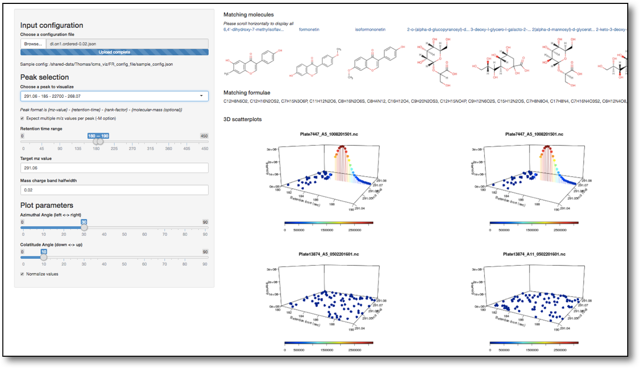
This module lets you setup a visualization server that allows comparative display of UntargetedMetabolomics analysis.
R version > 3.1
- If needed, clone a new version of the
actrepository
mkdir lcms-viz
git clone [email protected]:20n/act.git lcms-viz/
- Sync your lib directory as instructed here reachables/lib and install the Chemaxon license
cd lcms-viz/reachables/
rsync -azP LOC_LIB lib/
license CHEMAXON_LICENSE.cxl
At this point, you should be able to fully compile your SBT project.
Try performing a clean compilation and running tests on the project: sbt clean compile test.
The app needs to access outside resources, including the 20n logo, but more importantly some methods in the reachables project, such as the MS1 class for m/z values computation. By creating symlinks in the app's directory, we provide the path to these resources.
- Create a fat JAR with SBT
sbt assembly
- Create symlinks in the app directory:
cd src/main/r/LCMSDataVisualisation/
ln -s lcms-viz/reachables/target/scala-2.10/reachables-assembly-0.1.jar reachables-assembly-0.1.jar
ln -s lcms-viz/reachables/src/main/resources/20n.png 20nlogo
- Finally, make sure that the directory
data/mol-structure-cache/exists. This is where molecule structure images will be stored. TODO: add a symlink for this too!
- Install libraries netCDF and BOOST:
sudo apt-get install libnetcdf-dev
sudo apt-get install libboost-all-dev
You might have to upgrade to the latest version of R (e.g., in case "mzR" below fails). Search for "r install ubuntu" and follow instructions.
- Start R and install the required packages
sudo R
In the R console:
R.Version() # Should be > 3.1
install.packages(c("shiny", "rscala", "dplyr", "plot3D", "classInt", "jsonlite", "logging", "digest"))
library(rscala) # loads the "rscala" library
rscala::scalaInstall() # downloads and installs Scala
# Package "mzR" needs to be installed through the bioconductor package
source("https://bioconductor.org/biocLite.R") # try replacing http:// with https:// if it gives you an error
biocLite("mzR")In the R console:
library(shiny)
runApp(port = 9090, host = "0.0.0.0", launch.browser = FALSE)The app should be now accessible from http://hostname:9090
Try uploading the lcms-demo.json file for sample visualization.