plate (working plate)\n* Slot 2 \u2013 Wash buffer in 12 well reservoir (orange)\n* Slot 3 - Temperature module/96 well PCR plate (final plate)\n* Slot 4 - Tube rack/elution buffer in 15 mL conical tube (purple)\n* Slot 5 \u2013 Tiprack3 (Note: this tiprack is assigned for use in 3 wash steps. The tips are returned to the tiprack and reused and will not be discarded.) \n* Slot 6 \u2013 Tiprack4\n* Slot 7 - Tiprack1\n* Slot 8 - Tiprack2\n* Slot 9 \u2013 Liquid waste\n\n",
"description": "This protocol (Part 2) performs washing and elution on the OT2 as the second part of automated immunoprecipitation using Thermo Fisher Dynabeads Protein A or Protein G.\n\nThe user can determine the number of samples to be processed.\n\nThe samples (e.g. cell lysates), Dynabeads and the antibody specific for the protein of interest will be transferred to and mixed in a 96-well deepwell working plate on the OT2. The first part of the protocol completes at this point allowing the user to move the working plate to an agitation device, an antibody/target protein incubation period determined by the user. After incubation, the process proceeds with the second part of the protocol on the OT2. Dynabeads/antibody/target protein complex will be washed, eluted and heated to denature. The final product is ready for SDS-PAGE. \n\n",
diff --git a/protoBuilds/sci-idt-normalase/README.json b/protoBuilds/sci-idt-normalase/README.json
index 70caef05d1..42f049cf8c 100644
--- a/protoBuilds/sci-idt-normalase/README.json
+++ b/protoBuilds/sci-idt-normalase/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"IDT Normalase"
@@ -10,7 +10,7 @@
"internal": "sci-idt-normalase",
"labware": "\n2x Eppendorf 96 well plate full skirt\n1x Nest 96 well plate full skirt\n\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* IDT Normalase\n\n",
"deck-setup": "\n\n* Left: Initial Plate Setup, Right: After Moving the Sample plate to Pos 1 and Pooling the samples on a new plate on the\nThermo.\n\n# PLATE MOVING\nThe script requires manually transferring the sample plate between the Thermocycler and Magnet. NOTE: In the script\nthe two positions are handled as sample_plate_mag and sample_plate_thermo; during calibration use an empty plate of\nthe same labware as the sample plate on the magnet position to calibrate that position.\n\n# REAGENT PLATE\nPrepare the reagents in the Reagent Plate according to the table below. If available, prepare extra volume to account for\noverage. Following the instructions from the IDT Normalase protocols, reagent mixes for Norm II and Inactivation are\nmade for at least 24x samples even if fewer samples are run due to the low volumes.\n\n* [reagent plate](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/sci-idt-normalase/reagent+plate+setup.png)\n\n\n---\n\n",
"description": "This is the Normalase protocol that follows the Normalase PCR that is done after the usual fragmentation, End-Repair, A-tailing, and Ligation. The Ligation would have been performed with either a full length adapter ligation using the Normalase Reagent R5, or a stubby universal adapter followed by the barcoded stubby Normalase adapters in the PCR. This protocol follows that modified PCR that can be adapted into other NGS Library preps, before starting the prepared, amplified libraries should be cleaned and quantified, samples must be at least 5ul of 12nM for efficient Normalization. See the IDT Normalase protocol for more information.\n\n# EXAMPLE PROTOCOL SETUP\nThis is an example setup for 24 samples that will be pooled into 3 pools of 8 samples each. The first step is a reaction on the thermocycler, afterwards the plate is transferred to Pos 1 and a new plate for pooled samples and additional reactions goes on the thermocycler. This protocol will pool 5ul of each sample individually with a single channel pipette according to\nthe setup below. The sample setup is configured for 1, 2, or 3 columns of samples into 3 pools, but the configuration can be adjusted for different pooling amounts or number of pools.\n\n* \n\n* [c setup](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/sci-idt-normalase/example+protocol+csv.png)\n\n\nExplanation of complex parameters below:\n* `.CSV File`: Provide csv formatted as seen in documentation to specify how many samples per pool is desired. \n* `Number of Samples`: Specify number of samples for this run.\n* `Dry Run`: Yes will return tips, skip incubation times, shorten mix, for testing purposes.\n* `Use Modules?`: Yes will not require modules on the deck and will skip module steps, for testing purposes, if `Dry Run` is set to `Yes`, then this variable will automatically set itself to `No`.\n* `Use protocol specific z-offsets?`: Sets whether to use protocol specific z offsets for each tip and labware or no offsets aside from defaults\n* `Use NGS Magnetic Block?\"`: Sets whether there is the Magnetic Block on the Deck (for post-NGS Setup)\n* `P20 Single-Channel Mount`: Specify which mount (left or right) to mount the P20 single-channel pipette.\n* `P20 Multi-Channel Mount`: Specify which mount (left or right) to mount the P20 multi-channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/sci-idt-xgen-ez/README.json b/protoBuilds/sci-idt-xgen-ez/README.json
index 1d327c5f5b..793e5dd0ee 100644
--- a/protoBuilds/sci-idt-xgen-ez/README.json
+++ b/protoBuilds/sci-idt-xgen-ez/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"IDT xGEN EZ"
@@ -10,7 +10,7 @@
"internal": "sci-idt-xgen-ez",
"labware": "\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\n[* NEST 0.1 mL 96-Well PCR Plate, Full Skirt\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nOpentrons Aluminum Block Set\nOpentrons 20\u00b5L Filter Tips\nOpentrons 200\u00b5L Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* IDT xGEN EZ\n\n",
"deck-setup": "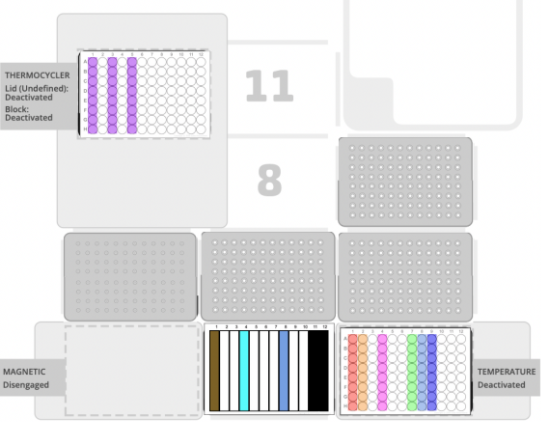\n\n",
"description": "This protocol performs the [IDT xGEN EZ kit](https://sfvideo.blob.core.windows.net/sitefinity/docs/default-source/protocol/xgen-dna-library-prep-ez-kit-and-xgen-dna-library-prep-ez-uni-kits-protocol.pdf?sfvrsn=57b1e007_8). This protocol uses the stubby adapter (Reagent W5) that is included in the xGEN EZ kit during ligation and the barcoding of samples is performed during PCR by using either xGEN UDIs or CDIs. The alternate protocol version is IDT xGEN EZ UNI that does the barcoding of samples at ligation instead. The Protocol steps and reagents are different between the two versions. See IDT\u2019s xGEN DNA Library Prep EZ Kit protocol for more information about sample barcode kit requirements.\n\nExplanation of complex parameters below:\n* `Number of Samples`: Samples are prepared as pictured below; with 100ng of input DNA in 50ul of Low EDTA. See IDT\u2019s xGEN DNA Library Prep EZ Kit protocol for more information about sample input requirements.\n\n\n* `Fragmentation Time (minutes)`: Minutes, Duration of the Fragmentation Step\n* `PCR Cycles`: Specify number of PCR cycles if performing PCR on deck.\n* `Dry Run`: Yes will return tips, skip incubation times, shorten mix, for testing purposes.\n* `Use Modules?`: Yes will not require modules on the deck and will skip module steps, for testing purposes, if `Dry Run` is set to `Yes`, then this variable will automatically set itself to `No`.\n* `Reuse Tips?`: No, NYI format for reusing tips\n* `Use protocol specific z-offsets?`: Sets whether to use protocol specific z offsets for each tip and labware or no offsets aside from defaults\n* `Include Fragmentation / End-Repair / A-Tailing Step?`: Specify whether to include this step below. This and all steps below allow you to customize where to start the protocol, run the protocol in parts over multiple days, choose whether to use Opentrons modules or off deck modules, etc.\n* `If yes, Fragmentation / End-Repair / A-Tailing on deck or off deck?`: Use this step on or off the deck.\n* `Include Ligation Step?`: Specify whether to include this step in this run.\n* `If yes, ligation Step on or off deck?`: Use this step on or off the deck.\n* `Include Post Ligation Step?`: Specify whether to include this step in this run.\n* `Include PCR Step?`: Specify whether to include this step in this run.\n* `If yes, PCR step on or off deck?`: Use this step on or off the deck.\n* `Include First Post PCR Step?`: Specify whether to include this step in this run.\n* `Include Second Post PCR Step?`: Specify whether to include this step in this run.\n* `P20 Multi-Channel Mount`: Specify which mount (left or right) to mount the P20 multi-channel pipette.\n* `P300 Multi-Channel Mount`: Specify which mount (left or right) to mount the P300 multi-channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/sci-idt-xgen-mc/README.json b/protoBuilds/sci-idt-xgen-mc/README.json
index dc7bd58954..7150a47b20 100644
--- a/protoBuilds/sci-idt-xgen-mc/README.json
+++ b/protoBuilds/sci-idt-xgen-mc/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"IDT xGEN MC"
@@ -10,7 +10,7 @@
"internal": "sci-idt-xgen-mc",
"labware": "\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\n[* NEST 0.1 mL 96-Well PCR Plate, Full Skirt\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\nOpentrons Aluminum Block Set\nOpentrons 20\u00b5L Filter Tips\nOpentrons 200\u00b5L Filter Tips\n\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* IDT xGEN MC\n\n",
"deck-setup": "\n\n",
"description": "This protocol performs the [IDT xGEN MC kit](https://sfvideo.blob.core.windows.net/sitefinity/docs/default-source/protocol/xgen-dna-library-prep-ez-kit-and-xgen-dna-library-prep-ez-uni-kits-protocol.pdf?sfvrsn=57b1e007_8). This protocol uses the stubby adapter (Reagent L3) that is included in the xGEN MC kit during ligation and the barcoding\nof samples is performed during PCR by using either xGEN UDIs or CDIs. The alternate protocol version is IDT xGEN MC\nUNI that does the barcoding of samples at ligation instead. The Protocol steps and reagents are different between the\ntwo versions. See IDT\u2019s xGEN DNA Library Prep MC Kit protocol for more information about sample barcode kit\nrequirements.\n\nThe script requires manually transferring the sample plate between the Thermocycler and Magnet 3 times. It starts on the\nThermocycler and needs to be moved to the Magnet for the post-ligation cleanup, and then moved to the Thermocycler\nfor PCR and then back to the Magnet for the post-PCR cleanup.\nNOTE: In the script the two positions are handled as magnetic sample plate and thermocycler sample plate; during calibration,\nuse an empty plate of the same labware as the sample plate on the magnet position to calibrate that position.\n\nExplanation of complex parameters below:\n* `Number of Samples`: Samples are prepared as pictured below; with 100ng of input DNA in 50ul of Low EDTA. See IDT\u2019s xGEN DNA Library Prep EZ Kit protocol for more information about sample input requirements.\n\n\n* `PCR Cycles`: Specify number of PCR cycles if performing PCR on deck.\n* `Dry Run`: Yes will return tips, skip incubation times, shorten mix, for testing purposes.\n* `Use Modules?`: Yes will not require modules on the deck and will skip module steps, for testing purposes, if `Dry Run` is set to `Yes`, then this variable will automatically set itself to `No`.\n* `Reuse Tips?`: No, NYI format for reusing tips\n* `Use protocol specific z-offsets?`: Sets whether to use protocol specific z offsets for each tip and labware or no offsets aside from defaults\n* `Include End-Repair / A-Tailing Step?`: Specify whether to include this step below. This and all steps below allow you to customize where to start the protocol, run the protocol in parts over multiple days, choose whether to use Opentrons modules or off deck modules, etc.\n* `If yes, End-Repair / A-Tailing on deck or off deck?`: Use this step on or off the deck.\n* `Include Ligation Step?`: Specify whether to include this step in this run.\n* `If yes, ligation Step on or off deck?`: Use this step on or off the deck.\n* `Include Post Ligation Step?`: Specify whether to include this step in this run.\n* `Include PCR Step?`: Specify whether to include this step in this run.\n* `If yes, PCR step on or off deck?`: Use this step on or off the deck.\n* `Include First Post PCR Step?`: Specify whether to include this step in this run.\n* `Include Second Post PCR Step?`: Specify whether to include this step in this run.\n* `P20 Multi-Channel Mount`: Specify which mount (left or right) to mount the P20 multi-channel pipette.\n* `P300 Multi-Channel Mount`: Specify which mount (left or right) to mount the P300 multi-channel pipette.\n\n---\n\n",
diff --git a/protoBuilds/sci-illumina-dna-prep/README.json b/protoBuilds/sci-illumina-dna-prep/README.json
index 34007e21c1..53df1f6679 100644
--- a/protoBuilds/sci-illumina-dna-prep/README.json
+++ b/protoBuilds/sci-illumina-dna-prep/README.json
@@ -18,7 +18,7 @@
"labware": "* [Nest 96 well plate full skirt 100 \u00b5L](https://shop.opentrons.com/nest-0-1-ml-96-well-pcr-plate-full-skirt/)\n* [NEST 2 mL 96-Well Deep Well Plate](https://shop.opentrons.com/nest-2-ml-96-well-deep-well-plate-v-bottom/)\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/nest-12-well-reservoirs-15-ml/)\n* [Opentrons aluminum block set](https://shop.opentrons.com/aluminum-block-set/)\n\n",
"modules": "* [Temperature Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Magnetic Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Illumina](https://www.illumina.com/)\n\n",
+ "partner": "[Illumina](https://www.illumina.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P300 multi-Channel (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n* [P20 multi-Channel (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n* [P10 multi-Channel](https://shop.opentrons.com/8-channel-electronic-pipette/)\n**Tips**\n* [Opentrons 20 \u00b5L filter tiprack](https://shop.opentrons.com/opentrons-20ul-filter-tips/)\n* [Opentrons 200 \u00b5L filter tiprack](https://shop.opentrons.com/opentrons-200ul-filter-tips/)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Prepare the thermocycler by setting the block temperature to 4 degrees, and the lid to 100 degrees.\n2. Add tagmentation mix to the samples\n3. User seals the plate and the protocol incubates the samples with the mix in the thermocycler, 55 degrees for 15 minutes.\n4. The thermocycler opens, and the user removes the seal.\n5. Add Tagmentation Stop Buffer to the samples.\n6. Seal and incubate the mix at 37 degrees for 15 minutes.\n7. User removes seal; remove the supernatant and wash the beads three times with Tagmentation Wash Buffer.\n8. Amplification of DNA: Addition of PCR mix and addition of barcodes.\n9. The protocol runs PCR protocol for 5 cycles. This takes 25 minutes to complete. The supernatant is transferred to columns 7, 9 and 11 depending on how many sample columns there are.\n10. Post-PCR cleanup using AMPure beads.\n11. The protocol performs two ethanol washes.\n12. RSB (resuspension buffer) is added to the bead wells and the supernatant is transferred to the output columns (8, 10, and 12).\n\n",
@@ -32,7 +32,7 @@
"Thermocycler Module"
],
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Illumina",
+ "partner": "Illumina\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP300 multi-Channel (GEN2)\nP20 multi-Channel (GEN2)\nP10 multi-Channel\nTips\nOpentrons 20 \u00b5L filter tiprack\nOpentrons 200 \u00b5L filter tiprack\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nPrepare the thermocycler by setting the block temperature to 4 degrees, and the lid to 100 degrees.\nAdd tagmentation mix to the samples\nUser seals the plate and the protocol incubates the samples with the mix in the thermocycler, 55 degrees for 15 minutes.\nThe thermocycler opens, and the user removes the seal.\nAdd Tagmentation Stop Buffer to the samples.\nSeal and incubate the mix at 37 degrees for 15 minutes.\nUser removes seal; remove the supernatant and wash the beads three times with Tagmentation Wash Buffer.\nAmplification of DNA: Addition of PCR mix and addition of barcodes.\nThe protocol runs PCR protocol for 5 cycles. This takes 25 minutes to complete. The supernatant is transferred to columns 7, 9 and 11 depending on how many sample columns there are.\nPost-PCR cleanup using AMPure beads.\nThe protocol performs two ethanol washes.\nRSB (resuspension buffer) is added to the bead wells and the supernatant is transferred to the output columns (8, 10, and 12).\n",
diff --git a/protoBuilds/sci-illumina-rna-enrichment-normalization/README.json b/protoBuilds/sci-illumina-rna-enrichment-normalization/README.json
index d904522723..d974a9001e 100644
--- a/protoBuilds/sci-illumina-rna-enrichment-normalization/README.json
+++ b/protoBuilds/sci-illumina-rna-enrichment-normalization/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Illumina RNA Prep with Enrichment"
@@ -10,7 +10,7 @@
"internal": "sci-illumina-rna-enrichment-normalization",
"labware": "\nNEST 0.1 mL 96-Well PCR Plate, Full Skirt\nOpentrons 4-in-1 Tube Rack Set with 15+50ml Falcon tube insert\nOpentrons 200\u00b5L Filter Tips\nOpentrons 20\u00b5L Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* Illumina RNA Prep with Enrichment\n\n",
"deck-setup": "\n\n\n---\n\n",
"description": "\nThis protocol performs normalization for the [Illimuina RNA Prep with Enrichment](https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/rna-prep-enrichment.html) protocol. Normalization is the method of adding buffer to a set of samples so that their concentrations are uniform. This is usually so that they can be treated uniformly in subsequent steps. For example, after NGS library prep of multiple samples, the samples are normalized to the same concentration, so that the same volume of each sample can be pooled, and each sample is represented in equal proportions. A requirement for this Normalization protocol is that the concentrations and volumes are known i.e. previously quantified by Picogreen and aliquoted into a plate at a set volume, and formatted\nin a .csv file as input.\n\nYou should upload a .csv file formatted in the following way, being sure to include the header line:\n\n```\nSample_Plate,Sample_well,InitialVol,InitialConc,TargetConc\nsample_plate,A1,20,2.3,2\nsample_plate,B1,20,3.99,2\nsample_plate,C1,20,4.39,2\nsample_plate,D1,20,3.95,2\nsample_plate,E1,20,4.16,2\nsample_plate,F1,20,3.81,2\nsample_plate,G1,20,3.96,2\nsample_plate,H1,20,3.41,2\n```\n\n---\n\n",
diff --git a/protoBuilds/sci-lucif-assay1-for4/README.json b/protoBuilds/sci-lucif-assay1-for4/README.json
index 322cc06b6d..3fdfd0dc84 100644
--- a/protoBuilds/sci-lucif-assay1-for4/README.json
+++ b/protoBuilds/sci-lucif-assay1-for4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "sci-lucif-assay1-for4",
"labware": "\nCorning 96 Well Plate 360 \u00b5L Flat #3650\nOpentrons 96 Tip Rack 300 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons HEPA Filter Module\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "The protocol performs liquid handling to prepare up to four 96-well plates of mammalian cells for luciferase reporter assay.\n\nThe luciferase reporter assay is used to investigate the regulation of expression of a gene of interest. This method relies on reporter cells created by stable or transient transfection of a promoter-driven, luciferase-expressing DNA. The bioluminescence proportional to luciferase expression can be quantified to assess the functional connection between the expression of the target gene which is controlled by this promoter, and the factor to be studied which may exhibit a regulatory effect on the cell behaviors at the transcription level. In this particular example, HeLa cells are stably or transiently transfected with an NF-kB promoter-driven, luciferase reporter construct, treated with an NF-kB activator phorbol 12-myristate 13-acetate (PMA), and activation of NF-kB assessed by measuring the luciferase enzymatic activity.\n\nCell suspension preparation:\nMammalian cervical cancer cell line HeLa (ATCC, Manassas, VA, USA) or HeLa-originated cell line with a sequence of NF-kB response element upstream of luciferase-expressing DNA integrated in the cell\u2019s genome (Cellomics Technology, Halethorpe, MD, USA) is maintained in Dulbecco\u2019s modified Eagle\u2019s medium (DMEM, Thermo Fisher Scientific, Waltham, MA, USA). containing 10% fetal bovine serum and 1% penicillin/streptomycin in humidified 5% CO2 at 37\u2009\u00b0C. HeLa cells are detached by using trypsin, resuspend in DMEM and poured into a NEST 12-well reservoir. To prepare one 96-well plate cell culture, for the assay that requires transient transfection to prepare reporter cells, resuspend 1/4 of cells collected from a confluent 100 mm tissue culture dish (~ 2.2 x 106 cells) in 11 mL DMEM. For the assay using the pre-engineered cell line stably expressing luciferase reporter, resuspend 1/2 of cells collected from a confluent 100 mm tissue culture dish (~ 4.4 x 106 cells) in 11 mL DMEM.\n\n\n\n",
diff --git a/protoBuilds/sci-lucif-assay1/README.json b/protoBuilds/sci-lucif-assay1/README.json
index 02e287d40f..7100d5f50b 100644
--- a/protoBuilds/sci-lucif-assay1/README.json
+++ b/protoBuilds/sci-lucif-assay1/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "sci-lucif-assay1",
"labware": "\nNEST 12 Well Reservoir 15 mL #360102\nCorning 96 Well Plate 360 \u00b5L Flat #3650\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "The protocol performs liquid handling to prepare a 96-well plate of mammalian cells for luciferase reporter assay.\n\nThe luciferase reporter assay is used to investigate the regulation of expression of a gene of interest. This method relies on reporter cells created by stable or transient transfection of a promoter-driven, luciferase-expressing DNA. The bioluminescence proportional to luciferase expression can be quantified to assess the functional connection between the expression of the target gene which is controlled by this promoter, and the factor to be studied which may exhibit a regulatory effect on the cell behaviors at the transcription level. In this particular example, HeLa cells are stably or transiently transfected with an NF-kB promoter-driven, luciferase reporter construct, treated with an NF-kB activator phorbol 12-myristate 13-acetate (PMA), and activation of NF-kB assessed by measuring the luciferase enzymatic activity.\n\nCell suspension preparation:\nMammalian cervical cancer cell line HeLa (ATCC, Manassas, VA, USA) or HeLa-originated cell line with a sequence of NF-kB response element upstream of luciferase-expressing DNA integrated in the cell\u2019s genome (Cellomics Technology, Halethorpe, MD, USA) is maintained in Dulbecco\u2019s modified Eagle\u2019s medium (DMEM, Thermo Fisher Scientific, Waltham, MA, USA). containing 10% fetal bovine serum and 1% penicillin/streptomycin in humidified 5% CO2 at 37\u2009\u00b0C. To prepare cell culture in a 96-well plate, HeLa cells are detached by using trypsin, resuspend in DMEM and poured into a NEST 12-well reservoir. For the assay that requires transient transfection to prepare reporter cells, resuspend 1/4 of cells collected from a confluent 100 mm tissue culture dish (~ 2.2 x 106 cells) in 11 mL DMEM. For the assay using the pre-engineered cell line stably expressing luciferase reporter, resuspend 1/2 of cells collected from a confluent 100 mm tissue culture dish (~ 4.4 x 106 cells) in 11 mL DMEM.\n\n\n",
diff --git a/protoBuilds/sci-lucif-assay2-for4/README.json b/protoBuilds/sci-lucif-assay2-for4/README.json
index 879fef6f9d..8a398727be 100644
--- a/protoBuilds/sci-lucif-assay2-for4/README.json
+++ b/protoBuilds/sci-lucif-assay2-for4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "sci-lucif-assay2-for4",
"labware": "\nCorning 96 Well Plate 360 \u00b5L Flat #3650\nOpentrons 96 Tip Rack 300 \u00b5L\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons HEPA Filter Module\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "The protocol performs liquid handling to conduct transient DNA transfection to develop reporter cells in a 96-well setting for luciferase reporter assay.\n\nThe luciferase reporter assay is used to investigate the regulation of expression of a gene of interest. This method relies on reporter cells created by stable or transient transfection of a promoter-driven, luciferase-expressing DNA. The bioluminescence proportional to luciferase expression can be quantified to assess the functional connection between the expression of the target gene which is controlled by this promoter, and the factor to be studied which may exhibit a regulatory effect on the cell behaviors at the transcription level. In this particular example, HeLa cells are stably or transiently transfected with an NF-kB promoter-driven, luciferase reporter construct, treated with an NF-kB activator phorbol 12-myristate 13-acetate (PMA), and activation of NF-kB assessed by measuring the luciferase enzymatic activity.\n\nTransfection reagent preparation:\nTo make the reagent for 100 reactions (sufficient for a 96-well plate), dilute 10 \u00b5L (0.1 \u00b5L per reaction) of NanoLuc Reporter Vector with NF-\u03baB Response Element (Promega, Madison, WI, USA) in 1 mL (10 \u00b5L per reaction) serum free DMEM (Thermo Fisher Scientific, Waltham, MA, USA) and then add 30 \u00b5L (0.3 \u00b5L per reaction) of FuGENE HD Transfection Reagent (Promega, Madison, WI, USA). After 10-minute incubation at room temperature, the transfection reagent/DNA mixture is brought up to 2 mL by adding serum free DMEM and then distributed to fill a column (250 \u00b5L per well, 8 well per column) of a NEST 96 Deep Well Plate.\n\n\n",
diff --git a/protoBuilds/sci-lucif-assay2/README.json b/protoBuilds/sci-lucif-assay2/README.json
index bd70da07e0..3efed0f511 100644
--- a/protoBuilds/sci-lucif-assay2/README.json
+++ b/protoBuilds/sci-lucif-assay2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Broad Category": [
"Specific Category"
@@ -10,7 +10,7 @@
"internal": "sci-lucif-assay2",
"labware": "\nOpentrons 24 Tube Rack with NEST 2 mL Snapcap\nCorning 96 Well Plate 360 \u00b5L Flat #3650\nOpentrons 96 Tip Rack 300 \u00b5L\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Broad Category\n\t* Specific Category\n\n\n",
"deck-setup": "\n\n\n\n",
"description": "The protocol performs liquid handling to conduct transient DNA transfection to develop reporter cells in a 96-well setting for luciferase reporter assay. Multiple transfection master mixes can be used (e.g., different DNA vectors or various reagent concentrations are tested), and the user determines how many columns of cells (12 columns in a 96-well plate, 8 wells per column) are subjected to a master mix.\n\nThe luciferase reporter assay is used to investigate the regulation of expression of a gene of interest. This method relies on reporter cells created by stable or transient transfection of a promoter-driven, luciferase-expressing DNA. The bioluminescence proportional to luciferase expression can be quantified to assess the functional connection between the expression of the target gene which is controlled by this promoter, and the factor to be studied which may exhibit a regulatory effect on the cell behaviors at the transcription level. In this particular example, HeLa cells are stably or transiently transfected with an NF-kB promoter-driven, luciferase reporter construct, treated with an NF-kB activator phorbol 12-myristate 13-acetate (PMA), and activation of NF-kB assessed by measuring the luciferase enzymatic activity.\n\nTransfection reagent preparation:\nThe table below lists examples of transfection master mix preparation. To make the reagent for 100 reactions (sufficient for a 96-well plate), dilute 10 uL (0.1 uL per reaction) of NanoLuc Reporter Vector with NF-\u03baB Response Element (Promega, Madison, WI, USA) in 1 mL (10 uL per reaction) serum free DMEM (Thermo Fisher Scientific, Waltham, MA, USA) and then add 30 uL (0.3 uL per reaction) of FuGENE HD Transfection Reagent (Promega, Madison, WI, USA). After 10-minute incubation at room temperature, the total volume of this transfection reagent/DNA mixture is brought up to 2 mL by adding serum free DMEM. All master mixes are kept in 2 mL microcentrifuge tubes with snap cap, loaded on Opentrons 24 Tube Rack.\n\n\n",
diff --git a/protoBuilds/sci-lucif-assay3-for4/README.json b/protoBuilds/sci-lucif-assay3-for4/README.json
index 58e4724164..0f968d27c7 100644
--- a/protoBuilds/sci-lucif-assay3-for4/README.json
+++ b/protoBuilds/sci-lucif-assay3-for4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "sci-lucif-assay3-for4",
"labware": "\nCorning 96 Well Plate 360 \u00b5L Flat #3650\nOpentrons 96 Tip Rack 300 \u00b5L\nNEST 1 Well Reservoir 195 mL #360103\nOpentrons HEPA Filter Module\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "The protocol performs liquid handling to apply test articles (e.g., serial dilutions of drug candidates) to the reporter cells cultured in 96-well plates.\n\nThe luciferase reporter assay is used to investigate the regulation of expression of a gene of interest. This method relies on reporter cells created by stable or transient transfection of a promoter-driven, luciferase-expressing DNA. The bioluminescence proportional to luciferase expression can be quantified to assess the functional connection between the expression of the target gene which is controlled by this promoter, and the factor to be studied which may exhibit a regulatory effect on the cell behaviors at the transcription level. In this particular example, HeLa cells are stably or transiently transfected with an NF-kB promoter-driven, luciferase reporter construct, treated with an NF-kB activator phorbol 12-myristate 13-acetate (PMA), and activation of NF-kB assessed by measuring the luciferase enzymatic activity.\n\nReagent preparation:\nTest articles are diluted in DMEM at the concentrations of interest and pre-loaded in a Corning 96 Well Plate 360 \u00b5L Flat (150 \u03bcL per well).\n\n\n",
diff --git a/protoBuilds/sci-lucif-assay3/README.json b/protoBuilds/sci-lucif-assay3/README.json
index c26f16fe13..9506d58757 100644
--- a/protoBuilds/sci-lucif-assay3/README.json
+++ b/protoBuilds/sci-lucif-assay3/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "sci-lucif-assay3",
"labware": "\nOpentrons 96 Tip Rack 300 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nCorning 96 Well Plate 360 \u00b5L Flat #3650\nNEST 1 Well Reservoir 195 mL #360103\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "\nThe protocol performs liquid handling to apply test articles (e.g., serial dilutions of drug candidates) to the reporter cells cultured in a 96-well plate.\n\nThe luciferase reporter assay is used to investigate the regulation of expression of a gene of interest. This method relies on reporter cells created by stable or transient transfection of a promoter-driven, luciferase-expressing DNA. The bioluminescence proportional to luciferase expression can be quantified to assess the functional connection between the expression of the target gene which is controlled by this promoter, and the factor to be studied which may exhibit a regulatory effect on the cell behaviors at the transcription level. In this particular example, HeLa cells are stably or transiently transfected with an NF-kB promoter-driven, luciferase reporter construct, treated with an NF-kB activator phorbol 12-myristate 13-acetate (PMA), and activation of NF-kB assessed by measuring the luciferase enzymatic activity.\n\nReagent preparation:\nPBS used for washing (50 uL per sample, 6.5 mL sufficient for 96 samples) is filled in the first well of a NEST 1 Well Reservoir 195 mL. Test articles are diluted in DMEM at the concentrations of interest and pre-loaded in a Corning 96 Well Plate 360 \u00b5L Flat (150 uL per well).\n\n\n",
diff --git a/protoBuilds/sci-lucif-assay4-for4/README.json b/protoBuilds/sci-lucif-assay4-for4/README.json
index af66901ceb..57ae63bc2b 100644
--- a/protoBuilds/sci-lucif-assay4-for4/README.json
+++ b/protoBuilds/sci-lucif-assay4-for4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "sci-lucif-assay4-for4",
"labware": "\nOpentrons 96 Tip Rack 300 \u00b5L\nCorning 96 Well Plate 360 \u00b5L Flat #3650\nNEST 12 Well Reservoir 15 mL #360102\nNEST 1 Well Reservoir 195 mL #360103\nOpentrons HEPA Filter Module\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "The protocol performs liquid handling for reporter cell lysis and luciferase-catalyzed chemical reaction in 96-well plates, ready for bioluminescence measurement by a microplate reader.\n\nThe luciferase reporter assay is used to investigate the regulation of expression of a gene of interest. This method relies on reporter cells created by stable or transient transfection of a promoter-driven, luciferase-expressing DNA. The bioluminescence proportional to luciferase expression can be quantified to assess the functional connection between the expression of the target gene which is controlled by this promoter, and the factor to be studied which may exhibit a regulatory effect on the cell behaviors at the transcription level. In this particular example, HeLa cells are stably or transiently transfected with an NF-kB promoter-driven, luciferase reporter construct, treated with an NF-kB activator phorbol 12-myristate 13-acetate (PMA), and activation of NF-kB assessed by measuring the luciferase enzymatic activity.\n\nReagent preparation:\nThe luciferase assay kit purchased from Promega (Madison, WI, USA) contains all reagents for cell lysis and luminescent signal development. To prepare luciferase assay reagent, the lyophilized luciferase assay substrate is dissolved in luciferase assay buffer per manufacturer\u2019s instructions. Cell culture lysis reagent (5X) provided in the assay kit is diluted to working concentration. For a full 96-well plate, 11 mL luciferase assay reagent (100 \u03bcL per sample) and 4.6 mL lysis reagent (30 \u03bcL per sample) are recommended. These 2 reagents are filled in a NEST 12 Well Reservoir, and PBS for washing (50 \u03bcL per sample, 30 mL sufficient for up to four 96-well plates) filled in a NEST 1 Well Reservoir. \n\n\n",
diff --git a/protoBuilds/sci-lucif-assay4/README.json b/protoBuilds/sci-lucif-assay4/README.json
index a296c82950..c53a33d120 100644
--- a/protoBuilds/sci-lucif-assay4/README.json
+++ b/protoBuilds/sci-lucif-assay4/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -10,7 +10,7 @@
"internal": "sci-lucif-assay4",
"labware": "\nOpentrons 96 Tip Rack 300 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nCorning 96 Well Plate 360 \u00b5L Flat #3650\nNEST 1 Well Reservoir 195 mL #360103\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n\n",
"deck-setup": "\n\n\n",
"description": "The protocol performs liquid handling for reporter cell lysis and luciferase-catalyzed chemical reaction in a 96-well plate, ready for bioluminescence measurement by a microplate reader.\n\nThe luciferase reporter assay is used to investigate the regulation of expression of a gene of interest. This method relies on reporter cells created by stable or transient transfection of a promoter-driven, luciferase-expressing DNA. The bioluminescence proportional to luciferase expression can be quantified to assess the functional connection between the expression of the target gene which is controlled by this promoter, and the factor to be studied which may exhibit a regulatory effect on the cell behaviors at the transcription level. In this particular example, HeLa cells are stably or transiently transfected with an NF-kB promoter-driven, luciferase reporter construct, treated with an NF-kB activator phorbol 12-myristate 13-acetate (PMA), and activation of NF-kB assessed by measuring the luciferase enzymatic activity.\n\nReagent preparation:\nThe luciferase assay kit purchased from Promega (Madison, WI, USA) contains all reagents for cell lysis and luminescent signal development. To prepare luciferase assay reagent, the lyophilized luciferase assay substrate is dissolved in luciferase assay buffer per manufacturer\u2019s instructions. Cell culture lysis reagent (5X) provided in the assay kit is diluted to working concentration. For a full 96-well plate, 11 mL luciferase assay reagent (100 uL per sample) and 4.6 mL lysis reagent (30 uL per sample) are recommended. These 2 reagents and PBS for washing (50 uL per sample, 6.5 mL sufficient for 96 samples) are filled in a NEST 1 Well Reservoir 195 mL.\n\n\n",
diff --git a/protoBuilds/sci-macherey-nagel-nucleomag/README.json b/protoBuilds/sci-macherey-nagel-nucleomag/README.json
index 601cc9e331..63be472b59 100644
--- a/protoBuilds/sci-macherey-nagel-nucleomag/README.json
+++ b/protoBuilds/sci-macherey-nagel-nucleomag/README.json
@@ -6,15 +6,15 @@
]
},
"deck-setup": "\nTip rack on Slot 4 is used for tip parking if selected.\n\n",
- "description": "Your OT-2 can fully automate the entire NucleoMag\u00ae Virus Viral DNA/RNA Isolation.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below:\n\nExplanation of complex parameters below:\n Number of samples: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n Deepwell type: Specify which well plate will be mounted on the magnetic module.\n Reservoir Type: Specify which reservoir will be employed.\n Starting Volume: Specify starting volume of sample (ul).\n* `: Specify the volume of binding buffer to use (ul).\n*Elution Volume: Specify elution volume (ul).\n*Park Tips: Specify whether to park tips or drop tips.\n*P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n",
+ "description": "Your OT-2 can fully automate the entire NucleoMag\u00ae Virus Viral DNA/RNA Isolation.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below:\n\nExplanation of complex parameters below:\n Number of samples: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n Deepwell type: Specify which well plate will be mounted on the magnetic module.\n Reservoir Type: Specify which reservoir will be employed.\n Starting Volume: Specify starting volume of sample (ul).\n Binding Buffer Volume: Specify the volume of binding buffer to use (ul).\n Elution Volume: Specify elution volume (ul).\n Park Tips: Specify whether to park tips or drop tips.\n P300 Multi Channel Pipette Mount: Specify whether the P300 multi channel pipette will be on the left or right mount.\n",
"internal": "sci-promega-magazorb-dna-mini-prep-kit",
"labware": "\nNEST 96 Wellplate 2mL\nUSA Scientific 96 Wellplate 2.4mL\nNEST 12 Reservoir 15mL\nUSA Scientific 12 Reservoir 22mL\nOpentrons 200uL Filter Tips\nOpentrons 96 Aluminum block Nest Wellplate 100ul\n",
"markdown": {
- "author": "[Opentrons (verified)](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-macherey-nagel-nucleomag). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons (verified)](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-macherey-nagel-nucleomag). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* DNA Extraction\n\n",
"deck-setup": "\n* Tip rack on Slot 4 is used for tip parking if selected.\n\n\n\n",
- "description": "Your OT-2 can fully automate the entire NucleoMag\u00ae Virus Viral DNA/RNA Isolation.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below:\n\n\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n* `Deepwell type`: Specify which well plate will be mounted on the magnetic module.\n* `Reservoir Type`: Specify which reservoir will be employed.\n* `Starting Volume`: Specify starting volume of sample (ul).\n* ``: Specify the volume of binding buffer to use (ul).\n* `Elution Volume`: Specify elution volume (ul).\n* `Park Tips`: Specify whether to park tips or drop tips.\n* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n\n---\n\n",
- "internal": "sci-promega-magazorb-dna-mini-prep-kit",
+ "description": "Your OT-2 can fully automate the entire NucleoMag\u00ae Virus Viral DNA/RNA Isolation.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below:\n\n\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n* `Deepwell type`: Specify which well plate will be mounted on the magnetic module.\n* `Reservoir Type`: Specify which reservoir will be employed.\n* `Starting Volume`: Specify starting volume of sample (ul).\n* `Binding Buffer Volume`: Specify the volume of binding buffer to use (ul).\n* `Elution Volume`: Specify elution volume (ul).\n* `Park Tips`: Specify whether to park tips or drop tips.\n* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n\n---\n\n",
+ "internal": "sci-promega-magazorb-dna-mini-prep-kit\n",
"labware": "* [NEST 96 Wellplate 2mL](https://shop.opentrons.com/collections/lab-plates/products/nest-0-2-ml-96-well-deep-well-plate-v-bottom)\n* [USA Scientific 96 Wellplate 2.4mL](https://labware.opentrons.com/?category=wellPlate)\n* [NEST 12 Reservoir 15mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [USA Scientific 12 Reservoir 22mL](https://labware.opentrons.com/?category=reservoir)\n* [Opentrons 200uL Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 96 Aluminum block Nest Wellplate 100ul](https://labware.opentrons.com/opentrons_96_aluminumblock_nest_wellplate_100ul?category=aluminumBlock)\n\n",
"modules": "* [Magnetic Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
diff --git a/protoBuilds/sci-mag-bind-blood-tissue-kit/README.json b/protoBuilds/sci-mag-bind-blood-tissue-kit/README.json
index f4b5070a14..1e5bdf5ce2 100644
--- a/protoBuilds/sci-mag-bind-blood-tissue-kit/README.json
+++ b/protoBuilds/sci-mag-bind-blood-tissue-kit/README.json
@@ -6,15 +6,15 @@
]
},
"deck-setup": "\nTip rack on Slot 4 is used for tip parking if selected.\n\n",
- "description": "Your OT-2 can fully automate the entire Mag-Bind\u00ae Blood & Tissue DNA HDQ 96 Kit.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below:\n\nExplanation of complex parameters below:\n Number of samples: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n Deepwell type: Specify which well plate will be mounted on the magnetic module.\n Reservoir Type: Specify which reservoir will be employed.\n Starting Volume: Specify starting volume of sample (ul).\n* `: Specify the volume of binding buffer to use (ul).\n*Elution Volume: Specify elution volume (ul).\n*Park Tips: Specify whether to park tips or drop tips.\n*P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n",
+ "description": "Your OT-2 can fully automate the entire Mag-Bind\u00ae Blood & Tissue DNA HDQ 96 Kit.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below:\n\nExplanation of complex parameters below:\n Number of samples: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n Deepwell type: Specify which well plate will be mounted on the magnetic module.\n Reservoir Type: Specify which reservoir will be employed.\n Starting Volume: Specify starting volume of sample (ul).\n Binding Buffer Volume: Specify the volume of binding buffer to use (ul).\n Elution Volume: Specify elution volume (ul).\n Park Tips: Specify whether to park tips or drop tips.\n P300 Multi Channel Pipette Mount: Specify whether the P300 multi channel pipette will be on the left or right mount.\n",
"internal": "sci-mag-bind-blood-tissue-kit",
"labware": "\nNEST 96 Wellplate 2mL\nUSA Scientific 96 Wellplate 2.4mL\nNEST 12 Reservoir 15mL\nUSA Scientific 12 Reservoir 22mL\nOpentrons 200uL Filter Tips\nOpentrons 96 Aluminum block Nest Wellplate 100ul\n",
"markdown": {
- "author": "[Opentrons (verified)](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-mag-bind-blood-tissue-kit). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons (verified)](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-mag-bind-blood-tissue-kit). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* DNA Extraction\n\n",
"deck-setup": "\n* Tip rack on Slot 4 is used for tip parking if selected.\n\n\n\n",
- "description": "Your OT-2 can fully automate the entire Mag-Bind\u00ae Blood & Tissue DNA HDQ 96 Kit.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below:\n\n\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n* `Deepwell type`: Specify which well plate will be mounted on the magnetic module.\n* `Reservoir Type`: Specify which reservoir will be employed.\n* `Starting Volume`: Specify starting volume of sample (ul).\n* ``: Specify the volume of binding buffer to use (ul).\n* `Elution Volume`: Specify elution volume (ul).\n* `Park Tips`: Specify whether to park tips or drop tips.\n* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n\n---\n\n",
- "internal": "sci-mag-bind-blood-tissue-kit",
+ "description": "Your OT-2 can fully automate the entire Mag-Bind\u00ae Blood & Tissue DNA HDQ 96 Kit.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below:\n\n\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n* `Deepwell type`: Specify which well plate will be mounted on the magnetic module.\n* `Reservoir Type`: Specify which reservoir will be employed.\n* `Starting Volume`: Specify starting volume of sample (ul).\n* `Binding Buffer Volume`: Specify the volume of binding buffer to use (ul).\n* `Elution Volume`: Specify elution volume (ul).\n* `Park Tips`: Specify whether to park tips or drop tips.\n* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n\n---\n\n",
+ "internal": "sci-mag-bind-blood-tissue-kit\n",
"labware": "* [NEST 96 Wellplate 2mL](https://shop.opentrons.com/collections/lab-plates/products/nest-0-2-ml-96-well-deep-well-plate-v-bottom)\n* [USA Scientific 96 Wellplate 2.4mL](https://labware.opentrons.com/?category=wellPlate)\n* [NEST 12 Reservoir 15mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [USA Scientific 12 Reservoir 22mL](https://labware.opentrons.com/?category=reservoir)\n* [Opentrons 200uL Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 96 Aluminum block Nest Wellplate 100ul](https://labware.opentrons.com/opentrons_96_aluminumblock_nest_wellplate_100ul?category=aluminumBlock)\n\n",
"modules": "* [Magnetic Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
diff --git a/protoBuilds/sci-mgi-mgieasy-nucleic-acid-extraction-kit/README.json b/protoBuilds/sci-mgi-mgieasy-nucleic-acid-extraction-kit/README.json
index a46230699e..41939bb2ef 100644
--- a/protoBuilds/sci-mgi-mgieasy-nucleic-acid-extraction-kit/README.json
+++ b/protoBuilds/sci-mgi-mgieasy-nucleic-acid-extraction-kit/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"MGI Kit"
@@ -9,7 +9,7 @@
"description": "This protocol automates the magnetic bead-based purification of nucleic acids from throat swabs or bronchoalveolar lavage for up to 96 samples using the MGI MGIEasy Nucleic Acid Extraction Kit.\nLinks:\n* MGI MGIEasy Nucleic Acid Extraction Kit\nThis protocol was developed to perform the following steps using the MGI MGIEasy Nucleic Acid Extraction Kit: Sample lysis and bead binding, three wash steps, then elution of purified nucleic acids from the beads.",
"internal": "sci-mgi-mgieasy-nucleic-acid-extraction-kit",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * MGI Kit\n\n",
"deck-setup": "\n\n* Opentrons 200 ul filter tips (Deck Slots 4, 5, 7, 8, 10, 11)\n* Opentrons Temperature Module (cooled to 4 degrees C) with elution plate opentrons_96_aluminumblock_nest_wellplate_100ul (Deck Slot 1)\n* Reagent Reservoirs nest_12_reservoir_15ml (Deck Slots 2, 3)\n* Liquid Waste Reservoir nest_1_reservoir_195ml (Deck Slot 9)\n* Opentrons Magnetic Module with deep well plate nest_96_wellplate_2ml_deep (Deck Slot 6)\n\n\n\n* For sample traceability and consistency, samples are mapped directly from the magnetic extraction plate to the elution plate (well A1 is transferred to well A1, B1 to B1, etc.)\n* If the tip parking feature is used (recommended), the protocol will conserve tips between reagent additiona and removal. Tips will be temporarily stored in tiprack wells corresponding to the well of the sample they access (tip parked in A1 will only be used for sample in A1). If not using the tip parking feature, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for higher throughput runs.\n\n",
"description": "\nThis protocol automates the magnetic bead-based purification of nucleic acids from throat swabs or bronchoalveolar lavage for up to 96 samples using the [MGI MGIEasy Nucleic Acid Extraction Kit](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/sci-mgi-mgieasy-nucleic-acid-extraction-kit/MGIEasy+Nucleic+Acid+Extraction+Kit+User+Manual.pdf).\n\nLinks:\n* [MGI MGIEasy Nucleic Acid Extraction Kit](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/sci-mgi-mgieasy-nucleic-acid-extraction-kit/MGIEasy+Nucleic+Acid+Extraction+Kit+User+Manual.pdf)\n\nThis protocol was developed to perform the following steps using the MGI MGIEasy Nucleic Acid Extraction Kit: Sample lysis and bead binding, three wash steps, then elution of purified nucleic acids from the beads.\n\n",
diff --git a/protoBuilds/sci-neb-next-ultra/README.json b/protoBuilds/sci-neb-next-ultra/README.json
index 3d038e534a..352dd3c890 100644
--- a/protoBuilds/sci-neb-next-ultra/README.json
+++ b/protoBuilds/sci-neb-next-ultra/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"NEBNext Ultra II"
@@ -10,7 +10,7 @@
"internal": "sci-neb-next-ultra",
"labware": "\nNEST 96-Well PCR Plates\nNEST 12-Well Reservoir or NEST 96-Well Deepwell Plate (for more information, please see this guide)\nOpentrons 200\u00b5L Filter Tips\nOpentrons 20\u00b5L Filter Tips\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t* NEBNext Ultra II\n\n",
"deck-setup": "Please see our guide for a detailed explanation of deck setup. [Link to guide](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/sci-neb-next-ultra/NEBNext+Ultra+II+-+Protocol+Library+Readme.pdf)\n\n",
"description": "This protocol automates the [NEBNext\u00ae Ultra\u2122 II DNA Library Prep Kit for Illumina\u00ae](https://www.neb.com/products/e7645-nebnext-ultra-ii-dna-library-prep-kit-for-illumina#Product%20Information) on the OT-2.\n\nThis protocol was developed internally by Opentrons for use on the OT-2 and provides several options. Users can select how many samples to prepare, whether or not to use on-deck modules (like our thermocycler), and whether or not to perform certain steps in the process. Additionally, users can select to enable a \"Test Mode\" that is great for performing a dry run of the protocol.\n\nFor a more detailed explanation of this protocol, including deck setup and reagent setup, please see this guide: [link to guide](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/sci-neb-next-ultra/NEBNext+Ultra+II+-+Protocol+Library+Readme.pdf)\n\nExplanation of complex parameters below:\n* **Number of Samples**: Select the number of samples to prepare\n* **Test Mode (dry run)**: Select whether to perform a dry run (Yes) instead of a sample run\n* **Use Modules**: Select whether or not you are using our Magnetic Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) and [Temperature Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* **Reuse Tips**: Select whether or not to reuse tips for certain steps (selecting yes will reuse tips in a sterile way, saving on the amount of tips used and eliminating the need to replace tips during the run)\n* **Use Thermocycler**: Select whether or not you will use our on-deck GEN1 thermocycler module. If not using (select \"No\"), the user will be prompted to move the sample plate to an off-deck thermocycler.\n* **Use Opentrons Offsets**: Select whether to use Z offsets used by the Opentrons Science Team (\"Yes\") or use the default Z heights of the labware (\"No\")\n* **Perform ERAT Step**: Select whether or not to perform the ERAT step.\n* **Perform Ligation Step**: Select whether or not to perform the Ligation step.\n* **Perform Post-Ligation Step**: Select whether or not to perform the Post-Ligtion step.\n* **Perform PCR Step**: Select whether or not to perform the PCR step.\n* **Perform Post-PCR Step**: Select whether or not to perform the Post-PCR step.\n\n---\n\n",
diff --git a/protoBuilds/sci-omegabiotek-extraction-fa/README.json b/protoBuilds/sci-omegabiotek-extraction-fa/README.json
index 60d494e828..85ad17bc52 100644
--- a/protoBuilds/sci-omegabiotek-extraction-fa/README.json
+++ b/protoBuilds/sci-omegabiotek-extraction-fa/README.json
@@ -10,7 +10,7 @@
"internal": "sci-omegabiotek-extraction",
"labware": "\nNEST 96 Wellplate 2mL\nUSA Scientific 96 Wellplate 2.4mL\nNEST 12 Reservoir 15mL\nUSA Scientific 12 Reservoir 22mL\nOpentrons 96 tiprack 300ul\nOpentrons 96 Aluminum block Nest Wellplate 100ul\n",
"markdown": {
- "author": "[Opentrons (verified)](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-omegabiotek-extraction-fa). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons (verified)](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-omegabiotek-extraction-fa). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* RNA Extraction\n\n",
"deck-setup": "\n\n\n",
"description": "After lysing samples, your OT-2 can fully automate the entire Omega Bio-tek Mag-Bind\u00ae Blood & Tissue DNA HDQ 96 Kit. Buffer systems tailored specifically for each type of starting material are added to samples to undergo lysis. Samples are then mixed with HDQ Binding Buffer and Mag-Bind\u00ae Particles HDQ to bind magnetic beads to DNA. DNA is eluted in the Elution Buffer after rapid wash steps.\n\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below: \n\n\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n* `Deepwell type`: Specify which well plate will be mounted on the magnetic module.\n* `Reservoir Type`: Specify which reservoir will be employed.\n* `Starting Volume`: Specify starting volume of sample (ul).\n* `Elution Volume`: Specify elution volume (ul).\n* `Park Tips`: Specify whether to park tips or drop tips.\n* `Mag Deck Generation`: Specify whether GEN1 or GEN2 magnetic module will be used.\n* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n\n\n---\n\n",
diff --git a/protoBuilds/sci-omegabiotek-extraction/README.json b/protoBuilds/sci-omegabiotek-extraction/README.json
index f3260a78cc..b42fe0c0ef 100644
--- a/protoBuilds/sci-omegabiotek-extraction/README.json
+++ b/protoBuilds/sci-omegabiotek-extraction/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons (verified)",
+ "author": "Opentrons (verified)\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"RNA Extraction"
@@ -10,7 +10,7 @@
"internal": "sci-omegabiotek-extraction",
"labware": "\nNEST 96 Wellplate 2mL\nUSA Scientific 96 Wellplate 2.4mL\nNEST 12 Reservoir 15mL\nUSA Scientific 12 Reservoir 22mL\nOpentrons 96 tiprack 300ul\nOpentrons 96 Aluminum block Nest Wellplate 100ul\n",
"markdown": {
- "author": "[Opentrons (verified)](https://opentrons.com/)\n\n",
+ "author": "[Opentrons (verified)](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* RNA Extraction\n\n",
"deck-setup": "\n\n\n",
"description": "After lysing samples, your OT-2 can fully automate the entire Omega Bio-tek Mag-Bind\u00ae Blood & Tissue DNA HDQ 96 Kit. Buffer systems tailored specifically for each type of starting material are added to samples to undergo lysis. Samples are then mixed with HDQ Binding Buffer and Mag-Bind\u00ae Particles HDQ to bind magnetic beads to DNA. DNA is eluted in the Elution Buffer after rapid wash steps.\n\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below: \n\n\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n* `Deepwell type`: Specify which well plate will be mounted on the magnetic module.\n* `Reservoir Type`: Specify which reservoir will be employed.\n* `Starting Volume`: Specify starting volume of sample (ul).\n* `Elution Volume`: Specify elution volume (ul).\n* `Park Tips`: Specify whether to park tips or drop tips.\n* `Mag Deck Generation`: Specify whether GEN1 or GEN2 magnetic module will be used.\n* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n\n\n---\n\n",
diff --git a/protoBuilds/sci-omegabiotek-magbind-temp/README.json b/protoBuilds/sci-omegabiotek-magbind-temp/README.json
index b9f600a7fc..dfefee649f 100644
--- a/protoBuilds/sci-omegabiotek-magbind-temp/README.json
+++ b/protoBuilds/sci-omegabiotek-magbind-temp/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons (verified)",
+ "author": "Opentrons (verified)\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"RNA Extraction"
@@ -10,7 +10,7 @@
"internal": "sci-omegabiotek-magbind",
"labware": "\nNEST 96 Wellplate 2mL\nNEST 1 Reservoir 195mL\nNEST 12 Reservoir 15mL\nOpentrons 96 tiprack 300ul\nOpentrons 96 Aluminum block Nest Wellplate 100ul\n",
"markdown": {
- "author": "[Opentrons (verified)](https://opentrons.com/)\n\n",
+ "author": "[Opentrons (verified)](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* RNA Extraction\n\n",
"deck-setup": "Tip rack on Slot 5 is used for tip parking if selected. If not tip parking, place [200ul Opentrons Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips) in Slot 5.\n\n\n",
"description": "Your OT-2 can automate the Mag-Bind\u00ae Viral DNA/RNA 96 Kit. Please see the kit description below found on the [kit website]((https://www.omegabiotek.com/product/mag-bind-viral-dna-rna-96-kit/):\n\n\"Mag-Bind\u00ae Viral DNA/RNA Kit is designed for the rapid and reliable isolation of viral RNA and viral DNA from serum, swabs, plasma, saliva, and other body fluids. The Mag-Bind\u00ae magnetic beads technology enables purification of high-quality nucleic acids that are free of proteins, nucleases, and other impurities. In addition to easily being adapted with automated systems, this procedure can also be scaled up or down depending on the amount of starting sample. The purified nucleic acids are ready for direct use in downstream applications such as amplification or other enzymatic reactions.\"\n\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below: \n\n\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n* `Starting Volume`: Specify starting volume of sample (ul).\n* `Elution Volume`: Specify elution volume (ul).\n* `Park Tips`: Specify whether to park tips or drop tips.\n* `Mag Deck Generation`: Specify whether GEN1 or GEN2 magnetic module will be used.\n* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n\n\n---\n\n",
diff --git a/protoBuilds/sci-omegabiotek-magbind-total-rna-96/README.json b/protoBuilds/sci-omegabiotek-magbind-total-rna-96/README.json
index dcff40d8ef..80df1ecf89 100644
--- a/protoBuilds/sci-omegabiotek-magbind-total-rna-96/README.json
+++ b/protoBuilds/sci-omegabiotek-magbind-total-rna-96/README.json
@@ -10,7 +10,7 @@
"internal": "sci-omegabiotek-magbind-total-rna-96",
"labware": "\nNEST 96 Wellplate 2mL\nUSA Scientific 96 Wellplate 2.4mL\nNEST 12 Reservoir 15mL\nUSA Scientific 12 Reservoir 22mL\nOpentrons 96 tiprack 300ul\nOpentrons 96 Aluminum block Nest Wellplate 100ul\n",
"markdown": {
- "author": "[Opentrons (verified)](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-omegabiotek-magbind-total-rna-96). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons (verified)](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-omegabiotek-magbind-total-rna-96). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* RNA Extraction\n\n",
"deck-setup": "\n\n\nSaliva: add 200uL of saliva\n\nBacteria culture: spin down 200uL of culture, wash once in PBS, resuspend in 200uL of chilled PBS\n\n200uL of sample + 200uL of lysis buffer. Mix thoroughly, add to deep well plate\n\nDnase 1 treatment: 49uL of buffer + 1uL of DNAse 1 per sample.\n\n\n\n\n---\n\n",
"description": "Your OT-2 can automate the Mag-Bind\u00ae Viral DNA/RNA 96 Kit. Please see the kit description below found on the [kit website](https://www.omegabiotek.com/product/mag-bind-total-rna-96-kit/):\n\n\"The Mag-Bind\u00ae Total RNA 96 Kit provides a novel technology for total RNA isolation. This kit allows the rapid and reliable isolation of high-quality total cellular RNA and viral RNA from a wide variety of cells and tissues. Unlike column-based systems, the binding of nucleic acids to magnetic particles occurs in solution resulting in increased binding kinetics and binding efficiency. Particles are also completely re-suspended during the wash steps of the purification protocol, which improves the removal of contaminants and increases nucleic acid purity. Mag-Bind\u00ae Total RNA 96 Kit procedure can be fully automated with most robotic workstations.\"\n\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below: \n\n\n\n\n\n\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n* `Deepwell type`: Specify which well plate will be mounted on the magnetic module.\n* `Reservoir Type`: Specify which reservoir will be employed.\n* `Starting Volume`: Specify starting volume of sample (ul).\n* `Binding Buffer Volume`: Specify binding buffer volume (ul).\n* `Wash Volumes`: Specify each of the three wash volumes (ul).\n* `Elution Volume`: Specify elution volume (ul).\n* `Settling Time`: Specify settling time for beads (minutes).\n* `Mag Deck Generation`: Specify whether GEN1 or GEN2 magnetic module will be used.\n* `Park Tips`: Specify whether to park tips or drop tips.\n* `Track Tips`: Specify whether to track tips between runs (starting with fresh tips or pick up from last runs tips).\n* `Flash`: Specify whether to flash OT-2 lights when the protocol runs out of tips, prompting the user to replenish tips.\n* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n\n\n---\n\n",
diff --git a/protoBuilds/sci-omegabiotek-magbind/README.json b/protoBuilds/sci-omegabiotek-magbind/README.json
index 5458a603f7..9713283f28 100644
--- a/protoBuilds/sci-omegabiotek-magbind/README.json
+++ b/protoBuilds/sci-omegabiotek-magbind/README.json
@@ -10,7 +10,7 @@
"internal": "sci-omegabiotek-magbind",
"labware": "\nNEST 96 Wellplate 2mL\nNEST 1 Reservoir 195mL\nNEST 12 Reservoir 15mL\nOpentrons 96 tiprack 300ul\nOpentrons 96 Aluminum block Nest Wellplate 100ul\n",
"markdown": {
- "author": "[Opentrons (verified)](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-omegabiotek-magbind). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons (verified)](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-omegabiotek-magbind). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* RNA Extraction\n\n",
"deck-setup": "Tip rack on Slot 5 is used for tip parking if selected. If not tip parking, place [200ul Opentrons Filter Tip Rack](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips) in Slot 5.\n\n\n",
"description": "Your OT-2 can automate the Mag-Bind\u00ae Viral DNA/RNA 96 Kit. Please see the kit description below found on the [kit website]((https://www.omegabiotek.com/product/mag-bind-viral-dna-rna-96-kit/):\n\n\"Mag-Bind\u00ae Viral DNA/RNA Kit is designed for the rapid and reliable isolation of viral RNA and viral DNA from serum, swabs, plasma, saliva, and other body fluids. The Mag-Bind\u00ae magnetic beads technology enables purification of high-quality nucleic acids that are free of proteins, nucleases, and other impurities. In addition to easily being adapted with automated systems, this procedure can also be scaled up or down depending on the amount of starting sample. The purified nucleic acids are ready for direct use in downstream applications such as amplification or other enzymatic reactions.\"\n\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below: \n\n\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n* `Starting Volume`: Specify starting volume of sample (ul).\n* `Elution Volume`: Specify elution volume (ul).\n* `Park Tips`: Specify whether to park tips or drop tips.\n* `Mag Deck Generation`: Specify whether GEN1 or GEN2 magnetic module will be used.\n* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n\n\n---\n\n",
diff --git a/protoBuilds/sci-phytip-neutralization/README.json b/protoBuilds/sci-phytip-neutralization/README.json
index df2549f8bb..6fe66d4ffd 100644
--- a/protoBuilds/sci-phytip-neutralization/README.json
+++ b/protoBuilds/sci-phytip-neutralization/README.json
@@ -17,7 +17,7 @@
"internal": "sci-phytip-neutralization\n",
"labware": "* Thermo Scientific 96 Well Plate V Bottom 450 uL #249944/249946\n* [Opentrons 96 Tip Rack 300 \u00b5L](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons 15 Tube Rack with NEST 15 mL Conical](https://shop.opentrons.com/collections/opentrons-tips/products/tube-rack-set-1)\n\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n\n",
- "partner": "[Biotage](https://www.biotage.com/)\n\n\n",
+ "partner": "[Biotage](https://www.biotage.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [Opentrons P300 Single Channel Electronic Pipette (GEN2)](https://shop.opentrons.com/single-channel-electronic-pipette-p20/)\n\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit \"Run\".\n\n\n",
"protocol-steps": "1. Pick up a tip (slot 6)\n2. Transfer neutralization buffer (slot 10) to each well containing the eluate (slot 11)\n\n\n",
@@ -25,7 +25,7 @@
"title": "Phytip Protein A, ProPlus, ProPlus LX Columns - Neutralization"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Biotage",
+ "partner": "Biotage\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nOpentrons P300 Single Channel Electronic Pipette (GEN2)\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit \"Run\".\n",
"protocol-steps": "\nPick up a tip (slot 6)\nTransfer neutralization buffer (slot 10) to each well containing the eluate (slot 11)\n",
diff --git a/protoBuilds/sci-phytip-plate-prep/README.json b/protoBuilds/sci-phytip-plate-prep/README.json
index fa740817b6..761c6753c0 100644
--- a/protoBuilds/sci-phytip-plate-prep/README.json
+++ b/protoBuilds/sci-phytip-plate-prep/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Protein Purification": [
"Protein A, Pro Plus, Pro Plus LX Columns"
@@ -10,7 +10,7 @@
"internal": "sci-phytip-plate-prep",
"labware": "\nThermo Scientific 96 Well Plate V Bottom 450 uL #249944/249946\nOpentrons 96 Tip Rack 300 \u00b5L\nNEST 12 Well Reservoir 15 mL #360102\nOpentrons Fixed Trash\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Protein Purification\n\t* Protein A, Pro Plus, Pro Plus LX Columns\n\n\n",
"deck-setup": "Slot 5 \u2013 96-well V-bottom plate \u2013 1st wash\nSlot 6 - Tiprack1\nSlot 7 - 96-well V-bottom plate - 2nd wash\nSlot 8 \u2013 12-well reagent stock reservoir\n* Green \u2013 equilibration buffer (well #1 and well #2)\n* Blue \u2013 wash buffer 1 (well #3 and well #4)\n* Pink \u2013 wash buffer 2 (well #5 and well #6)\n* Purple \u2013 elution buffer (well #7 and well #8)\n\nSlot 9 96-well V-bottom plate - equilibration\nSlot 11 - 96-well V-bottom plate - elution\n\n\n\n\n",
"description": "This protocol (Plate Prep) performs pipetting to transfer reagents (equilibration buffer, wash buffer 1, wash buffer 2 and elution buffer) from a 12-well reagent stock reservoir to 96-well V-bottom plates (the reagent plates) on the OT-2. These reagent plates are used for the protein purification protocol of Phytip\u00ae Protein A, ProPlus or ProPlus LX Columns.\n\nThe protocol is developed to prepare sufficient reagents to process up to 96 samples (a full 96-well plate).\n\n\n\n",
diff --git a/protoBuilds/sci-phytip-protein-A/README.json b/protoBuilds/sci-phytip-protein-A/README.json
index 4be81b1862..e6bc5008bf 100644
--- a/protoBuilds/sci-phytip-protein-A/README.json
+++ b/protoBuilds/sci-phytip-protein-A/README.json
@@ -17,7 +17,7 @@
"internal": "sci-phytip-protein-A\n",
"labware": "* [Thermo Fisher Nunc\u2122 96-Well Polypropylene Storage Microplates](https://www.thermofisher.com/order/catalog/product/249946?SID=srch-hj-249946)\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/nest-12-well-reservoirs-15-ml/)\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/opentrons-300ul-tips-1000-refills/)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Biotage](https://www.biotage.com/)\n\n",
+ "partner": "[Biotage](https://www.biotage.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P300 8-Channel GEN2](https://opentrons.com/pipettes/)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Equilibration buffer from reagent stock reservoir (slot 8) is transferred to the \u201cEquilibration\u201d plate (slot 9) by the 8-channel pipette (200 uL per well).\n2. Wash buffer 1 from reagent stock reservoir (slot 8) is transferred to the \u201c1st wash\u201d plate (slot 5) by the 8-channel pipette (200 uL per well).\n3. Wash buffer 2 from reagent stock reservoir (slot 8) is transferred to the \u201c2nd wash\u201d plate (slot 7) by the 8-channel pipette (200 uL per well).\n4. Elution buffer from reagent stock reservoir (slot 8) is transferred to the \u201cElution\u201d plate (slot 11) by the 8-channel pipette (80 uL per well).\n\n\n",
@@ -26,7 +26,7 @@
"title": "Phytip Protein A, ProPlus, ProPlus LX Columns - Plate Prep"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Biotage",
+ "partner": "Biotage\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP300 8-Channel GEN2\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nEquilibration buffer from reagent stock reservoir (slot 8) is transferred to the \u201cEquilibration\u201d plate (slot 9) by the 8-channel pipette (200 uL per well).\nWash buffer 1 from reagent stock reservoir (slot 8) is transferred to the \u201c1st wash\u201d plate (slot 5) by the 8-channel pipette (200 uL per well).\nWash buffer 2 from reagent stock reservoir (slot 8) is transferred to the \u201c2nd wash\u201d plate (slot 7) by the 8-channel pipette (200 uL per well).\nElution buffer from reagent stock reservoir (slot 8) is transferred to the \u201cElution\u201d plate (slot 11) by the 8-channel pipette (80 uL per well).\n",
diff --git a/protoBuilds/sci-phytip-protein-puri/README.json b/protoBuilds/sci-phytip-protein-puri/README.json
index 5efa3490c2..6f31de25e0 100644
--- a/protoBuilds/sci-phytip-protein-puri/README.json
+++ b/protoBuilds/sci-phytip-protein-puri/README.json
@@ -17,7 +17,7 @@
"internal": "sci-phytip-protein-puri\n",
"labware": "* [Thermo Fisher Nunc\u2122 96-Well Polypropylene Storage Microplates](https://www.thermofisher.com/order/catalog/product/249946?SID=srch-hj-249946)\n* [Empty Opentrons 300\u00b5L Tip Rack for PhyTip\u00ae Columns](https://shop.opentrons.com/opentrons-300ul-tips-1000-refills/)\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Biotage](https://www.biotage.com/)\n\n",
+ "partner": "[Biotage](https://www.biotage.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [P300 8-Channel GEN2](https://opentrons.com/pipettes/)\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. Pick up PhyTip\u00ae Columns (slot 3).\n2. Equilibrate \u2013 passing equilibration buffer (slot 9) over the resin bed for 4 cycles. In each cycle, the pipetting is programmed to aspirate and dispense 180 uL at 240 uL/min.\n3. Capture \u2013 pass the sample (slot 2) over the resin bed for 8 cycles. In each cycle, the pipetting is programmed to aspirate and dispense 180 uL at 240 uL/min.\n4. Wash \u2013 pass wash buffer 1 (slot 5) over the resin bed for 2 cycles. In each cycle, the pipetting is programmed to aspirate and dispense 180 uL at 480 uL/min.\n5. Wash again \u2013 pass wash buffer 2 (slot 7) over the resin bed for 2\ncycles. In each cycle, the pipetting is programmed to aspirate and\ndispense 180 uL at 480 uL/min.\n6. Elute - pass elution buffer (slot 11) over the resin bed for 4 cycles. In each cycle, the pipetting is programmed to aspirate and dispense 130 uL at 240 uL/min.\n\n\n",
@@ -26,7 +26,7 @@
"title": "Phytip Protein A, ProPlus, ProPlus LX Columns - Protein Purification"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Biotage",
+ "partner": "Biotage\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nP300 8-Channel GEN2\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nPick up PhyTip\u00ae Columns (slot 3).\nEquilibrate \u2013 passing equilibration buffer (slot 9) over the resin bed for 4 cycles. In each cycle, the pipetting is programmed to aspirate and dispense 180 uL at 240 uL/min.\nCapture \u2013 pass the sample (slot 2) over the resin bed for 8 cycles. In each cycle, the pipetting is programmed to aspirate and dispense 180 uL at 240 uL/min.\nWash \u2013 pass wash buffer 1 (slot 5) over the resin bed for 2 cycles. In each cycle, the pipetting is programmed to aspirate and dispense 180 uL at 480 uL/min.\nWash again \u2013 pass wash buffer 2 (slot 7) over the resin bed for 2\ncycles. In each cycle, the pipetting is programmed to aspirate and\ndispense 180 uL at 480 uL/min.\nElute - pass elution buffer (slot 11) over the resin bed for 4 cycles. In each cycle, the pipetting is programmed to aspirate and dispense 130 uL at 240 uL/min.\n",
diff --git a/protoBuilds/sci-pierce-ninta-magnetic-beads-part-2/README.json b/protoBuilds/sci-pierce-ninta-magnetic-beads-part-2/README.json
index a6509a2d2b..021be36c4a 100644
--- a/protoBuilds/sci-pierce-ninta-magnetic-beads-part-2/README.json
+++ b/protoBuilds/sci-pierce-ninta-magnetic-beads-part-2/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Protein Purification": [
"Thermo Fisher Pierce Ni-NTA Magnetic Agarose Beads"
@@ -9,7 +9,7 @@
"description": "This protocol (Part 2) performs washing and elution on the OT2 as the second part of automated process of His-tagged protein purification using Pierce Ni-NTA Magnetic Agarose Beads.\nThe user can determine the number of samples to be processed, and if desired, a second elution step can be performed to increase recovery.\nThe sample containing His-tagged protein (target protein) and Ni-NTA magnetic beads (beads) will be transferred to and mixed in a 96-well deepwell working plate on the OT2. The first part of the protocol completes at this point allowing the user to move the working plate to an agitation device, a beads/target protein incubation period determined by the user. After incubation, the process proceeds with the second part of the protocol on the OT2. Beads/target protein complex will be washed, and target protein eluted for further analysis.\nLinks:\n* Opentrons Protocol part-1: Pierce NiNTA Magnetic Beads\n\nOpentrons Protocol part-2: Pierce NiNTA Magnetic Beads: part-2\n\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nWash buffer (50 mM sodium phosphate, 300 mM sodium chloride, 40 mM imidazole, pH 7.4)\n\nElution buffer (50 mM sodium phosphate, 300 mM sodium chloride, 300 mM imidazole, pH 7.4)\n\n\nOpentrons OT-2\n\n\nOpentrons OT-2 Run App (Version 3.19.0 or later)\n\n\nMagnetic Module GEN2\n\n\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\n\nNEST 1-Well Reservoirs, 195 mL\n\nNEST 12-Well Reservoirs, 15 mL\n\n\nOpentrons 300\u00b5L Tips\n\n\nP300 8-Channel GEN2\n\n\n\n",
"internal": "sci-pierce-ninta-magnetic-beads-part-2",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Protein Purification\n * Thermo Fisher Pierce Ni-NTA Magnetic Agarose Beads\n\n",
"deck-setup": "\n* Slot 1 - Magnetic module/sample + beads (red) in 96 well deepwell plate (working plate)\n* Slot 2 \u2013 Elution buffer in 12 well reservoir (orange)\n* Slot 3 - 96 well deepwell plate (final plate)\n* Slot 4 \u2013 Wash buffer in 12 well reservoir (purple)\n* Slot 5 \u2013 Tiprack1 (Note: this tiprack is assigned for use in wash steps. The tips are returned to the tiprack and reused and will not be discarded.)\n* Slot 6 \u2013 Tiprack2 (Note: this tiprack is assigned for use in elution steps. The tips are returned to the tiprack and reused and will not be discarded.)\n\n\n\n* Slot 7 \u2013 Tiprack3\n* Slot 8 \u2013 Liquid waste\n\n\n\nWash buffer: 500 uL per sample per wash, i.e. 1,000 uL for 2 washes per sample (Note: Each well should contain at least 8,000 uL wash buffer, sufficient for 2 washing runs of 8 samples)\nElution buffer: 250 uL per sample per wash or 500 uL for 2 elutions per sample (Note: each well should contain at least 2,000 uL for 1 elution (or 4,000 uL for 2 elutions) of 8 samples)\n\n\n\n",
"description": "\nThis protocol (Part 2) performs washing and elution on the OT2 as the second part of automated process of His-tagged protein purification using Pierce Ni-NTA Magnetic Agarose Beads.\n\nThe user can determine the number of samples to be processed, and if desired, a second elution step can be performed to increase recovery.\n\nThe sample containing His-tagged protein (target protein) and Ni-NTA magnetic beads (beads) will be transferred to and mixed in a 96-well deepwell working plate on the OT2. The first part of the protocol completes at this point allowing the user to move the working plate to an agitation device, a beads/target protein incubation period determined by the user. After incubation, the process proceeds with the second part of the protocol on the OT2. Beads/target protein complex will be washed, and target protein eluted for further analysis.\n\nLinks:\n* [Opentrons Protocol part-1: Pierce NiNTA Magnetic Beads](https://protocols.opentrons.com/protocol/sci-pierce-ninta-magnetic-beads)\n\n* [Opentrons Protocol part-2: Pierce NiNTA Magnetic Beads: part-2](https://protocols.opentrons.com/protocol/sci-pierce-ninta-magnetic-beads-part-2)\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* Wash buffer (50 mM sodium phosphate, 300 mM sodium chloride, 40 mM imidazole, pH 7.4)\n* Elution buffer (50 mM sodium phosphate, 300 mM sodium chloride, 300 mM imidazole, pH 7.4)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n\n* [Magnetic Module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n\n* [NEST 2 mL 96-Well Deep Well Plate, V Bottom](https://shop.opentrons.com/nest-2-ml-96-well-deep-well-plate-v-bottom/)\n* [NEST 1-Well Reservoirs, 195 mL](https://shop.opentrons.com/nest-1-well-reservoirs-195-ml/)\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/nest-12-well-reservoirs-15-ml/)\n\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/opentrons-300ul-tips-1000-refills/)\n\n* [P300 8-Channel GEN2](https://opentrons.com/pipettes/)\n\n---\n\n\n",
diff --git a/protoBuilds/sci-pierce-ninta-magnetic-beads/README.json b/protoBuilds/sci-pierce-ninta-magnetic-beads/README.json
index 80fc409877..9b085943c3 100644
--- a/protoBuilds/sci-pierce-ninta-magnetic-beads/README.json
+++ b/protoBuilds/sci-pierce-ninta-magnetic-beads/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Protein Purification": [
"Thermo Fisher Pierce Ni-NTA Magnetic Agarose Beads"
@@ -9,7 +9,7 @@
"description": "This protocol (Part 1) performs pipetting and mixing of reagents and samples on the OT2 as the first part of automated process of His-tagged protein purification using Pierce Ni-NTA Magnetic Agarose Beads.\nThe user can determine the number of samples to be processed, and if desired, a second elution step can be performed to increase recovery.\nThe sample containing His-tagged protein (target protein) and Ni-NTA magnetic beads (beads) will be transferred to and mixed in a 96-well deepwell working plate on the OT2. The first part of the protocol completes at this point allowing the user to move the working plate to an agitation device, a beads/target protein incubation period determined by the user. After incubation, the process proceeds with the second part of the protocol on the OT2. Beads/target protein complex will be washed, and target protein eluted for further analysis.\nLinks:\n* Opentrons Protocol part-1: Pierce NiNTA Magnetic Beads\n\nOpentrons Protocol part-2: Pierce NiNTA Magnetic Beads: part-2\n\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nThermo Fisher Pierce Ni-NTA Magnetic Agarose Beads\n\nEquilibration buffer (Equilibration buffer (50 mM sodium phosphate, 300 mM sodium chloride, 20 mM imidazole, pH 7.4)\n\n\nOpentrons OT-2\n\n\nOpentrons OT-2 Run App (Version 3.19.0 or later)\n\n\nMagnetic Module GEN2\n\n\nNEST 2 mL 96-Well Deep Well Plate, V Bottom\n\nNEST 1-Well Reservoirs, 195 mL\nNEST 12-Well Reservoirs, 15 mL\n4-in-1 Tube Rack Set\n\nNEST 15 mL Centrifuge Tube\n\n\nOpentrons 300\u00b5L Tips\n\n\nP300 Single-Channel GEN2\n\nP300 8-Channel GEN2\n\n\n",
"internal": "sci-pierce-ninta-magnetic-beads",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Protein Purification\n * Thermo Fisher Pierce Ni-NTA Magnetic Agarose Beads\n\n",
"deck-setup": "\n* Slot 1 - Magnetic module/96 deepwell plate (working plate)\n* Slot 4 \u2013 Equilibration buffer in 12 well reservoir (blue)\n* Slot 5 - Tube rack/beads in 15 mL conical tube (Green)\n* Slot 7 - Samples in 96 well deepwell plate (red)\n* Slot 8 \u2013 Liquid waste\n* Slot 9 \u2013 Tiprack1\n* Slot 10 \u2013 Tiprack2\n\n\n\n* Beads: 50 uL per sample\n* Equilibration buffer: 450 uL and 500 uL per sample\n\n\n\n\n",
"description": "\nThis protocol (Part 1) performs pipetting and mixing of reagents and samples on the OT2 as the first part of automated process of His-tagged protein purification using Pierce Ni-NTA Magnetic Agarose Beads.\n\nThe user can determine the number of samples to be processed, and if desired, a second elution step can be performed to increase recovery.\n\nThe sample containing His-tagged protein (target protein) and Ni-NTA magnetic beads (beads) will be transferred to and mixed in a 96-well deepwell working plate on the OT2. The first part of the protocol completes at this point allowing the user to move the working plate to an agitation device, a beads/target protein incubation period determined by the user. After incubation, the process proceeds with the second part of the protocol on the OT2. Beads/target protein complex will be washed, and target protein eluted for further analysis.\n\nLinks:\n* [Opentrons Protocol part-1: Pierce NiNTA Magnetic Beads](https://protocols.opentrons.com/protocol/sci-pierce-ninta-magnetic-beads)\n\n* [Opentrons Protocol part-2: Pierce NiNTA Magnetic Beads: part-2](https://protocols.opentrons.com/protocol/sci-pierce-ninta-magnetic-beads-part-2)\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Thermo Fisher Pierce Ni-NTA Magnetic Agarose Beads](https://www.thermofisher.com/order/catalog/product/78606)\n* Equilibration buffer (Equilibration buffer (50 mM sodium phosphate, 300 mM sodium chloride, 20 mM imidazole, pH 7.4)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n\n* [Magnetic Module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n\n* [NEST 2 mL 96-Well Deep Well Plate, V Bottom](https://shop.opentrons.com/nest-2-ml-96-well-deep-well-plate-v-bottom/)\n* [NEST 1-Well Reservoirs, 195 mL](https://shop.opentrons.com/nest-1-well-reservoirs-195-ml/)\n* [NEST 12-Well Reservoirs, 15 mL](https://shop.opentrons.com/nest-12-well-reservoirs-15-ml/)\n* [4-in-1 Tube Rack Set](https://shop.opentrons.com/4-in-1-tube-rack-set/)\n* [NEST 15 mL Centrifuge Tube](https://shop.opentrons.com/nest-15-ml-centrifuge-tube/)\n\n* [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/opentrons-300ul-tips-1000-refills/)\n\n* [P300 Single-Channel GEN2](https://opentrons.com/pipettes/)\n* [P300 8-Channel GEN2](https://opentrons.com/pipettes/)\n\n---\n\n\n",
diff --git a/protoBuilds/sci-promega-magazorb-dna-mini-prep-kit/README.json b/protoBuilds/sci-promega-magazorb-dna-mini-prep-kit/README.json
index 099dac92de..29da383070 100644
--- a/protoBuilds/sci-promega-magazorb-dna-mini-prep-kit/README.json
+++ b/protoBuilds/sci-promega-magazorb-dna-mini-prep-kit/README.json
@@ -6,14 +6,14 @@
]
},
"deck-setup": "\nTip rack on Slot 4 is used for tip parking if selected.\n\n",
- "description": "Your OT-2 can fully automate the entire Promega MagaZorb\u00ae DNA Mini-Prep Kit.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below: \n\nExplanation of complex parameters below:\n Number of samples: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n Deepwell type: Specify which well plate will be mounted on the magnetic module.\n Reservoir Type: Specify which reservoir will be employed.\n Starting Volume: Specify starting volume of sample (ul).\n* `: Specify the volume of binding buffer to use (ul).\n*Elution Volume: Specify elution volume (ul).\n*Park Tips: Specify whether to park tips or drop tips.\n*P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n",
+ "description": "Your OT-2 can fully automate the entire Promega MagaZorb\u00ae DNA Mini-Prep Kit.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below: \n\nExplanation of complex parameters below:\n Number of samples: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n Deepwell type: Specify which well plate will be mounted on the magnetic module.\n Reservoir Type: Specify which reservoir will be employed.\n Starting Volume: Specify starting volume of sample (ul).\n Binding buffer volume: Specify the volume of binding buffer to use (ul).\n Elution Volume: Specify elution volume (ul).\n Park Tips: Specify whether to park tips or drop tips.\n P300 Multi Channel Pipette Mount: Specify whether the P300 multi channel pipette will be on the left or right mount.\n",
"internal": "sci-promega-magazorb-dna-mini-prep-kit",
"labware": "\nNEST 96 Wellplate 2mL\nUSA Scientific 96 Wellplate 2.4mL\nNEST 12 Reservoir 15mL\nUSA Scientific 12 Reservoir 22mL\nOpentrons 200uL Filter Tips\nOpentrons 96 Aluminum block Nest Wellplate 100ul\n",
"markdown": {
- "author": "[Opentrons (verified)](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-promega-magazorb-dna-mini-prep-kit). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons (verified)](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-promega-magazorb-dna-mini-prep-kit). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* DNA Extraction\n\n",
"deck-setup": "\n* Tip rack on Slot 4 is used for tip parking if selected.\n\n\n\n",
- "description": "Your OT-2 can fully automate the entire Promega MagaZorb\u00ae DNA Mini-Prep Kit.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below: \n\n\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n* `Deepwell type`: Specify which well plate will be mounted on the magnetic module.\n* `Reservoir Type`: Specify which reservoir will be employed.\n* `Starting Volume`: Specify starting volume of sample (ul).\n* ``: Specify the volume of binding buffer to use (ul).\n* `Elution Volume`: Specify elution volume (ul).\n* `Park Tips`: Specify whether to park tips or drop tips.\n* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n\n\n---\n\n",
+ "description": "Your OT-2 can fully automate the entire Promega MagaZorb\u00ae DNA Mini-Prep Kit.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below: \n\n\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n* `Deepwell type`: Specify which well plate will be mounted on the magnetic module.\n* `Reservoir Type`: Specify which reservoir will be employed.\n* `Starting Volume`: Specify starting volume of sample (ul).\n* `Binding buffer volume`: Specify the volume of binding buffer to use (ul).\n* `Elution Volume`: Specify elution volume (ul).\n* `Park Tips`: Specify whether to park tips or drop tips.\n* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n\n\n---\n\n",
"internal": "sci-promega-magazorb-dna-mini-prep-kit\n",
"labware": "* [NEST 96 Wellplate 2mL](https://shop.opentrons.com/collections/lab-plates/products/nest-0-2-ml-96-well-deep-well-plate-v-bottom)\n* [USA Scientific 96 Wellplate 2.4mL](https://labware.opentrons.com/?category=wellPlate)\n* [NEST 12 Reservoir 15mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [USA Scientific 12 Reservoir 22mL](https://labware.opentrons.com/?category=reservoir)\n* [Opentrons 200uL Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n* [Opentrons 96 Aluminum block Nest Wellplate 100ul](https://labware.opentrons.com/opentrons_96_aluminumblock_nest_wellplate_100ul?category=aluminumBlock)\n\n",
"modules": "* [Magnetic Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n\n",
diff --git a/protoBuilds/sci-promega-magnesil-total-rna-mini-isolation-system/README.json b/protoBuilds/sci-promega-magnesil-total-rna-mini-isolation-system/README.json
index 122d7db9b0..98af7738f4 100644
--- a/protoBuilds/sci-promega-magnesil-total-rna-mini-isolation-system/README.json
+++ b/protoBuilds/sci-promega-magnesil-total-rna-mini-isolation-system/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Promega Kit"
@@ -9,7 +9,7 @@
"description": "This protocol automates the paramagnetic bead-based purification of DNA-free total RNA for up to 96 samples using the Promega Magnesil Total RNA Mini Isolation System.\nLinks:\n* Promega Magnesil Total RNA Mini Isolation System\nThis protocol was developed to perform the following steps using the Promega Magnesil Total RNA Mini Isolation System: Bead binding, a wash step, DNAseI treatment and DNase inactivation, two additional washes followed by elution.",
"internal": "sci-promega-magnesil-total-rna-mini-isolation-system",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Promega Kit\n\n",
"deck-setup": "\n\n* Opentrons 200 ul filter tips (Deck Slots 2, 4, 5, 7, 8, 10, 11)\n* Opentrons Temperature Module (cooled to 4 degrees C) with elution plate opentrons_96_aluminumblock_nest_wellplate_100ul (Deck Slot 1)\n* Reagent Reservoir nest_12_reservoir_15ml (Deck Slot 3)\n* Liquid Waste Reservoir nest_1_reservoir_195ml (Deck Slot 9)\n* Opentrons Magnetic Module with deep well plate nest_96_wellplate_2ml_deep (Deck Slot 6)\n\n\n\n\n\n\n\n\n\n",
"description": "\nThis protocol automates the paramagnetic bead-based purification of DNA-free total RNA for up to 96 samples using the [Promega Magnesil Total RNA Mini Isolation System](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/sci-promega-magnesil-total-rna-mini-isolation-system/MagneSil+Total+RNA+mini-Isolation+System+TB328.pdf).\n\nLinks:\n* [Promega Magnesil Total RNA Mini Isolation System](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/sci-promega-magnesil-total-rna-mini-isolation-system/MagneSil+Total+RNA+mini-Isolation+System+TB328.pdf)\n\nThis protocol was developed to perform the following steps using the Promega Magnesil Total RNA Mini Isolation System: Bead binding, a wash step, DNAseI treatment and DNase inactivation, two additional washes followed by elution.\n\n",
diff --git a/protoBuilds/sci-roche-hyperprep/README.json b/protoBuilds/sci-roche-hyperprep/README.json
index 96d85263ce..77de763a59 100644
--- a/protoBuilds/sci-roche-hyperprep/README.json
+++ b/protoBuilds/sci-roche-hyperprep/README.json
@@ -18,7 +18,7 @@
"labware": "* NEST 12 Well Reservoir 195mL\n* Eppendorf 96 well plate full skirt\n* Nest 96 well plate full skirt\n* Opentrons 20ul Filter Tips\n* Opentrons 200ul Filter Tips\n\n",
"modules": "* [Temperature Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Magnetic Module (GEN2)](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module)\n\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Partner Name](partner website link)\n\n",
+ "partner": "[Partner Name](partner website link)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* Opentrons P300 Multi-Channel Pipette GEN2\n* Opentrons P20 Multi-Channel Pipette GEN2\n* Opentrons P300 Multi-Channel Pipette GEN1\n* Opentrons P10 Multi-Channel Pipette GEN1\n\n\n",
"process": "1. Input your protocol parameters above.\n2. Download your protocol and unzip if needed.\n3. Upload your custom labware to the [OT App](https://opentrons.com/ot-app) by navigating to `More` > `Custom Labware` > `Add Labware`, and selecting your labware files (.json extensions) if needed.\n4. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n5. Set up your deck according to the deck map.\n6. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n7. Hit 'Run'.\n\n",
"protocol-steps": "1. This section should consist of a numerical outline of the protocol steps, somewhat analogous to the steps outlined by the user in their custom protocol submission.\n2. example step: Samples are transferred from the source tuberacks on slots 1-2 to the PCR plate on slot 3, down columns and then across rows.\n3. example step: Waste is removed from each sample on the magnetic module, ensuring the bead pellets are not contacted by the pipette tips.\n\n",
@@ -32,7 +32,7 @@
"Thermocycler Module"
],
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Partner Name",
+ "partner": "Partner Name\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nOpentrons P300 Multi-Channel Pipette GEN2\nOpentrons P20 Multi-Channel Pipette GEN2\nOpentrons P300 Multi-Channel Pipette GEN1\nOpentrons P10 Multi-Channel Pipette GEN1\n",
"process": "\nInput your protocol parameters above.\nDownload your protocol and unzip if needed.\nUpload your custom labware to the OT App by navigating to More > Custom Labware > Add Labware, and selecting your labware files (.json extensions) if needed.\nUpload your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit 'Run'.\n",
"protocol-steps": "\nThis section should consist of a numerical outline of the protocol steps, somewhat analogous to the steps outlined by the user in their custom protocol submission.\nexample step: Samples are transferred from the source tuberacks on slots 1-2 to the PCR plate on slot 3, down columns and then across rows.\nexample step: Waste is removed from each sample on the magnetic module, ensuring the bead pellets are not contacted by the pipette tips.\n",
diff --git a/protoBuilds/sci-tecan-cortisol-saliva-elisa/README.json b/protoBuilds/sci-tecan-cortisol-saliva-elisa/README.json
index 18bbd4ddca..2f3df19c8d 100644
--- a/protoBuilds/sci-tecan-cortisol-saliva-elisa/README.json
+++ b/protoBuilds/sci-tecan-cortisol-saliva-elisa/README.json
@@ -1,5 +1,5 @@
{
- "author": "Boren Lin",
+ "author": "Boren Lin\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Proteins": [
"Proteins"
@@ -10,7 +10,7 @@
"internal": "sci-tecan-cortisol-saliva-elisa",
"labware": "\nTecan ELISA 96-well on Heater Shaker #446469\nThermo Scientific 96 Well Plate V Bottom 450 uL #249944/249946\nNEST 96 Deepwell Plate 2mL #503001\nOpentrons 96 Tip Rack 300 \u00b5L\nNEST 1 Well Reservoir 195 mL #360103\n",
"markdown": {
- "author": "Boren Lin\n\n\n",
+ "author": "Boren Lin\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Proteins\n\t* Proteins\n\n\n",
"deck-setup": "[deck](https://drive.google.com/open?id=19lOAqOhCQwh45I_KiY92_RhT9Nj_6pUF)\n\n\n",
"description": "Tecan\u2019s Cortisol Saliva ELISA (REF 30172091, M\u00e4nnedorf, Switzerland) is an enzyme immunoassay for the quantitative determination of free cortisol in human saliva. It is a competitive ELISA which the target antigen (free cortisol in specimen) competes with a reference antigen (fixed amount of HRP-labelled cortisol) for binding the anti-cortisol antibody pre-coated on the plate. A substrate for HRP is then added into the mixture to produce the photometric signal. The more the target antigen is present in the sample, the less the reference antigen can be captured by the antibody and the weaker the signal produced; therefore, the intensity of the developed color is inversely proportional to the amount of the target antigen in the sample.\n\nThe protocol performs transferring of the samples and reference antigen (Enzyme Conjugate) into the ELISA plate (Microtiter Plate, 8-well strips coated with rabbit anti-cortisol antibody) to allow the antibody to bind to the antigen. After a 2-hour incubation and washing, TMB Substrate Solution is transferred into the ELISA plate to develop photometric signal which can be measured on a microplate reader at 450 nm.\n\nMaterials provided in the kit:\n1.\tRabbit anti-cortisol antibody coated microtiter plate (8-well strip x12)\n2.\tHRP-conjugated cortisol (Enzyme Conjugate)\n3.\tCortisol Standards and Positive Controls\n4.\tTMB Substrate Solution \n5.\tTMB Stop Solution\n6.\tWash Buffer (10X)\n\nOther materials required:\n1.\tSlit seal (BioChromato, Fujisawa, Japan)\n2.\tNEST 1 Well Reservoir 195 mL \n3.\tNEST 96 Deep Well Plate 2mL\n4.\tOpentrons Tip Rack, 300 \u00b5L\n5.\tNunc 96-Well Polypropylene Storage Microplates (Cat. #: 249947, Thermo Fisher, Waltham, MA, USA) or compatible \n6.\tMicroplate Reader\n\nPipettes and Modules\n1.\tOpentrons P300 8-Channel Pipette (GEN2)\n2.\tHeater Shaker Module (GEN1) with Aluminum Adaptor for 96-well Flat Plate\n\nDeck setup:\nPlease see the deck layout as the general guideline for labware setup. \n1.\tThe ELISA plate is sealed with a slit seal and placed on the Heater Shaker Module (Slot 3). \n2.\tSamples or sample dilutions are pre-loaded in the Nunc 96-Well Polypropylene Storage Microplates, the SAMPLES plate (Slot 1).\n3.\tThe REAGENTS plate (Slot 4) is filled with Enzyme Conjugate in Column 1, TMB Substrate Solution in Column 2, and TMB Stop Solution in Column 3\nNote: See Reagent Step for reagent plate preparation\n4.\tFor WASH (Slot 5), sufficient wash buffer is filled in each well for 4 washes of 250 \u03bcL.\n5.\tThree boxes of Opentrons 96 tips (Slot 7, 10, and 11) are sufficient to run a full 96-well ELISA plate (12 8-well strips). \n6.\tThe WASTE plate is loaded at Slot 9.\n\n\n\n",
diff --git a/protoBuilds/sci-thermofisher-magmax/README.json b/protoBuilds/sci-thermofisher-magmax/README.json
index 86bd1c1d20..4cd269c221 100644
--- a/protoBuilds/sci-thermofisher-magmax/README.json
+++ b/protoBuilds/sci-thermofisher-magmax/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Thermofisher MagMAX"
@@ -10,7 +10,7 @@
"internal": "sci-thermofisher-magmax",
"labware": "\nNEST 96 Wellplate 2mL\nUSA Scientific 96 Wellplate 2.4mL\nNEST 12 Reservoir 15mL\nUSA Scientific 12 Reservoir 22mL\nOpentrons 200uL Filter Tips\nOpentrons 96 Aluminum block Nest Wellplate 100ul\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Thermofisher MagMAX\n\n",
"deck-setup": "\n\n\n",
"description": "Your OT-2 can fully automate the entire Thermofisher MagMAX Kit.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below: \n\n\n\nLysed samples should be loaded on the magnetic module in a NEST or USA Scientific 96-deepwell plate. For reagent layout in the 2 12-channel reservoirs used in this protocol, please see \"Setup\" below.\n\nFor sample traceability and consistency, samples are mapped directly from the magnetic extraction plate (magnetic module, slot 4) to the elution PCR plate (temperature module, slot 1). Magnetic extraction plate well A1 is transferred to elution PCR plate A1, extraction plate well B1 to elution plate B1, ..., D2 to D2, etc.\n\nExplanation of complex parameters below:\n* `park tips`: If set to `yes` (recommended), the protocol will conserve tips between reagent addition and removal. Tips will be stored in the wells of an empty rack corresponding to the well of the sample that they access (tip parked in A1 of the empty rack will only be used for sample A1, tip parked in B1 only used for sample B1, etc.). If set to `no`, tips will always be used only once, and the user will be prompted to manually refill tipracks mid-protocol for high throughput runs.\n* `track tips across protocol runs`: If set to `yes`, tip racks will be assumed to be in the same state that they were in the previous run. For example, if one completed protocol run accessed tips through column 5 of the 3rd tiprack, the next run will access tips starting at column 6 of the 3rd tiprack. If set to `no`, tips will be picked up from column 1 of the 1st tiprack.\n* `flash`: If set to `yes`, the robot rail lights will flash during any automatic pauses in the protocol. If set to `no`, the lights will not flash.\n\n---\n\n",
diff --git a/protoBuilds/sci-zymo-directzol-magbead/README.json b/protoBuilds/sci-zymo-directzol-magbead/README.json
index 6c393c39a5..f904402cc7 100644
--- a/protoBuilds/sci-zymo-directzol-magbead/README.json
+++ b/protoBuilds/sci-zymo-directzol-magbead/README.json
@@ -10,7 +10,7 @@
"internal": "sci-zymo-directzol-magbead",
"labware": "\nNEST 96 Wellplate 2mL\nUSA Scientific 96 Wellplate 2.4mL\nNEST 12 Reservoir 15mL\nUSA Scientific 12 Reservoir 22mL\nOpentrons 200uL Filter Tips\nOpentrons 96 Aluminum block Nest Wellplate 100ul\n",
"markdown": {
- "author": "[Opentrons (verified)](https://opentrons.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-zymo-directzol-magbead). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "author": "[Opentrons (verified)](https://opentrons.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-zymo-directzol-magbead). This page won\u2019t be available after January 31st, 2024.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* RNA Extraction\n\n",
"deck-setup": "\n* Tip rack on Slot 4 is used for tip parking if selected.\n\n\n\n",
"description": "Your OT-2 can fully automate the entire Zymo Research Direct-zol\u2122-96 MagBead RNA Kit.\nResults of the Opentrons Science team's internal testing of this protocol on the OT-2 are shown below: \n\n\n\n\n\nExplanation of complex parameters below:\n* `Number of samples`: Specify the number of samples this run (1-96 and divisible by 8, i.e. whole columns at a time).\n* `Deepwell type`: Specify which well plate will be mounted on the magnetic module.\n* `Reservoir Type`: Specify which reservoir will be employed.\n* `Starting Volume`: Specify starting volume of sample (ul).\n* `Elution Volume`: Specify elution volume (ul).\n* `Park Tips`: Specify whether to park tips or drop tips.\n* `Mag Deck Generation`: Specify whether GEN1 or GEN2 magnetic module will be used.\n* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.\n\n\n---\n\n",
diff --git a/protoBuilds/sci-zymo-quick-dna-rna-magbead/README.json b/protoBuilds/sci-zymo-quick-dna-rna-magbead/README.json
index be4722347b..e3bba4eac7 100644
--- a/protoBuilds/sci-zymo-quick-dna-rna-magbead/README.json
+++ b/protoBuilds/sci-zymo-quick-dna-rna-magbead/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Zymo Kit"
@@ -9,7 +9,7 @@
"description": "This protocol automates the magnetic bead-based purification of DNA-free total RNA (if default protocol parameters are used at the time of protocol download to include the DNAseI step) or alternatively both DNA and total RNA from the same starting sample (both DNA and RNA in a combined single sample, if protocol parameters are used at the time of protocol download to skip the DNAseI step) for up to 96 samples using the Zymo Quick-DNA-RNA MagBead Kit.\nLinks:\n* Zymo Quick-DNA-RNA MagBead Kit\nThis protocol was developed to perform the following steps using the Zymo Quick-DNA-RNA MagBead Kit: Bead binding, 4 wash steps, DNAseI treatment (if preparing DNA-free total RNA), stop and elution. The default output sample is DNA-free total RNA. A protocol parameter (below) can be used at the time of protocol download to skip the DNAseI step for an output sample containing combined DNA and total RNA. Additional protocol parameters can be used at the time of protocol download to skip or include any of these steps (for testing purposes, for a partially manual and partially automated process, for a custom sequence of steps etc.)",
"internal": "sci-zymo-quick-dna-rna-magbead",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n * Zymo Kit\n\n",
"deck-setup": "\n\n* Opentrons 200 ul filter tips (Deck Slots 5, 7, 8, 10, 11)\n* empty Opentrons 200 ul filter tip box for parking tips (Deck Slot 4)\n* Opentrons Temperature Module (cooled to 4 degrees C) with elution plate opentrons_96_aluminumblock_nest_wellplate_100ul (Deck Slot 1)\n* Reagent Reservoirs nest_12_reservoir_15ml (Deck Slots 2, 3)\n* Liquid Waste Reservoir nest_1_reservoir_195ml (Deck Slot 9)\n* Opentrons Magnetic Module with deep well plate nest_96_wellplate_2ml_deep (Deck Slot 6)\n\n\n\n\n\n",
"description": "\nThis protocol automates the magnetic bead-based purification of DNA-free total RNA (if default protocol parameters are used at the time of protocol download to include the DNAseI step) or alternatively both DNA and total RNA from the same starting sample (both DNA and RNA in a combined single sample, if protocol parameters are used at the time of protocol download to skip the DNAseI step) for up to 96 samples using the [Zymo Quick-DNA-RNA MagBead Kit](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/sci-zymo-quick-dna-rna-magbead/_r2130_r2131_quick-dna_rna_magbead.pdf).\n\nLinks:\n* [Zymo Quick-DNA-RNA MagBead Kit](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/sci-zymo-quick-dna-rna-magbead/_r2130_r2131_quick-dna_rna_magbead.pdf)\n\nThis protocol was developed to perform the following steps using the Zymo Quick-DNA-RNA MagBead Kit: Bead binding, 4 wash steps, DNAseI treatment (if preparing DNA-free total RNA), stop and elution. The default output sample is DNA-free total RNA. A protocol parameter (below) can be used at the time of protocol download to skip the DNAseI step for an output sample containing combined DNA and total RNA. Additional protocol parameters can be used at the time of protocol download to skip or include any of these steps (for testing purposes, for a partially manual and partially automated process, for a custom sequence of steps etc.)\n\n",
diff --git a/protoBuilds/speaker-test/README.json b/protoBuilds/speaker-test/README.json
index e336925a79..77785275e0 100644
--- a/protoBuilds/speaker-test/README.json
+++ b/protoBuilds/speaker-test/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Getting Started": [
"OT-2 Demo"
@@ -9,7 +9,7 @@
"internal": "speaker-test",
"labware": "\nOpentrons Tip Rack\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Getting Started\n\t* OT-2 Demo\n\n\n",
"description": "This is a simple protocol to illustrate the speaker functionality of the OT-2!\n\nWith this protocol, the robot will pick up and drop a tip. Following this simple command, a song will begin to play. Using this as a template, adding a sound to your protocol should be fairly easy.\n\nThis protocol allows you to select the pipette, mount, and tip rack you would like to use for this demonstration. The only labware needed will be the tip rack and this should be loaded on deck slot 1.\n\n\n",
"internal": "speaker-test\n",
diff --git a/protoBuilds/standard-biotools-da-192/README.json b/protoBuilds/standard-biotools-da-192/README.json
index d07c622ebd..4e091b8add 100644
--- a/protoBuilds/standard-biotools-da-192/README.json
+++ b/protoBuilds/standard-biotools-da-192/README.json
@@ -17,14 +17,14 @@
"internal": "standard-biotools-da-192\n",
"labware": "* Standard BioTools 192.24 Dynamic Array IFC\n* [Opentrons 96 Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set)\n* [NEST 96-Well PCR Plate, 100\u00b5L](https://shop.opentrons.com/nest-0-1-ml-96-well-pcr-plate-full-skirt/)\n* VWR 96 Well Plate 200 \u00b5L PCR (89218-290)\n* [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/4-in-1-tube-rack-set/)\n* Eppendorf 1.5 mL Safe-Lock Snapcap\n* [Opentrons 20\u00b5L Filter Tips](https://shop.opentrons.com/opentrons-20ul-filter-tips/)\n* [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/opentrons-200ul-filter-tips/)\n\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n\n",
- "partner": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "partner": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [Opentrons P20 8-Channel Pipette (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n* [Opentrons P300 Single-Channel Pipette (GEN2)](https://shop.opentrons.com/single-channel-electronic-pipette-p20/)\n\n\n",
"process": "1. Download your protocol and unzip if needed.\n2. Import your labware file(s) (.json extension) to the [OT App](https://opentrons.com/ot-app) in the `labware` tab, if needed.\n3. Import your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n4. Set up your deck according to the deck map.\n5. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/s/article/How-positional-calibration-works-on-the-OT-2).\n6. Hit \"Run\".\n\n\n",
"protocol-steps": "1. Transfer Actuation Fluid to IFC\n2. Transfer Pressure Fluid to IFC\n3. Transfer assays to IFC\n4. Transfer samples to IFC\n\n\n*NOTE*: Mapping from the sample / assay plates to the IFC matches the mapping defined by the workflow selected in Preparation: Step 1\n\n\n\n",
"title": "Standard Biotools Dynamic Array 192.24: Load 4 uL"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Opentrons",
+ "partner": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nOpentrons P20 8-Channel Pipette (GEN2)\nOpentrons P300 Single-Channel Pipette (GEN2)\n",
"process": "\nDownload your protocol and unzip if needed.\nImport your labware file(s) (.json extension) to the OT App in the labware tab, if needed.\nImport your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit \"Run\".\n",
"protocol-steps": "\nTransfer Actuation Fluid to IFC\nTransfer Pressure Fluid to IFC\nTransfer assays to IFC\nTransfer samples to IFC\n\n\nNOTE: Mapping from the sample / assay plates to the IFC matches the mapping defined by the workflow selected in Preparation: Step 1",
diff --git a/protoBuilds/standard-biotools-da-48/README.json b/protoBuilds/standard-biotools-da-48/README.json
index bab0fd4fd0..8457896791 100644
--- a/protoBuilds/standard-biotools-da-48/README.json
+++ b/protoBuilds/standard-biotools-da-48/README.json
@@ -17,14 +17,14 @@
"internal": "standard-biotools-da-48\n",
"labware": "* Standard BioTools 48.48 Dynamic Array IFC\n* [Opentrons 96 Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set)\n* [NEST 96-Well PCR Plate, 100\u00b5L](https://shop.opentrons.com/nest-0-1-ml-96-well-pcr-plate-full-skirt/)\n* [Opentrons 20\u00b5L Filter Tips](https://shop.opentrons.com/opentrons-20ul-filter-tips/)\n\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n\n",
- "partner": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "partner": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [Opentrons P20 8-Channel Pipette (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n\n\n",
"process": "1. Download your protocol and unzip if needed.\n2. Import your labware file(s) (.json extension) to the [OT App](https://opentrons.com/ot-app) in the `labware` tab, if needed.\n3. Import your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n4. Set up your deck according to the deck map.\n5. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/s/article/How-positional-calibration-works-on-the-OT-2).\n6. Hit \"Run\".\n\n\n",
"protocol-steps": "1. Transfer assays to left side of IFC\n2. Transfer samples to right side of IFC\n\n*NOTE*: Mapping from the sample / assay plates to the IFC matches the mapping defined by the workflow selected in Preparation: Step 1\n\n\n\n",
"title": "Standard BioTools Dynamic Array 48.48: Load 4 uL"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Opentrons",
+ "partner": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nOpentrons P20 8-Channel Pipette (GEN2)\n",
"process": "\nDownload your protocol and unzip if needed.\nImport your labware file(s) (.json extension) to the OT App in the labware tab, if needed.\nImport your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit \"Run\".\n",
"protocol-steps": "\nTransfer assays to left side of IFC\nTransfer samples to right side of IFC\n\nNOTE: Mapping from the sample / assay plates to the IFC matches the mapping defined by the workflow selected in Preparation: Step 1\n",
diff --git a/protoBuilds/standard-biotools-da-96/README.json b/protoBuilds/standard-biotools-da-96/README.json
index dea2d09a23..0eabf7e5f4 100644
--- a/protoBuilds/standard-biotools-da-96/README.json
+++ b/protoBuilds/standard-biotools-da-96/README.json
@@ -17,14 +17,14 @@
"internal": "standard-biotools-da-96\n",
"labware": "* Standard BioTools 96.96 Dynamic Array IFC\n* [Opentrons 96 Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set)\n* [NEST 96-Well PCR Plate, 100\u00b5L](https://shop.opentrons.com/nest-0-1-ml-96-well-pcr-plate-full-skirt/)\n* [Opentrons 20\u00b5L Filter Tips](https://shop.opentrons.com/opentrons-20ul-filter-tips/)\n\n\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n\n",
- "partner": "[Opentrons](https://opentrons.com/)\n\n\n",
+ "partner": "[Opentrons](https://opentrons.com/)\n\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"pipettes": "* [Opentrons P20 8-Channel Pipette (GEN2)](https://shop.opentrons.com/8-channel-electronic-pipette/)\n\n\n",
"process": "1. Download your protocol and unzip if needed.\n2. Import your labware file(s) (.json extension) to the [OT App](https://opentrons.com/ot-app) in the `labware` tab, if needed.\n3. Import your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.\n4. Set up your deck according to the deck map.\n5. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/s/article/How-positional-calibration-works-on-the-OT-2).\n6. Hit \"Run\".\n\n\n",
"protocol-steps": "1. Transfer assays to left side of IFC\n2. Transfer samples to right side of IFC\n\n*NOTE*: Mapping from the sample / assay plates to the IFC matches the mapping defined by the workflow selected in Preparation: Step 1\n\n\n\n",
"title": "Standard BioTools Dynamic Array 96.96: Load 4 uL"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Opentrons",
+ "partner": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"pipettes": "\nOpentrons P20 8-Channel Pipette (GEN2)\n",
"process": "\nDownload your protocol and unzip if needed.\nImport your labware file(s) (.json extension) to the OT App in the labware tab, if needed.\nImport your protocol file (.py extension) to the OT App in the Protocol tab.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit \"Run\".\n",
"protocol-steps": "\nTransfer assays to left side of IFC\nTransfer samples to right side of IFC\n\nNOTE: Mapping from the sample / assay plates to the IFC matches the mapping defined by the workflow selected in Preparation: Step 1\n",
diff --git a/protoBuilds/swift-2s-turbo-pt1/README.json b/protoBuilds/swift-2s-turbo-pt1/README.json
index 02f5801c9c..805fb2b0fb 100644
--- a/protoBuilds/swift-2s-turbo-pt1/README.json
+++ b/protoBuilds/swift-2s-turbo-pt1/README.json
@@ -13,7 +13,7 @@
"description": "\n\nPart 1 of 3: Enzymatic Prep & Ligation\n\n\nWith this protocol, your [OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2) can perform the [Swift 2S Turbo DNA Library Kit](https://swiftbiosci.com/swift-2s-turbo-dna-library-kits/). For more information about the Swift 2S Turbo Kit and the [Swift 2S Turbo Unique Dual Indexing Primer Kit](https://shop.opentrons.com/products/swift-2s-turbo-unique-dual-indexing-primer-kit-96-rxns?_pos=1&_sid=f1fb599e7&_ss=r) on the OT-2, please see our Application Note here: [Rapid high quality next generation sequencing library preparation with Swift 2S Turbo DNA Library Kits on the Opentrons OT-2](https://opentrons-landing-img.s3.amazonaws.com/bundles/swift_automated_ngs_application_note.pdf)\n\n\nIn this part of the protocol, your OT-2 will complete the enzymatic prep portion and the initial steps of the ligation portion prior to adding your samples to a thermocycler, as described in the [Swift 2S Turbo Kit Guide](https://swiftbiosci.com/wp-content/uploads/2019/11/PRT-001-2S-Turbo-DNA-Library-Kit-Rev-1.pdf).\n\n\nAt the completion of this step, you will add your samples to the thermocycler. Once the thermocycler step is complete, continue with [Part 2 of the protocol](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt2).\n\n\nLinks:\n* [Part 1: Enzymatic Prep & Ligation](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt1)\n* [Part 2: Ligation Clean-Up & PCR Prep](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt2)\n* [Part 3: Final Clean-Up](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt3)\n\n\n\n**Note:** This workflow replaces the Reagent K2 in the Enzymatic Prep Master Mix with Reagent DE in order to reduce the risk of over fragmentation. For more information, please see [this note.](https://swiftbiosci.com/wp-content/uploads/2019/12/PRT-022-Swift-Deceleration-Module-Rev-1.pdf)\n\n---\n\n\nTo purchase consumables, labware, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n\n**Attention**: You can now purchase all of the consumables needed to run this protocol by [clicking here](https://shop.opentrons.com/products/ngs-library-prep-workstation-consumables-refill).\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Swift 2S Turbo DNA Library Kit](https://swiftbiosci.com/swift-2s-turbo-dna-library-kits/)\n* [Swift 2S Turbo Unique Dual Indexing Primer Kit](https://shop.opentrons.com/products/swift-2s-turbo-unique-dual-indexing-primer-kit-96-rxns?_pos=1&_sid=f1fb599e7&_ss=r)\n* [Omega Mag-Bind TotalPure NGS Kit](https://shop.opentrons.com/collections/verified-reagents/products/mag-bind-total-pure-ngs)\n* [Opentrons Temperature Module with Aluminum Block Set](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) or Opentrons P50 Single-Channel Pipette*\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [NEST 2mL Tubes](https://shop.opentrons.com/collections/tubes/products/nest-2-0-ml-sample-vial)\n* Samples, resuspended in Low EDTA TE, bringing the total volume to 16\u00b5L\n* PCR Strip(s), *optional*\n\n\n\\*Opentrons now sells the P20 Single-Channel Pipette in place of the P50 Single-Channel Pipette. If you have the P50 Single-Channel Pipette, you can use it for this protocol.\n\n\nFull setup for the entire protocol:\n\n\n\n\n\n\n\n\n---\n\n\n\n*Specific to Part 1 of 3*\n\n\nSlot 1: [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) (or PCR Strips) on top of [96-Well Aluminum Block](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set) with samples\n* 8 Samples: Column 1\n* 16 Samples: Columns 1 & 2\n* 24 Samples: Columns 1, 2, & 3\n\nSlot 3: [Opentrons Temperature Module with 24-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) and [NEST 2mL Tubes](https://shop.opentrons.com/collections/tubes/products/nest-2-0-ml-sample-vial) with master mixes (for more information on master mixes, [click here](https://opentrons-protocol-library-website.s3.amazonaws.com/Technical+Notes/swift-2s-turbo-pt1/Swift+2S+Turbo+Master+Mix+Volumes.xlsx))\n* A1: Enzymatic Prep Master Mix\n* A2: Ligation Master Mix\n* A3: (**used in Part 2**) PCR Master Mix\n* B1: (**used in Part 2**) Indexing Reagent 1 (in original container); loaded sequentially (Reagent 2 - B2; Reagent 3 - B3...)\n\n\nSlot 5: [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n\n\n\n\n__Using the customizations fields, below set up your protocol.__\n* **Pipette and Tip Type**: Select which pipette (P50 Single-Channel or P20 Single-Channel) and corresponding tips to be used for this protocol. **The pipette should be attached to the left mount.**\n* **Number of Samples**: Specify the number of samples (8, 16 or 24) you'd like to run.\n\n\n\n\n",
"internal": "Swift-2S-Turbo-pt1\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Swift Biosciences](https://swiftbiosci.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/swift-2s-turbo-pt1). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "partner": "[Swift Biosciences](https://swiftbiosci.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/swift-2s-turbo-pt1). This page won\u2019t be available after January 31st, 2024.\n\n",
"process": "\n1. Input your protocol parameters.\n2. Download your protocol.\n3. Upload your protocol into the [OT App](https://opentrons.com/ot-app).\n4. Set up your deck according to the deck map.\n5. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n6. Hit \"Run\".\n\n",
"robot": "* [OT-2](https://opentrons.com/ot-2)\n\n",
"title": "Swift 2S Turbo DNA Library Kit Protocol: Part 1/3 - Enzymatic Prep & Ligation"
diff --git a/protoBuilds/swift-2s-turbo-pt2/README.json b/protoBuilds/swift-2s-turbo-pt2/README.json
index f4731eb8c2..6c20b882a1 100644
--- a/protoBuilds/swift-2s-turbo-pt2/README.json
+++ b/protoBuilds/swift-2s-turbo-pt2/README.json
@@ -13,13 +13,13 @@
"description": "\n\nPart 2 of 3: Ligation Clean-Up & PCR Prep\n\n\nWith this protocol, your [OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2) can perform the [Swift 2S Turbo DNA Library Kit](https://swiftbiosci.com/swift-2s-turbo-dna-library-kits/). For more information about the Swift 2S Turbo Kit and the [Swift 2S Turbo Unique Dual Indexing Primer Kit](https://shop.opentrons.com/products/swift-2s-turbo-unique-dual-indexing-primer-kit-96-rxns?_pos=1&_sid=f1fb599e7&_ss=r) on the OT-2, please see our Application Note here: [Rapid high quality next generation sequencing library preparation with Swift 2S Turbo DNA Library Kits on the Opentrons OT-2](https://opentrons-landing-img.s3.amazonaws.com/bundles/swift_automated_ngs_application_note.pdf)\n\n\nIn this part of the protocol, your OT-2 will complete the ligation process that was begun in [Part 1](http://develop.protocols.opentrons.com/protocol/swift-2s-turbo-pt1) and complete the indexing portion up to the thermocycling step (step 18) as outlined in the [Swift 2S Turbo Kit Guide](https://swiftbiosci.com/wp-content/uploads/2019/11/PRT-001-2S-Turbo-DNA-Library-Kit-Rev-1.pdf).\n\n\nAt the completion of this step, you will add your samples to the thermocycler. Once the thermocycler step is complete, continue with [Part 3 of the protocol](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt3).\n\n\nLinks:\n* [Part 1: Enzymatic Prep & Ligation](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt1)\n* [Part 2: Ligation Clean-Up & PCR Prep](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt2)\n* [Part 3: Final Clean-Up](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt3)\n\n\n\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n\n**Attention**: You can now purchase all of the consumables needed to run this protocol by [clicking here](https://shop.opentrons.com/products/ngs-library-prep-workstation-consumables-refill).\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Swift 2S Turbo DNA Library Kit](https://swiftbiosci.com/swift-2s-turbo-dna-library-kits/)\n[Swift 2S Turbo Unique Dual Indexing Primer Kit](https://shop.opentrons.com/products/swift-2s-turbo-unique-dual-indexing-primer-kit-96-rxns?_pos=1&_sid=f1fb599e7&_ss=r)\n* [Omega Mag-Bind TotalPure NGS Kit](https://shop.opentrons.com/collections/verified-reagents/products/mag-bind-total-pure-ngs)\n* [Opentrons Temperature Module with Aluminum Block Set](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) or Opentrons P50 Single-Channel Pipette\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [NEST 2mL Tubes](https://shop.opentrons.com/collections/tubes/products/nest-2-0-ml-sample-vial)\n* Samples, user collected from [Part 1](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt1)\n* PCR Strip(s), *optional*\n\n\nFull setup for the entire protocol:\n\n\n\n\n\n\n---\n\n\n*Specific to Part 2 of 3*\n\n\nSlot 1: [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) (or PCR Strips) on top of [96-Well Aluminum Block](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set) with samples the user has collected from [Part 1](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt1)\n* 8 Samples: Column 1\n* 16 Samples: Columns 1 & 2\n* 24 Samples: Columns 1, 2, & 3\n\nSlot 2: [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* A1: Magnetic Beads\n* A3: 80% Ethanol Solution, Freshly Prepared\n* A4: 80% Ethanol Solution, Freshly Prepared\n* A6: Low EDTA TE Buffer\n\nSlot 3: [Opentrons Temperature Module with 24-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) and [NEST 2mL Tubes](https://shop.opentrons.com/collections/tubes/products/nest-2-0-ml-sample-vial) with master mixes (for more information on master mixes, [click here](https://opentrons-protocol-library-website.s3.amazonaws.com/Technical+Notes/swift-2s-turbo-pt1/Swift+2S+Turbo+Master+Mix+Volumes.xlsx)) and indices (if automating index addition)\n* A1: (**used in Part 1**) Enzymatic Prep Master Mix\n* A2: (**used in Part 1**) Ligation Master Mix\n* A3: PCR Master Mix\n* B1: Indexing Reagent 1 (in original container); loaded sequentially (Reagent 2 - B2; Reagent 3 - B3...)\n*Note*: if the user is running 24 samples and chooses to automate indices addition, the robot will pause after addition of the first 16 indices to allow user to replace them with the final 8 indices.\n\nSlot 4: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n\nSlot 5: [Opentrons Tips for Single-Channel Pipette](https://shop.opentrons.com/collections/opentrons-tips)\n\n\nSlot 6: [Opentrons Tips for P300 8-Channel Pipette](https://shop.opentrons.com/collections/opentrons-tips)\n\n\nSlot 9: [Opentrons Tips for P300 8-Channel Pipette](https://shop.opentrons.com/collections/opentrons-tips)\n\n\n\n**Using the customizations fields, below set up your protocol.**\n* **Pipette and Tip Type**: Select which pipette (P50 Single-Channel or P20 Single-Channel) and corresponding tips to be used for this protocol. **The pipette should be attached to the left mount.**\n* **P300 8-Channel Pipette Tip Type**: Select which tips (filter/non-filter) to be used for this protocol.\n* **Number of Samples**: Specify the number of samples (8, 16, or 24) you'd like to run.\n* **Magdeck Generation**: Specify whether using Generation 1 or Generation 2 magdeck.\n* **Automate Indexing**: Specify whether the indices should be added to the samples with the OT-2, or manually.\n\n\n\n\n",
"internal": "Swift-2S-Turbo-pt2\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Swift Biosciences](https://swiftbiosci.com/)\n\n",
+ "partner": "[Swift Biosciences](https://swiftbiosci.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"process": "\n1. Input your protocol parameters.\n2. Download your protocol.\n3. Upload your protocol into the [OT App](https://opentrons.com/ot-app).\n4. Set up your deck according to the deck map.\n5. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n6. Hit \"Run\".\n\n",
"robot": "* [OT-2](https://opentrons.com/ot-2)\n\n",
"title": "Swift 2S Turbo DNA Library Kit Protocol: Part 2/3 - Ligation Clean-Up & PCR Prep"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Swift Biosciences",
+ "partner": "Swift Biosciences\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"process": "\nInput your protocol parameters.\nDownload your protocol.\nUpload your protocol into the OT App.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit \"Run\".\n",
"robot": [
"OT-2"
diff --git a/protoBuilds/swift-2s-turbo-pt3/README.json b/protoBuilds/swift-2s-turbo-pt3/README.json
index 101018be1e..81cd5a80fc 100644
--- a/protoBuilds/swift-2s-turbo-pt3/README.json
+++ b/protoBuilds/swift-2s-turbo-pt3/README.json
@@ -13,13 +13,13 @@
"description": "\n\nPart 3 of 3: Final Clean-Up\n\n\nWith this protocol, your [OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2) can perform the [Swift 2S Turbo DNA Library Kit](https://swiftbiosci.com/swift-2s-turbo-dna-library-kits/). For more information about the Swift 2S Turbo Kit and the [Swift 2S Turbo Unique Dual Indexing Primer Kit](https://shop.opentrons.com/products/swift-2s-turbo-unique-dual-indexing-primer-kit-96-rxns?_pos=1&_sid=f1fb599e7&_ss=r) on the OT-2, please see our Application Note here: [Rapid high quality next generation sequencing library preparation with Swift 2S Turbo DNA Library Kits on the Opentrons OT-2](https://opentrons-landing-img.s3.amazonaws.com/bundles/swift_automated_ngs_application_note.pdf)\n\n\nIn this part of the protocol, your OT-2 will complete the final bead clean-up of the protocol as outlined in the [Swift 2S Turbo Kit Guide](https://swiftbiosci.com/wp-content/uploads/2019/11/PRT-001-2S-Turbo-DNA-Library-Kit-Rev-1.pdf).\n\n\nAt the completion of this step, you will have 20\u03bcl of elution from each sample that contains the final library.\n\nLinks:\n* [Part 1: Enzymatic Prep & Ligation](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt1)\n* [Part 2: Ligation Clean-Up & PCR Prep](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt2)\n* [Part 3: Final Clean-Up](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt3)\n\n\n\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n\n**Attention**: You can now purchase all of the consumables needed to run this protocol by [clicking here](https://shop.opentrons.com/products/ngs-library-prep-workstation-consumables-refill).\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Swift 2S Turbo DNA Library Kit](https://swiftbiosci.com/swift-2s-turbo-dna-library-kits/)\n[Swift 2S Turbo Unique Dual Indexing Primer Kit](https://shop.opentrons.com/products/swift-2s-turbo-unique-dual-indexing-primer-kit-96-rxns?_pos=1&_sid=f1fb599e7&_ss=r)\n* [Omega Mag-Bind TotalPure NGS Kit](https://shop.opentrons.com/collections/verified-reagents/products/mag-bind-total-pure-ngs)\n* [Opentrons Temperature Module with Aluminum Block Set](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) or Opentrons P50 Single-Channel Pipette\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [NEST 2mL Tubes](https://shop.opentrons.com/collections/tubes/products/nest-2-0-ml-sample-vial)\n* Samples, user collected from [Part 2](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt2)\n* PCR Strip(s), *optional*\n\n\nFull setup for the entire protocol:\n\n\n\n\n\n\n\n---\n\n\n*Specific to Part 3 of 3*\n\n\nSlot 1: [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) (or PCR Strips) on top of [96-Well Aluminum Block](https://shop.opentrons.com/collections/racks-and-adapters/products/aluminum-block-set) with samples the user has collected from [Part 2](https://protocols.opentrons.com/protocol/swift-2s-turbo-pt2)\n* 8 Samples: Column 4\n* 16 Samples: Columns 4 & 5\n* 24 Samples: Columns 4, 5, & 6\n\n\nSlot 2: [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* A1: Magnetic Beads\n* A4: 80% Ethanol Solution, Freshly Prepared\n* A6: Low EDTA TE Buffer\n\n\nSlot 4: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n\n\nSlot 6: [Opentrons Tips for P300 8-Channel Pipette](https://shop.opentrons.com/collections/opentrons-tips)\n\n\nSlot 9: [Opentrons Tips for P300 8-Channel Pipette](https://shop.opentrons.com/collections/opentrons-tips)\n\n\n\n**Using the customizations fields, below set up your protocol.**\n* **P300 8-Channel Pipette Tip Type**: Select which tips (filter/non-filter) to be used for this protocol.\n* **Number of Samples**: Specify the number of samples (8, 16, or 24) you'd like to run.\n* **Magdeck Generation**: Specify whether using Generation 1 or Generation 2 magdeck. \n\n\n__Note:__ The final elution will be transferred to column 7 (and column 8 and column 9, if running 16 samples or 24 samples, respectively) of the PCR plate (or strips) in slot 1.\n\n\n",
"internal": "Swift-2S-Turbo-pt3\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Swift Biosciences](https://swiftbiosci.com/)\n\n",
+ "partner": "[Swift Biosciences](https://swiftbiosci.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"process": "\n1. Input your protocol parameters.\n2. Download your protocol.\n3. Upload your protocol into the [OT App](https://opentrons.com/ot-app).\n4. Set up your deck according to the deck map.\n5. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n6. Hit \"Run\".\n\n",
"robot": "* [OT-2](https://opentrons.com/ot-2)\n\n",
"title": "Swift 2S Turbo DNA Library Kit Protocol: Part 3/3 - Final Clean-Up"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Swift Biosciences",
+ "partner": "Swift Biosciences\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"process": "\nInput your protocol parameters.\nDownload your protocol.\nUpload your protocol into the OT App.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit \"Run\".\n",
"robot": [
"OT-2"
diff --git a/protoBuilds/swift-fully-automated-custom/README.json b/protoBuilds/swift-fully-automated-custom/README.json
index 0fd2d6e926..3bb253a5b7 100644
--- a/protoBuilds/swift-fully-automated-custom/README.json
+++ b/protoBuilds/swift-fully-automated-custom/README.json
@@ -13,13 +13,13 @@
"description": "\n\nThis protocol is a custom modification to the [Swift 2S Turbo DNA Library Kit Protocol: Fully Automated](https://protocols.opentrons.com/protocol/swift-fully-automated) already found in the Opentrons Protocol Library. This version of the protocol features lowers aspiration flow rates to avoid bead pick-up, longer incubation times, and less volume of supernatant removed. This version also features both generations of the magnetic module. If running 24 samples with this protocol, the protocol will stop when it runs out of tips, prompting the user to replace tip racks.\n\nWith this protocol, your [OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2) can **fully automate** the entire [Swift 2S Turbo DNA Library Kit](https://swiftbiosci.com/swift-2s-turbo-dna-library-kits/). Simply press start and your OT-2 can automate this entire workflow without any hands-on requirement - from enzymatic prep to sequence ready libraries! Up to 16 libraries can be prepared in under 3 hours.\n\n\nFor more information about the Swift 2S Turbo Kit and the [Swift 2S Turbo Unique Dual Indexing Primer Kit](https://shop.opentrons.com/products/swift-2s-turbo-unique-dual-indexing-primer-kit-96-rxns?_pos=1&_sid=f1fb599e7&_ss=r) on the OT-2, please see our Application Note here: [Rapid high quality next generation sequencing library preparation with Swift 2S Turbo DNA Library Kits on the Opentrons OT-2](https://opentrons-landing-img.s3.amazonaws.com/bundles/swift_automated_ngs_application_note.pdf)\n\n\n\n---\n\n\nTo purchase consumables, labware, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n\n**Attention**: You can now purchase all of the consumables needed to run this protocol by [clicking here](https://shop.opentrons.com/products/ngs-library-prep-workstation-consumables-refill).\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Swift 2S Turbo DNA Library Kit](https://swiftbiosci.com/swift-2s-turbo-dna-library-kits/)\n* [Swift 2S Turbo Unique Dual Indexing Primer Kit](https://shop.opentrons.com/products/swift-2s-turbo-unique-dual-indexing-primer-kit-96-rxns?_pos=1&_sid=f1fb599e7&_ss=r)\n* [Omega Mag-Bind TotalPure NGS Kit](https://shop.opentrons.com/collections/verified-reagents/products/mag-bind-total-pure-ngs)\n* [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module)\n* [Opentrons Temperature Module with Aluminum Block Set](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) or Opentrons P50 Single-Channel Pipette*\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [NEST 2mL Tubes](https://shop.opentrons.com/collections/tubes/products/nest-2-0-ml-sample-vial)\n* Samples, resuspended in Low EDTA TE, bringing the total volume to 19.5\u00b5L\n\n\n\n\\*Opentrons now sells the P20 Single-Channel Pipette in place of the P50 Single-Channel Pipette. If you have the P50 Single-Channel Pipette, you can use it for this protocol.\n\n\nFull setup for the entire protocol:\n\n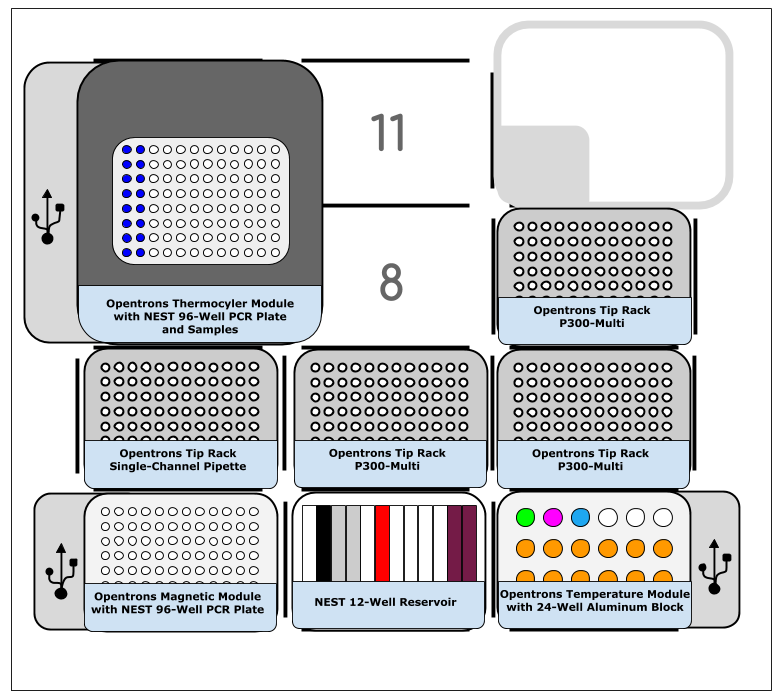\n\n\n\n\n\n\n---\n\n\n\nSlot 1: [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) on [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n\n\nSlot 2: [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml) with Reagents\n* A2: Magnetic Beads; recommended volume: 3-4mL, 4-5mL if running 24 samples.\n* A3: 80% Ethanol Solution, Freshly Prepared; recommended volume: 9-10mL, max allowable reservoir well volume if running 24 samples (watch for overflowing if the multi-channel pipette dunks).\n* A4: 80% Ethanol Solution, Freshly Prepared (if running 16 samples); recommended volume: 9-10mL, max allowable reservoir well volume if running 24 samples (watch for overflowing if the multi-channel pipette dunks).\n* A6: Low EDTA TE Buffer; recommended volume: ~3mL\n\n\nSlot 3: [Opentrons Temperature Module with 24-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) and [NEST 2mL Tubes](https://shop.opentrons.com/collections/tubes/products/nest-2-0-ml-sample-vial) with master mixes (for more information on master mixes, [click here](https://opentrons-protocol-library-website.s3.amazonaws.com/Technical+Notes/swift-fully-automated-custom/Swift+2s+Turbo+Fully+Automated+MasterMix+Volumes+(1).xlsx)) and indices (if automating index addition)\n* A1: Enzymatic Prep Master Mix\n* A2: Ligation Master Mix\n* A3: PCR Master Mix\n* B1: Indexing Reagent 1 (in original container); loaded sequentially (Reagent 2 - B2; Reagent 3 - B3...)\n\n\nSlot 4: [Opentrons Tips for Single-Channel Pipette](https://shop.opentrons.com/collections/opentrons-tips)\n\n\nSlot 5: [Opentrons Tips for P300 8-Channel Pipette](https://shop.opentrons.com/collections/opentrons-tips)\n\n\nSlot 6: [Opentrons Tips for P300 8-Channel Pipette](https://shop.opentrons.com/collections/opentrons-tips)\n\n\nSlot 9: [Opentrons Tips for P300 8-Channel Pipette](https://shop.opentrons.com/collections/opentrons-tips)\n\n\n\nSlot 7/8/10/11: [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module) with samples in a [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* 8 Samples: Column 1\n* 16 Samples: Columns 1 & 2\n\n\n\n__Using the customizations fields, below set up your protocol.__\n* **Number of Samples**: Specify the number of samples (8, 16, 24) you'd like to run.\n* **Pipette and Tip Type**: Select which pipette (P50 Single-Channel or P20 Single-Channel) and corresponding tips to be used for this protocol. **The pipette should be attached to the left mount.**\n* **P300 8-Channel Pipette Tip Type**: Select which tips (Filter/Non-Filter) for P300 8-Channel Pipette\n* **Automate Indexing**: Specify whether the indices should be added to the samples with the OT-2, or manually.\n* **Number of PCR Cycles**: See suggested cycles [here](https://opentrons-protocol-library-website.s3.amazonaws.com/Technical+Notes/swift-fully-automated-custom/Swift+Fully+Automated+-+PCR+Cycles+Recommendation.xlsx). See Swift 2S Turbo manual for more detailed information.\n* **Fragmentation Time**: Fragmentation time varies depending on *1)* Lot number of the kit and *2)* whether the desired insert size is 200bp or 350bp. Please refer to the manual and Lot number on your kit for more information.\n\n\n\n**Note**: The final 20\u00b5L elution will be transferred to Column 3 if running 8 samples, Columns 5 & 6 if running 16 samples, and Columns 5, 6, and 7 if running 24 samples of the PCR plate on the Thermocycler\n\n",
"internal": "swift-fully-automated-custom\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Swift Biosciences](https://swiftbiosci.com/)\n\n",
+ "partner": "[Swift Biosciences](https://swiftbiosci.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"process": "\n1. Input your protocol parameters.\n2. Download your protocol.\n3. Upload your protocol into the [OT App](https://opentrons.com/ot-app).\n4. Set up your deck according to the deck map.\n5. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n6. Hit \"Run\".\n\n",
"robot": "* [OT-2](https://opentrons.com/ot-2)\n\n",
"title": "Swift 2S Turbo DNA Library Kit Protocol: Fully Automated (Custom)"
},
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the Troubleshooting Survey.",
- "partner": "Swift Biosciences",
+ "partner": "Swift Biosciences\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"process": "\nInput your protocol parameters.\nDownload your protocol.\nUpload your protocol into the OT App.\nSet up your deck according to the deck map.\nCalibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our support articles.\nHit \"Run\".\n",
"robot": [
"OT-2"
diff --git a/protoBuilds/swift-fully-automated/README.json b/protoBuilds/swift-fully-automated/README.json
index df8d2dab71..249d2e11fe 100644
--- a/protoBuilds/swift-fully-automated/README.json
+++ b/protoBuilds/swift-fully-automated/README.json
@@ -13,7 +13,7 @@
"description": "\n\n\nWith this protocol, your [OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2) can **fully automate** the entire [Swift 2S Turbo DNA Library Kit](https://swiftbiosci.com/swift-2s-turbo-dna-library-kits/). Simply press start and your OT-2 can automate this entire workflow without any hands-on requirement - from enzymatic prep to sequence ready libraries! Up to 16 libraries can be prepared in under 3 hours.\n\n\nFor more information about the Swift 2S Turbo Kit and the [Swift 2S Turbo Unique Dual Indexing Primer Kit](https://shop.opentrons.com/products/swift-2s-turbo-unique-dual-indexing-primer-kit-96-rxns?_pos=1&_sid=f1fb599e7&_ss=r) on the OT-2, please see our Application Note here: [Rapid high quality next generation sequencing library preparation with Swift 2S Turbo DNA Library Kits on the Opentrons OT-2](https://opentrons-landing-img.s3.amazonaws.com/bundles/swift_automated_ngs_application_note.pdf)\n\n**Update (April 12, 2021)**: This protocol has been updated to fix an issue with the final elution destination when selecting 16 samples.\n**Update (April 13, 2021)**: This protocol has been updated to accommodate the GEN2 Magnetic Module and fix an issue when using 200\u00b5L filter tips.\n**Update (September 2, 2021)**: This protocol has been updated to allow for the [GEN2 P300 8-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette) and has fixed an issue when using 16 samples that caused the 8-channel pipette to go to the wrong source wells during step 1220.\n\n\n---\n\n\nTo purchase consumables, labware, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n\n**Attention**: You can now purchase all of the consumables needed to run this protocol by [clicking here](https://shop.opentrons.com/products/ngs-library-prep-workstation-consumables-refill).\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Swift 2S Turbo DNA Library Kit](https://swiftbiosci.com/swift-2s-turbo-dna-library-kits/)\n* [Swift 2S Turbo Unique Dual Indexing Primer Kit](https://shop.opentrons.com/products/swift-2s-turbo-unique-dual-indexing-primer-kit-96-rxns?_pos=1&_sid=f1fb599e7&_ss=r)\n* [Omega Mag-Bind TotalPure NGS Kit](https://shop.opentrons.com/collections/verified-reagents/products/mag-bind-total-pure-ngs)\n* [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module)\n* [Opentrons Temperature Module with Aluminum Block Set](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette) or Opentrons P50 Single-Channel Pipette*\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-robot/products/8-channel-electronic-pipette)\n* [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [NEST 2mL Tubes](https://shop.opentrons.com/collections/tubes/products/nest-2-0-ml-sample-vial)\n* Samples, resuspended in Low EDTA TE, bringing the total volume to 19.5\u00b5L\n\n\n\n\\*Opentrons now sells the P20 Single-Channel Pipette in place of the P50 Single-Channel Pipette. If you have the P50 Single-Channel Pipette, you can use it for this protocol.\n\n\nFull setup for the entire protocol:\n\n\n\n\n\n\n\n\n---\n\n\n\nSlot 1: [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt) on [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n\n\nSlot 2: [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml) with Reagents\n* A2: Magnetic Beads; recommended volume: 3-4mL\n* A3: 80% Ethanol Solution, Freshly Prepared; recommended volume: 9-10mL\n* A4: 80% Ethanol Solution, Freshly Prepared (if running 16 samples); recommended volume: 9-10mL\n* A6: Low EDTA TE Buffer; recommended volume: 3mL\n\n\nSlot 3: [Opentrons Temperature Module with 24-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) and [NEST 2mL Tubes](https://shop.opentrons.com/collections/tubes/products/nest-2-0-ml-sample-vial) with master mixes (for more information on master mixes, [click here](https://opentrons-protocol-library-website.s3.amazonaws.com/Technical+Notes/swift-fully-automated/Swift+2s+Turbo+Fully+Automated+MasterMix+Volumes.xlsx)) and indices (if automating index addition)\n* A1: Enzymatic Prep Master Mix\n* A2: Ligation Master Mix\n* A3: PCR Master Mix\n* B1: Indexing Reagent 1 (in original container); loaded sequentially (Reagent 2 - B2; Reagent 3 - B3...)\n\n\nSlot 4: [Opentrons Tips for Single-Channel Pipette](https://shop.opentrons.com/collections/opentrons-tips)\n\n\nSlot 5: [Opentrons Tips for P300 8-Channel Pipette](https://shop.opentrons.com/collections/opentrons-tips)\n\n\nSlot 6: [Opentrons Tips for P300 8-Channel Pipette](https://shop.opentrons.com/collections/opentrons-tips)\n\n\nSlot 9: [Opentrons Tips for P300 8-Channel Pipette](https://shop.opentrons.com/collections/opentrons-tips)\n\n\n\nSlot 7/8/10/11: [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module) with samples in a [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* 8 Samples: Column 1\n* 16 Samples: Columns 1 & 2\n\n\n\n__Using the customizations fields, below set up your protocol.__\n* **Number of Samples**: Specify the number of samples (8 or 16) you'd like to run.\n* **Pipette and Tip Type**: Select which pipette (P50 Single-Channel or P20 Single-Channel) and corresponding tips to be used for this protocol. **The pipette should be attached to the left mount.**\n* **P300 8-Channel Pipette Tip Type**: Select which tips (Filter/Non-Filter) for P300 8-Channel Pipette\n* **Automate Indexing**: Specify whether the indices should be added to the samples with the OT-2, or manually.\n* **Number of PCR Cycles**: See suggested cycles [here](https://opentrons-protocol-library-website.s3.amazonaws.com/Technical+Notes/swift-fully-automated/Swift+Fully+Automated+-+PCR+Cycles+Recommendation.xlsx). See Swift 2S Turbo manual for more detailed information.\n* **Fragmentation Time**: Fragmentation time varies depending on *1)* Lot number of the kit and *2)* whether the desired insert size is 200bp or 350bp. Please refer to the manual and Lot number on your kit for more information.\n\n\n\n**Note**: The final 20\u00b5L elution will be transferred to Column 3 if running 8 samples or Columns 5 & 6 if running 16 samples of the PCR plate on the Thermocycler\n\n",
"internal": "swift-fully-automated\n",
"notes": "If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).\n\n",
- "partner": "[Swift Biosciences](https://swiftbiosci.com/)\n\n***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/swift-fully-automated). This page won\u2019t be available after January 31st, 2024.***\n\n",
+ "partner": "[Swift Biosciences](https://swiftbiosci.com/)\n\n# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/swift-fully-automated). This page won\u2019t be available after January 31st, 2024.\n\n",
"process": "\n1. Input your protocol parameters.\n2. Download your protocol.\n3. Upload your protocol into the [OT App](https://opentrons.com/ot-app).\n4. Set up your deck according to the deck map.\n5. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).\n6. Hit \"Run\".\n\n",
"robot": "* [OT-2](https://opentrons.com/ot-2)\n\n",
"title": "Swift 2S Turbo DNA Library Kit Protocol: Fully Automated"
diff --git a/protoBuilds/testdrive/README.json b/protoBuilds/testdrive/README.json
index 93ad8f13d7..01a81793d2 100644
--- a/protoBuilds/testdrive/README.json
+++ b/protoBuilds/testdrive/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Getting Started": [
"OT-2 Guided Walk-through"
@@ -10,7 +10,7 @@
"internal": "testdrive",
"labware": "\nAll well plates found in our labware library can be used in this protocol.\nAll Opentrons tip racks found in our labware library can be used in this protocol.\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Getting Started\n\t* OT-2 Guided Walk-through\n\n",
"deck-setup": "\n\n",
"description": "Learn the OT-2 in under 10 minutes! This protocol will walk you through most of the OT-2 capabilities which include but are not limited to: touch tip, blow out, return tip, and distribute functions (for all functions covered in this protocol, see `Protocol Steps` section below). \n\nUnderstand what each OT-2 function does, and see it in real time before incorporating it into your own biology workflow.\n\nExplanation of complex parameters below:\n* `Well Plate`: Select which well plate will be loaded onto the deck.\n* `Pipette`: Select which single-channel pipette will be loaded onto the deck.\n* `Pipette Tips`: Select which pipette tips will accommodate the pipette you selected above.\n`Pipette Mount`: Select which mount your pipette will be hosted on.\n---\n\n",
diff --git a/protoBuilds/thermocycler-csv/README.json b/protoBuilds/thermocycler-csv/README.json
index c54ce05b49..dc64dc8192 100644
--- a/protoBuilds/thermocycler-csv/README.json
+++ b/protoBuilds/thermocycler-csv/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Thermocycler": [
"Thermocycler with CSV"
@@ -8,7 +8,7 @@
"description": "This is a demo protocol, intended for using the Opentrons Thermocycler Module as a thermocycler. This protocol does not involve any liquid handling, so no pipettes are required to run this protocol. If you are interested in a custom protocol that uses the thermocycler to your lab's specific needs, you can request one from our Applications Engineering Team, using the Protocol Request Form.\nWith this protocol, you can create steps for a a custom thermocycler profile.\nAdditionally, you can specify the lid temperature, sample volume per well, and a final hold temperature that will stay set until you're ready to move the samples.\nFor more information about the Opentrons Thermocycler Module, please see the Thermocycler Module white paper or our Thermocycler Module support article.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Thermocycler Module\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nThe Thermocycler Module should be loaded into its default position in slots 7 and 10 (with some overhang in slots 8 and 11).\nBefore running the protocol with samples, use the button on top of the Thermocycler to open the lid (if closed) and place your sample plate into the Thermocycler. If needed, place an Opentrons Thermocycler Seal on the lid. You do not need to close the lid before running the protocol.\nHow to setup your Thermocycler profiles\n1. Download the template file here\n2. Open the .csv file in a your spreadsheet editor of choice (Google Sheets, Excel, Numbers, etc.)\n3. Export the edited file as .csv\n4. Upload the new file in the field \"profile .csv file\" below.",
"internal": "thermocycler",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Thermocycler\n\t* Thermocycler with CSV\n\n\n",
"description": "\nThis is a demo protocol, intended for using the [Opentrons Thermocycler Module](https://shop.opentrons.com/products/thermocycler-module) as a thermocycler. This protocol does not involve any liquid handling, so no pipettes are required to run this protocol. If you are interested in a custom protocol that uses the thermocycler to your lab's specific needs, you can request one from our Applications Engineering Team, using the [Protocol Request Form](https://opentrons-protocol-dev.paperform.co/).\n\n\nWith this protocol, you can create steps for a a custom thermocycler profile.\n\n\nAdditionally, you can specify the lid temperature, sample volume per well, and a final hold temperature that will stay set until you're ready to move the samples.\n\n\nFor more information about the [Opentrons Thermocycler Module](https://shop.opentrons.com/products/thermocycler-module), please see the [Thermocycler Module white paper](https://opentrons.com/publications/Opentrons-Thermocycler-Module-White-Paper.pdf) or our [Thermocycler Module support article](https://support.opentrons.com/en/articles/3469797-thermocycler-module).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Thermocycler Module](https://shop.opentrons.com/products/thermocycler-module)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n\n---\n\n\nThe Thermocycler Module should be loaded into its default position in slots 7 and 10 (with some overhang in slots 8 and 11).\n\n\nBefore running the protocol with samples, use the button on top of the Thermocycler to open the lid (if closed) and place your sample plate into the Thermocycler. If needed, place an [Opentrons Thermocycler Seal](https://shop.opentrons.com/products/thermocycler-seals) on the lid. You do not need to close the lid before running the protocol.\n\n\n**How to setup your Thermocycler profiles**\n1. Download the template file [here](https://opentrons-protocol-library-website.s3.amazonaws.com/custom-README-images/thermocycler/ex.csv)\n2. Open the `.csv` file in a your spreadsheet editor of choice (Google Sheets, Excel, Numbers, etc.)\n3. Export the edited file as `.csv`\n4. Upload the new file in the field \"profile .csv file\" below.\n\n",
"internal": "thermocycler\n",
diff --git a/protoBuilds/thermocycler/README.json b/protoBuilds/thermocycler/README.json
index cd9d85ad7e..26e067ca23 100644
--- a/protoBuilds/thermocycler/README.json
+++ b/protoBuilds/thermocycler/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Getting Started": [
"Thermocycler Example"
@@ -8,7 +8,7 @@
"description": "This is a demo protocol, intended for using the Opentrons Thermocycler Module as a thermocycler. This protocol does not involve any liquid handling, so no pipettes are required to run this protocol. If you are interested in a custom protocol that uses the thermocycler to your lab's specific needs, you can request one from our Applications Engineering Team, using the Protocol Request Form.\nWith this protocol, you can specify the traditional thermocycler parameters (denaturation temperature/time, annealation temperature/time, elongation temperature/time).\nAdditionally, you can specify the lid temperature, sample volume per well, an initialization temperature/time, a final elongation temperature/time, and a final hold temperature that will stay set until you're ready to move the samples.\nFor more information about the Opentrons Thermocycler Module, please see the Thermocycler Module white paper or our Thermocycler Module support article.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.15.0 or later)\nOpentrons Thermocycler Module\n\nFor more detailed information on compatible labware, please visit our Labware Library.\n\n\nThe Thermocycler Module should be loaded into its default position in slots 7 and 10 (with some overhang in slots 8 and 11).\nBefore running the protocol with samples, use the button on top of the Thermocycler to open the lid (if closed) and place your sample plate into the Thermocycler. If needed, place an Opentrons Thermocycler Seal on the lid. You do not need to close the lid before running the protocol.\nUsing the customizations fields, below set up your thermocycler protocol.\n Sample Volume per Well: Specify how much volume will be in each well for greater accuracy\n Lid Temperature: Specify temperature of lid. We recommend a higher temperature to prevent condensation.\nInitialization\n Initialization Temperature: Specify the temperature during the initialization phase (will not be cycled)\n Initializatin Time: Specify the time during the initialization phase (will not be cycled)\nThermocycling\n Denaturation Temperature: Specify the temperature during the denaturation phase (will be cycled)\n Denaturation Time: Specify the time during the denaturation phase (will be cycled)\n Annealation Temperature: Specify the temperature during the annealation phase (will be cycled)\n Annealation Time: Specify the time during the annealtion phase (will be cycled)\n Elongation Temperature: Specify the temperature during the elongation phase (will be cycled)\n Elongation Time: Specify the time during the elongation phase (will be cycled)\n* Number of Cycles: Specify how many cycles to run the Denaturation-Annealation-Elongation loop\nFinal Elongation and Hold\n Final Elongation Temperature: Specify the temperature during the final elongation phase (will not be cycled)\n Final Elongation Time: Specify the time during the final elongation phase (will not be cycled)\n* Final Hold Temperature: Specify the final temperature the thermocycler will hold indefinitely.",
"internal": "thermocycler",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Getting Started\n\t* Thermocycler Example\n\n\n",
"description": "\nThis is a demo protocol, intended for using the [Opentrons Thermocycler Module](https://shop.opentrons.com/products/thermocycler-module) as a thermocycler. This protocol does not involve any liquid handling, so no pipettes are required to run this protocol. If you are interested in a custom protocol that uses the thermocycler to your lab's specific needs, you can request one from our Applications Engineering Team, using the [Protocol Request Form](https://opentrons-protocol-dev.paperform.co/).\n\n\nWith this protocol, you can specify the traditional thermocycler parameters (denaturation temperature/time, annealation temperature/time, elongation temperature/time).\n\n\nAdditionally, you can specify the lid temperature, sample volume per well, an initialization temperature/time, a final elongation temperature/time, and a final hold temperature that will stay set until you're ready to move the samples.\n\n\nFor more information about the [Opentrons Thermocycler Module](https://shop.opentrons.com/products/thermocycler-module), please see the [Thermocycler Module white paper](https://opentrons.com/publications/Opentrons-Thermocycler-Module-White-Paper.pdf) or our [Thermocycler Module support article](https://support.opentrons.com/en/articles/3469797-thermocycler-module).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.15.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Thermocycler Module](https://shop.opentrons.com/products/thermocycler-module)\n\nFor more detailed information on compatible labware, please visit our [Labware Library](https://labware.opentrons.com/).\n\n\n\n---\n\n\nThe Thermocycler Module should be loaded into its default position in slots 7 and 10 (with some overhang in slots 8 and 11).\n\n\nBefore running the protocol with samples, use the button on top of the Thermocycler to open the lid (if closed) and place your sample plate into the Thermocycler. If needed, place an [Opentrons Thermocycler Seal](https://shop.opentrons.com/products/thermocycler-seals) on the lid. You do not need to close the lid before running the protocol.\n\n\n**Using the customizations fields, below set up your thermocycler protocol.**\n* Sample Volume per Well: Specify how much volume will be in each well for greater accuracy\n* Lid Temperature: Specify temperature of lid. We recommend a higher temperature to prevent condensation.\n\n*Initialization*\n* Initialization Temperature: Specify the temperature during the initialization phase (will not be cycled)\n* Initializatin Time: Specify the time during the initialization phase (will not be cycled)\n\n*Thermocycling*\n* Denaturation Temperature: Specify the temperature during the denaturation phase (will be cycled)\n* Denaturation Time: Specify the time during the denaturation phase (will be cycled)\n* Annealation Temperature: Specify the temperature during the annealation phase (will be cycled)\n* Annealation Time: Specify the time during the annealtion phase (will be cycled)\n* Elongation Temperature: Specify the temperature during the elongation phase (will be cycled)\n* Elongation Time: Specify the time during the elongation phase (will be cycled)\n* Number of Cycles: Specify how many cycles to run the Denaturation-Annealation-Elongation loop\n\n*Final Elongation and Hold*\n* Final Elongation Temperature: Specify the temperature during the final elongation phase (will not be cycled)\n* Final Elongation Time: Specify the time during the final elongation phase (will not be cycled)\n* Final Hold Temperature: Specify the final temperature the thermocycler will hold indefinitely.\n\n\n",
"internal": "thermocycler\n",
diff --git a/protoBuilds/user-upload_ZooMS_Protocol/README.json b/protoBuilds/user-upload_ZooMS_Protocol/README.json
index d264911cd1..182303c802 100644
--- a/protoBuilds/user-upload_ZooMS_Protocol/README.json
+++ b/protoBuilds/user-upload_ZooMS_Protocol/README.json
@@ -1,5 +1,5 @@
{
- "author": "University of York BioArchaeology Team",
+ "author": "University of York BioArchaeology Team\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Sample Prep": [
"Plate Filling"
@@ -9,7 +9,7 @@
"description": "A start to finish protocol for preparation of archaeological bone samples for analysis by MALDI-TOF.\nCurrently configured for 12 or 24 samples, depending on the amount of eppendorf racks available.\nProtocol can be adjusted for non-destructive analysis, and for NaOH wash to remove humic acids.\nSamples are to be laid out left to right (A1 to A12), top to bottom (then B1 to B12).\nThe protocol is split into 5 main steps, with the first two being optional:\n\nHydrochloric acid demineralisation (optional)\nSodium hydroxide wash (optional)\nGelatine extraction\nTrypsin digestion\nPeptide extraction\n\n",
"labware": "\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\nOpentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap\nThis tube rack is optional however, it allows throughput to double from 12 to 24!\n\n\nOpentrons 6 Tube Rack with Falcon 50 mL Conical\nOpentrons 96 Tip Rack 1000 \u00b5L\nOpentrons 96 Tip Rack 1000 \u00b5L\nOpentrons 96 Tip Rack 300 \u00b5L\nNEST 12 Well Reservoir 15 mL\nCorning 96 Well Plate 360 \u00b5L Flat\n",
"markdown": {
- "author": "[University of York BioArchaeology Team](https://www.york.ac.uk/archaeology/centres-facilities/bioarch/)\n\n",
+ "author": "[University of York BioArchaeology Team](https://www.york.ac.uk/archaeology/centres-facilities/bioarch/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Sample Prep\n\t* Plate Filling\n\n",
"deck-setup": "\n\nNote that the 12 well reservoir in slot 11 is simply used as an aqueous waste (AqWaste) container.\n\n* IMPORTANT:\n * If using one rack then the top two rows (A and B) are used for the \"second extract\" eppendorfs (where you place your sample to begin with) and bottom two rows are used for \"Extract\" exppendorfs (where the sample will be processed)\n * If using two racks then the top rack (default deck location: 8) is used to hold samples (labelled as SE) and the bottom rack (default deck location: 5) will hold the extracts (labelled EXT).\n\n",
"description": "A start to finish protocol for preparation of archaeological bone samples for analysis by MALDI-TOF.\nCurrently configured for 12 or 24 samples, depending on the amount of eppendorf racks available.\nProtocol can be adjusted for non-destructive analysis, and for NaOH wash to remove humic acids.\n\nSamples are to be laid out left to right (A1 to A12), top to bottom (then B1 to B12).\n\nThe protocol is split into 5 main steps, with the first two being optional:\n\n1. Hydrochloric acid demineralisation (optional)\n2. Sodium hydroxide wash (optional)\n3. Gelatine extraction\n4. Trypsin digestion\n5. Peptide extraction\n\n---\n\n",
diff --git a/protoBuilds/zymo-quick-96/README.json b/protoBuilds/zymo-quick-96/README.json
index 6d6d62934e..dcfe644bf0 100644
--- a/protoBuilds/zymo-quick-96/README.json
+++ b/protoBuilds/zymo-quick-96/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Zymo Kit"
@@ -10,7 +10,7 @@
"internal": "zymo-quick-96",
"labware": "\n(5) Opentrons 200\u00b5L Filter Tips (recommended) or Opentrons 300\u00b5L Tips\n(1) NEST 96-Well PCR Plate\n(2) NEST 12-Well Reservoir\n(2) NEST 1-Well Reservoir\n(1) VWR Deepwell Plate, 1mL\n",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Zymo Kit\n\n",
"deck-setup": "\n**Slot 1:** [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) with reagents\n**Slot 2:** [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 3:** [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n**Slot 4:**:[NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) with reagents\n**Slot 5:** [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 6:** [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 7:** [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with VWR Deepwell Plate loaded with 300\u00b5L of sample\n**Slot 8:** [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 9:** [Opentrons Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 10:** [NEST 1-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml), empty for liquid waste\n**Slot 11:** [NEST 1-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml), empty for liquid waste\n\n\n",
"description": "This protocol automates nucleic acid purification with the [Zymo Quick-DNA/RNA Viral MagBead Kit](https://www.zymoresearch.com/collections/quick-dna-rna-viral-kits/products/quick-dna-rna-viral-magbead) and is a modifed version of [this protocol](https://protocols.opentrons.com/protocol/zymo-quick). In this version, tips are conserved to allow for a fully automated run of 96 samples.\n\n**Update (July 24, 2021)**: Added a new parameter - *Number of Samples* - to allow user to specify the number of samples used per run.\n \n\nExplanation of complex parameters below:\n**Number of Samples (1-96)**: Specify how many samples are used\n**P300-Multi Generation**: Select which P300-Multi is being used\n**P300-Mult Mount**: Select which mount the pipette is attached to\n**Magnetic Module Generation**: Select which version of the Magnetic Module is used\n\n---\n\n",
diff --git a/protoBuilds/zymo-quick-custom/README.json b/protoBuilds/zymo-quick-custom/README.json
index 9777c8ac96..63f44631a7 100644
--- a/protoBuilds/zymo-quick-custom/README.json
+++ b/protoBuilds/zymo-quick-custom/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Zymo Kit"
@@ -8,7 +8,7 @@
"description": "This protocol automates nucleic acid purification with the Zymo Quick-DNA/RNA Viral MagBead Kit. This protocol is a continuation of the original Zymo Quick Extraction protocol which can be found in our Protocol Library here. More specifically, this protocol distributes PCR mastermix to the plate before the elute is distributed. \n\nUsing the P300 Multi-Channel Pipette and a Single-Channel Pipette, this protocol can accommodate up to 48 samples per run and can be configured to run any multiple of 8 up to 48. Starting with 400\u00b5L of sample in a deepwell plate, the entire process is automated and ends with 60\u00b5L of elution containing nucleic acid dispensed into a 96-well PCR plate.The elution is then ready for RT/qPCR, Next-Gen Sequencing, hybridization, etc.\n\nThis kit can be used for Station B (RNA extraction) of our COVID Workstation.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons Magnetic Module, GEN2\nOpentrons P300 Multi-Channel Pipette\n(5) Opentrons 200\u00b5L Filter Tips (recommended) or Opentrons 300\u00b5L Tips\nOpentrons P20 (or P10) Single-Channel Pipette\nOpentrons 20\u00b5L Filter Tips\nNEST 96-Deep Well Plate, 2mL\nNEST 96-Well PCR Plate\nNEST 12-Well Reservoir\nNEST 1-Well Reservoir\nOpentrons 4-in-1 Tube Rack\n1.5mL Centrifuge Tube\nZymo Quick-DNA/RNA Viral MagBead Kit\nSamples\n15mL Falcon Tube\n\n\n\nReagent Preparation\nPrepare all reagents according to Zymo manual.\n\n\n\n\nProteinase K: In 1.5mL centrifuge tube, placed in \"D1\" in Opentrons 4-in-1 Tube Rack with 24-slot top.\nViral DNA/RNA Buffer + MagBinding Beads: In slots 1, 2, and 3 in the 12-well reservoir. For every column (8 samples), 7mL of Viral DNA/RNA Buffer should be combined with 175\u00b5L of MagBinding Beads. Up to two columns (16 samples) worth of buffer+beads can be prepared in one Falcon Tube at a time. Each slot of the 12 well reservoir can accommodate volume for 2 columns/16 samples and should be loaded sequentially (ex. if running 24 samples, slot 1 would get 14mL buffer + 350\u00b5L beads, slot 2 would get 7mL buffer + 175\u00b5L beads, slot 3 would be empty).\nMagBead DNA/RNA Wash 1: In slots 4 and 5 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 4 would get 13.5mL, slot 5 would be empty).\nMagBead DNA/RNA Wash 2: In slots 6 and 7 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 6 would get 13.5mL, slot 7 would be empty).\nEthanol Wash 1: In slots 8 and 9 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 8 would get 13.5mL, slot 9 would be empty).\nEthanol Wash 2: In slots 10 and 11 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 10 would get 13.5mL, slot 11 would be empty).\nNuclease-Free Water: In slot 12 in the 12-well reservoir. To accommodate 24 samples or less, 4mL of water should be used. For more than 24 samples, 4.5-5mL of water can be used.\nPCR Mastermix: In tube D6 of the tube rack.\n\nDeck Layout\n\n\nSlot 1:: Opentrons 200\u00b5L Filter Tips\nSlot 2:: NEST 12-Well Reservoir with Zymo Kit Reagents\nSlot 3:: NEST 96-Well PCR Plate\nSlot 4:: Opentrons Magnetic Module with NEST 96-Deep Well Plate, 2mL loaded with 400\u00b5L of sample in odd columns\nSlot 5:: Opentrons 10/20\u00b5L Filter Tips\nSlot 6:: Opentrons 200\u00b5L Filter Tips\nSlot 7:: Opentrons 200\u00b5L Filter Tips\nSlot 8:: Opentrons 4-in-1 Tube Rack with Proteinase K in \"D1\"\nSlot 9:: Opentrons 200\u00b5L Filter Tips\nSlot 10:: Opentrons 200\u00b5L Filter Tips\nSlot 11:: NEST 1-Well Reservoir, empty for liquid waste\n\nNote about tips: Each column of samples (8) requires ten columns of tips for this extraction. For 48 samples, all the tips in the five tip racks will be used; if running less than 48 samples, less tips/tipracks will be used and the tipracks will be accessed in this slot order: 1, 6, 9, 7, 10.\n\nUsing the customizations field (below), set up your protocol.\n Number of Samples: Select the number of samples to run\n P300-Multi Generation: Select which mount to place the P300 Gen2 Multi Channel Pipette\n* Single Channel Pipette: Select which mount to place the P20 Gen2 Single Channel Pipette",
"internal": "zymo-quick-pcr",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Zymo Kit\n\n\n",
"description": "This protocol automates nucleic acid purification with the [Zymo Quick-DNA/RNA Viral MagBead Kit](https://www.zymoresearch.com/collections/quick-dna-rna-viral-kits/products/quick-dna-rna-viral-magbead). This protocol is a continuation of the original Zymo Quick Extraction protocol which can be found in our Protocol Library [here](https://protocols.opentrons.com/protocol/zymo-quick). More specifically, this protocol distributes PCR mastermix to the plate before the elute is distributed. \n\nUsing the P300 Multi-Channel Pipette and a Single-Channel Pipette, this protocol can accommodate up to 48 samples per run and can be configured to run any multiple of 8 up to 48. Starting with 400\u00b5L of sample in a deepwell plate, the entire process is automated and ends with 60\u00b5L of elution containing nucleic acid dispensed into a 96-well PCR plate.The elution is then ready for RT/qPCR, Next-Gen Sequencing, hybridization, etc.\n\nThis kit can be used for Station B (RNA extraction) of our [COVID Workstation](https://blog.opentrons.com/how-to-use-opentrons-to-test-for-covid-19/).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module, GEN2](https://opentrons.com/modules#magnetic)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* (5) [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips) (recommended) or [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons P20 (or P10) Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 20\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [NEST 96-Deep Well Plate, 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 1-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [1.5mL Centrifuge Tube](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes)\n* [Zymo Quick-DNA/RNA Viral MagBead Kit](https://www.zymoresearch.com/collections/quick-dna-rna-viral-kits/products/quick-dna-rna-viral-magbead)\n* Samples\n* 15mL Falcon Tube\n\n\n\n---\n\n\n**Reagent Preparation**\nPrepare all reagents according to Zymo manual.\n\n\n\n\n**Proteinase K**: In 1.5mL centrifuge tube, placed in \"D1\" in [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 24-slot top.\n**Viral DNA/RNA Buffer + MagBinding Beads**: In slots 1, 2, and 3 in the 12-well reservoir. For every column (8 samples), 7mL of Viral DNA/RNA Buffer should be combined with 175\u00b5L of MagBinding Beads. Up to two columns (16 samples) worth of buffer+beads can be prepared in one Falcon Tube at a time. Each slot of the 12 well reservoir can accommodate volume for 2 columns/16 samples and should be loaded sequentially (ex. if running 24 samples, slot 1 would get 14mL buffer + 350\u00b5L beads, slot 2 would get 7mL buffer + 175\u00b5L beads, slot 3 would be empty).\n**MagBead DNA/RNA Wash 1**: In slots 4 and 5 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 4 would get 13.5mL, slot 5 would be empty).\n**MagBead DNA/RNA Wash 2**: In slots 6 and 7 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 6 would get 13.5mL, slot 7 would be empty).\n**Ethanol Wash 1**: In slots 8 and 9 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 8 would get 13.5mL, slot 9 would be empty).\n**Ethanol Wash 2**: In slots 10 and 11 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 10 would get 13.5mL, slot 11 would be empty).\n**Nuclease-Free Water**: In slot 12 in the 12-well reservoir. To accommodate 24 samples or less, 4mL of water should be used. For more than 24 samples, 4.5-5mL of water can be used.\n**PCR Mastermix**: In tube D6 of the tube rack.\n\n**Deck Layout**\n\n\n**Slot 1:**: [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 2:**: [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) with Zymo Kit Reagents\n**Slot 3:**: [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n**Slot 4:**: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [NEST 96-Deep Well Plate, 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate) loaded with 400\u00b5L of sample in **odd columns**\n**Slot 5:**: [Opentrons 10/20\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n**Slot 6:**: [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 7:**: [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 8:**: [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with Proteinase K in \"D1\"\n**Slot 9:**: [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 10:**: [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 11:**: [NEST 1-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml), empty for liquid waste\n\n*Note about tips*: Each column of samples (8) requires ten columns of tips for this extraction. For 48 samples, all the tips in the five tip racks will be used; if running less than 48 samples, less tips/tipracks will be used and the tipracks will be accessed in this slot order: 1, 6, 9, 7, 10.\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Samples**: Select the number of samples to run\n* **P300-Multi Generation**: Select which mount to place the P300 Gen2 Multi Channel Pipette\n* **Single Channel Pipette**: Select which mount to place the P20 Gen2 Single Channel Pipette\n\n\n\n",
"internal": "zymo-quick-pcr\n",
diff --git a/protoBuilds/zymo-quick/README.json b/protoBuilds/zymo-quick/README.json
index c6b2692b81..e6a2579c75 100644
--- a/protoBuilds/zymo-quick/README.json
+++ b/protoBuilds/zymo-quick/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Nucleic Acid Extraction & Purification": [
"Zymo Kit"
@@ -8,7 +8,7 @@
"description": "This protocol automates nucleic acid purification with the Zymo Quick-DNA/RNA Viral MagBead Kit.\n\nUsing the P300 Multi-Channel Pipette and a Single-Channel Pipette, this protocol can accommodate up to 48 samples per run and can be configured to run any multiple of 8 up to 48. Starting with 400\u00b5L of sample in a deepwell plate, the entire process is automated and ends with 60\u00b5L of elution containing nucleic acid dispensed into a 96-well PCR plate. The elution is then ready for RT/qPCR, Next-Gen Sequencing, hybridization, etc.\n\nThis kit can be used for Station B (RNA extraction) of our COVID Workstation.\n\n\nTo purchase tips, reagents, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.19.0 or later)\nOpentrons Magnetic Module, GEN1\nOpentrons P300 Multi-Channel Pipette\n(5) Opentrons 200\u00b5L Filter Tips (recommended) or Opentrons 300\u00b5L Tips\nOpentrons P20 (or P10) Single-Channel Pipette\nOpentrons 10/20\u00b5L Filter Tips\nNEST 96-Deep Well Plate, 2mL\nNEST 96-Well PCR Plate\nNEST 12-Well Reservoir\nNEST 1-Well Reservoir\nOpentrons 4-in-1 Tube Rack\n1.5mL Centrifuge Tube\nZymo Quick-DNA/RNA Viral MagBead Kit\nSamples\n15mL Falcon Tube\n\n\n\nReagent Preparation\nPrepare all reagents according to Zymo manual.\n\n\n\n\nProteinase K: In 1.5mL centrifuge tube, placed in \"D1\" in Opentrons 4-in-1 Tube Rack with 24-slot top.\nViral DNA/RNA Buffer + MagBinding Beads: In slots 1, 2, and 3 in the 12-well reservoir. For every column (8 samples), 7mL of Viral DNA/RNA Buffer should be combined with 175\u00b5L of MagBinding Beads. Up to two columns (16 samples) worth of buffer+beads can be prepared in one Falcon Tube at a time. Each slot of the 12 well reservoir can accommodate volume for 2 columns/16 samples and should be loaded sequentially (ex. if running 24 samples, slot 1 would get 14mL buffer + 350\u00b5L beads, slot 2 would get 7mL buffer + 175\u00b5L beads, slot 3 would be empty).\nMagBead DNA/RNA Wash 1: In slots 4 and 5 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 4 would get 13.5mL, slot 5 would be empty).\nMagBead DNA/RNA Wash 2: In slots 6 and 7 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 6 would get 13.5mL, slot 7 would be empty).\nEthanol Wash 1: In slots 8 and 9 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 8 would get 13.5mL, slot 9 would be empty).\nEthanol Wash 2: In slots 10 and 11 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 10 would get 13.5mL, slot 11 would be empty).\nNuclease-Free Water: In slot 12 in the 12-well reservoir. To accommodate 24 samples or less, 4mL of water should be used. For more than 24 samples, 4.5-5mL of water can be used.\n\nDeck Layout\n\n\nSlot 1:: Opentrons 200\u00b5L Filter Tips\nSlot 2:: NEST 12-Well Reservoir with Zymo Kit Reagents\nSlot 3:: NEST 96-Well PCR Plate\nSlot 4:: Opentrons Magnetic Module with NEST 96-Deep Well Plate, 2mL loaded with 400\u00b5L of sample in odd columns\nSlot 5:: Opentrons 10/20\u00b5L Filter Tips\nSlot 6:: Opentrons 200\u00b5L Filter Tips\nSlot 7:: Opentrons 200\u00b5L Filter Tips\nSlot 8:: Opentrons 4-in-1 Tube Rack with Proteinase K in \"D1\"\nSlot 9:: Opentrons 200\u00b5L Filter Tips\nSlot 10:: Opentrons 200\u00b5L Filter Tips\nSlot 11:: NEST 1-Well Reservoir, empty for liquid waste\n\nNote about tips: Each column of samples (8) requires ten columns of tips for this extraction. For 48 samples, all the tips in the five tip racks will be used; if running less than 48 samples, less tips/tipracks will be used and the tipracks will be accessed in this slot order: 1, 6, 9, 7, 10.\n\nUsing the customizations field (below), set up your protocol.\n Number of Samples: Select the number of samples to run\n P300-Multi Generation: Select which generation P300-Multi is being used\n* Single Channel Pipette: Select which single channel pipette is being used",
"internal": "zymo-quick",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Nucleic Acid Extraction & Purification\n\t* Zymo Kit\n\n\n",
"description": "This protocol automates nucleic acid purification with the [Zymo Quick-DNA/RNA Viral MagBead Kit](https://www.zymoresearch.com/collections/quick-dna-rna-viral-kits/products/quick-dna-rna-viral-magbead).\n\nUsing the P300 Multi-Channel Pipette and a Single-Channel Pipette, this protocol can accommodate up to 48 samples per run and can be configured to run any multiple of 8 up to 48. Starting with 400\u00b5L of sample in a deepwell plate, the entire process is automated and ends with 60\u00b5L of elution containing nucleic acid dispensed into a 96-well PCR plate. The elution is then ready for RT/qPCR, Next-Gen Sequencing, hybridization, etc.\n\nThis kit can be used for Station B (RNA extraction) of our [COVID Workstation](https://blog.opentrons.com/how-to-use-opentrons-to-test-for-covid-19/).\n\n---\n\n\nTo purchase tips, reagents, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.19.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module, GEN1](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons P300 Multi-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* (5) [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips) (recommended) or [Opentrons 300\u00b5L Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-300ul-tips)\n* [Opentrons P20 (or P10) Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 10/20\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n* [NEST 96-Deep Well Plate, 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [NEST 1-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml)\n* [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1)\n* [1.5mL Centrifuge Tube](https://shop.opentrons.com/collections/verified-consumables/products/nest-microcentrifuge-tubes)\n* [Zymo Quick-DNA/RNA Viral MagBead Kit](https://www.zymoresearch.com/collections/quick-dna-rna-viral-kits/products/quick-dna-rna-viral-magbead)\n* Samples\n* 15mL Falcon Tube\n\n\n\n---\n\n\n**Reagent Preparation**\nPrepare all reagents according to Zymo manual.\n\n\n\n\n**Proteinase K**: In 1.5mL centrifuge tube, placed in \"D1\" in [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with 24-slot top.\n**Viral DNA/RNA Buffer + MagBinding Beads**: In slots 1, 2, and 3 in the 12-well reservoir. For every column (8 samples), 7mL of Viral DNA/RNA Buffer should be combined with 175\u00b5L of MagBinding Beads. Up to two columns (16 samples) worth of buffer+beads can be prepared in one Falcon Tube at a time. Each slot of the 12 well reservoir can accommodate volume for 2 columns/16 samples and should be loaded sequentially (ex. if running 24 samples, slot 1 would get 14mL buffer + 350\u00b5L beads, slot 2 would get 7mL buffer + 175\u00b5L beads, slot 3 would be empty).\n**MagBead DNA/RNA Wash 1**: In slots 4 and 5 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 4 would get 13.5mL, slot 5 would be empty).\n**MagBead DNA/RNA Wash 2**: In slots 6 and 7 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 6 would get 13.5mL, slot 7 would be empty).\n**Ethanol Wash 1**: In slots 8 and 9 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 8 would get 13.5mL, slot 9 would be empty).\n**Ethanol Wash 2**: In slots 10 and 11 in the 12-well reservoir. Each slot can accommodate enough volume for 24 samples (3 columns). Each slot would get 13.5mL if running 48 samples and would be scaled back if running less (ex. if running 24 samples, slot 10 would get 13.5mL, slot 11 would be empty).\n**Nuclease-Free Water**: In slot 12 in the 12-well reservoir. To accommodate 24 samples or less, 4mL of water should be used. For more than 24 samples, 4.5-5mL of water can be used.\n\n**Deck Layout**\n\n\n**Slot 1:**: [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 2:**: [NEST 12-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml) with Zymo Kit Reagents\n**Slot 3:**: [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n**Slot 4:**: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [NEST 96-Deep Well Plate, 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate) loaded with 400\u00b5L of sample in **odd columns**\n**Slot 5:**: [Opentrons 10/20\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-20ul-filter-tips)\n**Slot 6:**: [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 7:**: [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 8:**: [Opentrons 4-in-1 Tube Rack](https://shop.opentrons.com/collections/verified-labware/products/tube-rack-set-1) with Proteinase K in \"D1\"\n**Slot 9:**: [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 10:**: [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips/products/opentrons-200ul-filter-tips)\n**Slot 11:**: [NEST 1-Well Reservoir](https://shop.opentrons.com/collections/verified-labware/products/nest-1-well-reservoir-195-ml), empty for liquid waste\n\n*Note about tips*: Each column of samples (8) requires ten columns of tips for this extraction. For 48 samples, all the tips in the five tip racks will be used; if running less than 48 samples, less tips/tipracks will be used and the tipracks will be accessed in this slot order: 1, 6, 9, 7, 10.\n\n**Using the customizations field (below), set up your protocol.**\n* **Number of Samples**: Select the number of samples to run\n* **P300-Multi Generation**: Select which generation P300-Multi is being used\n* **Single Channel Pipette**: Select which single channel pipette is being used\n\n\n\n",
"internal": "zymo-quick\n",
diff --git a/protoBuilds/zymo-ribofree-cleanup/README.json b/protoBuilds/zymo-ribofree-cleanup/README.json
index 9d6c13b193..7258b64fe5 100644
--- a/protoBuilds/zymo-ribofree-cleanup/README.json
+++ b/protoBuilds/zymo-ribofree-cleanup/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Zymo RiboFree\u2122 Total RNA Library Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs Select-a-Size MagBead Clean-up for the Zymo-Seq RiboFree\u2122 Total RNA Library Prep. This protocol is meant to be run on a second OT-2 in conjunction with another OT-2 running the rest of the library prep (first strand cDNA synthesis, universal depletion, P7 and P5 adapter ligations, and library index PCR).\nSamples will be processed down columns and then across rows (A1, B1, C1, ... A2, B2, etc.). Thoroughly mix reagent tubes thoroughly by flicking or pipetting before starting. Briefly spin down and load onto the temperature module according to Setup below.\nThe user is prompted to replace tipracks mid-protocol when necessary (for > 76 samples).\nLinks:\n First Strand cDNA Synthesis and RiboFreeTM Universal Depletion (robot 1)\n P7 Adapter Ligation (robot 1)\n P5 Adapter Ligation (robot 1)\n Library Index PCR (robot 1)\n* Select-a-Size Magbead Clean-up (robot 2)\n\n\n\nOpentrons Temperature Module GEN2\nOpentrons Magnetic Module GEN2 for NEST 96-well PCR plate, full skirt\nNEST 12-channel reservoir 15ml\nOpentrons P20 and P300 GEN2 multi-channel pipettes\nOpentrons 20\u00b5l and 50/300\u00b5l tipracks\n\n\n\n12-channel reservoir (slot 2)\n",
"internal": "zymo-ribofree",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t*\tZymo RiboFree\u2122 Total RNA Library Prep\n\n",
"description": "This protocol performs Select-a-Size MagBead Clean-up for the [Zymo-Seq RiboFree\u2122 Total RNA Library Prep](https://files.zymoresearch.com/protocols/r3000_zymo-seq_ribofree_total_rna_library_kit.pdf). This protocol is meant to be run on a second OT-2 in conjunction with another OT-2 running the rest of the library prep (first strand cDNA synthesis, universal depletion, P7 and P5 adapter ligations, and library index PCR).\n\nSamples will be processed down columns and then across rows (A1, B1, C1, ... A2, B2, etc.). Thoroughly mix reagent tubes thoroughly by flicking or pipetting before starting. Briefly spin down and load onto the temperature module according to `Setup` below.\n\nThe user is prompted to replace tipracks mid-protocol when necessary (for > 76 samples).\n\nLinks:\n* [First Strand cDNA Synthesis and RiboFreeTM Universal Depletion (robot 1)](./zymo-ribofree-first-strand-cdna-synth-universal-depletion)\n* [P7 Adapter Ligation (robot 1)](./zymo-ribofree-p7-adapter-ligation)\n* [P5 Adapter Ligation (robot 1)](./zymo-ribofree-p5-adapter-ligation)\n* [Library Index PCR (robot 1)](./zymo-ribofree-library-index-pcr)\n* [Select-a-Size Magbead Clean-up (robot 2)](./zymo-ribofree-cleanup)\n\n---\n\n\n* [Opentrons Temperature Module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Opentrons Magnetic Module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) for [NEST 96-well PCR plate, full skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-channel reservoir 15ml](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [Opentrons P20 and P300 GEN2 multi-channel pipettes](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons 20\u00b5l and 50/300\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n\n---\n\n\n12-channel reservoir (slot 2) \n\n\n",
"internal": "zymo-ribofree\n",
diff --git a/protoBuilds/zymo-ribofree-first-strand-cdna-synth-universal-depletion/README.json b/protoBuilds/zymo-ribofree-first-strand-cdna-synth-universal-depletion/README.json
index e6a362f943..cfc17e3a74 100644
--- a/protoBuilds/zymo-ribofree-first-strand-cdna-synth-universal-depletion/README.json
+++ b/protoBuilds/zymo-ribofree-first-strand-cdna-synth-universal-depletion/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Zymo RiboFree\u2122 Total RNA Library Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs First-Strand cDNA Synthesis and RiboFreeTM Universal Depletion for the Zymo-Seq RiboFree\u2122 Total RNA Library Prep. This protocol is meant to be run on one OT-2 in conjunction with a second OT-2 running solely Select-a-Size MagBead Clean-up Protocol.\nSamples will be processed down columns and then across rows (A1, B1, C1, ... A2, B2, etc.). Thoroughly mix reagent tubes thoroughly by flicking or pipetting before starting. Briefly spin down and load onto the temperature module according to Setup below.\nThe user is prompted to replace tipracks mid-protocol when necessary (for > 76 samples).\nLinks:\n First Strand cDNA Synthesis and RiboFreeTM Universal Depletion (robot 1)\n P7 Adapter Ligation (robot 1)\n P5 Adapter Ligation (robot 1)\n Library Index PCR (robot 1)\n* Select-a-Size Magbead Clean-up (robot 2)\n\n\n\nOpentrons Thermocycler Module with NEST 96-well PCR plate, full skirt\nOpentrons Temperature Module GEN2 with 4x6 aluminum block for NEST 1.5ml screwcap reagent tubes\nNEST 12-channel reservoir 15ml\nOpentrons P20 GEN2 single-channel pipette\nOpentrons P20 GEN2 multi-channel pipette\nOpentrons 20\u00b5l tipracks\n\n\n\n4x6 aluminum block on temperature module (slot 1)\n\n12-channel reservoir (slot 2)\n",
"internal": "zymo-ribofree",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t*\tZymo RiboFree\u2122 Total RNA Library Prep\n\n",
"description": "This protocol performs First-Strand cDNA Synthesis and RiboFreeTM Universal Depletion for the [Zymo-Seq RiboFree\u2122 Total RNA Library Prep](https://files.zymoresearch.com/protocols/r3000_zymo-seq_ribofree_total_rna_library_kit.pdf). This protocol is meant to be run on one OT-2 in conjunction with a second OT-2 running solely Select-a-Size MagBead Clean-up Protocol.\n\nSamples will be processed down columns and then across rows (A1, B1, C1, ... A2, B2, etc.). Thoroughly mix reagent tubes thoroughly by flicking or pipetting before starting. Briefly spin down and load onto the temperature module according to `Setup` below.\n\nThe user is prompted to replace tipracks mid-protocol when necessary (for > 76 samples).\n\nLinks:\n* [First Strand cDNA Synthesis and RiboFreeTM Universal Depletion (robot 1)](./zymo-ribofree-first-strand-cdna-synth-universal-depletion)\n* [P7 Adapter Ligation (robot 1)](./zymo-ribofree-p7-adapter-ligation)\n* [P5 Adapter Ligation (robot 1)](./zymo-ribofree-p5-adapter-ligation)\n* [Library Index PCR (robot 1)](./zymo-ribofree-library-index-pcr)\n* [Select-a-Size Magbead Clean-up (robot 2)](./zymo-ribofree-cleanup)\n\n---\n\n\n* [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module) with [NEST 96-well PCR plate, full skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons Temperature Module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [4x6 aluminum block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) for [NEST 1.5ml screwcap reagent tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-1-5-ml-sample-vial)\n* [NEST 12-channel reservoir 15ml](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [Opentrons P20 GEN2 single-channel pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons P20 GEN2 multi-channel pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette)\n* [Opentrons 20\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n\n---\n\n\n4x6 aluminum block on temperature module (slot 1) \n\n\n12-channel reservoir (slot 2) \n\n\n",
"internal": "zymo-ribofree\n",
diff --git a/protoBuilds/zymo-ribofree-library-index-pcr/README.json b/protoBuilds/zymo-ribofree-library-index-pcr/README.json
index c559b254bb..a3c9e26218 100644
--- a/protoBuilds/zymo-ribofree-library-index-pcr/README.json
+++ b/protoBuilds/zymo-ribofree-library-index-pcr/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Zymo RiboFree\u2122 Total RNA Library Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs Library Index PCR for the Zymo-Seq RiboFree\u2122 Total RNA Library Prep. This protocol is meant to be run on one OT-2 in conjunction with a second OT-2 running solely Select-a-Size MagBead Clean-up Protocol.\nSamples will be processed down columns and then across rows (A1, B1, C1, ... A2, B2, etc.). Indexes on the UDI Primer plate will be transferred to the corresponding well of the sample plate on the thermocycler module. Thoroughly mix reagent tubes thoroughly by flicking or pipetting before starting. Briefly spin down and load onto the temperature module according to Setup below.\nLinks:\n First Strand cDNA Synthesis and RiboFreeTM Universal Depletion (robot 1)\n P7 Adapter Ligation (robot 1)\n P5 Adapter Ligation (robot 1)\n Library Index PCR (robot 1)\n* Select-a-Size Magbead Clean-up (robot 2)\n\n\n\nOpentrons Thermocycler Module with NEST 96-well PCR plate, full skirt\nOpentrons Temperature Module GEN2 with 4x6 aluminum block for NEST 1.5ml screwcap reagent tubes\nNEST 12-channel reservoir 15ml\nOpentrons P20 GEN2 single-channel pipette\nOpentrons P50 GEN1 multi-channel pipette\nOpentrons 20\u00b5l and 50/300\u00b5l tipracks\n\n\n\n4x6 aluminum block on temperature module (slot 1)\n\n12-channel reservoir (slot 2)\n",
"internal": "zymo-ribofree",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t*\tZymo RiboFree\u2122 Total RNA Library Prep\n\n",
"description": "This protocol performs Library Index PCR for the [Zymo-Seq RiboFree\u2122 Total RNA Library Prep](https://files.zymoresearch.com/protocols/r3000_zymo-seq_ribofree_total_rna_library_kit.pdf). This protocol is meant to be run on one OT-2 in conjunction with a second OT-2 running solely Select-a-Size MagBead Clean-up Protocol.\n\nSamples will be processed down columns and then across rows (A1, B1, C1, ... A2, B2, etc.). Indexes on the UDI Primer plate will be transferred to the corresponding well of the sample plate on the thermocycler module. Thoroughly mix reagent tubes thoroughly by flicking or pipetting before starting. Briefly spin down and load onto the temperature module according to `Setup` below.\n\nLinks:\n* [First Strand cDNA Synthesis and RiboFreeTM Universal Depletion (robot 1)](./zymo-ribofree-first-strand-cdna-synth-universal-depletion)\n* [P7 Adapter Ligation (robot 1)](./zymo-ribofree-p7-adapter-ligation)\n* [P5 Adapter Ligation (robot 1)](./zymo-ribofree-p5-adapter-ligation)\n* [Library Index PCR (robot 1)](./zymo-ribofree-library-index-pcr)\n* [Select-a-Size Magbead Clean-up (robot 2)](./zymo-ribofree-cleanup)\n\n---\n\n\n* [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module) with [NEST 96-well PCR plate, full skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons Temperature Module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [4x6 aluminum block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) for [NEST 1.5ml screwcap reagent tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-1-5-ml-sample-vial)\n* [NEST 12-channel reservoir 15ml](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [Opentrons P20 GEN2 single-channel pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons P50 GEN1 multi-channel pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/8-channel-electronic-pipette?variant=5984202457117)\n* [Opentrons 20\u00b5l and 50/300\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n\n---\n\n\n4x6 aluminum block on temperature module (slot 1) \n\n\n12-channel reservoir (slot 2) \n\n\n",
"internal": "zymo-ribofree\n",
diff --git a/protoBuilds/zymo-ribofree-p5-adapter-ligation/README.json b/protoBuilds/zymo-ribofree-p5-adapter-ligation/README.json
index 31cecdf9e2..9f1574cb84 100644
--- a/protoBuilds/zymo-ribofree-p5-adapter-ligation/README.json
+++ b/protoBuilds/zymo-ribofree-p5-adapter-ligation/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Zymo RiboFree\u2122 Total RNA Library Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs P5 Adapter Ligation for the Zymo-Seq RiboFree\u2122 Total RNA Library Prep. This protocol is meant to be run on one OT-2 in conjunction with a second OT-2 running solely Select-a-Size MagBead Clean-up Protocol.\nSamples will be processed down columns and then across rows (A1, B1, C1, ... A2, B2, etc.). Thoroughly mix reagent tubes thoroughly by flicking or pipetting before starting. Briefly spin down and load onto the temperature module according to Setup below.\nThe user is prompted to replace tipracks mid-protocol when necessary (for > 76 samples).\nLinks:\n First Strand cDNA Synthesis and RiboFreeTM Universal Depletion (robot 1)\n P7 Adapter Ligation (robot 1)\n P5 Adapter Ligation (robot 1)\n Library Index PCR (robot 1)\n* Select-a-Size Magbead Clean-up (robot 2)\n\n\n\nOpentrons Thermocycler Module with NEST 96-well PCR plate, full skirt\nOpentrons Temperature Module GEN2 with 4x6 aluminum block for NEST 1.5ml screwcap reagent tubes\nNEST 12-channel reservoir 15ml\nOpentrons P20 GEN2 single-channel pipette\nOpentrons P20 GEN2 multi-channel pipette\nOpentrons 20\u00b5l tipracks\n\n\n\n4x6 aluminum block on temperature module (slot 1)\n\n12-channel reservoir (slot 2)\n",
"internal": "zymo-ribofree",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t*\tZymo RiboFree\u2122 Total RNA Library Prep\n\n",
"description": "This protocol performs P5 Adapter Ligation for the [Zymo-Seq RiboFree\u2122 Total RNA Library Prep](https://files.zymoresearch.com/protocols/r3000_zymo-seq_ribofree_total_rna_library_kit.pdf). This protocol is meant to be run on one OT-2 in conjunction with a second OT-2 running solely Select-a-Size MagBead Clean-up Protocol.\n\nSamples will be processed down columns and then across rows (A1, B1, C1, ... A2, B2, etc.). Thoroughly mix reagent tubes thoroughly by flicking or pipetting before starting. Briefly spin down and load onto the temperature module according to `Setup` below.\n\nThe user is prompted to replace tipracks mid-protocol when necessary (for > 76 samples).\n\nLinks:\n* [First Strand cDNA Synthesis and RiboFreeTM Universal Depletion (robot 1)](./zymo-ribofree-first-strand-cdna-synth-universal-depletion)\n* [P7 Adapter Ligation (robot 1)](./zymo-ribofree-p7-adapter-ligation)\n* [P5 Adapter Ligation (robot 1)](./zymo-ribofree-p5-adapter-ligation)\n* [Library Index PCR (robot 1)](./zymo-ribofree-library-index-pcr)\n* [Select-a-Size Magbead Clean-up (robot 2)](./zymo-ribofree-cleanup)\n\n---\n\n\n* [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module) with [NEST 96-well PCR plate, full skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons Temperature Module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [4x6 aluminum block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) for [NEST 1.5ml screwcap reagent tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-1-5-ml-sample-vial)\n* [NEST 12-channel reservoir 15ml](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [Opentrons P20 GEN2 single-channel pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons P20 GEN2 multi-channel pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 20\u00b5l tipracks](https://shop.opentrons.com/collections/opentrons-tips)\n\n---\n\n\n4x6 aluminum block on temperature module (slot 1) \n\n\n12-channel reservoir (slot 2) \n\n\n",
"internal": "zymo-ribofree\n",
diff --git a/protoBuilds/zymo-ribofree-p7-adapter-ligation/README.json b/protoBuilds/zymo-ribofree-p7-adapter-ligation/README.json
index c4f4e38867..4e9a0f9344 100644
--- a/protoBuilds/zymo-ribofree-p7-adapter-ligation/README.json
+++ b/protoBuilds/zymo-ribofree-p7-adapter-ligation/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"NGS Library Prep": [
"Zymo RiboFree\u2122 Total RNA Library Prep"
@@ -8,7 +8,7 @@
"description": "This protocol performs P7 Adapter Ligation for the Zymo-Seq RiboFree\u2122 Total RNA Library Prep. This protocol is meant to be run on one OT-2 in conjunction with a second OT-2 running solely Select-a-Size MagBead Clean-up Protocol.\nSamples will be processed down columns and then across rows (A1, B1, C1, ... A2, B2, etc.). Thoroughly mix reagent tubes thoroughly by flicking or pipetting before starting. Briefly spin down and load onto the temperature module according to Setup below.\nThe user is prompted to replace tipracks mid-protocol when necessary (for > 76 samples).\nLinks:\n First Strand cDNA Synthesis and RiboFreeTM Universal Depletion (robot 1)\n P7 Adapter Ligation (robot 1)\n P5 Adapter Ligation (robot 1)\n Library Index PCR (robot 1)\n* Select-a-Size Magbead Clean-up (robot 2)\n\n\n\nOpentrons Thermocycler Module with NEST 96-well PCR plate, full skirt\nOpentrons Temperature Module GEN2 with 4x6 aluminum block for NEST 1.5ml screwcap reagent tubes\nNEST 12-channel reservoir 15ml\nOpentrons P20 GEN2 single-channel pipette\nOpentrons 20\u00b5l\n\n\n\n4x6 aluminum block on temperature module (slot 1)\n\n12-channel reservoir (slot 2)\n",
"internal": "zymo-ribofree",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* NGS Library Prep\n\t*\tZymo RiboFree\u2122 Total RNA Library Prep\n\n",
"description": "This protocol performs P7 Adapter Ligation for the [Zymo-Seq RiboFree\u2122 Total RNA Library Prep](https://files.zymoresearch.com/protocols/r3000_zymo-seq_ribofree_total_rna_library_kit.pdf). This protocol is meant to be run on one OT-2 in conjunction with a second OT-2 running solely Select-a-Size MagBead Clean-up Protocol.\n\nSamples will be processed down columns and then across rows (A1, B1, C1, ... A2, B2, etc.). Thoroughly mix reagent tubes thoroughly by flicking or pipetting before starting. Briefly spin down and load onto the temperature module according to `Setup` below.\n\nThe user is prompted to replace tipracks mid-protocol when necessary (for > 76 samples).\n\nLinks:\n* [First Strand cDNA Synthesis and RiboFreeTM Universal Depletion (robot 1)](./zymo-ribofree-first-strand-cdna-synth-universal-depletion)\n* [P7 Adapter Ligation (robot 1)](./zymo-ribofree-p7-adapter-ligation)\n* [P5 Adapter Ligation (robot 1)](./zymo-ribofree-p5-adapter-ligation)\n* [Library Index PCR (robot 1)](./zymo-ribofree-library-index-pcr)\n* [Select-a-Size Magbead Clean-up (robot 2)](./zymo-ribofree-cleanup)\n\n---\n\n\n* [Opentrons Thermocycler Module](https://shop.opentrons.com/collections/hardware-modules/products/thermocycler-module) with [NEST 96-well PCR plate, full skirt](https://shop.opentrons.com/collections/verified-labware/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [Opentrons Temperature Module GEN2](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with [4x6 aluminum block](https://shop.opentrons.com/collections/hardware-modules/products/aluminum-block-set) for [NEST 1.5ml screwcap reagent tubes](https://shop.opentrons.com/collections/verified-consumables/products/nest-1-5-ml-sample-vial)\n* [NEST 12-channel reservoir 15ml](https://shop.opentrons.com/collections/verified-labware/products/nest-12-well-reservoir-15-ml)\n* [Opentrons P20 GEN2 single-channel pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons 20\u00b5l](https://shop.opentrons.com/collections/opentrons-tips)\n\n---\n\n\n4x6 aluminum block on temperature module (slot 1) \n\n\n12-channel reservoir (slot 2) \n\n\n",
"internal": "zymo-ribofree\n",
diff --git a/protoBuilds/zymo-rna-extraction/README.json b/protoBuilds/zymo-rna-extraction/README.json
index 041c10137c..c89f4ad925 100644
--- a/protoBuilds/zymo-rna-extraction/README.json
+++ b/protoBuilds/zymo-rna-extraction/README.json
@@ -1,5 +1,5 @@
{
- "author": "Opentrons",
+ "author": "Opentrons\nOpentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. Submit a request to add this protocol to the new library.",
"categories": {
"Covid Workstation": [
"RNA Extraction"
@@ -8,7 +8,7 @@
"description": "This protocol automates the Zymo Quick-DNA/RNA Viral MagBead Extraction Kit. This kit is designed for high-throughput purification of viral DNA and/or RNA and the isolated nucleic acids are ready for all downstream applications, such as RT-qPCR, Next-Gen Sequencing, and many others.\n\nThis kit can be used for Station B (RNA extraction) as part of our COVID Workstation.\n\n\nTo purchase consumables, labware, or pipettes, please visit our online store or contact our sales team at info@opentrons.com\n\nOpentrons OT-2\nOpentrons OT-2 Run App (Version 3.16.0 or later)\nOpentrons Magnetic Module\nOpentrons Temperature Module with 96-Well Aluminum Block\nOpentrons P20 Single-Channel Pipette\nOpentrons P300 8-Channel Pipette, Gen 2\nOpentrons 200\u00b5L Filter Tips\nOpentrons 20\u00b5L Filter Tips\nOpentrons 4-in-1 Tube Rack with 24-Well Top\nNEST 1.5mL Microcentrifuge Tubes\nNEST 96-Deepwell Plate, 2mL\nNEST 96-Well PCR Plate\nNEST 12-Well Reservoir, 15mL\nNEST 1-Well Reservoir, 195mL\nZymo Quick-DNA/RNA Viral MagBead Kit\nEthanol, 95-100%\nSample, 400\u00b5L per well\n\n\n\nNote - Before running this protocol, make sure you clean the OT-2. For more information on cleaning the OT-2, check out this guide.\n\nThe setup for this protocol varies slightly, depending on the number of samples the user selects.\n\nLabware Setup\n\nSlot 2: NEST 12-Well Reservoir, 15mL with reagents (see below)\n\nSlot 3: Opentrons Temperature Module with 96-Well Aluminum Block with clean and empty NEST 96-Well PCR Plate\n\nSlot 4: Opentrons Magnetic Module with NEST 96-Deepwell Plate, 2mL, containing samples\nSamples should be loaded in odd-numbered columns only\n\nSlot 5: Opentrons 20\u00b5L Filter Tips\n\nSlot 8: Opentrons 4-in-1 Tube Rack with 24-Well Top, containing reagents in NEST 1.5mL Microcentrifuge Tubes (see below)\n\nOpentrons 200\u00b5L Filter Tips\n if 8 samples, Slot 1\n if 16 samples, Slots 1 and 6\n if 24 samples, Slots 1, 6, and 9\n if 32 samples, Slots 1, 6, 7, and 9\n if 40 or 48 samples, Slots 1, 6, 7, 9, and 10\n\n\nReagent Setup\n\nOpentrons 4-in-1 Tube Rack with 24-Well Top\nD1: Proteinase K, 4\u00b5L per sample\nD6: Extraction Control Spike-In, if using\n\nNEST 12-Well Reservoir, 15mL\nFor every 8 samples, the following reagents are needed:\n 7mL Zymo Viral DNA/RNA Buffer + 350\u00b5L of Zymo MagBeads\n 4.5mL of Zymo Wash Buffer 1\n 4.5mL of Zymo Wash Buffer 2\n 4.5mL of Ethanol (Ethanol Wash 1)\n* 4.5mL Ethanol (Ethanol Wash 2)\n\nSlot 1: Zymo Viral DNA/RNA Buffer + Zymo MagBeads, 8/16 samples\nSlot 2: Zymo Viral DNA/RNA Buffer + Zymo MagBeads, 24/32 samples\nSlot 3: Zymo Viral DNA/RNA Buffer + Zymo MagBeads, 40/48 samples\nSlot 4: Zymo Wash Buffer 1, 8/16/24 samples\nSlot 5: Zymo Wash Buffer 1, 32/40/48 samples\nSlot 6: Zymo Wash Buffer 2, 8/16/24 samples\nSlot 7: Zymo Wash Buffer 2, 32/40/48 samples\nSlot 8: Ethanol (Ethanol Wash 1), 8/16/24 samples\nSlot 9: Ethanol (Ethanol Wash 1), 32/40/48 samples\nSlot 10: Ethanol (Ethanol Wash 2), 8/16/24 samples\nSlot 11: Ethanol (Ethanol Wash 2), 32/40/48 samples\nSlot 12: Nuclease-Free Water, 3.5-4mL\n\nFull Deck Layout:\n\nUsing the customizations fields, below set up your protocol.\n Number of Samples: Specify the number of samples you'd like to run.\n Amount of Spike-in: Specify how much control RNA (or other spike-in) should be added to each well. If not using a spike-in, leave as 0.\n* Return tips after wash step: Specify whether to drop tips into trash bin after use, or to replace in tip rack for easy disposal.",
"internal": "zymo-rna-extraction",
"markdown": {
- "author": "[Opentrons](https://opentrons.com/)\n\n",
+ "author": "[Opentrons](https://opentrons.com/)\n\n\n# Opentrons has launched a new Protocol Library. This page won\u2019t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.\n\n",
"categories": "* Covid Workstation\n\t* RNA Extraction\n\n\n",
"description": "This protocol automates the [Zymo Quick-DNA/RNA Viral MagBead Extraction Kit](https://www.zymoresearch.com/collections/quick-dna-rna-viral-kits/products/quick-dna-rna-viral-magbead). This kit is designed for high-throughput purification of viral DNA and/or RNA and the isolated nucleic acids are ready for all downstream applications, such as RT-qPCR, Next-Gen Sequencing, and many others.
\n
\nThis kit can be used for Station B (RNA extraction) as part of our [COVID Workstation](https://blog.opentrons.com/how-to-use-opentrons-to-test-for-covid-19/).\n\n\n\n---\n\n\nTo purchase consumables, labware, or pipettes, please visit our [online store](https://shop.opentrons.com/) or contact our sales team at [info@opentrons.com](mailto:info@opentrons.com)\n\n* [Opentrons OT-2](https://shop.opentrons.com/collections/ot-2-robot/products/ot-2)\n* [Opentrons OT-2 Run App (Version 3.16.0 or later)](https://opentrons.com/ot-app/)\n* [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck)\n* [Opentrons Temperature Module with 96-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck)\n* [Opentrons P20 Single-Channel Pipette](https://shop.opentrons.com/collections/ot-2-pipettes/products/single-channel-electronic-pipette)\n* [Opentrons P300 8-Channel Pipette, Gen 2](https://opentrons.com/pipettes/)\n* [Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons 20\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips)\n* [Opentrons 4-in-1 Tube Rack with 24-Well Top](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)\n* [NEST 1.5mL Microcentrifuge Tubes](https://shop.opentrons.com/collections/tubes/products/nest-microcentrifuge-tubes)\n* [NEST 96-Deepwell Plate, 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate)\n* [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)\n* [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)\n* [NEST 1-Well Reservoir, 195mL](https://labware.opentrons.com/nest_1_reservoir_195ml?category=reservoir)\n* [Zymo Quick-DNA/RNA Viral MagBead Kit](https://www.zymoresearch.com/collections/quick-dna-rna-viral-kits/products/quick-dna-rna-viral-magbead)\n* Ethanol, 95-100%\n* Sample, 400\u00b5L per well\n\n\n---\n\n\nNote - Before running this protocol, make sure you clean the OT-2. For more information on cleaning the OT-2, check out [this guide](https://support.opentrons.com/en/articles/1795522-cleaning-your-ot-2).
\n
\nThe setup for this protocol varies slightly, depending on the number of samples the user selects.
\n
\n**Labware Setup**
\n
\nSlot 2: [NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml) with reagents (see below)
\n
\nSlot 3: [Opentrons Temperature Module with 96-Well Aluminum Block](https://shop.opentrons.com/collections/hardware-modules/products/tempdeck) with clean and empty [NEST 96-Well PCR Plate](https://shop.opentrons.com/collections/lab-plates/products/nest-0-1-ml-96-well-pcr-plate-full-skirt)
\n
\nSlot 4: [Opentrons Magnetic Module](https://shop.opentrons.com/collections/hardware-modules/products/magdeck) with [NEST 96-Deepwell Plate, 2mL](https://labware.opentrons.com/nest_96_wellplate_2ml_deep?category=wellPlate), containing samples*
\n*Samples should be loaded in odd-numbered columns only*
\n
\nSlot 5: [Opentrons 20\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips)
\n
\nSlot 8: [Opentrons 4-in-1 Tube Rack with 24-Well Top](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1), containing reagents in [NEST 1.5mL Microcentrifuge Tubes](https://shop.opentrons.com/collections/tubes/products/nest-microcentrifuge-tubes) (see below)
\n
\n[Opentrons 200\u00b5L Filter Tips](https://shop.opentrons.com/collections/opentrons-tips)
\n* if 8 samples, Slot 1\n* if 16 samples, Slots 1 and 6\n* if 24 samples, Slots 1, 6, and 9\n* if 32 samples, Slots 1, 6, 7, and 9\n* if 40 or 48 samples, Slots 1, 6, 7, 9, and 10\n\n
\n
\n**Reagent Setup**
\n
\n[Opentrons 4-in-1 Tube Rack with 24-Well Top](https://shop.opentrons.com/collections/racks-and-adapters/products/tube-rack-set-1)
\nD1: *Proteinase K*, 4\u00b5L per sample
\nD6: *Extraction Control Spike-In*, if using
\n
\n[NEST 12-Well Reservoir, 15mL](https://shop.opentrons.com/collections/reservoirs/products/nest-12-well-reservoir-15-ml)
\nFor every 8 samples, the following reagents are needed:\n* 7mL Zymo Viral DNA/RNA Buffer + 350\u00b5L of Zymo MagBeads\n* 4.5mL of Zymo Wash Buffer 1\n* 4.5mL of Zymo Wash Buffer 2\n* 4.5mL of Ethanol (Ethanol Wash 1)\n* 4.5mL Ethanol (Ethanol Wash 2)\n\n
\n\nSlot 1: Zymo Viral DNA/RNA Buffer + Zymo MagBeads, 8/16 samples
\nSlot 2: Zymo Viral DNA/RNA Buffer + Zymo MagBeads, 24/32 samples
\nSlot 3: Zymo Viral DNA/RNA Buffer + Zymo MagBeads, 40/48 samples
\nSlot 4: Zymo Wash Buffer 1, 8/16/24 samples
\nSlot 5: Zymo Wash Buffer 1, 32/40/48 samples
\nSlot 6: Zymo Wash Buffer 2, 8/16/24 samples
\nSlot 7: Zymo Wash Buffer 2, 32/40/48 samples
\nSlot 8: Ethanol (Ethanol Wash 1), 8/16/24 samples
\nSlot 9: Ethanol (Ethanol Wash 1), 32/40/48 samples
\nSlot 10: Ethanol (Ethanol Wash 2), 8/16/24 samples
\nSlot 11: Ethanol (Ethanol Wash 2), 32/40/48 samples
\nSlot 12: Nuclease-Free Water, 3.5-4mL
\n
\n**Full Deck Layout:**\n\n\n\n\n__Using the customizations fields, below set up your protocol.__\n* **Number of Samples**: Specify the number of samples you'd like to run.\n* **Amount of Spike-in**: Specify how much control RNA (or other spike-in) should be added to each well. If not using a spike-in, leave as 0.\n* **Return tips after wash step**: Specify whether to drop tips into trash bin after use, or to replace in tip rack for easy disposal.\n\n\n\n\n\n",
"internal": "zymo-rna-extraction\n",
diff --git a/protocols/00222e-dd/README.md b/protocols/00222e-dd/README.md
index 7e81d8e4d8..7c02f0a6a3 100644
--- a/protocols/00222e-dd/README.md
+++ b/protocols/00222e-dd/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sterile Workflows
* Cell Culture
diff --git a/protocols/00222e-seed/README.md b/protocols/00222e-seed/README.md
index 94741b126f..a1b0c4cb84 100644
--- a/protocols/00222e-seed/README.md
+++ b/protocols/00222e-seed/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sterile Workflows
* Seeding
diff --git a/protocols/00222e/README.md b/protocols/00222e/README.md
index f133b863c1..2b43194403 100644
--- a/protocols/00222e/README.md
+++ b/protocols/00222e/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Serial Dilution and Screening
diff --git a/protocols/002af3/README.md b/protocols/002af3/README.md
index edb80f4e62..5ba62f5512 100644
--- a/protocols/002af3/README.md
+++ b/protocols/002af3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/0045f1/README.md b/protocols/0045f1/README.md
index 78c12d3acb..dd7c6d55e9 100644
--- a/protocols/0045f1/README.md
+++ b/protocols/0045f1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/007992/README.md b/protocols/007992/README.md
index e647e34d37..8d0b29c8a4 100644
--- a/protocols/007992/README.md
+++ b/protocols/007992/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Viral RNA
diff --git a/protocols/00a214-protocol-1/README.md b/protocols/00a214-protocol-1/README.md
index 18602cdad7..7a411e9997 100644
--- a/protocols/00a214-protocol-1/README.md
+++ b/protocols/00a214-protocol-1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Seeding Plate
diff --git a/protocols/00a214-protocol-2/README.md b/protocols/00a214-protocol-2/README.md
index 5f105752ae..e51768e544 100644
--- a/protocols/00a214-protocol-2/README.md
+++ b/protocols/00a214-protocol-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Staining
diff --git a/protocols/00a214-protocol-3/README.md b/protocols/00a214-protocol-3/README.md
index 33ba3d5940..fc22fa6cb2 100644
--- a/protocols/00a214-protocol-3/README.md
+++ b/protocols/00a214-protocol-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Staining
diff --git a/protocols/00a214-protocol-4/README.md b/protocols/00a214-protocol-4/README.md
index 98a323dde9..dba1217bba 100644
--- a/protocols/00a214-protocol-4/README.md
+++ b/protocols/00a214-protocol-4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Staining
diff --git a/protocols/00a214-protocol-5/README.md b/protocols/00a214-protocol-5/README.md
index 5b1922b3fa..c34750b5f6 100644
--- a/protocols/00a214-protocol-5/README.md
+++ b/protocols/00a214-protocol-5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Staining
diff --git a/protocols/00a577/README.md b/protocols/00a577/README.md
index 4625c84bfc..ba9b869a7a 100644
--- a/protocols/00a577/README.md
+++ b/protocols/00a577/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Purification
* MP Biomedicals magGENic Plant DNA Kit
diff --git a/protocols/00a6e5/README.md b/protocols/00a6e5/README.md
index 3d1ab85a30..31e162e6d9 100644
--- a/protocols/00a6e5/README.md
+++ b/protocols/00a6e5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/00bbd7/README.md b/protocols/00bbd7/README.md
index d09e15133d..74b088839a 100644
--- a/protocols/00bbd7/README.md
+++ b/protocols/00bbd7/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/00c517-pt2/README.md b/protocols/00c517-pt2/README.md
index 0a10687a8d..5a03385a14 100644
--- a/protocols/00c517-pt2/README.md
+++ b/protocols/00c517-pt2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/00c517/README.md b/protocols/00c517/README.md
index 5d15c98d1d..d58cd1d64e 100644
--- a/protocols/00c517/README.md
+++ b/protocols/00c517/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/00f7b1_part10/README.md b/protocols/00f7b1_part10/README.md
index 65ace30bc4..6af7046375 100644
--- a/protocols/00f7b1_part10/README.md
+++ b/protocols/00f7b1_part10/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext® Ultra™ II Directional RNA Library Prep
diff --git a/protocols/00f7b1_part2/README.md b/protocols/00f7b1_part2/README.md
index 1d99eaad91..97fae35386 100644
--- a/protocols/00f7b1_part2/README.md
+++ b/protocols/00f7b1_part2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext® Ultra™ II Directional RNA Library Prep
diff --git a/protocols/00f7b1_part3/README.md b/protocols/00f7b1_part3/README.md
index ae49c46da0..34504cc4c7 100644
--- a/protocols/00f7b1_part3/README.md
+++ b/protocols/00f7b1_part3/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext® Ultra™ II Directional RNA Library Prep
diff --git a/protocols/00f7b1_part4/README.md b/protocols/00f7b1_part4/README.md
index 3be5660865..c1afa9941c 100644
--- a/protocols/00f7b1_part4/README.md
+++ b/protocols/00f7b1_part4/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext® Ultra™ II Directional RNA Library Prep
diff --git a/protocols/00f7b1_part5/README.md b/protocols/00f7b1_part5/README.md
index b9de26ff56..e4aa3a8989 100644
--- a/protocols/00f7b1_part5/README.md
+++ b/protocols/00f7b1_part5/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext® Ultra™ II Directional RNA Library Prep
diff --git a/protocols/00f7b1_part6/README.md b/protocols/00f7b1_part6/README.md
index 0942846d1d..5aa75ff17a 100644
--- a/protocols/00f7b1_part6/README.md
+++ b/protocols/00f7b1_part6/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext® Ultra™ II Directional RNA Library Prep
diff --git a/protocols/00f7b1_part7/README.md b/protocols/00f7b1_part7/README.md
index 1f7a7532d2..20f6670791 100644
--- a/protocols/00f7b1_part7/README.md
+++ b/protocols/00f7b1_part7/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II Directional RNA Library Prep Kit for Illumina
diff --git a/protocols/00f7b1_part8/README.md b/protocols/00f7b1_part8/README.md
index 2bb69f247f..3d79077312 100644
--- a/protocols/00f7b1_part8/README.md
+++ b/protocols/00f7b1_part8/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext® Ultra™ II Directional RNA Library Prep
diff --git a/protocols/00f7b1_part9/README.md b/protocols/00f7b1_part9/README.md
index 9cf43517c6..960fb5471d 100644
--- a/protocols/00f7b1_part9/README.md
+++ b/protocols/00f7b1_part9/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II Directional RNA Library Prep Kit for Illumina
diff --git a/protocols/010526/README.md b/protocols/010526/README.md
index e18f72c1b0..715478aa96 100644
--- a/protocols/010526/README.md
+++ b/protocols/010526/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Restriction Digest
diff --git a/protocols/010d6c/README.md b/protocols/010d6c/README.md
index 2dc17b26cb..f513bec1f6 100644
--- a/protocols/010d6c/README.md
+++ b/protocols/010d6c/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0175a4/README.md b/protocols/0175a4/README.md
index 1e438a601a..a5f56b7a92 100644
--- a/protocols/0175a4/README.md
+++ b/protocols/0175a4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Pooling
diff --git a/protocols/017860/README.md b/protocols/017860/README.md
index e51e97b534..ad715f118e 100644
--- a/protocols/017860/README.md
+++ b/protocols/017860/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* CSV Transfer
diff --git a/protocols/019a7d/README.md b/protocols/019a7d/README.md
index bb48487caf..c44297abbf 100644
--- a/protocols/019a7d/README.md
+++ b/protocols/019a7d/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Gibson Prep
diff --git a/protocols/019a7d_part_2/README.md b/protocols/019a7d_part_2/README.md
index fe7c86b241..5a1017322d 100644
--- a/protocols/019a7d_part_2/README.md
+++ b/protocols/019a7d_part_2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Gibson Prep
diff --git a/protocols/01a6b9/README.md b/protocols/01a6b9/README.md
index aaa4bf9846..4815e37ad8 100644
--- a/protocols/01a6b9/README.md
+++ b/protocols/01a6b9/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sterile Workflows
* Cell Culture
diff --git a/protocols/01b4a2/README.md b/protocols/01b4a2/README.md
index a5fe31c0c7..47c1a529aa 100644
--- a/protocols/01b4a2/README.md
+++ b/protocols/01b4a2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/022548-2/README.md b/protocols/022548-2/README.md
index adeb953910..a7bc50c258 100644
--- a/protocols/022548-2/README.md
+++ b/protocols/022548-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* DNA Extraction
diff --git a/protocols/022548/README.md b/protocols/022548/README.md
index 4fae21ddf7..5b901d566c 100644
--- a/protocols/022548/README.md
+++ b/protocols/022548/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* DNA Extraction
diff --git a/protocols/022a99/README.md b/protocols/022a99/README.md
index e469d70720..4f224b1c29 100644
--- a/protocols/022a99/README.md
+++ b/protocols/022a99/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
- Sample Prep
diff --git a/protocols/029f5b/README.md b/protocols/029f5b/README.md
index b76fdfafdb..4ada8aeb13 100644
--- a/protocols/029f5b/README.md
+++ b/protocols/029f5b/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* SAMPLE PREP
* Custom Sample Transfer
diff --git a/protocols/02b308/README.md b/protocols/02b308/README.md
index e8a8c8b346..d8d0d7a75d 100644
--- a/protocols/02b308/README.md
+++ b/protocols/02b308/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* qPCR Prep
diff --git a/protocols/02pnzp/README.md b/protocols/02pnzp/README.md
index 5682ab49e8..d5a0a4cfcc 100644
--- a/protocols/02pnzp/README.md
+++ b/protocols/02pnzp/README.md
@@ -2,6 +2,9 @@
Michael Fichtner
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* DNA/RNA
* DNA/RNA
diff --git a/protocols/030dd8-amplify/README.md b/protocols/030dd8-amplify/README.md
index cebe2c8f16..9385e4f1b1 100644
--- a/protocols/030dd8-amplify/README.md
+++ b/protocols/030dd8-amplify/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Broad Category
* Specific Category
diff --git a/protocols/030dd8-amplify2/README.md b/protocols/030dd8-amplify2/README.md
index ae881392d8..86443397d1 100644
--- a/protocols/030dd8-amplify2/README.md
+++ b/protocols/030dd8-amplify2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Broad Category
* Specific Category
diff --git a/protocols/030dd8-anneal/README.md b/protocols/030dd8-anneal/README.md
index 6ec7936f6b..5c53ca236c 100644
--- a/protocols/030dd8-anneal/README.md
+++ b/protocols/030dd8-anneal/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Broad Category
* Specific Category
diff --git a/protocols/030dd8-cleanup/README.md b/protocols/030dd8-cleanup/README.md
index d8d87d6619..e068114732 100644
--- a/protocols/030dd8-cleanup/README.md
+++ b/protocols/030dd8-cleanup/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Broad Category
* Specific Category
diff --git a/protocols/030dd8-synthesize/README.md b/protocols/030dd8-synthesize/README.md
index 48b88d1c26..43ad6fefcc 100644
--- a/protocols/030dd8-synthesize/README.md
+++ b/protocols/030dd8-synthesize/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Broad Category
* Specific Category
diff --git a/protocols/030dd8-tagment/README.md b/protocols/030dd8-tagment/README.md
index 8bff48d260..c31ac334c2 100644
--- a/protocols/030dd8-tagment/README.md
+++ b/protocols/030dd8-tagment/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Broad Category
* Specific Category
diff --git a/protocols/030dd8/README.md b/protocols/030dd8/README.md
index d2e6fdbb46..f664b62e3e 100644
--- a/protocols/030dd8/README.md
+++ b/protocols/030dd8/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Broad Category
* Specific Category
diff --git a/protocols/036016/README.md b/protocols/036016/README.md
index 754e9acc93..dbcb53f7cb 100644
--- a/protocols/036016/README.md
+++ b/protocols/036016/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/039ecb/README.md b/protocols/039ecb/README.md
index 609bac9617..82a418314e 100644
--- a/protocols/039ecb/README.md
+++ b/protocols/039ecb/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext® Ultra™ II FS DNA Library Prep Kit for Illumina
diff --git a/protocols/03f9cd/README.md b/protocols/03f9cd/README.md
index cf4db212f2..394d83ef60 100644
--- a/protocols/03f9cd/README.md
+++ b/protocols/03f9cd/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/041adf/README.md b/protocols/041adf/README.md
index 6a13be6be5..253a641f34 100644
--- a/protocols/041adf/README.md
+++ b/protocols/041adf/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Screening
diff --git a/protocols/04318b/README.md b/protocols/04318b/README.md
index 3a16af27b4..ab7a20dcfa 100644
--- a/protocols/04318b/README.md
+++ b/protocols/04318b/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Serial Dilution
diff --git a/protocols/0451f3/README.md b/protocols/0451f3/README.md
index 11c01dff67..bc816b5c5f 100644
--- a/protocols/0451f3/README.md
+++ b/protocols/0451f3/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Purification & Extraction
* Zymo Zyppy™-96 Plasmid MagBead Miniprep
diff --git a/protocols/04552f/README.md b/protocols/04552f/README.md
index 7d6ed8aa69..0c94654269 100644
--- a/protocols/04552f/README.md
+++ b/protocols/04552f/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/0479ad-cp/README.md b/protocols/0479ad-cp/README.md
index fd23ebfe1d..3697c75d03 100644
--- a/protocols/0479ad-cp/README.md
+++ b/protocols/0479ad-cp/README.md
@@ -5,6 +5,9 @@
[Breakthrough Genomics](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/0479ad-part-2/README.md b/protocols/0479ad-part-2/README.md
index fa24941c23..781851efc0 100644
--- a/protocols/0479ad-part-2/README.md
+++ b/protocols/0479ad-part-2/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II RNA Library Prep Kit for Illumina
diff --git a/protocols/0479ad-part-3/README.md b/protocols/0479ad-part-3/README.md
index f2a8fae609..9dcc42461c 100644
--- a/protocols/0479ad-part-3/README.md
+++ b/protocols/0479ad-part-3/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II RNA Library Prep Kit for Illumina
diff --git a/protocols/0479ad-part-4/README.md b/protocols/0479ad-part-4/README.md
index 5405e04927..61e1fc37ed 100644
--- a/protocols/0479ad-part-4/README.md
+++ b/protocols/0479ad-part-4/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II RNA Library Prep Kit for Illumina
diff --git a/protocols/0479ad-part-5/README.md b/protocols/0479ad-part-5/README.md
index 9e5db88053..13fe2ee02f 100644
--- a/protocols/0479ad-part-5/README.md
+++ b/protocols/0479ad-part-5/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II RNA Library Prep Kit for Illumina
diff --git a/protocols/0479ad/README.md b/protocols/0479ad/README.md
index 059bb35aeb..500e413cf1 100644
--- a/protocols/0479ad/README.md
+++ b/protocols/0479ad/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II RNA Library Prep Kit for Illumina
diff --git a/protocols/04bec0/README.md b/protocols/04bec0/README.md
index bc7e009761..48ac55e495 100644
--- a/protocols/04bec0/README.md
+++ b/protocols/04bec0/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/04eeb1-part-2/README.md b/protocols/04eeb1-part-2/README.md
index 803ebc52d7..4e7d2659ed 100644
--- a/protocols/04eeb1-part-2/README.md
+++ b/protocols/04eeb1-part-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina COVIDSeq Test
diff --git a/protocols/04eeb1-part-3/README.md b/protocols/04eeb1-part-3/README.md
index 3251dc603b..a26e3e52e8 100644
--- a/protocols/04eeb1-part-3/README.md
+++ b/protocols/04eeb1-part-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina COVIDSeq Test
diff --git a/protocols/04eeb1-part-4/README.md b/protocols/04eeb1-part-4/README.md
index 1013866e75..11b61d7316 100644
--- a/protocols/04eeb1-part-4/README.md
+++ b/protocols/04eeb1-part-4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina COVIDSeq Test
diff --git a/protocols/04eeb1-part-5/README.md b/protocols/04eeb1-part-5/README.md
index 40e29830cc..c7a6c2a591 100644
--- a/protocols/04eeb1-part-5/README.md
+++ b/protocols/04eeb1-part-5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina COVIDSeq Test
diff --git a/protocols/04eeb1-part-6/README.md b/protocols/04eeb1-part-6/README.md
index 454a6ccdb7..ae48443576 100644
--- a/protocols/04eeb1-part-6/README.md
+++ b/protocols/04eeb1-part-6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina COVIDSeq Test
diff --git a/protocols/04eeb1-part-7/README.md b/protocols/04eeb1-part-7/README.md
index 886473ede5..bad69f5f54 100644
--- a/protocols/04eeb1-part-7/README.md
+++ b/protocols/04eeb1-part-7/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina COVIDSeq Test
diff --git a/protocols/04eeb1/README.md b/protocols/04eeb1/README.md
index 14aa21a0b6..240e7c1807 100644
--- a/protocols/04eeb1/README.md
+++ b/protocols/04eeb1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina COVIDSeq Test
diff --git a/protocols/04f310/README.md b/protocols/04f310/README.md
index ec9464aa39..ce908e3b84 100644
--- a/protocols/04f310/README.md
+++ b/protocols/04f310/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/04f4e7/README.md b/protocols/04f4e7/README.md
index c09e1dc2b7..a64dc3db64 100644
--- a/protocols/04f4e7/README.md
+++ b/protocols/04f4e7/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Purification
diff --git a/protocols/04fed3/README.md b/protocols/04fed3/README.md
index 57feefe162..8b06ef8b7c 100644
--- a/protocols/04fed3/README.md
+++ b/protocols/04fed3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/051557/README.md b/protocols/051557/README.md
index 0ba3bb22af..388d2ab31b 100644
--- a/protocols/051557/README.md
+++ b/protocols/051557/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Standard Curve
diff --git a/protocols/0523c2/README.md b/protocols/0523c2/README.md
index 9a321535f8..89b766727d 100644
--- a/protocols/0523c2/README.md
+++ b/protocols/0523c2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/052d03/README.md b/protocols/052d03/README.md
index 6ba9d9849f..341df2fcac 100644
--- a/protocols/052d03/README.md
+++ b/protocols/052d03/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR
diff --git a/protocols/0530d8-pt2/README.md b/protocols/0530d8-pt2/README.md
index 97847b4bb3..8b638f136d 100644
--- a/protocols/0530d8-pt2/README.md
+++ b/protocols/0530d8-pt2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* DNA Extraction
diff --git a/protocols/0530d8/README.md b/protocols/0530d8/README.md
index 6bc2b780ed..b7b40c5756 100644
--- a/protocols/0530d8/README.md
+++ b/protocols/0530d8/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* DNA Extraction
diff --git a/protocols/053386-pt2/README.md b/protocols/053386-pt2/README.md
index e6289b6263..c9ad9e9e6e 100644
--- a/protocols/053386-pt2/README.md
+++ b/protocols/053386-pt2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/053386-pt3/README.md b/protocols/053386-pt3/README.md
index e500b75f6b..e8359bfe73 100644
--- a/protocols/053386-pt3/README.md
+++ b/protocols/053386-pt3/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/053386/README.md b/protocols/053386/README.md
index 8770b8477c..30bd88b50a 100644
--- a/protocols/053386/README.md
+++ b/protocols/053386/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0556be-bmda/README.md b/protocols/0556be-bmda/README.md
index 35114bbb3b..2134159d8b 100644
--- a/protocols/0556be-bmda/README.md
+++ b/protocols/0556be-bmda/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/0556be-covid-mm-qc-v3/README.md b/protocols/0556be-covid-mm-qc-v3/README.md
index d7765c6814..e3063bc7b4 100644
--- a/protocols/0556be-covid-mm-qc-v3/README.md
+++ b/protocols/0556be-covid-mm-qc-v3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0556be-covid-mm-qc/README.md b/protocols/0556be-covid-mm-qc/README.md
index 794ef977b3..f6ef7c8b9d 100644
--- a/protocols/0556be-covid-mm-qc/README.md
+++ b/protocols/0556be-covid-mm-qc/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0556be-covid-ntc/README.md b/protocols/0556be-covid-ntc/README.md
index 282788ea29..202665b847 100644
--- a/protocols/0556be-covid-ntc/README.md
+++ b/protocols/0556be-covid-ntc/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/055b94/README.md b/protocols/055b94/README.md
index 933dc49008..3853ec65b9 100644
--- a/protocols/055b94/README.md
+++ b/protocols/055b94/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/055f76/README.md b/protocols/055f76/README.md
index 005deb132f..3c7a1800d8 100644
--- a/protocols/055f76/README.md
+++ b/protocols/055f76/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/056543/README.md b/protocols/056543/README.md
index 94e6eb717f..6709b41acc 100644
--- a/protocols/056543/README.md
+++ b/protocols/056543/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Compound Plating
diff --git a/protocols/056f47/README.md b/protocols/056f47/README.md
index c4a0edd13d..362eb43369 100644
--- a/protocols/056f47/README.md
+++ b/protocols/056f47/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Serial Dilution
diff --git a/protocols/058124/README.md b/protocols/058124/README.md
index 093836ec5a..6df3babc08 100644
--- a/protocols/058124/README.md
+++ b/protocols/058124/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Serial Dilution
diff --git a/protocols/059126/README.md b/protocols/059126/README.md
index 4feff80d2a..9af968981b 100644
--- a/protocols/059126/README.md
+++ b/protocols/059126/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Reagent Addition
diff --git a/protocols/05f673/README.md b/protocols/05f673/README.md
index ee6cd53104..79f1e1fcc5 100644
--- a/protocols/05f673/README.md
+++ b/protocols/05f673/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/05fb0f/README.md b/protocols/05fb0f/README.md
index d10bb1c926..2eb3618ef6 100644
--- a/protocols/05fb0f/README.md
+++ b/protocols/05fb0f/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Assay
diff --git a/protocols/060c4f/README.md b/protocols/060c4f/README.md
index 7934b94d43..8b3d5a3d26 100644
--- a/protocols/060c4f/README.md
+++ b/protocols/060c4f/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/063611-part-2/README.md b/protocols/063611-part-2/README.md
index 0bc97851b3..715d7b31b3 100644
--- a/protocols/063611-part-2/README.md
+++ b/protocols/063611-part-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/063611/README.md b/protocols/063611/README.md
index e6ba07b130..4e88cecdb9 100644
--- a/protocols/063611/README.md
+++ b/protocols/063611/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/06ab09/README.md b/protocols/06ab09/README.md
index b4a1e1be2b..4199fa4e2b 100644
--- a/protocols/06ab09/README.md
+++ b/protocols/06ab09/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/06da40/README.md b/protocols/06da40/README.md
index a91a5bdbbc..26dda564f9 100644
--- a/protocols/06da40/README.md
+++ b/protocols/06da40/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization & PCR Prep
diff --git a/protocols/06e5b6/README.md b/protocols/06e5b6/README.md
index a89480dbe3..2a7ebb1e3f 100644
--- a/protocols/06e5b6/README.md
+++ b/protocols/06e5b6/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Crystallization
diff --git a/protocols/072463/README.md b/protocols/072463/README.md
index 11bdb7417e..9ddc8ea9f9 100644
--- a/protocols/072463/README.md
+++ b/protocols/072463/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/075bee/README.md b/protocols/075bee/README.md
index a30c4e55fb..acfbb02175 100644
--- a/protocols/075bee/README.md
+++ b/protocols/075bee/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/076f67/README.md b/protocols/076f67/README.md
index 28c6e2ebc9..aeb5a7b812 100644
--- a/protocols/076f67/README.md
+++ b/protocols/076f67/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/079db2/README.md b/protocols/079db2/README.md
index 016f8d7e3f..700781795a 100644
--- a/protocols/079db2/README.md
+++ b/protocols/079db2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/07c65a/README.md b/protocols/07c65a/README.md
index 6f1289b583..d68ba7d5c7 100644
--- a/protocols/07c65a/README.md
+++ b/protocols/07c65a/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/07caf7/README.md b/protocols/07caf7/README.md
index 8d69685c5d..517f2fc0aa 100644
--- a/protocols/07caf7/README.md
+++ b/protocols/07caf7/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Mass Spec
diff --git a/protocols/08207f/README.md b/protocols/08207f/README.md
index 0ba751255a..3f9918da00 100644
--- a/protocols/08207f/README.md
+++ b/protocols/08207f/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Normalization
* Normalization and Pooling
diff --git a/protocols/0845ab/README.md b/protocols/0845ab/README.md
index 0a3cfdd24e..d26fa459cc 100644
--- a/protocols/0845ab/README.md
+++ b/protocols/0845ab/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0849c2/README.md b/protocols/0849c2/README.md
index 10c25a15ea..a885b98030 100644
--- a/protocols/0849c2/README.md
+++ b/protocols/0849c2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Extraction
diff --git a/protocols/08878b/README.md b/protocols/08878b/README.md
index c7687ffc76..5e6e2f0d44 100644
--- a/protocols/08878b/README.md
+++ b/protocols/08878b/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/089108/README.md b/protocols/089108/README.md
index 30baad69b5..94fa5c691e 100644
--- a/protocols/089108/README.md
+++ b/protocols/089108/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NUCLEIC ACID EXTRACTION & PURIFICATION
* Nucleic Acid Purification
diff --git a/protocols/08aeaa/README.md b/protocols/08aeaa/README.md
index 9b100d3bf2..41d707584e 100644
--- a/protocols/08aeaa/README.md
+++ b/protocols/08aeaa/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/08bbaf/README.md b/protocols/08bbaf/README.md
index c61508a0af..5611044681 100644
--- a/protocols/08bbaf/README.md
+++ b/protocols/08bbaf/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Standard Curve
diff --git a/protocols/08e3eb/README.md b/protocols/08e3eb/README.md
index fb5aea089a..1e886ddae5 100644
--- a/protocols/08e3eb/README.md
+++ b/protocols/08e3eb/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/08fd01-pt2/README.md b/protocols/08fd01-pt2/README.md
index 260ceed97d..6181857083 100644
--- a/protocols/08fd01-pt2/README.md
+++ b/protocols/08fd01-pt2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/08fd01-pt3/README.md b/protocols/08fd01-pt3/README.md
index 6c55cdca70..b92ea55838 100644
--- a/protocols/08fd01-pt3/README.md
+++ b/protocols/08fd01-pt3/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/08fd01/README.md b/protocols/08fd01/README.md
index 6e92322185..3d2a0b3a71 100644
--- a/protocols/08fd01/README.md
+++ b/protocols/08fd01/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/0909e6/README.md b/protocols/0909e6/README.md
index c73f0859c4..24fb01436a 100644
--- a/protocols/0909e6/README.md
+++ b/protocols/0909e6/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sterile Workflows
* Cell Culture
diff --git a/protocols/090c2e/README.md b/protocols/090c2e/README.md
index 21be984df3..00c478eaff 100644
--- a/protocols/090c2e/README.md
+++ b/protocols/090c2e/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
- Nucleic Acid Purification
diff --git a/protocols/0944e4/README.md b/protocols/0944e4/README.md
index 3d72f927b0..a89977891e 100644
--- a/protocols/0944e4/README.md
+++ b/protocols/0944e4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Ampure XP
diff --git a/protocols/0961d2-part1/README.md b/protocols/0961d2-part1/README.md
index 6a56fa0fbb..9e0f5ecbbb 100644
--- a/protocols/0961d2-part1/README.md
+++ b/protocols/0961d2-part1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* plexWell LP384
diff --git a/protocols/0961d2-part2/README.md b/protocols/0961d2-part2/README.md
index e2a13edc9a..aa664cb7fc 100644
--- a/protocols/0961d2-part2/README.md
+++ b/protocols/0961d2-part2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* plexWell LP384
diff --git a/protocols/0961d2-part3/README.md b/protocols/0961d2-part3/README.md
index d79763851d..0598984752 100644
--- a/protocols/0961d2-part3/README.md
+++ b/protocols/0961d2-part3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* plexWell LP384
diff --git a/protocols/0961d2-part4/README.md b/protocols/0961d2-part4/README.md
index 289b7c4013..63d7639652 100644
--- a/protocols/0961d2-part4/README.md
+++ b/protocols/0961d2-part4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* plexWell LP384
diff --git a/protocols/0966ae/README.md b/protocols/0966ae/README.md
index e621ff9522..97b1ed9c7c 100644
--- a/protocols/0966ae/README.md
+++ b/protocols/0966ae/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sterile Workflows
* Cell Culture
diff --git a/protocols/09cdbe/README.md b/protocols/09cdbe/README.md
index 750c2c089c..703f15be7f 100644
--- a/protocols/09cdbe/README.md
+++ b/protocols/09cdbe/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Consolidation
diff --git a/protocols/0a23c6/README.md b/protocols/0a23c6/README.md
index cc3cb51193..5835f59aa8 100644
--- a/protocols/0a23c6/README.md
+++ b/protocols/0a23c6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* qPCR setup
diff --git a/protocols/0a25f2/README.md b/protocols/0a25f2/README.md
index 26030c8f0e..0b8e986442 100644
--- a/protocols/0a25f2/README.md
+++ b/protocols/0a25f2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Tube Filling
diff --git a/protocols/0a33e8/README.md b/protocols/0a33e8/README.md
index 5744ed2b73..cb7bc20b33 100644
--- a/protocols/0a33e8/README.md
+++ b/protocols/0a33e8/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/0a9b58/README.md b/protocols/0a9b58/README.md
index 2ca41342cf..04f30b911a 100644
--- a/protocols/0a9b58/README.md
+++ b/protocols/0a9b58/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0aa51a/README.md b/protocols/0aa51a/README.md
index b795bc6dfc..64c3c6e7ce 100644
--- a/protocols/0aa51a/README.md
+++ b/protocols/0aa51a/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* Complete PCR Workflow
diff --git a/protocols/0add76/README.md b/protocols/0add76/README.md
index 2851ed0ea5..cc65834915 100644
--- a/protocols/0add76/README.md
+++ b/protocols/0add76/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0aee8a/README.md b/protocols/0aee8a/README.md
index c7ef871b25..7c61711643 100644
--- a/protocols/0aee8a/README.md
+++ b/protocols/0aee8a/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Serial Dilution
diff --git a/protocols/0b83b7/README.md b/protocols/0b83b7/README.md
index 0bc945b2a3..15da3ae4f2 100644
--- a/protocols/0b83b7/README.md
+++ b/protocols/0b83b7/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Cleanup
diff --git a/protocols/0b97ae-protocol-2A/README.md b/protocols/0b97ae-protocol-2A/README.md
index f54003a51a..50fc774841 100644
--- a/protocols/0b97ae-protocol-2A/README.md
+++ b/protocols/0b97ae-protocol-2A/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS LIBRARY PREP
* QIASeq FastSelect
diff --git a/protocols/0b97ae-protocol-2B/README.md b/protocols/0b97ae-protocol-2B/README.md
index 011929b476..6f6bc29c66 100644
--- a/protocols/0b97ae-protocol-2B/README.md
+++ b/protocols/0b97ae-protocol-2B/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS LIBRARY PREP
* QIASeq FastSelect
diff --git a/protocols/0b97ae-protocol-3B/README.md b/protocols/0b97ae-protocol-3B/README.md
index 2fc3530ce8..fea5438536 100644
--- a/protocols/0b97ae-protocol-3B/README.md
+++ b/protocols/0b97ae-protocol-3B/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS LIBRARY PREP
* QIASeq FastSelect
diff --git a/protocols/0b97ae/README.md b/protocols/0b97ae/README.md
index 9cec277a96..8331fd929e 100644
--- a/protocols/0b97ae/README.md
+++ b/protocols/0b97ae/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* SAMPLE PREP
* Normalization
diff --git a/protocols/0b98cc/README.md b/protocols/0b98cc/README.md
index dd279ece45..2150141c2d 100644
--- a/protocols/0b98cc/README.md
+++ b/protocols/0b98cc/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Serial Dilution
diff --git a/protocols/0ba998-2/README.md b/protocols/0ba998-2/README.md
index 31e416a5b6..c26a64eabf 100644
--- a/protocols/0ba998-2/README.md
+++ b/protocols/0ba998-2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep
diff --git a/protocols/0ba998-3/README.md b/protocols/0ba998-3/README.md
index ab1d0f3f40..5c6de0ed4e 100644
--- a/protocols/0ba998-3/README.md
+++ b/protocols/0ba998-3/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep
diff --git a/protocols/0ba998-4/README.md b/protocols/0ba998-4/README.md
index d00d44d11c..9fae99f81e 100644
--- a/protocols/0ba998-4/README.md
+++ b/protocols/0ba998-4/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep
diff --git a/protocols/0ba998/README.md b/protocols/0ba998/README.md
index 6287186409..f58710bda6 100644
--- a/protocols/0ba998/README.md
+++ b/protocols/0ba998/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep
diff --git a/protocols/0bd707/README.md b/protocols/0bd707/README.md
index 1b4af42c28..5c1d71ff75 100644
--- a/protocols/0bd707/README.md
+++ b/protocols/0bd707/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Aliquoting
diff --git a/protocols/0be1bf/README.md b/protocols/0be1bf/README.md
index 75209648fa..ccf2323bce 100644
--- a/protocols/0be1bf/README.md
+++ b/protocols/0be1bf/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Aliquoting
diff --git a/protocols/0bf4f4-pt2/README.md b/protocols/0bf4f4-pt2/README.md
index 14089578c2..6aca50f97a 100644
--- a/protocols/0bf4f4-pt2/README.md
+++ b/protocols/0bf4f4-pt2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep
diff --git a/protocols/0bf4f4-pt3/README.md b/protocols/0bf4f4-pt3/README.md
index c175c3a699..a7fcd341b6 100644
--- a/protocols/0bf4f4-pt3/README.md
+++ b/protocols/0bf4f4-pt3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep
diff --git a/protocols/0bf4f4/README.md b/protocols/0bf4f4/README.md
index 27b086c15f..bd7c38e191 100644
--- a/protocols/0bf4f4/README.md
+++ b/protocols/0bf4f4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep
diff --git a/protocols/0c24ca/README.md b/protocols/0c24ca/README.md
index 3c599c7bd0..8e8e51d20f 100644
--- a/protocols/0c24ca/README.md
+++ b/protocols/0c24ca/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0c3e45/README.md b/protocols/0c3e45/README.md
index de4f224535..2a5ee9cfc7 100644
--- a/protocols/0c3e45/README.md
+++ b/protocols/0c3e45/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0ca318-pt2/README.md b/protocols/0ca318-pt2/README.md
index 8b21899ac0..ae4e907788 100644
--- a/protocols/0ca318-pt2/README.md
+++ b/protocols/0ca318-pt2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR
diff --git a/protocols/0ca318/README.md b/protocols/0ca318/README.md
index 09354ad7fc..b1bd1cae72 100644
--- a/protocols/0ca318/README.md
+++ b/protocols/0ca318/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR
diff --git a/protocols/0cded6-amplify/README.md b/protocols/0cded6-amplify/README.md
index 938d03b360..361995b53c 100644
--- a/protocols/0cded6-amplify/README.md
+++ b/protocols/0cded6-amplify/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina COVIDSeq
diff --git a/protocols/0cded6-amplify2/README.md b/protocols/0cded6-amplify2/README.md
index f9131ae676..d236f0c513 100644
--- a/protocols/0cded6-amplify2/README.md
+++ b/protocols/0cded6-amplify2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina COVIDSeq
diff --git a/protocols/0cded6-cleanup/README.md b/protocols/0cded6-cleanup/README.md
index 6ea30c4334..06a06cf52e 100644
--- a/protocols/0cded6-cleanup/README.md
+++ b/protocols/0cded6-cleanup/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina COVIDSeq
diff --git a/protocols/0cded6-synthesize/README.md b/protocols/0cded6-synthesize/README.md
index 70992047bc..296040c4a9 100644
--- a/protocols/0cded6-synthesize/README.md
+++ b/protocols/0cded6-synthesize/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina COVIDSeq
diff --git a/protocols/0cded6-tagment/README.md b/protocols/0cded6-tagment/README.md
index 423bd197b8..cea10d5e57 100644
--- a/protocols/0cded6-tagment/README.md
+++ b/protocols/0cded6-tagment/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina COVIDSeq
diff --git a/protocols/0cded6/README.md b/protocols/0cded6/README.md
index 7a5c65229b..7c432d2199 100644
--- a/protocols/0cded6/README.md
+++ b/protocols/0cded6/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina COVIDSeq
diff --git a/protocols/0d2950/README.md b/protocols/0d2950/README.md
index e41d17d6e0..f06ecc25ee 100644
--- a/protocols/0d2950/README.md
+++ b/protocols/0d2950/README.md
@@ -6,6 +6,9 @@
### Partner
[appliedbiosystems](https://www.thermofisher.com/content/dam/LifeTech/Documents/PDFs/clinical/taqpath-COVID-19-combo-kit-full-instructions-for-use.pdf)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0d868e-part2/README.md b/protocols/0d868e-part2/README.md
index 66d2ac6879..49608eff2b 100644
--- a/protocols/0d868e-part2/README.md
+++ b/protocols/0d868e-part2/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Generic
diff --git a/protocols/0d868e/README.md b/protocols/0d868e/README.md
index 4a538b4324..554d4cc873 100644
--- a/protocols/0d868e/README.md
+++ b/protocols/0d868e/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Generic
diff --git a/protocols/0dda91/README.md b/protocols/0dda91/README.md
index c7fc92fe33..4cb5a52abf 100644
--- a/protocols/0dda91/README.md
+++ b/protocols/0dda91/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Pooling
diff --git a/protocols/0df680/README.md b/protocols/0df680/README.md
index bfd63aac91..7ea39ff948 100644
--- a/protocols/0df680/README.md
+++ b/protocols/0df680/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/0e39fc/README.md b/protocols/0e39fc/README.md
index 3f9b1d9359..41bbba1b43 100644
--- a/protocols/0e39fc/README.md
+++ b/protocols/0e39fc/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/0e525c/README.md b/protocols/0e525c/README.md
index ac8aa4c903..94e5b3b2fd 100644
--- a/protocols/0e525c/README.md
+++ b/protocols/0e525c/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/0e696e/README.md b/protocols/0e696e/README.md
index 3ac31e777a..dcfa0ecab7 100644
--- a/protocols/0e696e/README.md
+++ b/protocols/0e696e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/0e7175-pt2/README.md b/protocols/0e7175-pt2/README.md
index 045b76b2c3..54efc99025 100644
--- a/protocols/0e7175-pt2/README.md
+++ b/protocols/0e7175-pt2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0e7175/README.md b/protocols/0e7175/README.md
index 01c7bf229d..ff6f04c1c5 100644
--- a/protocols/0e7175/README.md
+++ b/protocols/0e7175/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0e8475/README.md b/protocols/0e8475/README.md
index 01f8d8e143..415daeb306 100644
--- a/protocols/0e8475/README.md
+++ b/protocols/0e8475/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina GUIDE-seq
diff --git a/protocols/0e8e3/README.md b/protocols/0e8e3/README.md
index 58b56156ec..df4039ebab 100644
--- a/protocols/0e8e3/README.md
+++ b/protocols/0e8e3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/0eaeb5/README.md b/protocols/0eaeb5/README.md
index 23aedd68f6..a467790533 100644
--- a/protocols/0eaeb5/README.md
+++ b/protocols/0eaeb5/README.md
@@ -3,6 +3,9 @@
## Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS
* Paragon Genomics CleanPlex NGS Panel Kit
diff --git a/protocols/0edc68/README.md b/protocols/0edc68/README.md
index 9c3c277f9e..3bbc9b0be1 100644
--- a/protocols/0edc68/README.md
+++ b/protocols/0edc68/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Purification
diff --git a/protocols/0ef96f/README.md b/protocols/0ef96f/README.md
index 6307799d11..0df6451c4c 100644
--- a/protocols/0ef96f/README.md
+++ b/protocols/0ef96f/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/0f3b60/README.md b/protocols/0f3b60/README.md
index 7a4a87eba1..75df82fed9 100644
--- a/protocols/0f3b60/README.md
+++ b/protocols/0f3b60/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0f409e/README.md b/protocols/0f409e/README.md
index 3165256ac3..922d5e4326 100644
--- a/protocols/0f409e/README.md
+++ b/protocols/0f409e/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0f4405/README.md b/protocols/0f4405/README.md
index 1fc4efb55f..63eb79587c 100644
--- a/protocols/0f4405/README.md
+++ b/protocols/0f4405/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext® Ultra™ II DNA Library Prep Kit for Illumina®
diff --git a/protocols/0f5985/README.md b/protocols/0f5985/README.md
index 6f806a8d82..a5e812e3e8 100644
--- a/protocols/0f5985/README.md
+++ b/protocols/0f5985/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Blood Sample
diff --git a/protocols/0f5c31/README.md b/protocols/0f5c31/README.md
index e9cefe38d1..bf32edbd13 100644
--- a/protocols/0f5c31/README.md
+++ b/protocols/0f5c31/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteiomics
* TPP
diff --git a/protocols/0f7910/README.md b/protocols/0f7910/README.md
index 33abdd3ab8..0a0a51c9e7 100644
--- a/protocols/0f7910/README.md
+++ b/protocols/0f7910/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0fa015/README.md b/protocols/0fa015/README.md
index 972592ac67..21cc60f7aa 100644
--- a/protocols/0fa015/README.md
+++ b/protocols/0fa015/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/0fd3dd/README.md b/protocols/0fd3dd/README.md
index 3674de85f9..a3fba34536 100644
--- a/protocols/0fd3dd/README.md
+++ b/protocols/0fd3dd/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PROTEINS & PROTEOMICS
* ELISA
diff --git a/protocols/0fdc01/README.md b/protocols/0fdc01/README.md
index 7150753f21..8b160a9a02 100644
--- a/protocols/0fdc01/README.md
+++ b/protocols/0fdc01/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/0ff0c3/README.md b/protocols/0ff0c3/README.md
index 4f754c6e69..6846ac9ca3 100644
--- a/protocols/0ff0c3/README.md
+++ b/protocols/0ff0c3/README.md
@@ -4,6 +4,9 @@ Nucleic Acid Purification
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Purification
diff --git a/protocols/1033g1/README.md b/protocols/1033g1/README.md
index 41f76b19b9..924706d738 100644
--- a/protocols/1033g1/README.md
+++ b/protocols/1033g1/README.md
@@ -5,6 +5,9 @@
Vasudha Nair
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Cell and Tissue Culture
* Cell and Tissue Culture
diff --git a/protocols/10bf60-station-B/README.md b/protocols/10bf60-station-B/README.md
index b0b3a59409..fb712a3320 100644
--- a/protocols/10bf60-station-B/README.md
+++ b/protocols/10bf60-station-B/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Covid Workstation
* RNA Extraction
diff --git a/protocols/10bf60-station-C/README.md b/protocols/10bf60-station-C/README.md
index 10f64a641d..3858c6246e 100644
--- a/protocols/10bf60-station-C/README.md
+++ b/protocols/10bf60-station-C/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/111210-part-10/README.md b/protocols/111210-part-10/README.md
index 5b57ae8d29..93f3d0fbbf 100644
--- a/protocols/111210-part-10/README.md
+++ b/protocols/111210-part-10/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung RNA Fusion UMI Panel Kit
diff --git a/protocols/111210-part-2/README.md b/protocols/111210-part-2/README.md
index 8756c4e8bc..6a8812a685 100644
--- a/protocols/111210-part-2/README.md
+++ b/protocols/111210-part-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung RNA Fusion UMI Panel Kit
diff --git a/protocols/111210-part-3/README.md b/protocols/111210-part-3/README.md
index 2320fc30c7..fd6629ac0f 100644
--- a/protocols/111210-part-3/README.md
+++ b/protocols/111210-part-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung RNA Fusion UMI Panel Kit
diff --git a/protocols/111210-part-4/README.md b/protocols/111210-part-4/README.md
index 56cdd30df8..82f20b23dc 100644
--- a/protocols/111210-part-4/README.md
+++ b/protocols/111210-part-4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung RNA Fusion UMI Panel Kit
diff --git a/protocols/111210-part-5/README.md b/protocols/111210-part-5/README.md
index ed218fe8f2..f680829b59 100644
--- a/protocols/111210-part-5/README.md
+++ b/protocols/111210-part-5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung RNA Fusion UMI Panel Kit
diff --git a/protocols/111210-part-6/README.md b/protocols/111210-part-6/README.md
index 3c9e6f29e8..89d4781ba7 100644
--- a/protocols/111210-part-6/README.md
+++ b/protocols/111210-part-6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung RNA Fusion UMI Panel Kit
diff --git a/protocols/111210-part-7/README.md b/protocols/111210-part-7/README.md
index 64b478c869..0a6054224a 100644
--- a/protocols/111210-part-7/README.md
+++ b/protocols/111210-part-7/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung RNA Fusion UMI Panel Kit
diff --git a/protocols/111210-part-8/README.md b/protocols/111210-part-8/README.md
index c88fbc95cb..78ea51b2c9 100644
--- a/protocols/111210-part-8/README.md
+++ b/protocols/111210-part-8/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung RNA Fusion UMI Panel Kit
diff --git a/protocols/111210-part-9/README.md b/protocols/111210-part-9/README.md
index 006d412077..e04fea7049 100644
--- a/protocols/111210-part-9/README.md
+++ b/protocols/111210-part-9/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung DNA UMI Panel Kit
diff --git a/protocols/111210/README.md b/protocols/111210/README.md
index 1a45ff484e..f27f3fe914 100644
--- a/protocols/111210/README.md
+++ b/protocols/111210/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung RNA Fusion UMI Panel Kit
diff --git a/protocols/115d98/README.md b/protocols/115d98/README.md
index cb9df036b7..a25d5516cb 100644
--- a/protocols/115d98/README.md
+++ b/protocols/115d98/README.md
@@ -6,6 +6,9 @@
### Partner
[BasisDx](https://www.basisdx.org/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Aliquoting
diff --git a/protocols/1185d3/README.md b/protocols/1185d3/README.md
index 8163617235..e56f2fa452 100644
--- a/protocols/1185d3/README.md
+++ b/protocols/1185d3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* MagMAX Nucleic Acid Isolation Kit
diff --git a/protocols/118e8c/README.md b/protocols/118e8c/README.md
index d4bcdbc3ee..05ee7d5fa4 100644
--- a/protocols/118e8c/README.md
+++ b/protocols/118e8c/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/11bb6a-part-2/README.md b/protocols/11bb6a-part-2/README.md
index c7ae45ec9d..62be95ebf6 100644
--- a/protocols/11bb6a-part-2/README.md
+++ b/protocols/11bb6a-part-2/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext
diff --git a/protocols/11bb6a-part-3/README.md b/protocols/11bb6a-part-3/README.md
index 5720778e3b..66e8112f04 100644
--- a/protocols/11bb6a-part-3/README.md
+++ b/protocols/11bb6a-part-3/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext
diff --git a/protocols/11bb6a-part-4/README.md b/protocols/11bb6a-part-4/README.md
index 0134cab645..4b0c1259df 100644
--- a/protocols/11bb6a-part-4/README.md
+++ b/protocols/11bb6a-part-4/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext
diff --git a/protocols/11bb6a-part-5/README.md b/protocols/11bb6a-part-5/README.md
index b7e876c9e1..c9f18e03e1 100644
--- a/protocols/11bb6a-part-5/README.md
+++ b/protocols/11bb6a-part-5/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext
diff --git a/protocols/11bb6a-part-6/README.md b/protocols/11bb6a-part-6/README.md
index 23d3165448..a31adbb7cc 100644
--- a/protocols/11bb6a-part-6/README.md
+++ b/protocols/11bb6a-part-6/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext
diff --git a/protocols/11bb6a-part-7/README.md b/protocols/11bb6a-part-7/README.md
index 8ace51520a..2a69b14084 100644
--- a/protocols/11bb6a-part-7/README.md
+++ b/protocols/11bb6a-part-7/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext
diff --git a/protocols/11bb6a-part-8/README.md b/protocols/11bb6a-part-8/README.md
index 34bafb0b62..ba4e85062a 100644
--- a/protocols/11bb6a-part-8/README.md
+++ b/protocols/11bb6a-part-8/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext
diff --git a/protocols/11bb6a-part-9/README.md b/protocols/11bb6a-part-9/README.md
index 809a3548a0..75c11b51ff 100644
--- a/protocols/11bb6a-part-9/README.md
+++ b/protocols/11bb6a-part-9/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext
diff --git a/protocols/11bb6a/README.md b/protocols/11bb6a/README.md
index e2e73f9de2..09b7ab0705 100644
--- a/protocols/11bb6a/README.md
+++ b/protocols/11bb6a/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext
diff --git a/protocols/11beaa/README.md b/protocols/11beaa/README.md
index 5ebd43c12b..5d94f29370 100644
--- a/protocols/11beaa/README.md
+++ b/protocols/11beaa/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/1211/README.md b/protocols/1211/README.md
index 3d94029b1d..b9423e5736 100644
--- a/protocols/1211/README.md
+++ b/protocols/1211/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/121d15-2-384/README.md b/protocols/121d15-2-384/README.md
index bf598bd844..cd75bf4041 100644
--- a/protocols/121d15-2-384/README.md
+++ b/protocols/121d15-2-384/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/121d15-2-96-Greiner-1000/README.md b/protocols/121d15-2-96-Greiner-1000/README.md
index 98d99f3f6e..056ff66a3b 100644
--- a/protocols/121d15-2-96-Greiner-1000/README.md
+++ b/protocols/121d15-2-96-Greiner-1000/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/121d15-2-96-Greiner-500/README.md b/protocols/121d15-2-96-Greiner-500/README.md
index d3526f3427..064da3004d 100644
--- a/protocols/121d15-2-96-Greiner-500/README.md
+++ b/protocols/121d15-2-96-Greiner-500/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/121d15-2-96-Irish-2200/README.md b/protocols/121d15-2-96-Irish-2200/README.md
index 7cb5e70cf4..51c67eb9c7 100644
--- a/protocols/121d15-2-96-Irish-2200/README.md
+++ b/protocols/121d15-2-96-Irish-2200/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/121d15-3/README.md b/protocols/121d15-3/README.md
index e2e51d0c2b..89d83f55bb 100644
--- a/protocols/121d15-3/README.md
+++ b/protocols/121d15-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/121d15-4/README.md b/protocols/121d15-4/README.md
index 57719fbd13..c9310d964e 100644
--- a/protocols/121d15-4/README.md
+++ b/protocols/121d15-4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/121d15-5/README.md b/protocols/121d15-5/README.md
index c62b72b6df..7f8ca5916a 100644
--- a/protocols/121d15-5/README.md
+++ b/protocols/121d15-5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/121d15-6/README.md b/protocols/121d15-6/README.md
index a8f8c45861..8a05748e74 100644
--- a/protocols/121d15-6/README.md
+++ b/protocols/121d15-6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/121d15-7-2ml-15ml-aliquot/README.md b/protocols/121d15-7-2ml-15ml-aliquot/README.md
index 744ca52a16..778ba8f805 100644
--- a/protocols/121d15-7-2ml-15ml-aliquot/README.md
+++ b/protocols/121d15-7-2ml-15ml-aliquot/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Pooling
diff --git a/protocols/121d15-7-2ml-15ml-pool/README.md b/protocols/121d15-7-2ml-15ml-pool/README.md
index 744ca52a16..778ba8f805 100644
--- a/protocols/121d15-7-2ml-15ml-pool/README.md
+++ b/protocols/121d15-7-2ml-15ml-pool/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Pooling
diff --git a/protocols/121d15-7-2ml-2ml-aliquot/README.md b/protocols/121d15-7-2ml-2ml-aliquot/README.md
index 8f9a36740f..60a7f99f37 100644
--- a/protocols/121d15-7-2ml-2ml-aliquot/README.md
+++ b/protocols/121d15-7-2ml-2ml-aliquot/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Aliquoting
diff --git a/protocols/121d15-7-2ml-2ml-pool/README.md b/protocols/121d15-7-2ml-2ml-pool/README.md
index ffdc029435..3c98ac247e 100644
--- a/protocols/121d15-7-2ml-2ml-pool/README.md
+++ b/protocols/121d15-7-2ml-2ml-pool/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Pooling
diff --git a/protocols/121d15-repeat/README.md b/protocols/121d15-repeat/README.md
index 57e690cf9d..4b657da312 100644
--- a/protocols/121d15-repeat/README.md
+++ b/protocols/121d15-repeat/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/121d15/README.md b/protocols/121d15/README.md
index f53d6c5854..5e92aee8f6 100644
--- a/protocols/121d15/README.md
+++ b/protocols/121d15/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/12213d/README.md b/protocols/12213d/README.md
index d9aa70b00d..8f384c26e2 100644
--- a/protocols/12213d/README.md
+++ b/protocols/12213d/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/131de0/README.md b/protocols/131de0/README.md
index 5d734ad513..870c2d7dbc 100644
--- a/protocols/131de0/README.md
+++ b/protocols/131de0/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/139815/README.md b/protocols/139815/README.md
index ca365ed96a..3f0c637680 100644
--- a/protocols/139815/README.md
+++ b/protocols/139815/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Consolidation
diff --git a/protocols/13ac78/README.md b/protocols/13ac78/README.md
index 9f027c2e11..c73e4600dc 100644
--- a/protocols/13ac78/README.md
+++ b/protocols/13ac78/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/13c0b8/README.md b/protocols/13c0b8/README.md
index d88bbd3248..c37ad4c2e3 100644
--- a/protocols/13c0b8/README.md
+++ b/protocols/13c0b8/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Staining Prep
diff --git a/protocols/13ea1e-part2/README.md b/protocols/13ea1e-part2/README.md
index b808d40d63..8cc85462d6 100644
--- a/protocols/13ea1e-part2/README.md
+++ b/protocols/13ea1e-part2/README.md
@@ -6,6 +6,9 @@
### Partner
[Partner Name](partner website link)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/13ea1e/README.md b/protocols/13ea1e/README.md
index e1601eea4e..d559cf1c9d 100644
--- a/protocols/13ea1e/README.md
+++ b/protocols/13ea1e/README.md
@@ -6,6 +6,9 @@
### Partner
[Partner Name](partner website link)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/13fd88/README.md b/protocols/13fd88/README.md
index 998170811d..c6a221f037 100644
--- a/protocols/13fd88/README.md
+++ b/protocols/13fd88/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Purification
diff --git a/protocols/141a1a/README.md b/protocols/141a1a/README.md
index 1f37c91ff8..f8e1d8c4a1 100644
--- a/protocols/141a1a/README.md
+++ b/protocols/141a1a/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cell-Free Gene Expression
diff --git a/protocols/1440ad/README.md b/protocols/1440ad/README.md
index 71123fcbac..6551890227 100644
--- a/protocols/1440ad/README.md
+++ b/protocols/1440ad/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* RNA Extraction
diff --git a/protocols/1464-2/README.md b/protocols/1464-2/README.md
index 3d79c006fb..97cece847e 100644
--- a/protocols/1464-2/README.md
+++ b/protocols/1464-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](http://www.opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* ELISA
diff --git a/protocols/1464-3/README.md b/protocols/1464-3/README.md
index cd8289c2fb..99e29fe1b5 100644
--- a/protocols/1464-3/README.md
+++ b/protocols/1464-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](http://www.opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* ELISA
diff --git a/protocols/1464/README.md b/protocols/1464/README.md
index bdf81837c7..fee28781ab 100644
--- a/protocols/1464/README.md
+++ b/protocols/1464/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](http://www.opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* ELISA
diff --git a/protocols/1477ea/README.md b/protocols/1477ea/README.md
index a3314b8b3d..9a7fcf2b09 100644
--- a/protocols/1477ea/README.md
+++ b/protocols/1477ea/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/1479/README.md b/protocols/1479/README.md
index 14785aad0e..a6b682bc3b 100644
--- a/protocols/1479/README.md
+++ b/protocols/1479/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/14b685/README.md b/protocols/14b685/README.md
index 4bf74b1614..6780bf70c9 100644
--- a/protocols/14b685/README.md
+++ b/protocols/14b685/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina 16S
diff --git a/protocols/154ec5/README.md b/protocols/154ec5/README.md
index 32507e0810..8bae3ae6df 100644
--- a/protocols/154ec5/README.md
+++ b/protocols/154ec5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/1577-v2/README.md b/protocols/1577-v2/README.md
index 89b51de23e..b0badd45a3 100644
--- a/protocols/1577-v2/README.md
+++ b/protocols/1577-v2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](http://www.opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/158d42/README.md b/protocols/158d42/README.md
index 4cadd8799d..3d3eb25ab8 100644
--- a/protocols/158d42/README.md
+++ b/protocols/158d42/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/15dc4c/README.md b/protocols/15dc4c/README.md
index e4e9d91c19..3501d98bd7 100644
--- a/protocols/15dc4c/README.md
+++ b/protocols/15dc4c/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* ELISA
diff --git a/protocols/1601a9/README.md b/protocols/1601a9/README.md
index 9868094f89..0d1a65aabd 100644
--- a/protocols/1601a9/README.md
+++ b/protocols/1601a9/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Illumina Beachip Amplification
diff --git a/protocols/1649b1/README.md b/protocols/1649b1/README.md
index 47e6c9b47a..228b0b90b6 100644
--- a/protocols/1649b1/README.md
+++ b/protocols/1649b1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Extraction
diff --git a/protocols/165a77/README.md b/protocols/165a77/README.md
index 46136ad88e..66a45f3c95 100644
--- a/protocols/165a77/README.md
+++ b/protocols/165a77/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* qPCR setup
diff --git a/protocols/16b3fb/README.md b/protocols/16b3fb/README.md
index 1bcf6d80e4..0f0adb4370 100644
--- a/protocols/16b3fb/README.md
+++ b/protocols/16b3fb/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/17cb2d/README.md b/protocols/17cb2d/README.md
index 34b9b88f88..998a1644b2 100644
--- a/protocols/17cb2d/README.md
+++ b/protocols/17cb2d/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/17d210-part-2/README.md b/protocols/17d210-part-2/README.md
index 2402dfee86..76bcfffa8b 100644
--- a/protocols/17d210-part-2/README.md
+++ b/protocols/17d210-part-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Verogen ForenSeq DNA Signature Prep Kit
diff --git a/protocols/17d210-part-3/README.md b/protocols/17d210-part-3/README.md
index 15ca8235f7..2b2afa46ed 100644
--- a/protocols/17d210-part-3/README.md
+++ b/protocols/17d210-part-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Verogen ForenSeq DNA Signature Prep Kit
diff --git a/protocols/17d210-part-4/README.md b/protocols/17d210-part-4/README.md
index 7016a785b2..b0ac60ce3a 100644
--- a/protocols/17d210-part-4/README.md
+++ b/protocols/17d210-part-4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Verogen ForenSeq DNA Signature Prep Kit
diff --git a/protocols/17d210-part-5/README.md b/protocols/17d210-part-5/README.md
index e8f3e1b36e..3ba27a56d8 100644
--- a/protocols/17d210-part-5/README.md
+++ b/protocols/17d210-part-5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Verogen ForenSeq DNA Signature Prep Kit
diff --git a/protocols/17d210/README.md b/protocols/17d210/README.md
index 29fdfbb1d4..ce77a99de9 100644
--- a/protocols/17d210/README.md
+++ b/protocols/17d210/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Verogen ForenSeq DNA Signature Prep Kit
diff --git a/protocols/18050b/README.md b/protocols/18050b/README.md
index 6a498d5f96..627f9d431d 100644
--- a/protocols/18050b/README.md
+++ b/protocols/18050b/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/18d6a1/README.md b/protocols/18d6a1/README.md
index fe1676f73a..2de95dbed4 100644
--- a/protocols/18d6a1/README.md
+++ b/protocols/18d6a1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Extraction
diff --git a/protocols/18e62e/README.md b/protocols/18e62e/README.md
index 196c8b75b8..e4b93eb7e2 100644
--- a/protocols/18e62e/README.md
+++ b/protocols/18e62e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/19313a/README.md b/protocols/19313a/README.md
index 46c88e2ca0..6db73710e5 100644
--- a/protocols/19313a/README.md
+++ b/protocols/19313a/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Purification
diff --git a/protocols/1980cd/README.md b/protocols/1980cd/README.md
index 3503672dd9..0fe0e044ae 100644
--- a/protocols/1980cd/README.md
+++ b/protocols/1980cd/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Plant DNA
diff --git a/protocols/199721/README.md b/protocols/199721/README.md
index 802f468fee..2b08f05a41 100644
--- a/protocols/199721/README.md
+++ b/protocols/199721/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Pooling
diff --git a/protocols/19fc32/README.md b/protocols/19fc32/README.md
index 50bf05e372..459ef9f8c6 100644
--- a/protocols/19fc32/README.md
+++ b/protocols/19fc32/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Clean Up
diff --git a/protocols/1a2343/README.md b/protocols/1a2343/README.md
index ccd66ad791..0afd205cc8 100644
--- a/protocols/1a2343/README.md
+++ b/protocols/1a2343/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/1adec6-2/README.md b/protocols/1adec6-2/README.md
index 7e90c383a8..61c8fc6519 100644
--- a/protocols/1adec6-2/README.md
+++ b/protocols/1adec6-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Assay
diff --git a/protocols/1adec6-3/README.md b/protocols/1adec6-3/README.md
index 1c99423098..fed535cf2e 100644
--- a/protocols/1adec6-3/README.md
+++ b/protocols/1adec6-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Assay
diff --git a/protocols/1adec6-4/README.md b/protocols/1adec6-4/README.md
index 04962f0266..36ab481641 100644
--- a/protocols/1adec6-4/README.md
+++ b/protocols/1adec6-4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Assay
diff --git a/protocols/1adec6-5/README.md b/protocols/1adec6-5/README.md
index d05c72f4da..fd5809a861 100644
--- a/protocols/1adec6-5/README.md
+++ b/protocols/1adec6-5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Assay
diff --git a/protocols/1adec6-6/README.md b/protocols/1adec6-6/README.md
index bd05268de4..af14e43aeb 100644
--- a/protocols/1adec6-6/README.md
+++ b/protocols/1adec6-6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Assay
diff --git a/protocols/1adec6-7/README.md b/protocols/1adec6-7/README.md
index 01beffa509..483ff3a0ff 100644
--- a/protocols/1adec6-7/README.md
+++ b/protocols/1adec6-7/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Assay
diff --git a/protocols/1adec6/README.md b/protocols/1adec6/README.md
index 95be50fb23..b8bacb5911 100644
--- a/protocols/1adec6/README.md
+++ b/protocols/1adec6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Assay
diff --git a/protocols/1af856/README.md b/protocols/1af856/README.md
index b3863dfbea..ecf3cbbd4b 100644
--- a/protocols/1af856/README.md
+++ b/protocols/1af856/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Lexogen
diff --git a/protocols/1b2788/README.md b/protocols/1b2788/README.md
index 9fbd4e5496..2f4c55c772 100644
--- a/protocols/1b2788/README.md
+++ b/protocols/1b2788/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* DNA Extraction
diff --git a/protocols/1bcd67/README.md b/protocols/1bcd67/README.md
index f91f09779f..8143eef3f1 100644
--- a/protocols/1bcd67/README.md
+++ b/protocols/1bcd67/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/1c086c/README.md b/protocols/1c086c/README.md
index ac6a28322e..daa45d6e6c 100644
--- a/protocols/1c086c/README.md
+++ b/protocols/1c086c/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Distribution
diff --git a/protocols/1c3611/README.md b/protocols/1c3611/README.md
index c888b970f7..a1c3851092 100644
--- a/protocols/1c3611/README.md
+++ b/protocols/1c3611/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/1c8468/README.md b/protocols/1c8468/README.md
index 7b40a178c8..da7f3b9a51 100644
--- a/protocols/1c8468/README.md
+++ b/protocols/1c8468/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/1ccd23-station-A/README.md b/protocols/1ccd23-station-A/README.md
index 67c82cf579..cd2066b2b7 100644
--- a/protocols/1ccd23-station-A/README.md
+++ b/protocols/1ccd23-station-A/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Covid Workstation
* Sample Plating
diff --git a/protocols/1ccd23-station-B/README.md b/protocols/1ccd23-station-B/README.md
index 196cdcfabe..65af1851e1 100644
--- a/protocols/1ccd23-station-B/README.md
+++ b/protocols/1ccd23-station-B/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons (verified)](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Covid Workstation
* RNA Extraction
diff --git a/protocols/1ccd23-station-C/README.md b/protocols/1ccd23-station-C/README.md
index fff69e62f4..f006e0993a 100644
--- a/protocols/1ccd23-station-C/README.md
+++ b/protocols/1ccd23-station-C/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Covid Workstation
* qPCR Setup
diff --git a/protocols/1ce6aa/README.md b/protocols/1ce6aa/README.md
index 564c099196..9749d7b1d7 100644
--- a/protocols/1ce6aa/README.md
+++ b/protocols/1ce6aa/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/1d37e5/README.md b/protocols/1d37e5/README.md
index cebded3036..28a2137d11 100644
--- a/protocols/1d37e5/README.md
+++ b/protocols/1d37e5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/1d6d1b/README.md b/protocols/1d6d1b/README.md
index 35256c926b..84ebede8e5 100644
--- a/protocols/1d6d1b/README.md
+++ b/protocols/1d6d1b/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Slide Spotting
diff --git a/protocols/1dbc24/README.md b/protocols/1dbc24/README.md
index 47732cd6b5..5b12c33f93 100644
--- a/protocols/1dbc24/README.md
+++ b/protocols/1dbc24/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/1dec68/README.md b/protocols/1dec68/README.md
index 520a7e338d..c61f0102e2 100644
--- a/protocols/1dec68/README.md
+++ b/protocols/1dec68/README.md
@@ -6,6 +6,9 @@
### Partner
[Partner Name](partner website link)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/1dfebe/README.md b/protocols/1dfebe/README.md
index 659421fee6..97018ef4d1 100644
--- a/protocols/1dfebe/README.md
+++ b/protocols/1dfebe/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/1e744e/README.md b/protocols/1e744e/README.md
index c7f004d8cf..fde556b680 100644
--- a/protocols/1e744e/README.md
+++ b/protocols/1e744e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/1eeb01/README.md b/protocols/1eeb01/README.md
index fa704c1077..cf4fc3fa42 100644
--- a/protocols/1eeb01/README.md
+++ b/protocols/1eeb01/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](http://www.opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Extraction
diff --git a/protocols/1ef67e/README.md b/protocols/1ef67e/README.md
index 041bddb126..fd43d49441 100644
--- a/protocols/1ef67e/README.md
+++ b/protocols/1ef67e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Assay
diff --git a/protocols/1f62ba/README.md b/protocols/1f62ba/README.md
index f8923d61f6..b545e4de7d 100644
--- a/protocols/1f62ba/README.md
+++ b/protocols/1f62ba/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/1f8b55/README.md b/protocols/1f8b55/README.md
index 1d754d2409..03739f83f8 100644
--- a/protocols/1f8b55/README.md
+++ b/protocols/1f8b55/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/1f96ca-part-2/README.md b/protocols/1f96ca-part-2/README.md
index c83fdbbab3..58036ea9f5 100644
--- a/protocols/1f96ca-part-2/README.md
+++ b/protocols/1f96ca-part-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* KAPA Hyper Plus 96rx, cat 07962428001 ROCHE
diff --git a/protocols/1f96ca/README.md b/protocols/1f96ca/README.md
index f42113180a..e75c161650 100644
--- a/protocols/1f96ca/README.md
+++ b/protocols/1f96ca/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* KAPA Hyper Plus 96rx, cat 07962428001 ROCHE
diff --git a/protocols/1fcf02/README.md b/protocols/1fcf02/README.md
index 118edf9dce..32b4e0109e 100644
--- a/protocols/1fcf02/README.md
+++ b/protocols/1fcf02/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/203136/README.md b/protocols/203136/README.md
index 9796a67a1f..ed5c1636b4 100644
--- a/protocols/203136/README.md
+++ b/protocols/203136/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Invitrogen ChargeSwitch
diff --git a/protocols/2082dd/README.md b/protocols/2082dd/README.md
index aee607a7b8..8d369015c5 100644
--- a/protocols/2082dd/README.md
+++ b/protocols/2082dd/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/211a24/README.md b/protocols/211a24/README.md
index 758a86e39c..ae64ffa12e 100644
--- a/protocols/211a24/README.md
+++ b/protocols/211a24/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Serial Dilution
diff --git a/protocols/211fe1/README.md b/protocols/211fe1/README.md
index e08149bf2a..e4301b559d 100644
--- a/protocols/211fe1/README.md
+++ b/protocols/211fe1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Mass Spec
diff --git a/protocols/21e4d8-pt1/README.md b/protocols/21e4d8-pt1/README.md
index 810c56f1d0..66ad14fa99 100644
--- a/protocols/21e4d8-pt1/README.md
+++ b/protocols/21e4d8-pt1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Twist Library Prep
diff --git a/protocols/21e4d8-pt2/README.md b/protocols/21e4d8-pt2/README.md
index ed09771a81..ab1a3526e7 100644
--- a/protocols/21e4d8-pt2/README.md
+++ b/protocols/21e4d8-pt2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Twist Library Prep
diff --git a/protocols/21e4d8-pt3/README.md b/protocols/21e4d8-pt3/README.md
index 3c833d33b0..17c7ba3760 100644
--- a/protocols/21e4d8-pt3/README.md
+++ b/protocols/21e4d8-pt3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Twist Library Prep
diff --git a/protocols/21u968/README.md b/protocols/21u968/README.md
index a425c49ebb..3eb25840a3 100644
--- a/protocols/21u968/README.md
+++ b/protocols/21u968/README.md
@@ -5,6 +5,9 @@
Iva Pitelkova
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* DNA/RNA
* DNA/RNA
diff --git a/protocols/226d24/README.md b/protocols/226d24/README.md
index c8baf59ec3..db93f63e74 100644
--- a/protocols/226d24/README.md
+++ b/protocols/226d24/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/234495/README.md b/protocols/234495/README.md
index 46e4008ba5..519431a80f 100644
--- a/protocols/234495/README.md
+++ b/protocols/234495/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Mass Spec
diff --git a/protocols/243973/README.md b/protocols/243973/README.md
index ef244c52e8..2b0cbfa973 100644
--- a/protocols/243973/README.md
+++ b/protocols/243973/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/245eb2/README.md b/protocols/245eb2/README.md
index feb8d78cc9..23274dd7ab 100644
--- a/protocols/245eb2/README.md
+++ b/protocols/245eb2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Extraction
diff --git a/protocols/25a03d/README.md b/protocols/25a03d/README.md
index bf13a365d1..8cb4b704fe 100644
--- a/protocols/25a03d/README.md
+++ b/protocols/25a03d/README.md
@@ -6,6 +6,9 @@
### Partner
[Partner Name](partner website link)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/269b21/README.md b/protocols/269b21/README.md
index 4fcbb23a50..6ea421b3db 100644
--- a/protocols/269b21/README.md
+++ b/protocols/269b21/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/26a414/README.md b/protocols/26a414/README.md
index 288733d7e2..8d3683a550 100644
--- a/protocols/26a414/README.md
+++ b/protocols/26a414/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/26c304/README.md b/protocols/26c304/README.md
index 1b2819a565..5b67529c4b 100644
--- a/protocols/26c304/README.md
+++ b/protocols/26c304/README.md
@@ -6,6 +6,9 @@
### Partner
[Partner Name](partner website link)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/272c63/README.md b/protocols/272c63/README.md
index c0d93879ff..a7a37c4323 100644
--- a/protocols/272c63/README.md
+++ b/protocols/272c63/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Tube Filling
diff --git a/protocols/274d2a/README.md b/protocols/274d2a/README.md
index 724314a38c..42bd7a2a41 100644
--- a/protocols/274d2a/README.md
+++ b/protocols/274d2a/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/274d2a). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/274d2a). This page won’t be available after January 31st, 2024.
## Categories
* Sample Prep
diff --git a/protocols/277a3d/README.md b/protocols/277a3d/README.md
index 2dbe017912..e6805732c8 100644
--- a/protocols/277a3d/README.md
+++ b/protocols/277a3d/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Oxford Nanopore 16S Barcoding Kit
diff --git a/protocols/280ea4/README.md b/protocols/280ea4/README.md
index 23a5cc4076..ea23686305 100644
--- a/protocols/280ea4/README.md
+++ b/protocols/280ea4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/2905a4/README.md b/protocols/2905a4/README.md
index 977f7e88d1..95b8e1b130 100644
--- a/protocols/2905a4/README.md
+++ b/protocols/2905a4/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
diff --git a/protocols/29225e-part-2/README.md b/protocols/29225e-part-2/README.md
index 80ed6d9eaa..f3b2101b4f 100644
--- a/protocols/29225e-part-2/README.md
+++ b/protocols/29225e-part-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/29225e/README.md b/protocols/29225e/README.md
index 07517b016d..3b7a0fb124 100644
--- a/protocols/29225e/README.md
+++ b/protocols/29225e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/29414b-ot/README.md b/protocols/29414b-ot/README.md
index 9b083c20e0..768267ba21 100644
--- a/protocols/29414b-ot/README.md
+++ b/protocols/29414b-ot/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Getting Started
* Calibration
diff --git a/protocols/29414b/README.md b/protocols/29414b/README.md
index f0a4df6e51..d1785f6bc5 100644
--- a/protocols/29414b/README.md
+++ b/protocols/29414b/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Getting Started
* Calibration
diff --git a/protocols/2942dd/README.md b/protocols/2942dd/README.md
index 7337da77b4..c05cdd323e 100644
--- a/protocols/2942dd/README.md
+++ b/protocols/2942dd/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/295c73/README.md b/protocols/295c73/README.md
index 495cc5754c..1b608908f4 100644
--- a/protocols/295c73/README.md
+++ b/protocols/295c73/README.md
@@ -1,5 +1,8 @@
# Accel-Amplicon® Plus EGFR Pathway Panel
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Accel-Amplicon Plus EGFR Pathway Panel
diff --git a/protocols/298069/README.md b/protocols/298069/README.md
index ed7d3ee41d..3ef17a71ff 100644
--- a/protocols/298069/README.md
+++ b/protocols/298069/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Aliquoting
diff --git a/protocols/29effa-buffer/README.md b/protocols/29effa-buffer/README.md
index b260b74379..8112f26cbb 100644
--- a/protocols/29effa-buffer/README.md
+++ b/protocols/29effa-buffer/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/29effa-mastermix/README.md b/protocols/29effa-mastermix/README.md
index 1e6c5c0875..d85f9acfaf 100644
--- a/protocols/29effa-mastermix/README.md
+++ b/protocols/29effa-mastermix/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/29f643/README.md b/protocols/29f643/README.md
index fb2e09f96b..ebcd7533c1 100644
--- a/protocols/29f643/README.md
+++ b/protocols/29f643/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* ELISA
diff --git a/protocols/2a370c/README.md b/protocols/2a370c/README.md
index 035fd837d3..a3348b2e63 100644
--- a/protocols/2a370c/README.md
+++ b/protocols/2a370c/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Omega Mag-Bind Bacterial DNA 96 Kit
diff --git a/protocols/2aee74-48-2/README.md b/protocols/2aee74-48-2/README.md
index dd64f15492..97db5250d5 100644
--- a/protocols/2aee74-48-2/README.md
+++ b/protocols/2aee74-48-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Olink® Target 48
diff --git a/protocols/2aee74-48-3/README.md b/protocols/2aee74-48-3/README.md
index 74053efaef..951ba5aa61 100644
--- a/protocols/2aee74-48-3/README.md
+++ b/protocols/2aee74-48-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Olink® Target 48
diff --git a/protocols/2aee74-48/README.md b/protocols/2aee74-48/README.md
index bf03b6a7f5..c1e2ae1681 100644
--- a/protocols/2aee74-48/README.md
+++ b/protocols/2aee74-48/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Olink® Target 48
diff --git a/protocols/2aee74-96-2/README.md b/protocols/2aee74-96-2/README.md
index 665989aa4d..20ccab837f 100644
--- a/protocols/2aee74-96-2/README.md
+++ b/protocols/2aee74-96-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Olink® Target 96
diff --git a/protocols/2aee74-96-3/README.md b/protocols/2aee74-96-3/README.md
index 38ef0b9f49..8b42020dfe 100644
--- a/protocols/2aee74-96-3/README.md
+++ b/protocols/2aee74-96-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Olink® Target 96
diff --git a/protocols/2aee74-96/README.md b/protocols/2aee74-96/README.md
index f1d3d1ba14..de45b37654 100644
--- a/protocols/2aee74-96/README.md
+++ b/protocols/2aee74-96/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Olink® Target 96
diff --git a/protocols/2b3642/README.md b/protocols/2b3642/README.md
index 52bbca4ca6..52b8bc11c2 100644
--- a/protocols/2b3642/README.md
+++ b/protocols/2b3642/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II DNA Library Preparation Kit for Illumina E7645S
diff --git a/protocols/2b9486/README.md b/protocols/2b9486/README.md
index 519454ca30..290b245008 100644
--- a/protocols/2b9486/README.md
+++ b/protocols/2b9486/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/2bba96/README.md b/protocols/2bba96/README.md
index 902f1cb3b0..bc8f81d4d2 100644
--- a/protocols/2bba96/README.md
+++ b/protocols/2bba96/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/2bdff3/README.md b/protocols/2bdff3/README.md
index bd82185a20..0ea8b85ff0 100644
--- a/protocols/2bdff3/README.md
+++ b/protocols/2bdff3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Pooling
diff --git a/protocols/2c19aa/README.md b/protocols/2c19aa/README.md
index e869e30337..37c6daddf6 100644
--- a/protocols/2c19aa/README.md
+++ b/protocols/2c19aa/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/2c62b7/README.md b/protocols/2c62b7/README.md
index ef781b5aa7..e7f362a21f 100644
--- a/protocols/2c62b7/README.md
+++ b/protocols/2c62b7/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Omega Mag-Bind Environmental DNA 96 Kit
diff --git a/protocols/2c81c0/README.md b/protocols/2c81c0/README.md
index b96fb860ef..67a3b71e51 100644
--- a/protocols/2c81c0/README.md
+++ b/protocols/2c81c0/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Featured
* Cherrypicking
diff --git a/protocols/2c83f7/README.md b/protocols/2c83f7/README.md
index 5fb2f5b900..7eb0d8aac2 100644
--- a/protocols/2c83f7/README.md
+++ b/protocols/2c83f7/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Zymo Kit
diff --git a/protocols/2cc59d/README.md b/protocols/2cc59d/README.md
index 0627993e5a..9b7c788b83 100644
--- a/protocols/2cc59d/README.md
+++ b/protocols/2cc59d/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/2d7126/README.md b/protocols/2d7126/README.md
index c863791696..3f260b92e3 100644
--- a/protocols/2d7126/README.md
+++ b/protocols/2d7126/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Slide Spotting
diff --git a/protocols/2d7d86/README.md b/protocols/2d7d86/README.md
index ee6dbbec3a..ccfa66a0a3 100644
--- a/protocols/2d7d86/README.md
+++ b/protocols/2d7d86/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* qpcr setup
diff --git a/protocols/2d9a0c/README.md b/protocols/2d9a0c/README.md
index 8ffcffcd42..ed2743eb8d 100644
--- a/protocols/2d9a0c/README.md
+++ b/protocols/2d9a0c/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* Complete PCR Workflow
diff --git a/protocols/2df9f8-pt2/README.md b/protocols/2df9f8-pt2/README.md
index 926087de6a..933a3712f5 100644
--- a/protocols/2df9f8-pt2/README.md
+++ b/protocols/2df9f8-pt2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/2df9f8-pt3/README.md b/protocols/2df9f8-pt3/README.md
index 8cae1d9746..d81e5e4f6e 100644
--- a/protocols/2df9f8-pt3/README.md
+++ b/protocols/2df9f8-pt3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/2df9f8-pt4/README.md b/protocols/2df9f8-pt4/README.md
index 2420a62054..b43b54bc6e 100644
--- a/protocols/2df9f8-pt4/README.md
+++ b/protocols/2df9f8-pt4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/2df9f8/README.md b/protocols/2df9f8/README.md
index 1d6f4b527c..9447519a01 100644
--- a/protocols/2df9f8/README.md
+++ b/protocols/2df9f8/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/2e84cc/README.md b/protocols/2e84cc/README.md
index 40bf487f46..f9c4d6bf48 100644
--- a/protocols/2e84cc/README.md
+++ b/protocols/2e84cc/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina Nextera XT
diff --git a/protocols/2ed4de-2/README.md b/protocols/2ed4de-2/README.md
index bbfcd627b9..e90c166adf 100644
--- a/protocols/2ed4de-2/README.md
+++ b/protocols/2ed4de-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Ribogreen Assay
diff --git a/protocols/2ed4de/README.md b/protocols/2ed4de/README.md
index c9b8926e6c..7fe717d0d2 100644
--- a/protocols/2ed4de/README.md
+++ b/protocols/2ed4de/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Ribogreen Assay
diff --git a/protocols/2ee5be/README.md b/protocols/2ee5be/README.md
index ef2505c0d7..d662b3b070 100644
--- a/protocols/2ee5be/README.md
+++ b/protocols/2ee5be/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/300073-2/README.md b/protocols/300073-2/README.md
index 2bc6fe3c76..58cff5916a 100644
--- a/protocols/300073-2/README.md
+++ b/protocols/300073-2/README.md
@@ -2,6 +2,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* PCR prep
diff --git a/protocols/300073/README.md b/protocols/300073/README.md
index ec264179d3..1f5c945824 100644
--- a/protocols/300073/README.md
+++ b/protocols/300073/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* PCR prep
diff --git a/protocols/30174a/README.md b/protocols/30174a/README.md
index 23f2f54b9b..93fa4366e3 100644
--- a/protocols/30174a/README.md
+++ b/protocols/30174a/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/30450c-part-2/README.md b/protocols/30450c-part-2/README.md
index f8a2683d23..315da0c44e 100644
--- a/protocols/30450c-part-2/README.md
+++ b/protocols/30450c-part-2/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/30450c/README.md b/protocols/30450c/README.md
index baa7670298..48f329bda6 100644
--- a/protocols/30450c/README.md
+++ b/protocols/30450c/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/313086-logixsmart-station-B/README.md b/protocols/313086-logixsmart-station-B/README.md
index cabec3488f..b19103f1e8 100644
--- a/protocols/313086-logixsmart-station-B/README.md
+++ b/protocols/313086-logixsmart-station-B/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Covid Workstation
* qPCR Setup
diff --git a/protocols/313086/README.md b/protocols/313086/README.md
index 821285f42d..368c871ab7 100644
--- a/protocols/313086/README.md
+++ b/protocols/313086/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Covid Workstation
* Sample Plating
diff --git a/protocols/315118/README.md b/protocols/315118/README.md
index c2bdc8eb9a..d68e3bf7a1 100644
--- a/protocols/315118/README.md
+++ b/protocols/315118/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Pooling
diff --git a/protocols/32207e/README.md b/protocols/32207e/README.md
index 5d753abe89..6bd8bb9d53 100644
--- a/protocols/32207e/README.md
+++ b/protocols/32207e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/3308b4/README.md b/protocols/3308b4/README.md
index 00cf9c8e4c..97f6531b58 100644
--- a/protocols/3308b4/README.md
+++ b/protocols/3308b4/README.md
@@ -2,6 +2,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/3359a5/README.md b/protocols/3359a5/README.md
index db1dd557f4..d75b7566cb 100644
--- a/protocols/3359a5/README.md
+++ b/protocols/3359a5/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/3359a5). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/3359a5). This page won’t be available after January 31st, 2024.
## Categories
* Featured
diff --git a/protocols/33900b/README.md b/protocols/33900b/README.md
index 35e62826b7..b9c9e90b38 100644
--- a/protocols/33900b/README.md
+++ b/protocols/33900b/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* MagneHis™ Protein Purification System
diff --git a/protocols/33a449/README.md b/protocols/33a449/README.md
index b78b11372c..d4f4c8a576 100644
--- a/protocols/33a449/README.md
+++ b/protocols/33a449/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* DNA Extraction
diff --git a/protocols/33b12a/README.md b/protocols/33b12a/README.md
index 81b5aa3e61..e10e3890ed 100644
--- a/protocols/33b12a/README.md
+++ b/protocols/33b12a/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/33y0f3/README.md b/protocols/33y0f3/README.md
index e79075d96b..98182e0397 100644
--- a/protocols/33y0f3/README.md
+++ b/protocols/33y0f3/README.md
@@ -5,6 +5,9 @@
Lachlan Munro
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Microbiology
* Microbiology
diff --git a/protocols/343001/README.md b/protocols/343001/README.md
index 29c1987221..661cbf96a5 100644
--- a/protocols/343001/README.md
+++ b/protocols/343001/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Zymo Extraction
diff --git a/protocols/35269b/README.md b/protocols/35269b/README.md
index 9d5256fe8e..7ce8a9b025 100644
--- a/protocols/35269b/README.md
+++ b/protocols/35269b/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Viral RNA
diff --git a/protocols/355b30/README.md b/protocols/355b30/README.md
index deb49cfdd3..84bbdbd307 100644
--- a/protocols/355b30/README.md
+++ b/protocols/355b30/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Pooling
diff --git a/protocols/357404/README.md b/protocols/357404/README.md
index 3ff0a2f795..9813f79118 100644
--- a/protocols/357404/README.md
+++ b/protocols/357404/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Microscopy Slide Antibody Staining
diff --git a/protocols/35b1a9/README.md b/protocols/35b1a9/README.md
index 4179280e03..20e23d6168 100644
--- a/protocols/35b1a9/README.md
+++ b/protocols/35b1a9/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/3607d5-2/README.md b/protocols/3607d5-2/README.md
index 4df64f3578..dc7991412a 100644
--- a/protocols/3607d5-2/README.md
+++ b/protocols/3607d5-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Extraction
diff --git a/protocols/3607d5-3/README.md b/protocols/3607d5-3/README.md
index 1bfd052c1d..ad9d0ee127 100644
--- a/protocols/3607d5-3/README.md
+++ b/protocols/3607d5-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Extraction
diff --git a/protocols/3607d5-4/README.md b/protocols/3607d5-4/README.md
index 9ad5b2a4ce..ff8e51fa40 100644
--- a/protocols/3607d5-4/README.md
+++ b/protocols/3607d5-4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/3607d5-5/README.md b/protocols/3607d5-5/README.md
index b6ad7e7994..2f8c60275a 100644
--- a/protocols/3607d5-5/README.md
+++ b/protocols/3607d5-5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/3607d5/README.md b/protocols/3607d5/README.md
index 826b173545..2bd74e160c 100644
--- a/protocols/3607d5/README.md
+++ b/protocols/3607d5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/3633ca/README.md b/protocols/3633ca/README.md
index 1fd6975f8b..d778a74d50 100644
--- a/protocols/3633ca/README.md
+++ b/protocols/3633ca/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/3662e6/README.md b/protocols/3662e6/README.md
index 41d2fcb76f..39bdd64373 100644
--- a/protocols/3662e6/README.md
+++ b/protocols/3662e6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](http://www.opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Serial Dilution
diff --git a/protocols/37190e/README.md b/protocols/37190e/README.md
index 3ca99f4d8d..d26cc6cb25 100644
--- a/protocols/37190e/README.md
+++ b/protocols/37190e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sterile Workflows
* Cell Culture
diff --git a/protocols/37de37/README.md b/protocols/37de37/README.md
index 287af802bb..575f48faba 100644
--- a/protocols/37de37/README.md
+++ b/protocols/37de37/README.md
@@ -6,6 +6,9 @@
### Partner
[Partner Name](partner website link)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/37ffa5/README.md b/protocols/37ffa5/README.md
index 7e2851813a..1765a21c09 100644
--- a/protocols/37ffa5/README.md
+++ b/protocols/37ffa5/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/384b23-plate-transfer/README.md b/protocols/384b23-plate-transfer/README.md
index 064a0f498f..358219a6b0 100644
--- a/protocols/384b23-plate-transfer/README.md
+++ b/protocols/384b23-plate-transfer/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/384b23-pooling/README.md b/protocols/384b23-pooling/README.md
index a66709348c..0b8a14ee5e 100644
--- a/protocols/384b23-pooling/README.md
+++ b/protocols/384b23-pooling/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Normalization
* Normalization and Pooling
diff --git a/protocols/384b23-tube-transfer/README.md b/protocols/384b23-tube-transfer/README.md
index 198716aa43..8d65a4feef 100644
--- a/protocols/384b23-tube-transfer/README.md
+++ b/protocols/384b23-tube-transfer/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/387961/README.md b/protocols/387961/README.md
index beaac4f987..c9707d55be 100644
--- a/protocols/387961/README.md
+++ b/protocols/387961/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/38efd4/README.md b/protocols/38efd4/README.md
index f0be96a5de..6bc44ad4c6 100644
--- a/protocols/38efd4/README.md
+++ b/protocols/38efd4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/), Ricardo Padua, Brandeis University
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/3955db/README.md b/protocols/3955db/README.md
index 136573ceb1..94b2ddf1a0 100644
--- a/protocols/3955db/README.md
+++ b/protocols/3955db/README.md
@@ -6,6 +6,9 @@
### Partner
[BasisDx](https://www.basisdx.org/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Pooling
diff --git a/protocols/39b4c7/README.md b/protocols/39b4c7/README.md
index c4318caea2..1de21d1e3b 100644
--- a/protocols/39b4c7/README.md
+++ b/protocols/39b4c7/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* qPCR setup
diff --git a/protocols/39bbbb/README.md b/protocols/39bbbb/README.md
index 478d1e1cfe..30cb4faaa3 100644
--- a/protocols/39bbbb/README.md
+++ b/protocols/39bbbb/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/3a49b6/README.md b/protocols/3a49b6/README.md
index 244d5284e4..86eaf5dd6d 100644
--- a/protocols/3a49b6/README.md
+++ b/protocols/3a49b6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/3b0db0/README.md b/protocols/3b0db0/README.md
index 26302c9c14..13031bce2a 100644
--- a/protocols/3b0db0/README.md
+++ b/protocols/3b0db0/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/3b3d2f/README.md b/protocols/3b3d2f/README.md
index 8185b0535c..18a3f2f5dc 100644
--- a/protocols/3b3d2f/README.md
+++ b/protocols/3b3d2f/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/3b3f1c/README.md b/protocols/3b3f1c/README.md
index 152c621bcb..2583919715 100644
--- a/protocols/3b3f1c/README.md
+++ b/protocols/3b3f1c/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/3bc25d/README.md b/protocols/3bc25d/README.md
index bfb5a9693c..e77e4d5957 100644
--- a/protocols/3bc25d/README.md
+++ b/protocols/3bc25d/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Nanopore
diff --git a/protocols/3c9aec/README.md b/protocols/3c9aec/README.md
index d7abf2bd73..e246a508ee 100644
--- a/protocols/3c9aec/README.md
+++ b/protocols/3c9aec/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sterile Workflows
* Cell Culture
diff --git a/protocols/3cf31f/README.md b/protocols/3cf31f/README.md
index 029babd3c0..8bac93fa1f 100644
--- a/protocols/3cf31f/README.md
+++ b/protocols/3cf31f/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Distribution
diff --git a/protocols/3d02be/README.md b/protocols/3d02be/README.md
index 60c0a51c0c..24e9010387 100644
--- a/protocols/3d02be/README.md
+++ b/protocols/3d02be/README.md
@@ -6,6 +6,9 @@
### Partner
[Partner Name](partner website link)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Aliquoting
diff --git a/protocols/3d942d/README.md b/protocols/3d942d/README.md
index f4b0102eb6..ffe38a13a7 100644
--- a/protocols/3d942d/README.md
+++ b/protocols/3d942d/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/3db190-manual/README.md b/protocols/3db190-manual/README.md
index e9c6ab622d..d5549e22dc 100644
--- a/protocols/3db190-manual/README.md
+++ b/protocols/3db190-manual/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/3db190/README.md b/protocols/3db190/README.md
index 310319bf3a..17d7f28796 100644
--- a/protocols/3db190/README.md
+++ b/protocols/3db190/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Featured
* PCR Prep
diff --git a/protocols/3e0bef/README.md b/protocols/3e0bef/README.md
index 4c83001f8b..0b30760bf0 100644
--- a/protocols/3e0bef/README.md
+++ b/protocols/3e0bef/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Omega Bio-Tek Mag-Bind Plant DNA DS Kit
diff --git a/protocols/3e3c9d-protocol-Adding-BTM/README.md b/protocols/3e3c9d-protocol-Adding-BTM/README.md
index 16782fa49a..3f72631589 100644
--- a/protocols/3e3c9d-protocol-Adding-BTM/README.md
+++ b/protocols/3e3c9d-protocol-Adding-BTM/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/3e3c9d-protocol-Adding-Water-RBC/README.md b/protocols/3e3c9d-protocol-Adding-Water-RBC/README.md
index b9bc7fb652..1310216ce9 100644
--- a/protocols/3e3c9d-protocol-Adding-Water-RBC/README.md
+++ b/protocols/3e3c9d-protocol-Adding-Water-RBC/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/3e3c9d-protocol-master-rbc-transfer/README.md b/protocols/3e3c9d-protocol-master-rbc-transfer/README.md
index 3e2572933a..cf48b15121 100644
--- a/protocols/3e3c9d-protocol-master-rbc-transfer/README.md
+++ b/protocols/3e3c9d-protocol-master-rbc-transfer/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/3e3c9d-protocols-adding-wistd-water/README.md b/protocols/3e3c9d-protocols-adding-wistd-water/README.md
index e4043ff422..75449becd6 100644
--- a/protocols/3e3c9d-protocols-adding-wistd-water/README.md
+++ b/protocols/3e3c9d-protocols-adding-wistd-water/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/3fad82-part-2/README.md b/protocols/3fad82-part-2/README.md
index ff0b94054c..19186fddbb 100644
--- a/protocols/3fad82-part-2/README.md
+++ b/protocols/3fad82-part-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II FS DNA Library Prep Kit for Illumina
diff --git a/protocols/3fad82-part-3/README.md b/protocols/3fad82-part-3/README.md
index 8d700a3ec3..de07834100 100644
--- a/protocols/3fad82-part-3/README.md
+++ b/protocols/3fad82-part-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II FS DNA Library Prep Kit for Illumina
diff --git a/protocols/3fad82/README.md b/protocols/3fad82/README.md
index c46682fd21..0d1a62723b 100644
--- a/protocols/3fad82/README.md
+++ b/protocols/3fad82/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II FS DNA Library Prep Kit for Illumina
diff --git a/protocols/3fb582/README.md b/protocols/3fb582/README.md
index d2d9318336..6365a6ceb1 100644
--- a/protocols/3fb582/README.md
+++ b/protocols/3fb582/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/400b8e/README.md b/protocols/400b8e/README.md
index 9e5edd2d36..0d6e7ceada 100644
--- a/protocols/400b8e/README.md
+++ b/protocols/400b8e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/407d5e/README.md b/protocols/407d5e/README.md
index f676b04ca3..e8dd524478 100644
--- a/protocols/407d5e/README.md
+++ b/protocols/407d5e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](http://www.opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Assay
diff --git a/protocols/412ec7/README.md b/protocols/412ec7/README.md
index 359d747ed9..7ffcae088a 100644
--- a/protocols/412ec7/README.md
+++ b/protocols/412ec7/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Serial Dilution
* Optional Temperature Module Serial Dilution
diff --git a/protocols/414aaa/README.md b/protocols/414aaa/README.md
index c9405341d8..854e34b195 100644
--- a/protocols/414aaa/README.md
+++ b/protocols/414aaa/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/4175de/README.md b/protocols/4175de/README.md
index 3b31e787be..0c2fcf9470 100644
--- a/protocols/4175de/README.md
+++ b/protocols/4175de/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/41ece2/README.md b/protocols/41ece2/README.md
index eaec027441..06db7ca1ba 100644
--- a/protocols/41ece2/README.md
+++ b/protocols/41ece2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](http://www.opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Serial Dilution
diff --git a/protocols/422b1e/README.md b/protocols/422b1e/README.md
index 00cabe1c0f..b61644c101 100644
--- a/protocols/422b1e/README.md
+++ b/protocols/422b1e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Titration
* Custom Titration
diff --git a/protocols/42d27b/README.md b/protocols/42d27b/README.md
index c15a2f762c..01de86093b 100644
--- a/protocols/42d27b/README.md
+++ b/protocols/42d27b/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Quant-iT dsDNA Kit
diff --git a/protocols/43163c/README.md b/protocols/43163c/README.md
index 73b9cc3949..cc7b9e2066 100644
--- a/protocols/43163c/README.md
+++ b/protocols/43163c/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Plant DNA
diff --git a/protocols/43d313-part-2/README.md b/protocols/43d313-part-2/README.md
index f7e638ee41..bec5cf6442 100644
--- a/protocols/43d313-part-2/README.md
+++ b/protocols/43d313-part-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/43d313-part-3/README.md b/protocols/43d313-part-3/README.md
index 8d94ad3ec0..450a5a2955 100644
--- a/protocols/43d313-part-3/README.md
+++ b/protocols/43d313-part-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/43d313-part-4/README.md b/protocols/43d313-part-4/README.md
index 425e34de9a..596928d542 100644
--- a/protocols/43d313-part-4/README.md
+++ b/protocols/43d313-part-4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/43d313-part-5/README.md b/protocols/43d313-part-5/README.md
index a2916d937b..99c265c61e 100644
--- a/protocols/43d313-part-5/README.md
+++ b/protocols/43d313-part-5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/43d313-part-6/README.md b/protocols/43d313-part-6/README.md
index e15df8bf8d..2077807c18 100644
--- a/protocols/43d313-part-6/README.md
+++ b/protocols/43d313-part-6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/43d313-part-7/README.md b/protocols/43d313-part-7/README.md
index 46b62dffbc..b247ada1bb 100644
--- a/protocols/43d313-part-7/README.md
+++ b/protocols/43d313-part-7/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/43d313-part-8/README.md b/protocols/43d313-part-8/README.md
index 9e68f6d746..56f4944b88 100644
--- a/protocols/43d313-part-8/README.md
+++ b/protocols/43d313-part-8/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/43d313-part-9/README.md b/protocols/43d313-part-9/README.md
index 358536adaa..042f57b5ca 100644
--- a/protocols/43d313-part-9/README.md
+++ b/protocols/43d313-part-9/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/43d313/README.md b/protocols/43d313/README.md
index 0687d27228..a4fccf6d5c 100644
--- a/protocols/43d313/README.md
+++ b/protocols/43d313/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/441cc8/README.md b/protocols/441cc8/README.md
index ae2d13d530..5322e5f86b 100644
--- a/protocols/441cc8/README.md
+++ b/protocols/441cc8/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* PCR Prep
diff --git a/protocols/4471ba/README.md b/protocols/4471ba/README.md
index 66ac79d873..633672aa2f 100644
--- a/protocols/4471ba/README.md
+++ b/protocols/4471ba/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/44981c/README.md b/protocols/44981c/README.md
index 4c638c36e5..dbc7788901 100644
--- a/protocols/44981c/README.md
+++ b/protocols/44981c/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](http://www.opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/44b1ac/README.md b/protocols/44b1ac/README.md
index 110339cd90..bce8b3411c 100644
--- a/protocols/44b1ac/README.md
+++ b/protocols/44b1ac/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* QIAseq Targeted RNAscan Panel for Illumina Instruments
diff --git a/protocols/44b43c/README.md b/protocols/44b43c/README.md
index d72f77c2be..3515c328b8 100644
--- a/protocols/44b43c/README.md
+++ b/protocols/44b43c/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/453b5a/README.md b/protocols/453b5a/README.md
index 5ed1aea43a..d31a9fd8d4 100644
--- a/protocols/453b5a/README.md
+++ b/protocols/453b5a/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/4568fa-2/README.md b/protocols/4568fa-2/README.md
index fec0617933..5808a13d74 100644
--- a/protocols/4568fa-2/README.md
+++ b/protocols/4568fa-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/4568fa/README.md b/protocols/4568fa/README.md
index a9481dcb0f..35600cfd76 100644
--- a/protocols/4568fa/README.md
+++ b/protocols/4568fa/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/459a55/README.md b/protocols/459a55/README.md
index 3dbdf4c297..a2c9642ec1 100644
--- a/protocols/459a55/README.md
+++ b/protocols/459a55/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Mass Spec
diff --git a/protocols/459cc2/README.md b/protocols/459cc2/README.md
index bc92305cb2..d24bc8bab2 100644
--- a/protocols/459cc2/README.md
+++ b/protocols/459cc2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Mass Spec
diff --git a/protocols/45a320/README.md b/protocols/45a320/README.md
index c384785181..e2e74ddbe1 100644
--- a/protocols/45a320/README.md
+++ b/protocols/45a320/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* RNA Extraction
diff --git a/protocols/470d8c/README.md b/protocols/470d8c/README.md
index a2d85cb142..224d5fa925 100644
--- a/protocols/470d8c/README.md
+++ b/protocols/470d8c/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/48648f/README.md b/protocols/48648f/README.md
index b27fbf03ea..1ff0ebab6c 100644
--- a/protocols/48648f/README.md
+++ b/protocols/48648f/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Purification
diff --git a/protocols/4883fc/README.md b/protocols/4883fc/README.md
index 6eb481d26b..19b4c2b428 100644
--- a/protocols/4883fc/README.md
+++ b/protocols/4883fc/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/4920bc/README.md b/protocols/4920bc/README.md
index 668013649f..7389b26c51 100644
--- a/protocols/4920bc/README.md
+++ b/protocols/4920bc/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Omega Mag-Bind® Total RNA 96 Kit
diff --git a/protocols/49b828/README.md b/protocols/49b828/README.md
index cebc73a9d5..dae7d060ef 100644
--- a/protocols/49b828/README.md
+++ b/protocols/49b828/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/49de51-pt1/README.md b/protocols/49de51-pt1/README.md
index ba53a02591..741a698403 100644
--- a/protocols/49de51-pt1/README.md
+++ b/protocols/49de51-pt1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Plant DNA
diff --git a/protocols/49de51-pt2/README.md b/protocols/49de51-pt2/README.md
index 56b3b9efa2..ee4ab5f1ca 100644
--- a/protocols/49de51-pt2/README.md
+++ b/protocols/49de51-pt2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Plant DNA
diff --git a/protocols/49de51/README.md b/protocols/49de51/README.md
index 6b88de722b..61e5261bb1 100644
--- a/protocols/49de51/README.md
+++ b/protocols/49de51/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Plant DNA
diff --git a/protocols/4a0be6/README.md b/protocols/4a0be6/README.md
index 1f8f3897d5..dbc59b9a9d 100644
--- a/protocols/4a0be6/README.md
+++ b/protocols/4a0be6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Tube Filling
diff --git a/protocols/4a5f32/README.md b/protocols/4a5f32/README.md
index 1ca5d95ed3..e0698b3ff8 100644
--- a/protocols/4a5f32/README.md
+++ b/protocols/4a5f32/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
diff --git a/protocols/4b4a80-adapter_ligation/README.md b/protocols/4b4a80-adapter_ligation/README.md
index dfb2309082..a14daf7f78 100644
--- a/protocols/4b4a80-adapter_ligation/README.md
+++ b/protocols/4b4a80-adapter_ligation/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L
diff --git a/protocols/4b4a80-fragmentation/README.md b/protocols/4b4a80-fragmentation/README.md
index 4aac3893fc..7ca8333091 100644
--- a/protocols/4b4a80-fragmentation/README.md
+++ b/protocols/4b4a80-fragmentation/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L
diff --git a/protocols/4b4a80-pcr_enrichment/README.md b/protocols/4b4a80-pcr_enrichment/README.md
index 26f1483140..d49e8d147d 100644
--- a/protocols/4b4a80-pcr_enrichment/README.md
+++ b/protocols/4b4a80-pcr_enrichment/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L
diff --git a/protocols/4b4a80-size_selection/README.md b/protocols/4b4a80-size_selection/README.md
index 5ff8766367..ff318adcec 100644
--- a/protocols/4b4a80-size_selection/README.md
+++ b/protocols/4b4a80-size_selection/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II FS DNA Library Prep Kit for Illumina E6177S/L
diff --git a/protocols/4b7268/README.md b/protocols/4b7268/README.md
index 50a0936463..6254cfec65 100644
--- a/protocols/4b7268/README.md
+++ b/protocols/4b7268/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Purification
diff --git a/protocols/4b76c0/README.md b/protocols/4b76c0/README.md
index fab858a864..87f6cde4f6 100644
--- a/protocols/4b76c0/README.md
+++ b/protocols/4b76c0/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/4bdd14/README.md b/protocols/4bdd14/README.md
index 13a421fac7..2b27d7f7ca 100644
--- a/protocols/4bdd14/README.md
+++ b/protocols/4bdd14/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/4c8861/README.md b/protocols/4c8861/README.md
index c59bed182d..a2dfeba771 100644
--- a/protocols/4c8861/README.md
+++ b/protocols/4c8861/README.md
@@ -6,6 +6,9 @@
### Partner
[Partner Name](partner website link)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/4d9b8b/README.md b/protocols/4d9b8b/README.md
index 4385f827d2..cad484cfca 100644
--- a/protocols/4d9b8b/README.md
+++ b/protocols/4d9b8b/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/4e4c0c/README.md b/protocols/4e4c0c/README.md
index 888c0349fb..5f20c89b79 100644
--- a/protocols/4e4c0c/README.md
+++ b/protocols/4e4c0c/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/4ee4d6-part2/README.md b/protocols/4ee4d6-part2/README.md
index 41b830a8b7..035f0fcf4f 100644
--- a/protocols/4ee4d6-part2/README.md
+++ b/protocols/4ee4d6-part2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep with Enrichment
diff --git a/protocols/4ee4d6/README.md b/protocols/4ee4d6/README.md
index 21282f11e6..0dc800185e 100644
--- a/protocols/4ee4d6/README.md
+++ b/protocols/4ee4d6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep with Enrichment
diff --git a/protocols/4f7a6f/README.md b/protocols/4f7a6f/README.md
index 1c0d1e01f7..95aab6460b 100644
--- a/protocols/4f7a6f/README.md
+++ b/protocols/4f7a6f/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/4fa62e/README.md b/protocols/4fa62e/README.md
index 445ac085b8..d369671215 100644
--- a/protocols/4fa62e/README.md
+++ b/protocols/4fa62e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/4fc750/README.md b/protocols/4fc750/README.md
index 133fcffc3a..476e767e5a 100644
--- a/protocols/4fc750/README.md
+++ b/protocols/4fc750/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Omega Mag-Bind® Blood & Tissue DNA 96 Kit
diff --git a/protocols/50486f-v2-part1/README.md b/protocols/50486f-v2-part1/README.md
index e9c136c177..bed7841cc4 100644
--- a/protocols/50486f-v2-part1/README.md
+++ b/protocols/50486f-v2-part1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/50486f-v2-part2/README.md b/protocols/50486f-v2-part2/README.md
index d4b7d1d647..f0233d8dd8 100644
--- a/protocols/50486f-v2-part2/README.md
+++ b/protocols/50486f-v2-part2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/50486f-v2-part3/README.md b/protocols/50486f-v2-part3/README.md
index 910956ebf1..181cfddb75 100644
--- a/protocols/50486f-v2-part3/README.md
+++ b/protocols/50486f-v2-part3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/50486f-v2-part4/README.md b/protocols/50486f-v2-part4/README.md
index 1543e577f6..a87cad12b0 100644
--- a/protocols/50486f-v2-part4/README.md
+++ b/protocols/50486f-v2-part4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/50486f-v2-part5/README.md b/protocols/50486f-v2-part5/README.md
index d88757278f..cd15144d05 100644
--- a/protocols/50486f-v2-part5/README.md
+++ b/protocols/50486f-v2-part5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/50c11b/README.md b/protocols/50c11b/README.md
index af22e09971..6dcf9fea9d 100644
--- a/protocols/50c11b/README.md
+++ b/protocols/50c11b/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/516336-part-2/README.md b/protocols/516336-part-2/README.md
index 34595c4aae..e35aeb56b9 100644
--- a/protocols/516336-part-2/README.md
+++ b/protocols/516336-part-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Oxford Nanopore Ligation Sequencing Kit
diff --git a/protocols/516336/README.md b/protocols/516336/README.md
index be999f1278..ef616175bb 100644
--- a/protocols/516336/README.md
+++ b/protocols/516336/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Oxford Nanopore Ligation Sequencing Kit
diff --git a/protocols/5191b6/README.md b/protocols/5191b6/README.md
index 2cce2815ca..d2652ee452 100644
--- a/protocols/5191b6/README.md
+++ b/protocols/5191b6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/51b9a5/README.md b/protocols/51b9a5/README.md
index 4325b5210a..baa35c5ac0 100644
--- a/protocols/51b9a5/README.md
+++ b/protocols/51b9a5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Agar Plating
diff --git a/protocols/51df08/README.md b/protocols/51df08/README.md
index 46ff5cd679..74893b6f69 100644
--- a/protocols/51df08/README.md
+++ b/protocols/51df08/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Assay
diff --git a/protocols/528c16/README.md b/protocols/528c16/README.md
index 6443c54af6..54cd3e2ae2 100644
--- a/protocols/528c16/README.md
+++ b/protocols/528c16/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/52d238/README.md b/protocols/52d238/README.md
index 7f97c3623d..ceab64410b 100644
--- a/protocols/52d238/README.md
+++ b/protocols/52d238/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEB Ultra II FS DNA Library Prep
diff --git a/protocols/53134e/README.md b/protocols/53134e/README.md
index 7d7e4629b1..a62631f1a2 100644
--- a/protocols/53134e/README.md
+++ b/protocols/53134e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Aliquoting
diff --git a/protocols/53e6bc_mastermix_creation/README.md b/protocols/53e6bc_mastermix_creation/README.md
index 079fca6775..efa64ee1e6 100644
--- a/protocols/53e6bc_mastermix_creation/README.md
+++ b/protocols/53e6bc_mastermix_creation/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Assay
diff --git a/protocols/53fec2/README.md b/protocols/53fec2/README.md
index 326d747392..42019f5a62 100644
--- a/protocols/53fec2/README.md
+++ b/protocols/53fec2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* Cleanup
diff --git a/protocols/543bf9/README.md b/protocols/543bf9/README.md
index 7a9b6426fd..b5f49dacf2 100644
--- a/protocols/543bf9/README.md
+++ b/protocols/543bf9/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Serial Dilution
diff --git a/protocols/5520f0/README.md b/protocols/5520f0/README.md
index e0f8fa11d5..2b079446c8 100644
--- a/protocols/5520f0/README.md
+++ b/protocols/5520f0/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
diff --git a/protocols/558d02/README.md b/protocols/558d02/README.md
index fa3f585f76..c11c9ab0e5 100644
--- a/protocols/558d02/README.md
+++ b/protocols/558d02/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Purification
diff --git a/protocols/559aa0/README.md b/protocols/559aa0/README.md
index 1cd9a0ff71..b6b5f3e4a4 100644
--- a/protocols/559aa0/README.md
+++ b/protocols/559aa0/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* qPCR setup
diff --git a/protocols/563624/README.md b/protocols/563624/README.md
index 9b2606ad4e..4bff3358f7 100644
--- a/protocols/563624/README.md
+++ b/protocols/563624/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Purification
diff --git a/protocols/5654c0/README.md b/protocols/5654c0/README.md
index e5906f6d64..e2cc9e594d 100644
--- a/protocols/5654c0/README.md
+++ b/protocols/5654c0/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/5689f5/README.md b/protocols/5689f5/README.md
index 477d9c3893..9b78da3e14 100644
--- a/protocols/5689f5/README.md
+++ b/protocols/5689f5/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Clean Up
diff --git a/protocols/56a6a1/README.md b/protocols/56a6a1/README.md
index 5dea95bd2f..1cfe08f236 100644
--- a/protocols/56a6a1/README.md
+++ b/protocols/56a6a1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins & Proteomics
* Assay
diff --git a/protocols/56b4ec/README.md b/protocols/56b4ec/README.md
index 68daea0199..f74dbe82ab 100644
--- a/protocols/56b4ec/README.md
+++ b/protocols/56b4ec/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/581011-pt2/README.md b/protocols/581011-pt2/README.md
index b7e53b2aa4..31b1162545 100644
--- a/protocols/581011-pt2/README.md
+++ b/protocols/581011-pt2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/581011/README.md b/protocols/581011/README.md
index 4d095b91ba..a6923e02d0 100644
--- a/protocols/581011/README.md
+++ b/protocols/581011/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/5829d7/README.md b/protocols/5829d7/README.md
index 3682352056..124dd765ba 100644
--- a/protocols/5829d7/README.md
+++ b/protocols/5829d7/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/58d57d/README.md b/protocols/58d57d/README.md
index 8425b892e1..d6b0947eeb 100644
--- a/protocols/58d57d/README.md
+++ b/protocols/58d57d/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/5a2a35/README.md b/protocols/5a2a35/README.md
index 252f1427ae..bc365b68c9 100644
--- a/protocols/5a2a35/README.md
+++ b/protocols/5a2a35/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/5a3797-protocol-1/README.md b/protocols/5a3797-protocol-1/README.md
index 1b7fbfa8cf..9cc679684b 100644
--- a/protocols/5a3797-protocol-1/README.md
+++ b/protocols/5a3797-protocol-1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/5a3797-protocol-2/README.md b/protocols/5a3797-protocol-2/README.md
index 28d701a503..090809873b 100644
--- a/protocols/5a3797-protocol-2/README.md
+++ b/protocols/5a3797-protocol-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/5a3797-protocol-3/README.md b/protocols/5a3797-protocol-3/README.md
index a311b95578..a6b16a533d 100644
--- a/protocols/5a3797-protocol-3/README.md
+++ b/protocols/5a3797-protocol-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/5a5767/README.md b/protocols/5a5767/README.md
index df46ed7979..3b0c5190b2 100644
--- a/protocols/5a5767/README.md
+++ b/protocols/5a5767/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/5b16ef-pt2/README.md b/protocols/5b16ef-pt2/README.md
index c58ccaaace..be7dd73ad6 100644
--- a/protocols/5b16ef-pt2/README.md
+++ b/protocols/5b16ef-pt2/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina Nextera Flex
diff --git a/protocols/5b16ef/README.md b/protocols/5b16ef/README.md
index 879efa92e8..ae18925df0 100644
--- a/protocols/5b16ef/README.md
+++ b/protocols/5b16ef/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina Nextera Flex
diff --git a/protocols/5b8174/README.md b/protocols/5b8174/README.md
index 99c265a312..8b2d65d73e 100644
--- a/protocols/5b8174/README.md
+++ b/protocols/5b8174/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/5c14ad/README.md b/protocols/5c14ad/README.md
index 9c486263b8..4b06a1c34e 100644
--- a/protocols/5c14ad/README.md
+++ b/protocols/5c14ad/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/5c24e2/README.md b/protocols/5c24e2/README.md
index 7dfb7a660b..f12ed76e95 100644
--- a/protocols/5c24e2/README.md
+++ b/protocols/5c24e2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/5c50d9/README.md b/protocols/5c50d9/README.md
index 876fb03bcd..ccffdfa3e3 100644
--- a/protocols/5c50d9/README.md
+++ b/protocols/5c50d9/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/5c7384-hhu/README.md b/protocols/5c7384-hhu/README.md
index 443614467c..0dbf6f3ef4 100644
--- a/protocols/5c7384-hhu/README.md
+++ b/protocols/5c7384-hhu/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Mass Spec
diff --git a/protocols/5c7384/README.md b/protocols/5c7384/README.md
index 443614467c..0dbf6f3ef4 100644
--- a/protocols/5c7384/README.md
+++ b/protocols/5c7384/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Mass Spec
diff --git a/protocols/5dcd88/README.md b/protocols/5dcd88/README.md
index 27a73dcbd4..22e7de9eb9 100644
--- a/protocols/5dcd88/README.md
+++ b/protocols/5dcd88/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/5eee17/README.md b/protocols/5eee17/README.md
index 8b4015a7a7..8bcc3866c7 100644
--- a/protocols/5eee17/README.md
+++ b/protocols/5eee17/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Dilution
diff --git a/protocols/5f37a2/README.md b/protocols/5f37a2/README.md
index e7fc2775c1..5e578090b5 100644
--- a/protocols/5f37a2/README.md
+++ b/protocols/5f37a2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* MagneSil
diff --git a/protocols/5f791f/README.md b/protocols/5f791f/README.md
index 2c083faef2..48a8c332aa 100644
--- a/protocols/5f791f/README.md
+++ b/protocols/5f791f/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep and Magnetic Bead Cleanup
diff --git a/protocols/5f7d18/README.md b/protocols/5f7d18/README.md
index ca31bc03a4..eee322a608 100644
--- a/protocols/5f7d18/README.md
+++ b/protocols/5f7d18/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* RNA Extraction
diff --git a/protocols/5fa647/README.md b/protocols/5fa647/README.md
index aac110615e..8ab6ebe421 100644
--- a/protocols/5fa647/README.md
+++ b/protocols/5fa647/README.md
@@ -6,6 +6,9 @@
### Partner
[Invitrogen](https://www.thermofisher.com/order/catalog/product/11752250#/11752250)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/6030b7/README.md b/protocols/6030b7/README.md
index 3f55f0d7bf..d69fdc5ae5 100644
--- a/protocols/6030b7/README.md
+++ b/protocols/6030b7/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Lexogen QuantSeq
diff --git a/protocols/60d861-2/README.md b/protocols/60d861-2/README.md
index e23c080a89..5e08f606c9 100644
--- a/protocols/60d861-2/README.md
+++ b/protocols/60d861-2/README.md
@@ -6,6 +6,9 @@
### Partner
[Partner Name](partner website link)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/60d861-3/README.md b/protocols/60d861-3/README.md
index e00f48af89..e48220b2d2 100644
--- a/protocols/60d861-3/README.md
+++ b/protocols/60d861-3/README.md
@@ -6,6 +6,9 @@
### Partner
[Partner Name](partner website link)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/60d861/README.md b/protocols/60d861/README.md
index adcf7f8935..fec6b43aad 100644
--- a/protocols/60d861/README.md
+++ b/protocols/60d861/README.md
@@ -6,6 +6,9 @@
### Partner
[Partner Name](partner website link)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/6151af/README.md b/protocols/6151af/README.md
index b18388e9a3..255bdd623d 100644
--- a/protocols/6151af/README.md
+++ b/protocols/6151af/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/62679a/README.md b/protocols/62679a/README.md
index e9f99a3eff..60b33b3931 100644
--- a/protocols/62679a/README.md
+++ b/protocols/62679a/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Serial Dilution
diff --git a/protocols/629f38/README.md b/protocols/629f38/README.md
index ebcd48922c..73b27c04e0 100644
--- a/protocols/629f38/README.md
+++ b/protocols/629f38/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Zymo Quick-DNA HMW MagBead Kit
diff --git a/protocols/6401f1/README.md b/protocols/6401f1/README.md
index 3958459051..f23572e062 100644
--- a/protocols/6401f1/README.md
+++ b/protocols/6401f1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/640a85/README.md b/protocols/640a85/README.md
index cd877a5a53..410b418531 100644
--- a/protocols/640a85/README.md
+++ b/protocols/640a85/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/642c73/README.md b/protocols/642c73/README.md
index e591b26367..cb63c9c904 100644
--- a/protocols/642c73/README.md
+++ b/protocols/642c73/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/649b67/README.md b/protocols/649b67/README.md
index 6efbdfc9f1..a87b960259 100644
--- a/protocols/649b67/README.md
+++ b/protocols/649b67/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Advanced cherrypicking
diff --git a/protocols/64c54a/README.md b/protocols/64c54a/README.md
index 6e16a44271..b41fb5ee7d 100644
--- a/protocols/64c54a/README.md
+++ b/protocols/64c54a/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Mass Spec
diff --git a/protocols/657ee9-2/README.md b/protocols/657ee9-2/README.md
index 9cf85cc5ca..5f49de8b5f 100644
--- a/protocols/657ee9-2/README.md
+++ b/protocols/657ee9-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Oncomine Focus Assay
diff --git a/protocols/657ee9-3/README.md b/protocols/657ee9-3/README.md
index b2f65db4d4..1e5694fac9 100644
--- a/protocols/657ee9-3/README.md
+++ b/protocols/657ee9-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Oncomine Focus Assay
diff --git a/protocols/657ee9-4/README.md b/protocols/657ee9-4/README.md
index cb2f9d6ffb..bf76d8341c 100644
--- a/protocols/657ee9-4/README.md
+++ b/protocols/657ee9-4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Oncomine Focus Assay
diff --git a/protocols/657ee9/README.md b/protocols/657ee9/README.md
index 41e767cb6d..5b942468f6 100644
--- a/protocols/657ee9/README.md
+++ b/protocols/657ee9/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Oncomine Focus Assay
diff --git a/protocols/659888/README.md b/protocols/659888/README.md
index 9e1f515859..c538d35775 100644
--- a/protocols/659888/README.md
+++ b/protocols/659888/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/65ed01/README.md b/protocols/65ed01/README.md
index 04915abd2c..abbdf8edef 100644
--- a/protocols/65ed01/README.md
+++ b/protocols/65ed01/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Purification
diff --git a/protocols/66aa48/README.md b/protocols/66aa48/README.md
index acb656be9e..7ee0090642 100644
--- a/protocols/66aa48/README.md
+++ b/protocols/66aa48/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/66e60f/README.md b/protocols/66e60f/README.md
index b72258f5e3..9f5923e6de 100644
--- a/protocols/66e60f/README.md
+++ b/protocols/66e60f/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/68ce98/README.md b/protocols/68ce98/README.md
index 65de982371..b3636235a2 100644
--- a/protocols/68ce98/README.md
+++ b/protocols/68ce98/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/69486f/README.md b/protocols/69486f/README.md
index f3defc91ac..08075abe96 100644
--- a/protocols/69486f/README.md
+++ b/protocols/69486f/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* KAPA HiFi
diff --git a/protocols/698b9e-part2/README.md b/protocols/698b9e-part2/README.md
index ef6edecbea..b458033618 100644
--- a/protocols/698b9e-part2/README.md
+++ b/protocols/698b9e-part2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/698b9e/README.md b/protocols/698b9e/README.md
index 2cfad9c752..f310a4d5c1 100644
--- a/protocols/698b9e/README.md
+++ b/protocols/698b9e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/69cf81-2/README.md b/protocols/69cf81-2/README.md
index ab240b8224..1709421228 100644
--- a/protocols/69cf81-2/README.md
+++ b/protocols/69cf81-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/69cf81/README.md b/protocols/69cf81/README.md
index 0cf640bdb2..34ade30e1a 100644
--- a/protocols/69cf81/README.md
+++ b/protocols/69cf81/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/69db93/README.md b/protocols/69db93/README.md
index 5e49f3a7dc..20aa5cdfd3 100644
--- a/protocols/69db93/README.md
+++ b/protocols/69db93/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* RNA Extraction
diff --git a/protocols/6a77d9/README.md b/protocols/6a77d9/README.md
index a6f7c7cc40..a7411cef54 100644
--- a/protocols/6a77d9/README.md
+++ b/protocols/6a77d9/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Slide Spotting
diff --git a/protocols/6a93a2-part2/README.md b/protocols/6a93a2-part2/README.md
index db89a3c43f..4922afada8 100644
--- a/protocols/6a93a2-part2/README.md
+++ b/protocols/6a93a2-part2/README.md
@@ -6,6 +6,9 @@
### Partner
[Swift Biosciences](https://swiftbiosci.com/protocols/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Swift Rapid RNA Library Kit
diff --git a/protocols/6a93a2-part3/README.md b/protocols/6a93a2-part3/README.md
index 468040d693..5185d26670 100644
--- a/protocols/6a93a2-part3/README.md
+++ b/protocols/6a93a2-part3/README.md
@@ -6,6 +6,9 @@
### Partner
[Swift Biosciences](https://swiftbiosci.com/protocols/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Swift Rapid RNA Library Kit
diff --git a/protocols/6a93a2-part4/README.md b/protocols/6a93a2-part4/README.md
index e2d486f822..10e78b334e 100644
--- a/protocols/6a93a2-part4/README.md
+++ b/protocols/6a93a2-part4/README.md
@@ -6,6 +6,9 @@
### Partner
[Swift Biosciences](https://swiftbiosci.com/protocols/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Swift Rapid RNA Library Kit
diff --git a/protocols/6a93a2-part5/README.md b/protocols/6a93a2-part5/README.md
index 0947d21083..9f37f4b2fb 100644
--- a/protocols/6a93a2-part5/README.md
+++ b/protocols/6a93a2-part5/README.md
@@ -6,6 +6,9 @@
### Partner
[Swift Biosciences](https://swiftbiosci.com/protocols/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Swift Rapid RNA Library Kit
diff --git a/protocols/6a93a2/README.md b/protocols/6a93a2/README.md
index 0dd0730ba2..ba6f44b4c7 100644
--- a/protocols/6a93a2/README.md
+++ b/protocols/6a93a2/README.md
@@ -6,6 +6,9 @@
### Partner
[Swift Biosciences](https://swiftbiosci.com/protocols/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Swift Rapid RNA Library Kit
diff --git a/protocols/6aee6e/README.md b/protocols/6aee6e/README.md
index fcf5787141..fb89ad855a 100644
--- a/protocols/6aee6e/README.md
+++ b/protocols/6aee6e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/6af807-amendment/README.md b/protocols/6af807-amendment/README.md
index a7a4c43d26..8bf12ae4f8 100644
--- a/protocols/6af807-amendment/README.md
+++ b/protocols/6af807-amendment/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/6af807/README.md b/protocols/6af807/README.md
index 9287a79870..d2b498a634 100644
--- a/protocols/6af807/README.md
+++ b/protocols/6af807/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/6b51bd/README.md b/protocols/6b51bd/README.md
index b483705b3d..412ebb003c 100644
--- a/protocols/6b51bd/README.md
+++ b/protocols/6b51bd/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/6b8c90-2/README.md b/protocols/6b8c90-2/README.md
index 46c68de518..74cb22f42e 100644
--- a/protocols/6b8c90-2/README.md
+++ b/protocols/6b8c90-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](http://www.opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/6b8c90-3/README.md b/protocols/6b8c90-3/README.md
index f37fab042a..3df03e171d 100644
--- a/protocols/6b8c90-3/README.md
+++ b/protocols/6b8c90-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](http://www.opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/6b8c90/README.md b/protocols/6b8c90/README.md
index 7a831ebf05..7bb2fee1bc 100644
--- a/protocols/6b8c90/README.md
+++ b/protocols/6b8c90/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](http://www.opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/6b9ee3/README.md b/protocols/6b9ee3/README.md
index 9178b29d5e..e35e3ef52b 100644
--- a/protocols/6b9ee3/README.md
+++ b/protocols/6b9ee3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/6bc986-part-2/README.md b/protocols/6bc986-part-2/README.md
index 13b1e2bed0..c98c997a9f 100644
--- a/protocols/6bc986-part-2/README.md
+++ b/protocols/6bc986-part-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/6bc986-part-3/README.md b/protocols/6bc986-part-3/README.md
index 21d3187a1e..60e893a09a 100644
--- a/protocols/6bc986-part-3/README.md
+++ b/protocols/6bc986-part-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/6bc986-part-4/README.md b/protocols/6bc986-part-4/README.md
index 5ae95f8730..0dbb3ed81b 100644
--- a/protocols/6bc986-part-4/README.md
+++ b/protocols/6bc986-part-4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/6bc986-part-5/README.md b/protocols/6bc986-part-5/README.md
index 4c8aecfe4b..bc26bd2a7f 100644
--- a/protocols/6bc986-part-5/README.md
+++ b/protocols/6bc986-part-5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/6bc986-part-6/README.md b/protocols/6bc986-part-6/README.md
index 3069e4e09a..a2bc18ed5a 100644
--- a/protocols/6bc986-part-6/README.md
+++ b/protocols/6bc986-part-6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/6bc986/README.md b/protocols/6bc986/README.md
index ffabe43a89..ab398dfc02 100644
--- a/protocols/6bc986/README.md
+++ b/protocols/6bc986/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/6be1f8/README.md b/protocols/6be1f8/README.md
index 7e8e9d07f9..ba3844fc69 100644
--- a/protocols/6be1f8/README.md
+++ b/protocols/6be1f8/README.md
@@ -6,6 +6,9 @@
### Partner
[Partner Name](partner website link)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/6c27ee/README.md b/protocols/6c27ee/README.md
index 64b69161e2..ee73831f64 100644
--- a/protocols/6c27ee/README.md
+++ b/protocols/6c27ee/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](http://www.opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/6c7447/README.md b/protocols/6c7447/README.md
index 086fed89ea..f43bd9148d 100644
--- a/protocols/6c7447/README.md
+++ b/protocols/6c7447/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/6cb818/README.md b/protocols/6cb818/README.md
index e4ad49932f..fe2fee0fcd 100644
--- a/protocols/6cb818/README.md
+++ b/protocols/6cb818/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Staining
diff --git a/protocols/6cd9d6/README.md b/protocols/6cd9d6/README.md
index 3a2b08c1dc..ab5cde5d94 100644
--- a/protocols/6cd9d6/README.md
+++ b/protocols/6cd9d6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/6d2e65/README.md b/protocols/6d2e65/README.md
index fe110db9f1..692a7f45d6 100644
--- a/protocols/6d2e65/README.md
+++ b/protocols/6d2e65/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Purification
diff --git a/protocols/6d3eda/README.md b/protocols/6d3eda/README.md
index bb8acab127..9b188c86e2 100644
--- a/protocols/6d3eda/README.md
+++ b/protocols/6d3eda/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Plant DNA
diff --git a/protocols/6d7eb7/README.md b/protocols/6d7eb7/README.md
index eb9ec9e2dc..f5153b4374 100644
--- a/protocols/6d7eb7/README.md
+++ b/protocols/6d7eb7/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/6d7fc3-part-2/README.md b/protocols/6d7fc3-part-2/README.md
index 035e8fe6d5..86358c4334 100644
--- a/protocols/6d7fc3-part-2/README.md
+++ b/protocols/6d7fc3-part-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung DNA UMI Panel Kit
diff --git a/protocols/6d7fc3-part-3/README.md b/protocols/6d7fc3-part-3/README.md
index 435307f1d5..6bd4c6497a 100644
--- a/protocols/6d7fc3-part-3/README.md
+++ b/protocols/6d7fc3-part-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung DNA UMI Panel Kit
diff --git a/protocols/6d7fc3-part-4/README.md b/protocols/6d7fc3-part-4/README.md
index c6fd2d7d46..213bd89baa 100644
--- a/protocols/6d7fc3-part-4/README.md
+++ b/protocols/6d7fc3-part-4/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung DNA UMI Panel Kit
diff --git a/protocols/6d7fc3-part-5/README.md b/protocols/6d7fc3-part-5/README.md
index 49aafc7602..bd20c3f71b 100644
--- a/protocols/6d7fc3-part-5/README.md
+++ b/protocols/6d7fc3-part-5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung DNA UMI Panel Kit
diff --git a/protocols/6d7fc3-part-6/README.md b/protocols/6d7fc3-part-6/README.md
index 8cd7dcc658..899f9e4040 100644
--- a/protocols/6d7fc3-part-6/README.md
+++ b/protocols/6d7fc3-part-6/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung DNA UMI Panel Kit
diff --git a/protocols/6d7fc3-part-7/README.md b/protocols/6d7fc3-part-7/README.md
index 15d1573c93..b482fa2180 100644
--- a/protocols/6d7fc3-part-7/README.md
+++ b/protocols/6d7fc3-part-7/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung DNA UMI Panel Kit
diff --git a/protocols/6d7fc3/README.md b/protocols/6d7fc3/README.md
index ea997dcdd5..1925d65cd8 100644
--- a/protocols/6d7fc3/README.md
+++ b/protocols/6d7fc3/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* GeneRead QIAact Lung DNA UMI Panel Kit
diff --git a/protocols/6d901d-2/README.md b/protocols/6d901d-2/README.md
index 256ca481b9..9f7048f896 100644
--- a/protocols/6d901d-2/README.md
+++ b/protocols/6d901d-2/README.md
@@ -6,6 +6,9 @@
### Partner
[AstraZeneca](https://www.astrazeneca.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/6d901d/README.md b/protocols/6d901d/README.md
index f6f568a4cd..86ac53a796 100644
--- a/protocols/6d901d/README.md
+++ b/protocols/6d901d/README.md
@@ -6,6 +6,9 @@
### Partner
[AstraZeneca](https://www.astrazeneca.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization with a multi-channel pipette substituting for a single-channel pipette
diff --git a/protocols/6e160e/README.md b/protocols/6e160e/README.md
index 6a22103327..078492c124 100644
--- a/protocols/6e160e/README.md
+++ b/protocols/6e160e/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/6e2509-v2/README.md b/protocols/6e2509-v2/README.md
index 7886f2711b..96236aabda 100644
--- a/protocols/6e2509-v2/README.md
+++ b/protocols/6e2509-v2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/6e2509/README.md b/protocols/6e2509/README.md
index 891df25efa..eb67426f47 100644
--- a/protocols/6e2509/README.md
+++ b/protocols/6e2509/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/6ec4dd-2/README.md b/protocols/6ec4dd-2/README.md
index a16171061f..79dc62492d 100644
--- a/protocols/6ec4dd-2/README.md
+++ b/protocols/6ec4dd-2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Clean Up
diff --git a/protocols/6ec4dd/README.md b/protocols/6ec4dd/README.md
index 74238bc10a..52d441623e 100644
--- a/protocols/6ec4dd/README.md
+++ b/protocols/6ec4dd/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/6f2d9b/README.md b/protocols/6f2d9b/README.md
index 48c5e94102..5b2979ec49 100644
--- a/protocols/6f2d9b/README.md
+++ b/protocols/6f2d9b/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Serial Dilution
diff --git a/protocols/6f4e2c/README.md b/protocols/6f4e2c/README.md
index c6f4ba9c9e..2d52a94bf9 100644
--- a/protocols/6f4e2c/README.md
+++ b/protocols/6f4e2c/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Blood Sample
diff --git a/protocols/6faa1e/README.md b/protocols/6faa1e/README.md
index a58a755544..589fc9fcc6 100644
--- a/protocols/6faa1e/README.md
+++ b/protocols/6faa1e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/6fe477-workflow-1/README.md b/protocols/6fe477-workflow-1/README.md
index a826395aef..1fa5e1b1a8 100644
--- a/protocols/6fe477-workflow-1/README.md
+++ b/protocols/6fe477-workflow-1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Purification
diff --git a/protocols/6fe477-workflow-2/README.md b/protocols/6fe477-workflow-2/README.md
index 5560b1f050..40c64c14e0 100644
--- a/protocols/6fe477-workflow-2/README.md
+++ b/protocols/6fe477-workflow-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Purification
diff --git a/protocols/6ff9e9/README.md b/protocols/6ff9e9/README.md
index 008d32a8e6..96f1c8e83e 100644
--- a/protocols/6ff9e9/README.md
+++ b/protocols/6ff9e9/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Extraction
diff --git a/protocols/701319/README.md b/protocols/701319/README.md
index d9e4cd35c2..c913fc8649 100644
--- a/protocols/701319/README.md
+++ b/protocols/701319/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Mass Spec
diff --git a/protocols/70567f/README.md b/protocols/70567f/README.md
index 1f74781cc9..de53332ed1 100644
--- a/protocols/70567f/README.md
+++ b/protocols/70567f/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Ampure XP
diff --git a/protocols/7062c9-2/README.md b/protocols/7062c9-2/README.md
index fe0433bba9..010aa4fab4 100644
--- a/protocols/7062c9-2/README.md
+++ b/protocols/7062c9-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Nucleic Acid Extraction
diff --git a/protocols/7062c9-aliquot/README.md b/protocols/7062c9-aliquot/README.md
index 328dd9e6cc..dca5b97a11 100644
--- a/protocols/7062c9-aliquot/README.md
+++ b/protocols/7062c9-aliquot/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Aliquoting
diff --git a/protocols/7062c9/README.md b/protocols/7062c9/README.md
index 949a64432f..5816ef6d00 100644
--- a/protocols/7062c9/README.md
+++ b/protocols/7062c9/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/71381b/README.md b/protocols/71381b/README.md
index e3d223706a..46a4e39593 100644
--- a/protocols/71381b/README.md
+++ b/protocols/71381b/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Zymo Kit
diff --git a/protocols/7363e4/README.md b/protocols/7363e4/README.md
index 40f1cdfce8..09554b4e54 100644
--- a/protocols/7363e4/README.md
+++ b/protocols/7363e4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Aliquoting
diff --git a/protocols/74312c/README.md b/protocols/74312c/README.md
index 20e3e4da85..73dfb0de64 100644
--- a/protocols/74312c/README.md
+++ b/protocols/74312c/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/743f54/README.md b/protocols/743f54/README.md
index 3911daa35b..affaa79414 100644
--- a/protocols/743f54/README.md
+++ b/protocols/743f54/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/74750e/README.md b/protocols/74750e/README.md
index 9e5786c659..e29444bab4 100644
--- a/protocols/74750e/README.md
+++ b/protocols/74750e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Serial Dilution
diff --git a/protocols/747d99-assay-1/README.md b/protocols/747d99-assay-1/README.md
index 489d95ce32..ffffb8de9b 100644
--- a/protocols/747d99-assay-1/README.md
+++ b/protocols/747d99-assay-1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Covid Workstation
* RNA Extraction
diff --git a/protocols/747d99-assay-2/README.md b/protocols/747d99-assay-2/README.md
index 43e077e554..6c119a8a80 100644
--- a/protocols/747d99-assay-2/README.md
+++ b/protocols/747d99-assay-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Covid Workstation
* RNA Extraction
diff --git a/protocols/74841a-2/README.md b/protocols/74841a-2/README.md
index 1cb40b423e..eec446c00f 100644
--- a/protocols/74841a-2/README.md
+++ b/protocols/74841a-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Swift Normalase Amplicon Panels (SNAP)
diff --git a/protocols/74841a/README.md b/protocols/74841a/README.md
index caedd97435..e27570c3d4 100644
--- a/protocols/74841a/README.md
+++ b/protocols/74841a/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Swift Normalase Amplicon Panels (SNAP)
diff --git a/protocols/74b303/README.md b/protocols/74b303/README.md
index 8ce3b33e1f..2fa83f99d4 100644
--- a/protocols/74b303/README.md
+++ b/protocols/74b303/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Normalization
* Normalization and Pooling
diff --git a/protocols/751127/README.md b/protocols/751127/README.md
index 7039ddfb60..80e1619e02 100644
--- a/protocols/751127/README.md
+++ b/protocols/751127/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/75cfa6/README.md b/protocols/75cfa6/README.md
index 56878071ff..7e727637f5 100644
--- a/protocols/75cfa6/README.md
+++ b/protocols/75cfa6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/76174b/README.md b/protocols/76174b/README.md
index cf385ea63f..fc31e69e6d 100644
--- a/protocols/76174b/README.md
+++ b/protocols/76174b/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/7663a6/README.md b/protocols/7663a6/README.md
index 858797ca1b..c4546f8055 100644
--- a/protocols/7663a6/README.md
+++ b/protocols/7663a6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/76ab0e/README.md b/protocols/76ab0e/README.md
index 31ca5bb6ab..bd860a6102 100644
--- a/protocols/76ab0e/README.md
+++ b/protocols/76ab0e/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/76ba23-bisulfite_conversion/README.md b/protocols/76ba23-bisulfite_conversion/README.md
index de2bf1aa54..a599166b97 100644
--- a/protocols/76ba23-bisulfite_conversion/README.md
+++ b/protocols/76ba23-bisulfite_conversion/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Zymoresearch: EZ DNA Methylation-Gold Kit
diff --git a/protocols/76ba23-pcr1/README.md b/protocols/76ba23-pcr1/README.md
index 3d89a3fce3..0b68b6a7cd 100644
--- a/protocols/76ba23-pcr1/README.md
+++ b/protocols/76ba23-pcr1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Zymoresearch: EZ DNA Methylation-Gold Kit
diff --git a/protocols/76ba23-pcr2/README.md b/protocols/76ba23-pcr2/README.md
index 6b1ad2a6f7..3062afb2bf 100644
--- a/protocols/76ba23-pcr2/README.md
+++ b/protocols/76ba23-pcr2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Zymoresearch: EZ DNA Methylation-Gold Kit
diff --git a/protocols/76ba23-pooling/README.md b/protocols/76ba23-pooling/README.md
index 14af90bbea..5097eed4d1 100644
--- a/protocols/76ba23-pooling/README.md
+++ b/protocols/76ba23-pooling/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Zymoresearch: EZ DNA Methylation-Gold Kit
diff --git a/protocols/76ba23/README.md b/protocols/76ba23/README.md
index 9b6fbc9c9f..7bf9a578dd 100644
--- a/protocols/76ba23/README.md
+++ b/protocols/76ba23/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Zymoresearch: EZ DNA Methylation-Gold Kit
diff --git a/protocols/76c562/README.md b/protocols/76c562/README.md
index d8ef46c1d7..255374c1f6 100644
--- a/protocols/76c562/README.md
+++ b/protocols/76c562/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/772041/README.md b/protocols/772041/README.md
index 13590f0924..0cd683543e 100644
--- a/protocols/772041/README.md
+++ b/protocols/772041/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/776039/README.md b/protocols/776039/README.md
index 47b32ffe38..400754c01e 100644
--- a/protocols/776039/README.md
+++ b/protocols/776039/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Consolidation
diff --git a/protocols/7805de-part-2/README.md b/protocols/7805de-part-2/README.md
index 96e2449aef..ec14de92e6 100644
--- a/protocols/7805de-part-2/README.md
+++ b/protocols/7805de-part-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II Directional Library Prep Kit for Illumina
diff --git a/protocols/7805de-part-3/README.md b/protocols/7805de-part-3/README.md
index 42b13eef02..bf5eae1535 100644
--- a/protocols/7805de-part-3/README.md
+++ b/protocols/7805de-part-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II Directional Library Prep Kit for Illumina
diff --git a/protocols/7805de-part-4/README.md b/protocols/7805de-part-4/README.md
index 5604053cf4..3b4901f454 100644
--- a/protocols/7805de-part-4/README.md
+++ b/protocols/7805de-part-4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II Directional Library Prep Kit for Illumina
diff --git a/protocols/7805de/README.md b/protocols/7805de/README.md
index 5b770d77c9..39e6ab32a5 100644
--- a/protocols/7805de/README.md
+++ b/protocols/7805de/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II Directional Library Prep Kit for Illumina
diff --git a/protocols/7855ef-part2/README.md b/protocols/7855ef-part2/README.md
index 0b65a897d8..d121b0102a 100644
--- a/protocols/7855ef-part2/README.md
+++ b/protocols/7855ef-part2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* AgriSeq HTS Library Kit
diff --git a/protocols/7855ef-part3/README.md b/protocols/7855ef-part3/README.md
index 9931ca898d..0d97af678f 100644
--- a/protocols/7855ef-part3/README.md
+++ b/protocols/7855ef-part3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* AgriSeq HTS Library Kit
diff --git a/protocols/7855ef-part4/README.md b/protocols/7855ef-part4/README.md
index 769152c12d..979245c241 100644
--- a/protocols/7855ef-part4/README.md
+++ b/protocols/7855ef-part4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* AgriSeq HTS Library Kit
diff --git a/protocols/7855ef-plate-part2/README.md b/protocols/7855ef-plate-part2/README.md
index 6f84bc04d3..6d0be08250 100644
--- a/protocols/7855ef-plate-part2/README.md
+++ b/protocols/7855ef-plate-part2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* AgriSeq HTS Library Kit
diff --git a/protocols/7855ef-plate-part3/README.md b/protocols/7855ef-plate-part3/README.md
index f9d591d264..4cabdf334a 100644
--- a/protocols/7855ef-plate-part3/README.md
+++ b/protocols/7855ef-plate-part3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* AgriSeq HTS Library Kit
diff --git a/protocols/7855ef-plate-part4/README.md b/protocols/7855ef-plate-part4/README.md
index 4604094274..e77ba965ae 100644
--- a/protocols/7855ef-plate-part4/README.md
+++ b/protocols/7855ef-plate-part4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* AgriSeq HTS Library Kit
diff --git a/protocols/7855ef-plate/README.md b/protocols/7855ef-plate/README.md
index 001be59fd0..74c64252f0 100644
--- a/protocols/7855ef-plate/README.md
+++ b/protocols/7855ef-plate/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* AgriSeq HTS Library Kit
diff --git a/protocols/7855ef/README.md b/protocols/7855ef/README.md
index 9d05e77e6a..ac4e2f5526 100644
--- a/protocols/7855ef/README.md
+++ b/protocols/7855ef/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* AgriSeq HTS Library Kit
diff --git a/protocols/78d33c-part-2/README.md b/protocols/78d33c-part-2/README.md
index b911eda91b..0fa0085018 100644
--- a/protocols/78d33c-part-2/README.md
+++ b/protocols/78d33c-part-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/78d33c-part-3/README.md b/protocols/78d33c-part-3/README.md
index 3bded090a1..d1b7d0d29f 100644
--- a/protocols/78d33c-part-3/README.md
+++ b/protocols/78d33c-part-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/78d33c-part-4/README.md b/protocols/78d33c-part-4/README.md
index 8b6c7f0dae..87b5e4f8ac 100644
--- a/protocols/78d33c-part-4/README.md
+++ b/protocols/78d33c-part-4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/78d33c-part-5/README.md b/protocols/78d33c-part-5/README.md
index e27679a036..da82a16846 100644
--- a/protocols/78d33c-part-5/README.md
+++ b/protocols/78d33c-part-5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/78d33c-part-6/README.md b/protocols/78d33c-part-6/README.md
index 22c7e1c5e6..7a771843e3 100644
--- a/protocols/78d33c-part-6/README.md
+++ b/protocols/78d33c-part-6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/78d33c/README.md b/protocols/78d33c/README.md
index b14e031d57..af0aba5516 100644
--- a/protocols/78d33c/README.md
+++ b/protocols/78d33c/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/796af8/README.md b/protocols/796af8/README.md
index 292fb56e47..e8efb7e54d 100644
--- a/protocols/796af8/README.md
+++ b/protocols/796af8/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEB Ultra II FS DNA Library Prep
diff --git a/protocols/797dee-normalization/README.md b/protocols/797dee-normalization/README.md
index 043939d800..3bb046303b 100644
--- a/protocols/797dee-normalization/README.md
+++ b/protocols/797dee-normalization/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/79e9a1-cleanup/README.md b/protocols/79e9a1-cleanup/README.md
index 8bf9f8313a..e97cdaf0d9 100644
--- a/protocols/79e9a1-cleanup/README.md
+++ b/protocols/79e9a1-cleanup/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
- NGS Library Prep
diff --git a/protocols/7a1ae8/README.md b/protocols/7a1ae8/README.md
index 24a1fd968c..285a6aba57 100644
--- a/protocols/7a1ae8/README.md
+++ b/protocols/7a1ae8/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Zymo Kit
diff --git a/protocols/7a3e4b/README.md b/protocols/7a3e4b/README.md
index e982849ec1..e14b3d3191 100644
--- a/protocols/7a3e4b/README.md
+++ b/protocols/7a3e4b/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Featured
* Cherrypicking
diff --git a/protocols/7aa3fd-library-pooling/README.md b/protocols/7aa3fd-library-pooling/README.md
index 717a42479c..506a4f205f 100644
--- a/protocols/7aa3fd-library-pooling/README.md
+++ b/protocols/7aa3fd-library-pooling/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/7aa3fd-library-quantification/README.md b/protocols/7aa3fd-library-quantification/README.md
index f0dc4b6fde..3cbd7337d3 100644
--- a/protocols/7aa3fd-library-quantification/README.md
+++ b/protocols/7aa3fd-library-quantification/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/7aa3fd-size-selection/README.md b/protocols/7aa3fd-size-selection/README.md
index a4f6e17b6f..cd6c44db04 100644
--- a/protocols/7aa3fd-size-selection/README.md
+++ b/protocols/7aa3fd-size-selection/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Custom
diff --git a/protocols/7aad4e/README.md b/protocols/7aad4e/README.md
index 28c76d797e..182379ccd3 100644
--- a/protocols/7aad4e/README.md
+++ b/protocols/7aad4e/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherry Picking
diff --git a/protocols/7ada78-pt2/README.md b/protocols/7ada78-pt2/README.md
index b8e61a15ec..8afcefa8f7 100644
--- a/protocols/7ada78-pt2/README.md
+++ b/protocols/7ada78-pt2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* Complete PCR Workflow
diff --git a/protocols/7ada78/README.md b/protocols/7ada78/README.md
index 96e43a27e9..03b7cca28d 100644
--- a/protocols/7ada78/README.md
+++ b/protocols/7ada78/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* Complete PCR Workflow
diff --git a/protocols/7cfbde/README.md b/protocols/7cfbde/README.md
index 68140c80fe..ca294cb3ec 100644
--- a/protocols/7cfbde/README.md
+++ b/protocols/7cfbde/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* RNA Extraction
diff --git a/protocols/7e3b31/README.md b/protocols/7e3b31/README.md
index a111615713..1af9bcd2d2 100644
--- a/protocols/7e3b31/README.md
+++ b/protocols/7e3b31/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina TruSeq Stranded mRNA Sample Prep
diff --git a/protocols/7e4c3e-1/README.md b/protocols/7e4c3e-1/README.md
index da9223ae51..ea168fd027 100644
--- a/protocols/7e4c3e-1/README.md
+++ b/protocols/7e4c3e-1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/7e4c3e-2/README.md b/protocols/7e4c3e-2/README.md
index 31dc139c83..3616cc36de 100644
--- a/protocols/7e4c3e-2/README.md
+++ b/protocols/7e4c3e-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/7e4c3e-3/README.md b/protocols/7e4c3e-3/README.md
index d114eb53c2..3363833f00 100644
--- a/protocols/7e4c3e-3/README.md
+++ b/protocols/7e4c3e-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/7e4c3e-4/README.md b/protocols/7e4c3e-4/README.md
index 1fed71e4a0..adc091ab6c 100644
--- a/protocols/7e4c3e-4/README.md
+++ b/protocols/7e4c3e-4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/7e4c3e-5/README.md b/protocols/7e4c3e-5/README.md
index 7b7e270eac..14709fae46 100644
--- a/protocols/7e4c3e-5/README.md
+++ b/protocols/7e4c3e-5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/7e4c3e-6/README.md b/protocols/7e4c3e-6/README.md
index 5ca2520381..4939166b20 100644
--- a/protocols/7e4c3e-6/README.md
+++ b/protocols/7e4c3e-6/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/7f0f89/README.md b/protocols/7f0f89/README.md
index c6cf679bd5..ad2bc57ff4 100644
--- a/protocols/7f0f89/README.md
+++ b/protocols/7f0f89/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/7f9595/README.md b/protocols/7f9595/README.md
index bf7952f8b3..e74bb7b06a 100644
--- a/protocols/7f9595/README.md
+++ b/protocols/7f9595/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Serial Dilution
diff --git a/protocols/7fc7b1/README.md b/protocols/7fc7b1/README.md
index d5ae0d13fa..a7a57453e0 100644
--- a/protocols/7fc7b1/README.md
+++ b/protocols/7fc7b1/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Featured
* Cherrypicking
diff --git a/protocols/85b0dc/README.md b/protocols/85b0dc/README.md
index 2a56c136af..ba4c1c0578 100644
--- a/protocols/85b0dc/README.md
+++ b/protocols/85b0dc/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* Syber Green Prep
diff --git a/protocols/8nhsa0-norm/README.md b/protocols/8nhsa0-norm/README.md
index 6baf71f639..c3b082427e 100644
--- a/protocols/8nhsa0-norm/README.md
+++ b/protocols/8nhsa0-norm/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/8nhsa0/README.md b/protocols/8nhsa0/README.md
index ee2ee8f6e5..729b71557f 100644
--- a/protocols/8nhsa0/README.md
+++ b/protocols/8nhsa0/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/9038af/README.md b/protocols/9038af/README.md
index ad312d0b13..00d0944567 100644
--- a/protocols/9038af/README.md
+++ b/protocols/9038af/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Library Prep
* Sequencing
diff --git a/protocols/925d07-cp/README.md b/protocols/925d07-cp/README.md
index ea7fddc91f..29f7845e71 100644
--- a/protocols/925d07-cp/README.md
+++ b/protocols/925d07-cp/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Aliquoting
diff --git a/protocols/925d07-pla/README.md b/protocols/925d07-pla/README.md
index 4b5d4212c4..b86a54c498 100644
--- a/protocols/925d07-pla/README.md
+++ b/protocols/925d07-pla/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/925d07-q/README.md b/protocols/925d07-q/README.md
index 1e37b841f8..53e81f83d3 100644
--- a/protocols/925d07-q/README.md
+++ b/protocols/925d07-q/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/925d07-v3/README.md b/protocols/925d07-v3/README.md
index 7511361610..44b5b0db28 100644
--- a/protocols/925d07-v3/README.md
+++ b/protocols/925d07-v3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/9778eb/README.md b/protocols/9778eb/README.md
index 7184d728d3..20a096fe6a 100644
--- a/protocols/9778eb/README.md
+++ b/protocols/9778eb/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* DNA Extraction
diff --git a/protocols/9778eb_mass_norm/README.md b/protocols/9778eb_mass_norm/README.md
index 9fe5bdab4e..ca3d02736d 100644
--- a/protocols/9778eb_mass_norm/README.md
+++ b/protocols/9778eb_mass_norm/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/9778eb_spri/README.md b/protocols/9778eb_spri/README.md
index 449a5835b7..c04e943e7a 100644
--- a/protocols/9778eb_spri/README.md
+++ b/protocols/9778eb_spri/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* SPRI Beads
diff --git a/protocols/979d28-normalization/README.md b/protocols/979d28-normalization/README.md
index c35481bc94..8e8c70a046 100644
--- a/protocols/979d28-normalization/README.md
+++ b/protocols/979d28-normalization/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/979d28-pcr/README.md b/protocols/979d28-pcr/README.md
index 2b04fe57c4..f8cbe7133d 100644
--- a/protocols/979d28-pcr/README.md
+++ b/protocols/979d28-pcr/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/979d28/README.md b/protocols/979d28/README.md
index 3a5f6e1ef5..c85137b6b5 100644
--- a/protocols/979d28/README.md
+++ b/protocols/979d28/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Omega Mag-Bind® Total RNA Isolation
diff --git a/protocols/9ot019/README.md b/protocols/9ot019/README.md
index 7793490963..c41d47fcc4 100644
--- a/protocols/9ot019/README.md
+++ b/protocols/9ot019/README.md
@@ -5,6 +5,9 @@
Tim Dobbs
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* General Liquid Handling
* General Liquid Handling
diff --git a/protocols/ML-normalization-cdna-library/README.md b/protocols/ML-normalization-cdna-library/README.md
index 2b28303bbb..ef1ca34545 100644
--- a/protocols/ML-normalization-cdna-library/README.md
+++ b/protocols/ML-normalization-cdna-library/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/ML-normalization/README.md b/protocols/ML-normalization/README.md
index 2b28303bbb..ef1ca34545 100644
--- a/protocols/ML-normalization/README.md
+++ b/protocols/ML-normalization/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Normalization
diff --git a/protocols/Opentrons_Logo/README.md b/protocols/Opentrons_Logo/README.md
index 62e2f78e2d..fb2bba11de 100644
--- a/protocols/Opentrons_Logo/README.md
+++ b/protocols/Opentrons_Logo/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/opentrons_logo). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/opentrons_logo). This page won’t be available after January 31st, 2024.
## Categories
* Getting Started
diff --git a/protocols/arcis-multi-test/README.md b/protocols/arcis-multi-test/README.md
index 67afc4a07a..e835841038 100644
--- a/protocols/arcis-multi-test/README.md
+++ b/protocols/arcis-multi-test/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Arcis Blood Kit
diff --git a/protocols/arcis-single-test/README.md b/protocols/arcis-single-test/README.md
index 9eb34eccaf..5ec653a530 100644
--- a/protocols/arcis-single-test/README.md
+++ b/protocols/arcis-single-test/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Arcis Blood Kit
diff --git a/protocols/batch-test-plating/README.md b/protocols/batch-test-plating/README.md
index 624ba773e9..33a424f00d 100644
--- a/protocols/batch-test-plating/README.md
+++ b/protocols/batch-test-plating/README.md
@@ -6,6 +6,9 @@
### Partner
[Facundo Rodriguez Goren]
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/bc-rnadvance-viral-feat/README.md b/protocols/bc-rnadvance-viral-feat/README.md
index 297d9ad6ad..bd0884062d 100644
--- a/protocols/bc-rnadvance-viral-feat/README.md
+++ b/protocols/bc-rnadvance-viral-feat/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Featured
* Beckman Coulter RNAdvance Viral
diff --git a/protocols/bc-rnadvance-viral/README.md b/protocols/bc-rnadvance-viral/README.md
index 310de1f240..77f004317a 100644
--- a/protocols/bc-rnadvance-viral/README.md
+++ b/protocols/bc-rnadvance-viral/README.md
@@ -6,6 +6,9 @@
### Partner
[Beckman Coulter Life Sciences](https://www.beckmancoulter.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Viral RNA
diff --git a/protocols/bms-illumina-dna-prep/README.md b/protocols/bms-illumina-dna-prep/README.md
index fd25fc40de..40034619e9 100644
--- a/protocols/bms-illumina-dna-prep/README.md
+++ b/protocols/bms-illumina-dna-prep/README.md
@@ -5,6 +5,9 @@
### Partner
[Illumina](https://www.illumina.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep
diff --git a/protocols/bpg-rna-extraction/README.md b/protocols/bpg-rna-extraction/README.md
index 73c8f42d63..aebe618bab 100644
--- a/protocols/bpg-rna-extraction/README.md
+++ b/protocols/bpg-rna-extraction/README.md
@@ -6,6 +6,9 @@
### Partner
[BP Genomics](https://bpgenomics.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Covid Workstation
* RNA Extraction
diff --git a/protocols/cepheid/README.md b/protocols/cepheid/README.md
index 60aad431a4..49c891f967 100644
--- a/protocols/cepheid/README.md
+++ b/protocols/cepheid/README.md
@@ -6,6 +6,9 @@
### Partner
[BasisDx](https://www.basisdx.org/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cepheid
diff --git a/protocols/cerillo/README.md b/protocols/cerillo/README.md
index 0850f33be1..2ad485e723 100644
--- a/protocols/cerillo/README.md
+++ b/protocols/cerillo/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/checkit/README.md b/protocols/checkit/README.md
index 18146f8c7a..1cce9beeff 100644
--- a/protocols/checkit/README.md
+++ b/protocols/checkit/README.md
@@ -6,6 +6,9 @@
### Partner

+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Featured
* Next Advance Checkit Go
diff --git a/protocols/cherrypicking/README.md b/protocols/cherrypicking/README.md
index d3cefadacd..c4f9d14476 100644
--- a/protocols/cherrypicking/README.md
+++ b/protocols/cherrypicking/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/cherrypicking). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/cherrypicking). This page won’t be available after January 31st, 2024.
## Categories
* Featured
diff --git a/protocols/cherrypicking_simple/README.md b/protocols/cherrypicking_simple/README.md
index 8756f6576a..fc226da631 100644
--- a/protocols/cherrypicking_simple/README.md
+++ b/protocols/cherrypicking_simple/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Cherrypicking
diff --git a/protocols/complete-pcr/README.md b/protocols/complete-pcr/README.md
index 918fb2e1bc..cbffaab294 100644
--- a/protocols/complete-pcr/README.md
+++ b/protocols/complete-pcr/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](http://www.opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* Complete PCR Workflow
diff --git a/protocols/covid19-rna-extraction/README.md b/protocols/covid19-rna-extraction/README.md
index 9af2bb0a7d..9dec7dd2d7 100644
--- a/protocols/covid19-rna-extraction/README.md
+++ b/protocols/covid19-rna-extraction/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Covid Workstation
* RNA Extraction
diff --git a/protocols/csv_transfer/README.md b/protocols/csv_transfer/README.md
index d88b6247c2..a58acec34e 100644
--- a/protocols/csv_transfer/README.md
+++ b/protocols/csv_transfer/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* CSV Transfer
diff --git a/protocols/customizable_serial_dilution_ot2/README.md b/protocols/customizable_serial_dilution_ot2/README.md
index e544d77342..21fdf4b889 100644
--- a/protocols/customizable_serial_dilution_ot2/README.md
+++ b/protocols/customizable_serial_dilution_ot2/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/customizable_serial_dilution_ot2). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/customizable_serial_dilution_ot2). This page won’t be available after January 31st, 2024.
## Categories
* Featured
@@ -18,17 +18,17 @@ With this protocol, you can do a simple serial dilution across a 96-well plate u

-***Example Setup***
+Example Setup
-This protocol uses the inputs you define for "***Dilution Factor***" and "***Total Mixing Volume***" to automatically infer the necessary transfer volume for each dilution across your plate. For a 1 in 3 dilution series across an entire plate, as seen above:
+This protocol uses the inputs you define for "Dilution Factor" and "Total Mixing Volume" to automatically infer the necessary transfer volume for each dilution across your plate. For a 1 in 3 dilution series across an entire plate, as seen above:
-- Start with your samples/reagents in Column 1 of your plate. In this example, you would pre-add 150 uL of concentrated sample to the first column of your 96-well plate.
--- Define a ***Total Mixing Volume*** of 150uL, a ***Dilution Factor*** of 3, and set ***Number of Dilutions*** = 11.
+-- Define a Total Mixing Volume of 150uL, a Dilution Factor of 3, and set Number of Dilutions = 11.
-- Your OT-2 will add 100uL of diluent to each empty well in your plate. Then it will transfer 50uL from Column 1 between each well/column in the plate.
--- "***Total mixing volume***" = transfer volume + diluent volume.
+-- "Total mixing volume" = transfer volume + diluent volume.
---
@@ -54,13 +54,13 @@ This protocol uses the inputs you define for "***Dilution Factor***" and "***Tot
## Process
1. Choose the pipette you want to use from the dropdown menu above and which side it is installed on the OT-2
2. Set your dilution factor.
- ***Example:*** If you want a 1:2 ratio of sample to total reaction volume, you would set your dilution factor to 2.
+ Example: If you want a 1:2 ratio of sample to total reaction volume, you would set your dilution factor to 2.
3. Set your number of dilutions (max is 11, 10 if using blank)
4. Set your total mixing volume. (Total mixing volume = transfer volume + diluent volume). Be careful to make sure this number does not exceed the volume capacity of your plate. To see how this number is used, scroll to the example above.
5. Set whether a blank will be made or not. The blank will be added to the first available column in the plate.
- ***NOTE:*** 10 dilutions is the max allowed when using a blank
+ NOTE: 10 dilutions is the max allowed when using a blank
6. Set your tip reuse strategy.
- ***Note:*** This defaults to no tip changes; adjust only if you want to change tips between each well.
+ Note: This defaults to no tip changes; adjust only if you want to change tips between each well.
7. Set your air gap, if desired. This will add a specified amount of air into the tip after aspiration
8. Download your customized OT-2 Serial Dilution protocol using the blue "Download" button.
9. Upload into the Opentrons Run App and follow the instructions there to set up your deck and proceed to run!
diff --git a/protocols/demo/README.md b/protocols/demo/README.md
index b265f254e5..574da2482b 100644
--- a/protocols/demo/README.md
+++ b/protocols/demo/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Getting Started
* OT-2 Demo
diff --git a/protocols/dinosaur/README.md b/protocols/dinosaur/README.md
index a7d2b5eb1c..6337b94c57 100644
--- a/protocols/dinosaur/README.md
+++ b/protocols/dinosaur/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/dinosaur). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/dinosaur). This page won’t be available after January 31st, 2024.
## Categories
* Getting Started
diff --git a/protocols/dynabeads_plate_prep_1/README.md b/protocols/dynabeads_plate_prep_1/README.md
index 95fcd12ed6..38e198b65b 100644
--- a/protocols/dynabeads_plate_prep_1/README.md
+++ b/protocols/dynabeads_plate_prep_1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Protein Purification
* Thermo Fisher Dynabeads™ Protein A
diff --git a/protocols/dynabeads_plate_prep_2/README.md b/protocols/dynabeads_plate_prep_2/README.md
index c0048504b7..828a7e3764 100644
--- a/protocols/dynabeads_plate_prep_2/README.md
+++ b/protocols/dynabeads_plate_prep_2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Protein Purification
* Thermo Fisher Dynabeads™ Protein A
diff --git a/protocols/dz1j39/README.md b/protocols/dz1j39/README.md
index 1c734a2e56..c1550d0a46 100644
--- a/protocols/dz1j39/README.md
+++ b/protocols/dz1j39/README.md
@@ -5,6 +5,9 @@
Opentrons
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Cell and Tissue Culture
* Cell and Tissue Culture
diff --git a/protocols/e54ada/README.md b/protocols/e54ada/README.md
index 100e4e71f6..504fe4ff82 100644
--- a/protocols/e54ada/README.md
+++ b/protocols/e54ada/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* DNA/RNA Extraction
diff --git a/protocols/e54ada_rt/README.md b/protocols/e54ada_rt/README.md
index 999d6cb39c..f45765e32e 100644
--- a/protocols/e54ada_rt/README.md
+++ b/protocols/e54ada_rt/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR Prep
* Reverse Transcriptase
diff --git a/protocols/e9ff8d/README.md b/protocols/e9ff8d/README.md
index 7048bef33c..4469f3d5a1 100644
--- a/protocols/e9ff8d/README.md
+++ b/protocols/e9ff8d/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* Pre-Amplification
diff --git a/protocols/e9ff8d_part2/README.md b/protocols/e9ff8d_part2/README.md
index ef6eee1cef..3a87e8c06e 100644
--- a/protocols/e9ff8d_part2/README.md
+++ b/protocols/e9ff8d_part2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Purification
* DNA Cleanup
diff --git a/protocols/e9ff8d_part3/README.md b/protocols/e9ff8d_part3/README.md
index 7355783b42..bdc01a42ff 100644
--- a/protocols/e9ff8d_part3/README.md
+++ b/protocols/e9ff8d_part3/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/e9ff8d_part4/README.md b/protocols/e9ff8d_part4/README.md
index 413a87c3eb..9622029007 100644
--- a/protocols/e9ff8d_part4/README.md
+++ b/protocols/e9ff8d_part4/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Purification
* DNA Cleanup
diff --git a/protocols/ff5763/README.md b/protocols/ff5763/README.md
index 511e05b6c3..b3a7685174 100644
--- a/protocols/ff5763/README.md
+++ b/protocols/ff5763/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep
diff --git a/protocols/ff5763_part2/README.md b/protocols/ff5763_part2/README.md
index fa98fd92f2..f1b26f5c9d 100644
--- a/protocols/ff5763_part2/README.md
+++ b/protocols/ff5763_part2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep
diff --git a/protocols/ff5763_part3/README.md b/protocols/ff5763_part3/README.md
index 1db5d788ff..ebae2c8a9e 100644
--- a/protocols/ff5763_part3/README.md
+++ b/protocols/ff5763_part3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep
diff --git a/protocols/ff5763_part4/README.md b/protocols/ff5763_part4/README.md
index ca5a841f3b..8bfb297c18 100644
--- a/protocols/ff5763_part4/README.md
+++ b/protocols/ff5763_part4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep
diff --git a/protocols/ff5763_part5/README.md b/protocols/ff5763_part5/README.md
index be8578c842..64d37ed86c 100644
--- a/protocols/ff5763_part5/README.md
+++ b/protocols/ff5763_part5/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep
diff --git a/protocols/generic_ngs_prep/README.md b/protocols/generic_ngs_prep/README.md
index 3a6b57268d..8202414280 100644
--- a/protocols/generic_ngs_prep/README.md
+++ b/protocols/generic_ngs_prep/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Generic
diff --git a/protocols/generic_pcr_prep_1/README.md b/protocols/generic_pcr_prep_1/README.md
index 33346bfe89..15ba76453f 100644
--- a/protocols/generic_pcr_prep_1/README.md
+++ b/protocols/generic_pcr_prep_1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* Generic Mastermix Assembly
diff --git a/protocols/generic_pcr_prep_2/README.md b/protocols/generic_pcr_prep_2/README.md
index f0713db09c..147ae55dc0 100644
--- a/protocols/generic_pcr_prep_2/README.md
+++ b/protocols/generic_pcr_prep_2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* Generic PCR Prep
diff --git a/protocols/generic_protein_purification/README.md b/protocols/generic_protein_purification/README.md
index ef70c9be1b..18bc99450e 100644
--- a/protocols/generic_protein_purification/README.md
+++ b/protocols/generic_protein_purification/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Protein Purification
* Generic Protein Purification
diff --git a/protocols/generic_station_A/README.md b/protocols/generic_station_A/README.md
index 73cc12834b..dfa04d531c 100644
--- a/protocols/generic_station_A/README.md
+++ b/protocols/generic_station_A/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Covid Workstation
* Sample Plating
diff --git a/protocols/generic_station_C/README.md b/protocols/generic_station_C/README.md
index 638312157c..fc40caf544 100644
--- a/protocols/generic_station_C/README.md
+++ b/protocols/generic_station_C/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Covid Workstation
* qPCR Setup
diff --git a/protocols/gna_octea/README.md b/protocols/gna_octea/README.md
index ff223c2b8b..10d86125de 100644
--- a/protocols/gna_octea/README.md
+++ b/protocols/gna_octea/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Covid
diff --git a/protocols/gna_octea_v2/README.md b/protocols/gna_octea_v2/README.md
index 423afc2bdc..60bb8adf6a 100644
--- a/protocols/gna_octea_v2/README.md
+++ b/protocols/gna_octea_v2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Covid
diff --git a/protocols/gsdx/README.md b/protocols/gsdx/README.md
index 8ab7985dcb..8c52412f61 100644
--- a/protocols/gsdx/README.md
+++ b/protocols/gsdx/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Screening
diff --git a/protocols/heater-shaker-beta/README.md b/protocols/heater-shaker-beta/README.md
index 6bce2123eb..6488f13eac 100644
--- a/protocols/heater-shaker-beta/README.md
+++ b/protocols/heater-shaker-beta/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Heater Shaker
* Heater Shaker Beta Test
diff --git a/protocols/i7g1ym/README.md b/protocols/i7g1ym/README.md
index 4b706e6cd6..915a4aa209 100644
--- a/protocols/i7g1ym/README.md
+++ b/protocols/i7g1ym/README.md
@@ -5,6 +5,9 @@
Tim Fallon
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* General Liquid Handling
* General Liquid Handling
diff --git a/protocols/illumina-nextera-XT-library-prep-part1/README.md b/protocols/illumina-nextera-XT-library-prep-part1/README.md
index 13f1352701..08debc37c9 100644
--- a/protocols/illumina-nextera-XT-library-prep-part1/README.md
+++ b/protocols/illumina-nextera-XT-library-prep-part1/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/illumina-nextera-xt-library-prep-part1). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/illumina-nextera-xt-library-prep-part1). This page won’t be available after January 31st, 2024.
## Categories
* Featured
diff --git a/protocols/illumina-nextera-XT-library-prep-part2/README.md b/protocols/illumina-nextera-XT-library-prep-part2/README.md
index 61c3579fce..305a27612a 100644
--- a/protocols/illumina-nextera-XT-library-prep-part2/README.md
+++ b/protocols/illumina-nextera-XT-library-prep-part2/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/illumina-nextera-xt-library-prep-part2). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/illumina-nextera-xt-library-prep-part2). This page won’t be available after January 31st, 2024.
## Categories
* Featured
diff --git a/protocols/illumina-nextera-XT-library-prep-part3/README.md b/protocols/illumina-nextera-XT-library-prep-part3/README.md
index ad80ea2071..0802814534 100644
--- a/protocols/illumina-nextera-XT-library-prep-part3/README.md
+++ b/protocols/illumina-nextera-XT-library-prep-part3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Featured
* NGS Library Prep: Illumina Nextera XT
diff --git a/protocols/illumina-nextera-XT-library-prep-part4/README.md b/protocols/illumina-nextera-XT-library-prep-part4/README.md
index 0a8d655876..0f7685001a 100644
--- a/protocols/illumina-nextera-XT-library-prep-part4/README.md
+++ b/protocols/illumina-nextera-XT-library-prep-part4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Featured
* NGS Library Prep: Illumina Nextera XT
diff --git a/protocols/macherey-nagel-nucleomag-DNA-microbiome/README.md b/protocols/macherey-nagel-nucleomag-DNA-microbiome/README.md
index 1be8715316..adf8b6af6c 100644
--- a/protocols/macherey-nagel-nucleomag-DNA-microbiome/README.md
+++ b/protocols/macherey-nagel-nucleomag-DNA-microbiome/README.md
@@ -8,7 +8,7 @@
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-dna-microbiome). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-dna-microbiome). This page won’t be available after January 31st, 2024.
## Categories
* Nucleic Acid Extraction & Purification
diff --git a/protocols/macherey-nagel-nucleomag-clean-up/README.md b/protocols/macherey-nagel-nucleomag-clean-up/README.md
index 8fa6dd92fe..97403c1b10 100644
--- a/protocols/macherey-nagel-nucleomag-clean-up/README.md
+++ b/protocols/macherey-nagel-nucleomag-clean-up/README.md
@@ -8,7 +8,7 @@
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-clean-up). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-clean-up). This page won’t be available after January 31st, 2024.
## Categories
* NGS Library Prep
diff --git a/protocols/macherey-nagel-nucleomag-dna-food/README.md b/protocols/macherey-nagel-nucleomag-dna-food/README.md
index cbb77c9dc9..e1ed5309d5 100644
--- a/protocols/macherey-nagel-nucleomag-dna-food/README.md
+++ b/protocols/macherey-nagel-nucleomag-dna-food/README.md
@@ -8,7 +8,7 @@
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-dna-food). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-dna-food). This page won’t be available after January 31st, 2024.
## Categories
* Nucleic Acid Extraction & Purification
diff --git a/protocols/macherey-nagel-nucleomag-pathogen/README.md b/protocols/macherey-nagel-nucleomag-pathogen/README.md
index 3b47b47cf6..2278a2ab69 100644
--- a/protocols/macherey-nagel-nucleomag-pathogen/README.md
+++ b/protocols/macherey-nagel-nucleomag-pathogen/README.md
@@ -8,6 +8,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* MACHEREY-NAGEL NucleoMag® Pathogen
diff --git a/protocols/macherey-nagel-nucleomag-rna/README.md b/protocols/macherey-nagel-nucleomag-rna/README.md
index 6b8304ce98..9c094d43a4 100644
--- a/protocols/macherey-nagel-nucleomag-rna/README.md
+++ b/protocols/macherey-nagel-nucleomag-rna/README.md
@@ -8,6 +8,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* MACHEREY-NAGEL NucleoMag® RNA
diff --git a/protocols/macherey-nagel-nucleomag-size-select/README.md b/protocols/macherey-nagel-nucleomag-size-select/README.md
index d30418f730..99aef0f3a6 100644
--- a/protocols/macherey-nagel-nucleomag-size-select/README.md
+++ b/protocols/macherey-nagel-nucleomag-size-select/README.md
@@ -8,6 +8,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* MACHEREY-NAGEL Double-Size Select
diff --git a/protocols/macherey-nagel-nucleomag-tissue/README.md b/protocols/macherey-nagel-nucleomag-tissue/README.md
index 4c3e4df054..3d28a2b2f6 100644
--- a/protocols/macherey-nagel-nucleomag-tissue/README.md
+++ b/protocols/macherey-nagel-nucleomag-tissue/README.md
@@ -8,7 +8,7 @@
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-tissue). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-tissue). This page won’t be available after January 31st, 2024.
## Categories
* Nucleic Acid Extraction & Purification
diff --git a/protocols/macherey-nagel-nucleomag-virus/README.md b/protocols/macherey-nagel-nucleomag-virus/README.md
index 2513798592..8c5fbc2715 100644
--- a/protocols/macherey-nagel-nucleomag-virus/README.md
+++ b/protocols/macherey-nagel-nucleomag-virus/README.md
@@ -8,7 +8,7 @@
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-virus). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/macherey-nagel-nucleomag-virus). This page won’t be available after January 31st, 2024.
## Categories
* Nucleic Acid Extraction & Purification
diff --git a/protocols/magnesil/README.md b/protocols/magnesil/README.md
index 0a32756d94..77c98b1799 100644
--- a/protocols/magnesil/README.md
+++ b/protocols/magnesil/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* MagneSil
diff --git a/protocols/nextera-flex-library-prep-amplify-tagmented-dna/README.md b/protocols/nextera-flex-library-prep-amplify-tagmented-dna/README.md
index da30ab8ac9..4a3fed59ec 100644
--- a/protocols/nextera-flex-library-prep-amplify-tagmented-dna/README.md
+++ b/protocols/nextera-flex-library-prep-amplify-tagmented-dna/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Nextera DNA Flex Library Prep
diff --git a/protocols/nextera-flex-library-prep-cherrypick-samples/README.md b/protocols/nextera-flex-library-prep-cherrypick-samples/README.md
index e320aa0d6a..463ffde7f0 100644
--- a/protocols/nextera-flex-library-prep-cherrypick-samples/README.md
+++ b/protocols/nextera-flex-library-prep-cherrypick-samples/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Nextera DNA Flex Library Prep
diff --git a/protocols/nextera-flex-library-prep-cleanup-libraries/README.md b/protocols/nextera-flex-library-prep-cleanup-libraries/README.md
index 4cd6751878..b8b414a7c6 100644
--- a/protocols/nextera-flex-library-prep-cleanup-libraries/README.md
+++ b/protocols/nextera-flex-library-prep-cleanup-libraries/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Nextera DNA Flex Library Prep
diff --git a/protocols/nextera-flex-library-prep-post-tag-cleanup/README.md b/protocols/nextera-flex-library-prep-post-tag-cleanup/README.md
index 7375e3a834..278b6e75a9 100644
--- a/protocols/nextera-flex-library-prep-post-tag-cleanup/README.md
+++ b/protocols/nextera-flex-library-prep-post-tag-cleanup/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Nextera DNA Flex Library Prep
diff --git a/protocols/nextera-flex-library-prep-tagment-dna/README.md b/protocols/nextera-flex-library-prep-tagment-dna/README.md
index 667193f7ad..0f90f16a9d 100644
--- a/protocols/nextera-flex-library-prep-tagment-dna/README.md
+++ b/protocols/nextera-flex-library-prep-tagment-dna/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Nextera DNA Flex Library Prep
diff --git a/protocols/normalization/README.md b/protocols/normalization/README.md
index 12ba60db43..ed2fba1d07 100644
--- a/protocols/normalization/README.md
+++ b/protocols/normalization/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/normalization). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/normalization). This page won’t be available after January 31st, 2024.
## Categories
* Featured
diff --git a/protocols/nucleic_acid_purification_with_magnetic_beads/README.md b/protocols/nucleic_acid_purification_with_magnetic_beads/README.md
index 44bc561cc0..fb41c92df1 100644
--- a/protocols/nucleic_acid_purification_with_magnetic_beads/README.md
+++ b/protocols/nucleic_acid_purification_with_magnetic_beads/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/nucleic_acid_purification_with_magnetic_beads). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/nucleic_acid_purification_with_magnetic_beads). This page won’t be available after January 31st, 2024.
## Categories
* Featured
diff --git a/protocols/omega-biotek-xpress/README.md b/protocols/omega-biotek-xpress/README.md
index 4f85132f30..d08e15345c 100644
--- a/protocols/omega-biotek-xpress/README.md
+++ b/protocols/omega-biotek-xpress/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Covid Workstation
* RNA Extraction
diff --git a/protocols/omega_biotek_magbind_totalpure_NGS/README.md b/protocols/omega_biotek_magbind_totalpure_NGS/README.md
index fd78e16b17..68f75ff1e8 100644
--- a/protocols/omega_biotek_magbind_totalpure_NGS/README.md
+++ b/protocols/omega_biotek_magbind_totalpure_NGS/README.md
@@ -6,7 +6,7 @@
### Partner
[Omega Bio-tek](http://omegabiotek.com/store/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/omega_biotek_magbind_totalpure_ngs). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/omega_biotek_magbind_totalpure_ngs). This page won’t be available after January 31st, 2024.
## Categories
* Featured
diff --git a/protocols/onsite-bms-48plate/README.md b/protocols/onsite-bms-48plate/README.md
index 3bed0bef6c..5b7e6fe45b 100644
--- a/protocols/onsite-bms-48plate/README.md
+++ b/protocols/onsite-bms-48plate/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/onsite-bms/README.md b/protocols/onsite-bms/README.md
index 1212ad27ef..693b2c6579 100644
--- a/protocols/onsite-bms/README.md
+++ b/protocols/onsite-bms/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/onsite-circlelabs/README.md b/protocols/onsite-circlelabs/README.md
index b813c05608..995730ed1a 100644
--- a/protocols/onsite-circlelabs/README.md
+++ b/protocols/onsite-circlelabs/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS LIBRARY PREP
* Custom
diff --git a/protocols/onsite-dmso/README.md b/protocols/onsite-dmso/README.md
index bc71c35d06..b0d8d8caef 100644
--- a/protocols/onsite-dmso/README.md
+++ b/protocols/onsite-dmso/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/onsite-ganda-1/README.md b/protocols/onsite-ganda-1/README.md
index c61dfc9e3f..575a115f1b 100644
--- a/protocols/onsite-ganda-1/README.md
+++ b/protocols/onsite-ganda-1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* rhAmpSeq Library Prep
diff --git a/protocols/onsite-ganda-2/README.md b/protocols/onsite-ganda-2/README.md
index d199a6515f..4872de0933 100644
--- a/protocols/onsite-ganda-2/README.md
+++ b/protocols/onsite-ganda-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* rhAmpSeq Library Prep
diff --git a/protocols/onsite-ganda-3/README.md b/protocols/onsite-ganda-3/README.md
index 9dd328b301..73e7bfccf0 100644
--- a/protocols/onsite-ganda-3/README.md
+++ b/protocols/onsite-ganda-3/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* rhAmpSeq Library Prep
diff --git a/protocols/onsite-ganda-4/README.md b/protocols/onsite-ganda-4/README.md
index 56ab1cf38f..ac7a2142c0 100644
--- a/protocols/onsite-ganda-4/README.md
+++ b/protocols/onsite-ganda-4/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* rhAmpSeq Library Prep
diff --git a/protocols/onsite_atila_nasal/README.md b/protocols/onsite_atila_nasal/README.md
index 9e656da1a1..a524466729 100644
--- a/protocols/onsite_atila_nasal/README.md
+++ b/protocols/onsite_atila_nasal/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/onsite_atila_saliva/README.md b/protocols/onsite_atila_saliva/README.md
index 95c5fc685a..155bb2241d 100644
--- a/protocols/onsite_atila_saliva/README.md
+++ b/protocols/onsite_atila_saliva/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/onsite_atila_saliva384_pt1/README.md b/protocols/onsite_atila_saliva384_pt1/README.md
index ab81018ad3..585197a8ca 100644
--- a/protocols/onsite_atila_saliva384_pt1/README.md
+++ b/protocols/onsite_atila_saliva384_pt1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/onsite_atila_saliva384_pt2/README.md b/protocols/onsite_atila_saliva384_pt2/README.md
index f0f48ff3d9..f1158fcfbe 100644
--- a/protocols/onsite_atila_saliva384_pt2/README.md
+++ b/protocols/onsite_atila_saliva384_pt2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* PCR Prep
diff --git a/protocols/onsite_egenesis/README.md b/protocols/onsite_egenesis/README.md
index 03fd954e76..b7414959e2 100644
--- a/protocols/onsite_egenesis/README.md
+++ b/protocols/onsite_egenesis/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/onsite_egenesis_part2/README.md b/protocols/onsite_egenesis_part2/README.md
index 31cdac9f77..28d2f0fb67 100644
--- a/protocols/onsite_egenesis_part2/README.md
+++ b/protocols/onsite_egenesis_part2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/onsite_galatea_mag_bind/README.md b/protocols/onsite_galatea_mag_bind/README.md
index e0f7b9d1ea..1ab4211dd1 100644
--- a/protocols/onsite_galatea_mag_bind/README.md
+++ b/protocols/onsite_galatea_mag_bind/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons (verified)](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* DNA Extraction
diff --git a/protocols/onsite_galatea_normalization/README.md b/protocols/onsite_galatea_normalization/README.md
index 191b2a2305..e12494b6b0 100644
--- a/protocols/onsite_galatea_normalization/README.md
+++ b/protocols/onsite_galatea_normalization/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/onsite_galatea_pre_normalization/README.md b/protocols/onsite_galatea_pre_normalization/README.md
index 3a11a6023a..9eb5741198 100644
--- a/protocols/onsite_galatea_pre_normalization/README.md
+++ b/protocols/onsite_galatea_pre_normalization/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/pcr_prep_part_1/README.md b/protocols/pcr_prep_part_1/README.md
index 5169565db8..f23df14e34 100644
--- a/protocols/pcr_prep_part_1/README.md
+++ b/protocols/pcr_prep_part_1/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons (verified)](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/pcr_prep_part_1). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/pcr_prep_part_1). This page won’t be available after January 31st, 2024.
## Categories
* PCR
diff --git a/protocols/pcr_prep_part_2/README.md b/protocols/pcr_prep_part_2/README.md
index 2d5a172a02..2932e362a3 100644
--- a/protocols/pcr_prep_part_2/README.md
+++ b/protocols/pcr_prep_part_2/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons (verified)](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/pcr_prep_part_2). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/pcr_prep_part_2). This page won’t be available after January 31st, 2024.
## Categories
* PCR
diff --git a/protocols/pcr_test_plan/README.md b/protocols/pcr_test_plan/README.md
index 6e0bd0a546..28443b189c 100644
--- a/protocols/pcr_test_plan/README.md
+++ b/protocols/pcr_test_plan/README.md
@@ -6,6 +6,9 @@
### Partner
[Stanford](https://www.stanford.edu/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Sequencing
diff --git a/protocols/pooling/README.md b/protocols/pooling/README.md
index 9d8f822065..9439b5b9da 100644
--- a/protocols/pooling/README.md
+++ b/protocols/pooling/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Pooling
diff --git a/protocols/primer_hydration/README.md b/protocols/primer_hydration/README.md
index e33471a280..369700286b 100644
--- a/protocols/primer_hydration/README.md
+++ b/protocols/primer_hydration/README.md
@@ -3,6 +3,9 @@
### Author
[REM Analytics](https://www.remanalytics.ch/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR Prep
* Primer hydration
diff --git a/protocols/promega-maxwell/README.md b/protocols/promega-maxwell/README.md
index 54b1ea625b..2b2497401b 100644
--- a/protocols/promega-maxwell/README.md
+++ b/protocols/promega-maxwell/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Viral RNA
diff --git a/protocols/quintara_onsite_nanopore/README.md b/protocols/quintara_onsite_nanopore/README.md
index a5ee79a454..f265c9970e 100644
--- a/protocols/quintara_onsite_nanopore/README.md
+++ b/protocols/quintara_onsite_nanopore/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Nanopore
diff --git a/protocols/quintara_onsite_nanopore_pooling/README.md b/protocols/quintara_onsite_nanopore_pooling/README.md
index 2712937fa4..dfc7dbbbae 100644
--- a/protocols/quintara_onsite_nanopore_pooling/README.md
+++ b/protocols/quintara_onsite_nanopore_pooling/README.md
@@ -4,6 +4,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Nanopore
diff --git a/protocols/quintara_onsite_part1/README.md b/protocols/quintara_onsite_part1/README.md
index 1b44d46352..f014ef9442 100644
--- a/protocols/quintara_onsite_part1/README.md
+++ b/protocols/quintara_onsite_part1/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/quintara_onsite_part1_384/README.md b/protocols/quintara_onsite_part1_384/README.md
index c620b7dba5..55305ede2f 100644
--- a/protocols/quintara_onsite_part1_384/README.md
+++ b/protocols/quintara_onsite_part1_384/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/quintara_onsite_part2/README.md b/protocols/quintara_onsite_part2/README.md
index 92477f0b26..79dfe15959 100644
--- a/protocols/quintara_onsite_part2/README.md
+++ b/protocols/quintara_onsite_part2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/quintara_onsite_part3/README.md b/protocols/quintara_onsite_part3/README.md
index 588e69bab2..9760c7535a 100644
--- a/protocols/quintara_onsite_part3/README.md
+++ b/protocols/quintara_onsite_part3/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/quintara_onsite_part4/README.md b/protocols/quintara_onsite_part4/README.md
index ae20988a73..251f7f7182 100644
--- a/protocols/quintara_onsite_part4/README.md
+++ b/protocols/quintara_onsite_part4/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/rem-pcr-plate/README.md b/protocols/rem-pcr-plate/README.md
index 8583736c80..63fb1a4456 100644
--- a/protocols/rem-pcr-plate/README.md
+++ b/protocols/rem-pcr-plate/README.md
@@ -3,6 +3,9 @@
### Author
[REM Analytics](https://www.remanalytics.ch/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR Prep
diff --git a/protocols/sci-amplex-red/README.md b/protocols/sci-amplex-red/README.md
index b0f7558c39..19ab9d853b 100644
--- a/protocols/sci-amplex-red/README.md
+++ b/protocols/sci-amplex-red/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Standard curve for Hydrogen Peroxide
* Measurement of peroxides/hydrogen peroxide from THP-1 cells
diff --git a/protocols/sci-bead-based-elisa-for-sars-cov-2-surrogate-virus-neutralization-test/README.md b/protocols/sci-bead-based-elisa-for-sars-cov-2-surrogate-virus-neutralization-test/README.md
index dc9763ccc1..6dc07f7d64 100644
--- a/protocols/sci-bead-based-elisa-for-sars-cov-2-surrogate-virus-neutralization-test/README.md
+++ b/protocols/sci-bead-based-elisa-for-sars-cov-2-surrogate-virus-neutralization-test/README.md
@@ -5,6 +5,9 @@
Boren Lin
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins
* Proteins
diff --git a/protocols/sci-dynabeads-ip-1/README.md b/protocols/sci-dynabeads-ip-1/README.md
index 5a393c993f..3de9c21593 100644
--- a/protocols/sci-dynabeads-ip-1/README.md
+++ b/protocols/sci-dynabeads-ip-1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Protein Purification
* ThermoFisher Dynabeads Protein A/G
diff --git a/protocols/sci-dynabeads-ip-2/README.md b/protocols/sci-dynabeads-ip-2/README.md
index 65fa9667b0..e7a3fc5f35 100644
--- a/protocols/sci-dynabeads-ip-2/README.md
+++ b/protocols/sci-dynabeads-ip-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Protein Purification
* ThermoFisher Dynabeads Protein A/G
diff --git a/protocols/sci-dynabeads-tube-1/README.md b/protocols/sci-dynabeads-tube-1/README.md
index 01a58e9658..2912d2a4a7 100644
--- a/protocols/sci-dynabeads-tube-1/README.md
+++ b/protocols/sci-dynabeads-tube-1/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Protein Purification
* Thermo Fisher Dynabeads™ Protein A/G
diff --git a/protocols/sci-dynabeads-tube-2/README.md b/protocols/sci-dynabeads-tube-2/README.md
index 9292504968..f4d478362e 100644
--- a/protocols/sci-dynabeads-tube-2/README.md
+++ b/protocols/sci-dynabeads-tube-2/README.md
@@ -3,6 +3,9 @@
### Author
Opentrons(https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Protein Purification
* Thermo Fisher Dynabeads™ Protein A/G
diff --git a/protocols/sci-idt-normalase/README.md b/protocols/sci-idt-normalase/README.md
index 6162c19b9e..f92065de60 100644
--- a/protocols/sci-idt-normalase/README.md
+++ b/protocols/sci-idt-normalase/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* IDT Normalase
diff --git a/protocols/sci-idt-xgen-ez/README.md b/protocols/sci-idt-xgen-ez/README.md
index 399cc67785..2465434b46 100644
--- a/protocols/sci-idt-xgen-ez/README.md
+++ b/protocols/sci-idt-xgen-ez/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* IDT xGEN EZ
diff --git a/protocols/sci-idt-xgen-mc/README.md b/protocols/sci-idt-xgen-mc/README.md
index 0aacd985ad..ebe39d2beb 100644
--- a/protocols/sci-idt-xgen-mc/README.md
+++ b/protocols/sci-idt-xgen-mc/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* IDT xGEN MC
diff --git a/protocols/sci-illumina-dna-prep/README.md b/protocols/sci-illumina-dna-prep/README.md
index a4c486e434..c1db9530fd 100644
--- a/protocols/sci-illumina-dna-prep/README.md
+++ b/protocols/sci-illumina-dna-prep/README.md
@@ -5,6 +5,9 @@
### Partner
[Illumina](https://www.illumina.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina DNA Prep
diff --git a/protocols/sci-illumina-rna-enrichment-normalization/README.md b/protocols/sci-illumina-rna-enrichment-normalization/README.md
index b23a9c05f7..f6ca7fb964 100644
--- a/protocols/sci-illumina-rna-enrichment-normalization/README.md
+++ b/protocols/sci-illumina-rna-enrichment-normalization/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Illumina RNA Prep with Enrichment
diff --git a/protocols/sci-lucif-assay1-for4/README.md b/protocols/sci-lucif-assay1-for4/README.md
index 1e2cd1eff8..51dd5c7bcd 100644
--- a/protocols/sci-lucif-assay1-for4/README.md
+++ b/protocols/sci-lucif-assay1-for4/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/sci-lucif-assay1/README.md b/protocols/sci-lucif-assay1/README.md
index c904e7af48..affbaffa40 100644
--- a/protocols/sci-lucif-assay1/README.md
+++ b/protocols/sci-lucif-assay1/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/sci-lucif-assay2-for4/README.md b/protocols/sci-lucif-assay2-for4/README.md
index 6aeee8b4b5..eda93a3d9d 100644
--- a/protocols/sci-lucif-assay2-for4/README.md
+++ b/protocols/sci-lucif-assay2-for4/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/sci-lucif-assay2/README.md b/protocols/sci-lucif-assay2/README.md
index 8a3b6b78c4..cb43cb09f5 100644
--- a/protocols/sci-lucif-assay2/README.md
+++ b/protocols/sci-lucif-assay2/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Broad Category
* Specific Category
diff --git a/protocols/sci-lucif-assay3-for4/README.md b/protocols/sci-lucif-assay3-for4/README.md
index 92f2fe89db..f22d0d9633 100644
--- a/protocols/sci-lucif-assay3-for4/README.md
+++ b/protocols/sci-lucif-assay3-for4/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/sci-lucif-assay3/README.md b/protocols/sci-lucif-assay3/README.md
index e217cd949f..41863d5a04 100644
--- a/protocols/sci-lucif-assay3/README.md
+++ b/protocols/sci-lucif-assay3/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/sci-lucif-assay4-for4/README.md b/protocols/sci-lucif-assay4-for4/README.md
index f9edadaba1..a8dd40bd81 100644
--- a/protocols/sci-lucif-assay4-for4/README.md
+++ b/protocols/sci-lucif-assay4-for4/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/sci-lucif-assay4/README.md b/protocols/sci-lucif-assay4/README.md
index fa8e8fc01a..d7b5e3d40e 100644
--- a/protocols/sci-lucif-assay4/README.md
+++ b/protocols/sci-lucif-assay4/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Sample Prep
* Plate Filling
diff --git a/protocols/sci-macherey-nagel-nucleomag/README.md b/protocols/sci-macherey-nagel-nucleomag/README.md
index 2dc21ad2ab..8dd64fcd56 100644
--- a/protocols/sci-macherey-nagel-nucleomag/README.md
+++ b/protocols/sci-macherey-nagel-nucleomag/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons (verified)](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-macherey-nagel-nucleomag). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-macherey-nagel-nucleomag). This page won’t be available after January 31st, 2024.
## Categories
* Nucleic Acid Extraction & Purification
@@ -20,7 +20,7 @@ Explanation of complex parameters below:
* `Deepwell type`: Specify which well plate will be mounted on the magnetic module.
* `Reservoir Type`: Specify which reservoir will be employed.
* `Starting Volume`: Specify starting volume of sample (ul).
-* ``: Specify the volume of binding buffer to use (ul).
+* `Binding Buffer Volume`: Specify the volume of binding buffer to use (ul).
* `Elution Volume`: Specify elution volume (ul).
* `Park Tips`: Specify whether to park tips or drop tips.
* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.
@@ -93,4 +93,4 @@ Explanation of complex parameters below:
If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).
###### Internal
-sci-promega-magazorb-dna-mini-prep-kit
\ No newline at end of file
+sci-promega-magazorb-dna-mini-prep-kit
diff --git a/protocols/sci-mag-bind-blood-tissue-kit/README.md b/protocols/sci-mag-bind-blood-tissue-kit/README.md
index 30d10e2231..07ca71348e 100644
--- a/protocols/sci-mag-bind-blood-tissue-kit/README.md
+++ b/protocols/sci-mag-bind-blood-tissue-kit/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons (verified)](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-mag-bind-blood-tissue-kit). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-mag-bind-blood-tissue-kit). This page won’t be available after January 31st, 2024.
## Categories
* Nucleic Acid Extraction & Purification
@@ -20,7 +20,7 @@ Explanation of complex parameters below:
* `Deepwell type`: Specify which well plate will be mounted on the magnetic module.
* `Reservoir Type`: Specify which reservoir will be employed.
* `Starting Volume`: Specify starting volume of sample (ul).
-* ``: Specify the volume of binding buffer to use (ul).
+* `Binding Buffer Volume`: Specify the volume of binding buffer to use (ul).
* `Elution Volume`: Specify elution volume (ul).
* `Park Tips`: Specify whether to park tips or drop tips.
* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.
@@ -93,4 +93,4 @@ Explanation of complex parameters below:
If you have any questions about this protocol, please contact the Protocol Development Team by filling out the [Troubleshooting Survey](https://protocol-troubleshooting.paperform.co/).
###### Internal
-sci-mag-bind-blood-tissue-kit
\ No newline at end of file
+sci-mag-bind-blood-tissue-kit
diff --git a/protocols/sci-mgi-mgieasy-nucleic-acid-extraction-kit/README.md b/protocols/sci-mgi-mgieasy-nucleic-acid-extraction-kit/README.md
index 98b59d7c1e..b953aff5d4 100644
--- a/protocols/sci-mgi-mgieasy-nucleic-acid-extraction-kit/README.md
+++ b/protocols/sci-mgi-mgieasy-nucleic-acid-extraction-kit/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* MGI Kit
diff --git a/protocols/sci-neb-next-ultra/README.md b/protocols/sci-neb-next-ultra/README.md
index 1459b45002..d5134a2d9f 100644
--- a/protocols/sci-neb-next-ultra/README.md
+++ b/protocols/sci-neb-next-ultra/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* NEBNext Ultra II
diff --git a/protocols/sci-omegabiotek-extraction-fa/README.md b/protocols/sci-omegabiotek-extraction-fa/README.md
index 074aa72aa2..5a42b5dbe8 100644
--- a/protocols/sci-omegabiotek-extraction-fa/README.md
+++ b/protocols/sci-omegabiotek-extraction-fa/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons (verified)](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-omegabiotek-extraction-fa). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-omegabiotek-extraction-fa). This page won’t be available after January 31st, 2024.
## Categories
* Nucleic Acid Extraction & Purification
diff --git a/protocols/sci-omegabiotek-extraction/README.md b/protocols/sci-omegabiotek-extraction/README.md
index b705dc97be..254df9077d 100644
--- a/protocols/sci-omegabiotek-extraction/README.md
+++ b/protocols/sci-omegabiotek-extraction/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons (verified)](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* RNA Extraction
diff --git a/protocols/sci-omegabiotek-magbind-temp/README.md b/protocols/sci-omegabiotek-magbind-temp/README.md
index f941e93dd2..0c73b98664 100644
--- a/protocols/sci-omegabiotek-magbind-temp/README.md
+++ b/protocols/sci-omegabiotek-magbind-temp/README.md
@@ -2,6 +2,9 @@
### Author
[Opentrons (verified)](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* RNA Extraction
diff --git a/protocols/sci-omegabiotek-magbind-total-rna-96/README.md b/protocols/sci-omegabiotek-magbind-total-rna-96/README.md
index ae7daf371d..906e5f615e 100644
--- a/protocols/sci-omegabiotek-magbind-total-rna-96/README.md
+++ b/protocols/sci-omegabiotek-magbind-total-rna-96/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons (verified)](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-omegabiotek-magbind-total-rna-96). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-omegabiotek-magbind-total-rna-96). This page won’t be available after January 31st, 2024.
## Categories
* Nucleic Acid Extraction & Purification
diff --git a/protocols/sci-omegabiotek-magbind/README.md b/protocols/sci-omegabiotek-magbind/README.md
index 21d8015a7d..d11dc2626e 100644
--- a/protocols/sci-omegabiotek-magbind/README.md
+++ b/protocols/sci-omegabiotek-magbind/README.md
@@ -2,7 +2,7 @@
### Author
[Opentrons (verified)](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-omegabiotek-magbind). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-omegabiotek-magbind). This page won’t be available after January 31st, 2024.
## Categories
* Nucleic Acid Extraction & Purification
diff --git a/protocols/sci-phytip-neutralization/README.md b/protocols/sci-phytip-neutralization/README.md
index 6ad762ab47..5cfc97ec1c 100644
--- a/protocols/sci-phytip-neutralization/README.md
+++ b/protocols/sci-phytip-neutralization/README.md
@@ -9,6 +9,9 @@
[Biotage](https://www.biotage.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Protein Purification
* ProPlus PhyTip® Columns
diff --git a/protocols/sci-phytip-plate-prep/README.md b/protocols/sci-phytip-plate-prep/README.md
index 80168fd759..9bf285a821 100644
--- a/protocols/sci-phytip-plate-prep/README.md
+++ b/protocols/sci-phytip-plate-prep/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Protein Purification
* Protein A, Pro Plus, Pro Plus LX Columns
diff --git a/protocols/sci-phytip-protein-A/README.md b/protocols/sci-phytip-protein-A/README.md
index cda407d9b9..b3a4c659b2 100644
--- a/protocols/sci-phytip-protein-A/README.md
+++ b/protocols/sci-phytip-protein-A/README.md
@@ -6,6 +6,9 @@
### Partner
[Biotage](https://www.biotage.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Protein Purification
* Protein A, Pro Plus, Pro Plus LX Columns
diff --git a/protocols/sci-phytip-protein-puri/README.md b/protocols/sci-phytip-protein-puri/README.md
index 72b8bb5ecf..0fc853ea0a 100644
--- a/protocols/sci-phytip-protein-puri/README.md
+++ b/protocols/sci-phytip-protein-puri/README.md
@@ -6,6 +6,9 @@
### Partner
[Biotage](https://www.biotage.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Protein Purification
* Protein A, Pro Plus, Pro Plus LX Columns
diff --git a/protocols/sci-pierce-ninta-magnetic-beads-part-2/README.md b/protocols/sci-pierce-ninta-magnetic-beads-part-2/README.md
index 5fd44a5193..20f20409d8 100644
--- a/protocols/sci-pierce-ninta-magnetic-beads-part-2/README.md
+++ b/protocols/sci-pierce-ninta-magnetic-beads-part-2/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Protein Purification
* Thermo Fisher Pierce Ni-NTA Magnetic Agarose Beads
diff --git a/protocols/sci-pierce-ninta-magnetic-beads/README.md b/protocols/sci-pierce-ninta-magnetic-beads/README.md
index cd50100ec9..cd6fb81013 100644
--- a/protocols/sci-pierce-ninta-magnetic-beads/README.md
+++ b/protocols/sci-pierce-ninta-magnetic-beads/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Protein Purification
* Thermo Fisher Pierce Ni-NTA Magnetic Agarose Beads
diff --git a/protocols/sci-promega-magazorb-dna-mini-prep-kit/README.md b/protocols/sci-promega-magazorb-dna-mini-prep-kit/README.md
index 9fe1287435..d193061751 100644
--- a/protocols/sci-promega-magazorb-dna-mini-prep-kit/README.md
+++ b/protocols/sci-promega-magazorb-dna-mini-prep-kit/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons (verified)](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-promega-magazorb-dna-mini-prep-kit). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-promega-magazorb-dna-mini-prep-kit). This page won’t be available after January 31st, 2024.
## Categories
* Nucleic Acid Extraction & Purification
@@ -20,7 +20,7 @@ Explanation of complex parameters below:
* `Deepwell type`: Specify which well plate will be mounted on the magnetic module.
* `Reservoir Type`: Specify which reservoir will be employed.
* `Starting Volume`: Specify starting volume of sample (ul).
-* ``: Specify the volume of binding buffer to use (ul).
+* `Binding buffer volume`: Specify the volume of binding buffer to use (ul).
* `Elution Volume`: Specify elution volume (ul).
* `Park Tips`: Specify whether to park tips or drop tips.
* `P300 Multi Channel Pipette Mount`: Specify whether the P300 multi channel pipette will be on the left or right mount.
diff --git a/protocols/sci-promega-magnesil-total-rna-mini-isolation-system/README.md b/protocols/sci-promega-magnesil-total-rna-mini-isolation-system/README.md
index a0b071ef0d..345e974913 100644
--- a/protocols/sci-promega-magnesil-total-rna-mini-isolation-system/README.md
+++ b/protocols/sci-promega-magnesil-total-rna-mini-isolation-system/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Promega Kit
diff --git a/protocols/sci-roche-hyperprep/README.md b/protocols/sci-roche-hyperprep/README.md
index 6cd5e7dde7..8403ac3b8e 100644
--- a/protocols/sci-roche-hyperprep/README.md
+++ b/protocols/sci-roche-hyperprep/README.md
@@ -6,6 +6,9 @@
### Partner
[Partner Name](partner website link)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Roche HyperPrep
diff --git a/protocols/sci-tecan-cortisol-saliva-elisa/README.md b/protocols/sci-tecan-cortisol-saliva-elisa/README.md
index c83a32d857..ce194a8e47 100644
--- a/protocols/sci-tecan-cortisol-saliva-elisa/README.md
+++ b/protocols/sci-tecan-cortisol-saliva-elisa/README.md
@@ -5,6 +5,9 @@
Boren Lin
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Proteins
* Proteins
diff --git a/protocols/sci-thermofisher-magmax/README.md b/protocols/sci-thermofisher-magmax/README.md
index 877d8cf572..b2b0b82738 100644
--- a/protocols/sci-thermofisher-magmax/README.md
+++ b/protocols/sci-thermofisher-magmax/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Thermofisher MagMAX
diff --git a/protocols/sci-zymo-directzol-magbead/README.md b/protocols/sci-zymo-directzol-magbead/README.md
index 151826be16..78970cb76d 100644
--- a/protocols/sci-zymo-directzol-magbead/README.md
+++ b/protocols/sci-zymo-directzol-magbead/README.md
@@ -3,7 +3,7 @@
### Author
[Opentrons (verified)](https://opentrons.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-zymo-directzol-magbead). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/sci-zymo-directzol-magbead). This page won’t be available after January 31st, 2024.
## Categories
* Nucleic Acid Extraction & Purification
diff --git a/protocols/sci-zymo-quick-dna-rna-magbead/README.md b/protocols/sci-zymo-quick-dna-rna-magbead/README.md
index 24fd1d059f..87f443517d 100644
--- a/protocols/sci-zymo-quick-dna-rna-magbead/README.md
+++ b/protocols/sci-zymo-quick-dna-rna-magbead/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Zymo Kit
diff --git a/protocols/speaker-test/README.md b/protocols/speaker-test/README.md
index 2d24949bb2..daa485937e 100644
--- a/protocols/speaker-test/README.md
+++ b/protocols/speaker-test/README.md
@@ -5,6 +5,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Getting Started
* OT-2 Demo
diff --git a/protocols/standard-biotools-da-192/README.md b/protocols/standard-biotools-da-192/README.md
index f66a34a31c..d780461f2a 100644
--- a/protocols/standard-biotools-da-192/README.md
+++ b/protocols/standard-biotools-da-192/README.md
@@ -7,6 +7,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* Standard BioTools
diff --git a/protocols/standard-biotools-da-48/README.md b/protocols/standard-biotools-da-48/README.md
index 122189584c..25ed11b3a9 100644
--- a/protocols/standard-biotools-da-48/README.md
+++ b/protocols/standard-biotools-da-48/README.md
@@ -8,6 +8,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* Standard BioTools
diff --git a/protocols/standard-biotools-da-96/README.md b/protocols/standard-biotools-da-96/README.md
index bc68b804bb..57a322a6f9 100644
--- a/protocols/standard-biotools-da-96/README.md
+++ b/protocols/standard-biotools-da-96/README.md
@@ -8,6 +8,9 @@
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* PCR
* Standard BioTools
diff --git a/protocols/swift-2s-turbo-pt1/README.md b/protocols/swift-2s-turbo-pt1/README.md
index 5dedfb721b..ff09ff2b86 100644
--- a/protocols/swift-2s-turbo-pt1/README.md
+++ b/protocols/swift-2s-turbo-pt1/README.md
@@ -6,7 +6,7 @@
### Partner
[Swift Biosciences](https://swiftbiosci.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/swift-2s-turbo-pt1). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/swift-2s-turbo-pt1). This page won’t be available after January 31st, 2024.
## Categories
* Featured
diff --git a/protocols/swift-2s-turbo-pt2/README.md b/protocols/swift-2s-turbo-pt2/README.md
index bd829ce524..616529edd5 100644
--- a/protocols/swift-2s-turbo-pt2/README.md
+++ b/protocols/swift-2s-turbo-pt2/README.md
@@ -6,6 +6,9 @@
### Partner
[Swift Biosciences](https://swiftbiosci.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Featured
* NGS Library Prep: Swift 2S Turbo
diff --git a/protocols/swift-2s-turbo-pt3/README.md b/protocols/swift-2s-turbo-pt3/README.md
index 0796117ce8..62c1cd91a2 100644
--- a/protocols/swift-2s-turbo-pt3/README.md
+++ b/protocols/swift-2s-turbo-pt3/README.md
@@ -6,6 +6,9 @@
### Partner
[Swift Biosciences](https://swiftbiosci.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Featured
* NGS Library Prep: Swift 2S Turbo
diff --git a/protocols/swift-fully-automated-custom/README.md b/protocols/swift-fully-automated-custom/README.md
index 6f279de83a..c8d01878aa 100644
--- a/protocols/swift-fully-automated-custom/README.md
+++ b/protocols/swift-fully-automated-custom/README.md
@@ -6,6 +6,9 @@
### Partner
[Swift Biosciences](https://swiftbiosci.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Swift 2S Turbo
diff --git a/protocols/swift-fully-automated/README.md b/protocols/swift-fully-automated/README.md
index 22fc9a3554..c818c20c5a 100644
--- a/protocols/swift-fully-automated/README.md
+++ b/protocols/swift-fully-automated/README.md
@@ -6,7 +6,7 @@
### Partner
[Swift Biosciences](https://swiftbiosci.com/)
-***Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/swift-fully-automated). This page won’t be available after January 31st, 2024.***
+# Opentrons has launched a new Protocol Library. You should use the [new page for this protocol](library.opentrons.com/p/swift-fully-automated). This page won’t be available after January 31st, 2024.
## Categories
* NGS Library Prep
diff --git a/protocols/testdrive/README.md b/protocols/testdrive/README.md
index 8cc58ea79b..e60c1c073f 100644
--- a/protocols/testdrive/README.md
+++ b/protocols/testdrive/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Getting Started
* OT-2 Guided Walk-through
diff --git a/protocols/thermocycler-csv/README.md b/protocols/thermocycler-csv/README.md
index ab4b419de1..bff0227188 100644
--- a/protocols/thermocycler-csv/README.md
+++ b/protocols/thermocycler-csv/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Thermocycler
* Thermocycler with CSV
diff --git a/protocols/thermocycler/README.md b/protocols/thermocycler/README.md
index 79477c5670..c8e39672a4 100644
--- a/protocols/thermocycler/README.md
+++ b/protocols/thermocycler/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Getting Started
* Thermocycler Example
diff --git a/protocols/user-upload_ZooMS_Protocol/README.md b/protocols/user-upload_ZooMS_Protocol/README.md
index f868b91e04..0f6c975547 100644
--- a/protocols/user-upload_ZooMS_Protocol/README.md
+++ b/protocols/user-upload_ZooMS_Protocol/README.md
@@ -1,89 +1,92 @@
-# Automated RoboZooMS Protocol
-
-### Author
-[University of York BioArchaeology Team](https://www.york.ac.uk/archaeology/centres-facilities/bioarch/)
-
-## Categories
-* Sample Prep
- * Plate Filling
-
-## Description
-A start to finish protocol for preparation of archaeological bone samples for analysis by MALDI-TOF.
-Currently configured for 12 or 24 samples, depending on the amount of eppendorf racks available.
-Protocol can be adjusted for non-destructive analysis, and for NaOH wash to remove humic acids.
-
-Samples are to be laid out left to right (A1 to A12), top to bottom (then B1 to B12).
-
-The protocol is split into 5 main steps, with the first two being optional:
-
-1. Hydrochloric acid demineralisation (optional)
-2. Sodium hydroxide wash (optional)
-3. Gelatine extraction
-4. Trypsin digestion
-5. Peptide extraction
-
----
-
-### Labware
- * [Opentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap](https://labware.opentrons.com/opentrons_24_tuberack_eppendorf_1.5ml_safelock_snapcap?category=tubeRack)
- * [Opentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap](https://labware.opentrons.com/opentrons_24_tuberack_eppendorf_1.5ml_safelock_snapcap?category=tubeRack)
- * This tube rack is optional however, it allows throughput to double from 12 to 24!
- * [Opentrons 6 Tube Rack with Falcon 50 mL Conical](https://labware.opentrons.com/opentrons_6_tuberack_falcon_50ml_conical?category=tubeRack)
- * [Opentrons 96 Tip Rack 1000 µL](https://labware.opentrons.com/opentrons_96_tiprack_1000ul?category=tipRack&manufacturer=Opentrons)
- * [Opentrons 96 Tip Rack 1000 µL](https://labware.opentrons.com/opentrons_96_tiprack_1000ul?category=tipRack&manufacturer=Opentrons)
- * [Opentrons 96 Tip Rack 300 µL](https://labware.opentrons.com/opentrons_96_tiprack_300ul?category=tipRack&manufacturer=Opentrons)
- * [NEST 12 Well Reservoir 15 mL](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir&manufacturer=NEST)
- * [Corning 96 Well Plate 360 µL Flat](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate&manufacturer=Corning)
-
-### Pipettes
- * P1000 Single GEN1
- * P300 Single GEN1
-
-### Reagents
-HCl and NaOH should be stored at 4°C prior to use.
- * 0.6 M Hydrochloric Acid (HCl)
- * 0.1 M Sodium Hydroxide (NaOH)
- * 50 mM Ammonium bicarbonate (AmBIC) buffer
- * 0.1% Trifluoroacetic acid (TFA) in ultrapure (UHQ) water (washing solution)
- * 0.1% TFA in 50:50 solution of UHQ water and acetonitrile (conditioning solution)
- * Trypsin solution (frozen and resuspended in 50 uL of re-suspension buffer as sold, as needed), made up to 10 mL with UHQ water
-
----
-
-### Deck Setup
-
-
-Note that the 12 well reservoir in slot 11 is simply used as an aqueous waste (AqWaste) container.
-
-* IMPORTANT:
- * If using one rack then the top two rows (A and B) are used for the "second extract" eppendorfs (where you place your sample to begin with) and bottom two rows are used for "Extract" exppendorfs (where the sample will be processed)
- * If using two racks then the top rack (default deck location: 8) is used to hold samples (labelled as SE) and the bottom rack (default deck location: 5) will hold the extracts (labelled EXT).
-
-### Reagent Setup
-
-
-
-
----
-
-### Protocol Steps
-Note: volumes shown here are defaults which can be changed if needed.
-Note: Manual steps indicated, followed by "automatically" when a robot takes over.
-
-1. (optional) Add 250 uL of 0.6 M HCl from "falcon tube well 0" into the 1.5 mL eppendorf tubes containg 10-30 mg of bone (pause and manual resume here after demineralisation 1-14 days).
-2. (optional) Remove acid from each eppendorf into AqWaste (reservoir trough 'A1'), add 250 uL 0.1 M of NaOH from "falcon tube well 2", manually vortex and centrifuge samples, then automatically remove NaOH from eppendorfs into AqWaste
-3. Distribute 100 uL of 50 mM AmBIC into samples, manually incubate for 1 hr at 65°C then centrifuge 1 min and place samples back, automatically transfer 50 uL of gelatinized sample into EXT eppendorfs.
-4. Distribute 1 uL of trypsin into EXT eppendorfs, mix, then manually incubate overnight at 37°C and place samples back, automatically add 50 uL of washing soltuion to quench trypsin.
-5. Using a regular 1000 uL pipette: prepare the zip-tip wellplate by placing 250 uL of conditioning solution and 350 uL of washing solution into well plate as above
-6. Using a filter-tip 300 uL pipette: slowly draw up conditioning solution twice (50 uL), wash the filter with washing solution twice (100 uL), draw sample (in EXT eppendorf) in and out 10 times, wash filter twice (100 uL), draw up conditioning solution (100 uL) and elute into third well, mixing 10 times (50 uL).
-
-If not spotting directly, store eluted samples at -20 °C.
-
-
-### Process
-1. Input your protocol parameters above.
-2. Download your protocol and unzip if needed.
-3. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.
-4. Set up your deck according to the deck map.
-5. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).
-6. Hit 'Run'.
+# Automated RoboZooMS Protocol
+
+### Author
+[University of York BioArchaeology Team](https://www.york.ac.uk/archaeology/centres-facilities/bioarch/)
+
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
+## Categories
+* Sample Prep
+ * Plate Filling
+
+## Description
+A start to finish protocol for preparation of archaeological bone samples for analysis by MALDI-TOF.
+Currently configured for 12 or 24 samples, depending on the amount of eppendorf racks available.
+Protocol can be adjusted for non-destructive analysis, and for NaOH wash to remove humic acids.
+
+Samples are to be laid out left to right (A1 to A12), top to bottom (then B1 to B12).
+
+The protocol is split into 5 main steps, with the first two being optional:
+
+1. Hydrochloric acid demineralisation (optional)
+2. Sodium hydroxide wash (optional)
+3. Gelatine extraction
+4. Trypsin digestion
+5. Peptide extraction
+
+---
+
+### Labware
+ * [Opentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap](https://labware.opentrons.com/opentrons_24_tuberack_eppendorf_1.5ml_safelock_snapcap?category=tubeRack)
+ * [Opentrons 24 Tube Rack with Eppendorf 1.5 mL Safe-Lock Snapcap](https://labware.opentrons.com/opentrons_24_tuberack_eppendorf_1.5ml_safelock_snapcap?category=tubeRack)
+ * This tube rack is optional however, it allows throughput to double from 12 to 24!
+ * [Opentrons 6 Tube Rack with Falcon 50 mL Conical](https://labware.opentrons.com/opentrons_6_tuberack_falcon_50ml_conical?category=tubeRack)
+ * [Opentrons 96 Tip Rack 1000 µL](https://labware.opentrons.com/opentrons_96_tiprack_1000ul?category=tipRack&manufacturer=Opentrons)
+ * [Opentrons 96 Tip Rack 1000 µL](https://labware.opentrons.com/opentrons_96_tiprack_1000ul?category=tipRack&manufacturer=Opentrons)
+ * [Opentrons 96 Tip Rack 300 µL](https://labware.opentrons.com/opentrons_96_tiprack_300ul?category=tipRack&manufacturer=Opentrons)
+ * [NEST 12 Well Reservoir 15 mL](https://labware.opentrons.com/nest_12_reservoir_15ml?category=reservoir&manufacturer=NEST)
+ * [Corning 96 Well Plate 360 µL Flat](https://labware.opentrons.com/corning_96_wellplate_360ul_flat?category=wellPlate&manufacturer=Corning)
+
+### Pipettes
+ * P1000 Single GEN1
+ * P300 Single GEN1
+
+### Reagents
+HCl and NaOH should be stored at 4°C prior to use.
+ * 0.6 M Hydrochloric Acid (HCl)
+ * 0.1 M Sodium Hydroxide (NaOH)
+ * 50 mM Ammonium bicarbonate (AmBIC) buffer
+ * 0.1% Trifluoroacetic acid (TFA) in ultrapure (UHQ) water (washing solution)
+ * 0.1% TFA in 50:50 solution of UHQ water and acetonitrile (conditioning solution)
+ * Trypsin solution (frozen and resuspended in 50 uL of re-suspension buffer as sold, as needed), made up to 10 mL with UHQ water
+
+---
+
+### Deck Setup
+
+
+Note that the 12 well reservoir in slot 11 is simply used as an aqueous waste (AqWaste) container.
+
+* IMPORTANT:
+ * If using one rack then the top two rows (A and B) are used for the "second extract" eppendorfs (where you place your sample to begin with) and bottom two rows are used for "Extract" exppendorfs (where the sample will be processed)
+ * If using two racks then the top rack (default deck location: 8) is used to hold samples (labelled as SE) and the bottom rack (default deck location: 5) will hold the extracts (labelled EXT).
+
+### Reagent Setup
+
+
+
+
+---
+
+### Protocol Steps
+Note: volumes shown here are defaults which can be changed if needed.
+Note: Manual steps indicated, followed by "automatically" when a robot takes over.
+
+1. (optional) Add 250 uL of 0.6 M HCl from "falcon tube well 0" into the 1.5 mL eppendorf tubes containg 10-30 mg of bone (pause and manual resume here after demineralisation 1-14 days).
+2. (optional) Remove acid from each eppendorf into AqWaste (reservoir trough 'A1'), add 250 uL 0.1 M of NaOH from "falcon tube well 2", manually vortex and centrifuge samples, then automatically remove NaOH from eppendorfs into AqWaste
+3. Distribute 100 uL of 50 mM AmBIC into samples, manually incubate for 1 hr at 65°C then centrifuge 1 min and place samples back, automatically transfer 50 uL of gelatinized sample into EXT eppendorfs.
+4. Distribute 1 uL of trypsin into EXT eppendorfs, mix, then manually incubate overnight at 37°C and place samples back, automatically add 50 uL of washing soltuion to quench trypsin.
+5. Using a regular 1000 uL pipette: prepare the zip-tip wellplate by placing 250 uL of conditioning solution and 350 uL of washing solution into well plate as above
+6. Using a filter-tip 300 uL pipette: slowly draw up conditioning solution twice (50 uL), wash the filter with washing solution twice (100 uL), draw sample (in EXT eppendorf) in and out 10 times, wash filter twice (100 uL), draw up conditioning solution (100 uL) and elute into third well, mixing 10 times (50 uL).
+
+If not spotting directly, store eluted samples at -20 °C.
+
+
+### Process
+1. Input your protocol parameters above.
+2. Download your protocol and unzip if needed.
+3. Upload your protocol file (.py extension) to the [OT App](https://opentrons.com/ot-app) in the `Protocol` tab.
+4. Set up your deck according to the deck map.
+5. Calibrate your labware, tiprack and pipette using the OT App. For calibration tips, check out our [support articles](https://support.opentrons.com/en/collections/1559720-guide-for-getting-started-with-the-ot-2).
+6. Hit 'Run'.
diff --git a/protocols/zymo-quick-96/README.md b/protocols/zymo-quick-96/README.md
index 2a100317c9..9d211d2f45 100644
--- a/protocols/zymo-quick-96/README.md
+++ b/protocols/zymo-quick-96/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Zymo Kit
diff --git a/protocols/zymo-quick-custom/README.md b/protocols/zymo-quick-custom/README.md
index a1d00b9805..7cc778060c 100644
--- a/protocols/zymo-quick-custom/README.md
+++ b/protocols/zymo-quick-custom/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Zymo Kit
diff --git a/protocols/zymo-quick/README.md b/protocols/zymo-quick/README.md
index fe54259aad..10ca35b5d7 100644
--- a/protocols/zymo-quick/README.md
+++ b/protocols/zymo-quick/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Nucleic Acid Extraction & Purification
* Zymo Kit
diff --git a/protocols/zymo-ribofree-cleanup/README.md b/protocols/zymo-ribofree-cleanup/README.md
index da2eaae25b..aa1cc5c3c7 100644
--- a/protocols/zymo-ribofree-cleanup/README.md
+++ b/protocols/zymo-ribofree-cleanup/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Zymo RiboFree™ Total RNA Library Prep
diff --git a/protocols/zymo-ribofree-first-strand-cdna-synth-universal-depletion/README.md b/protocols/zymo-ribofree-first-strand-cdna-synth-universal-depletion/README.md
index cdeddabdb1..07170ebd11 100644
--- a/protocols/zymo-ribofree-first-strand-cdna-synth-universal-depletion/README.md
+++ b/protocols/zymo-ribofree-first-strand-cdna-synth-universal-depletion/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Zymo RiboFree™ Total RNA Library Prep
diff --git a/protocols/zymo-ribofree-library-index-pcr/README.md b/protocols/zymo-ribofree-library-index-pcr/README.md
index ee83ea06de..427d92dd38 100644
--- a/protocols/zymo-ribofree-library-index-pcr/README.md
+++ b/protocols/zymo-ribofree-library-index-pcr/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Zymo RiboFree™ Total RNA Library Prep
diff --git a/protocols/zymo-ribofree-p5-adapter-ligation/README.md b/protocols/zymo-ribofree-p5-adapter-ligation/README.md
index 07c7a1f9ee..b2afb6295f 100644
--- a/protocols/zymo-ribofree-p5-adapter-ligation/README.md
+++ b/protocols/zymo-ribofree-p5-adapter-ligation/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Zymo RiboFree™ Total RNA Library Prep
diff --git a/protocols/zymo-ribofree-p7-adapter-ligation/README.md b/protocols/zymo-ribofree-p7-adapter-ligation/README.md
index 80c112dafc..71db1d92f8 100644
--- a/protocols/zymo-ribofree-p7-adapter-ligation/README.md
+++ b/protocols/zymo-ribofree-p7-adapter-ligation/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* NGS Library Prep
* Zymo RiboFree™ Total RNA Library Prep
diff --git a/protocols/zymo-rna-extraction/README.md b/protocols/zymo-rna-extraction/README.md
index 100a70402a..859222ceee 100644
--- a/protocols/zymo-rna-extraction/README.md
+++ b/protocols/zymo-rna-extraction/README.md
@@ -3,6 +3,9 @@
### Author
[Opentrons](https://opentrons.com/)
+
+# Opentrons has launched a new Protocol Library. This page won’t be available after January 31st, 2024. [Submit a request](https://docs.google.com/forms/d/e/1FAIpQLSdYYp9QCKow4nn0KlCVsMS3HX0eJ0N9O7-erajKvcpT0lWbSg/viewform) to add this protocol to the new library.
+
## Categories
* Covid Workstation
* RNA Extraction